Abstract
Irritable bowel syndrome (IBS) is a multifactorial and heterogeneous disorder estimated to affect over 10% of the Western population. A subset of the patients reports the start of the disease after an episode of gastroenteritis. The alterations in the intestinal microbiota of the post-infectious IBS (PI-IBS) patients were recently investigated in a British cohort and shown to differentiate from the healthy controls and resemble that of diarrhea-predominant IBS (IBS-D) patients. The altered 27 genus-like groups created a microbial signature, which could be used to objectively stratify patients and healthy controls. In this addendum, we combine the microbiota data derived from the British cohort with that of a recently reported Swedish PI-IBS cohort. Remarkably, robust and reproducible microbiota signatures were observed in these PI-IBS patients. We discuss these results with attention on the emerging role of microbiota in the classification, development and treatment of PI-IBS.
Key words: irritable bowel syndrome, IMD, microbiota, PI-IBS
Introduction
Irritable bowel syndrome (IBS) is a common gastrointestinal disorder affecting approximately 10–15% of the people in Western world.1 The patient group is very heterogeneous and characterized by abdominal pain, discomfort and abnormal bowel habits. The pathophysiology of IBS is unknown, it would appear that the due to the heterogeneous patient material there are multiple explanations for the symptom development. Currently, patients are subdivided according to their main symptom that have been proposed in the so called Rome criteria; IBS with diarrhea (IBS-D), IBS with constipation (IBS-C) or IBS with mixed bowel habits (IBS-M).2 No single unifying cause for IBS has been reported3 but several factors have been associated with the disease etiology, including genetic predisposition,4 anxiety and depression5 as well as female gender.6 Moreover, there is strong evidence indicating that changes in the intestinal microbiota and their function could be one of the contributing factors in the development of IBS. Several studies have reported differences in the intestinal microbiota composition of IBS patients and healthy controls, however the results show little overlap from study to study.7,8 This is likely caused by different methodological approaches in the analysis steps as well as the heterogeneity of the IBS patients. Regardless of the lack of coherent changes in the microbial composition, there is clear evidence for the role of the microbiota in the pathogenesis of IBS. For one, the patients report improved symptoms after consuming agents targeted to modify the intestinal microbiota, including probiotics,9 antibiotics10 or changing diet, such as avoidance of FODMAP carbohydrates.11 Moreover, the strongest predictor for IBS is a prior gastroenteritic episode, which will increase the risk of developing IBS by 7-fold.12 It has been estimated that about 10% of the IBS cases begin after an episode of gastroenteritis, causing post-infectious (PI) IBS, indicating a cause and effect relationship.13,14 This is in line with a previously proposed model in which an event, such as an infection, affects the intestinal microbiota and generates an alternative stable state.15,16 Recently, a number of bistable microbial groups, i.e. microbes with two stable homeostatic states, have been identified that could act as tipping points in this well known ecological process that is associated with resilience.17
In our recent study by Jalanka and colleagues18 the differences in the microbial composition of PI-IBS patients were addressed by analyzing the faecal microbiota of the study subjects with a previously validated and benchmarked phylogenetic microarray, detecting the abundance of over 1000 bacterial phylotypes found in the human intestine.19,20 The studied cohort consisted of 57 British subjects divided into 5 study groups; patients with a PI-IBS diagnosis (group 1, Rome II), patients whom 6 months after the gastroenteritic episode had a persisting bowel dysfunction, which however did not qualify for IBS (PI-BD) (group 2) or did not experience any bowel dysfunction anymore (PI-nonBD) (group 3). The results were benchmarked against IBS-D patients (group 4, Rome II) and healthy controls (group 5). Our aim was to determine the microbial compositional differences and to address associations between the faecal microbiota and the clinical features of IBS from patients with a varying degree of symptoms. As the development of IBS is multifactorial, we hypothesized that the observed symptoms and development of PI-IBS may arise from the interplay between the faecal microbiota, the host immune response, and psychological factors. Therefore, we analyzed in parallel the detailed records of the participants' psychological well-being, intestinal symptoms, systemic immunological markers as well as the levels of host gene expression on rectal mucosa. This integrated approach, addressing the associations between clinical phenotype and the faecal microbiota allowed us to address the complex relationships between the host and microbiota in the pathogenesis of IBS and PI-IBS.
Microbial Signatures and IMD
Since the intestinal microbiota is very heterogeneous and subject-specific, we hypothesized that instead of limiting the comparative microbiota analysis to individual taxa, it would be more relevant to search for a microbial signature that potentially differs between the patients and healthy controls. By using a novel bioinformatic approach (bootstrap aggregated RDA method; baggedRDA) we identified a combination of 27 genus-like bacterial groups that significantly separated healthy controls and patients with IBS symptoms. Moreover, we found that the microbial profile of the PI-IBS and PI-BD groups resembled those suffering from IBS-D. The major difference in the patients' microbiota was the 12-fold increase of Bacteroidetes phylum and the 35-fold decrease of Uncultured Clostridiales belonging to the Firmicutes phylum. Furthermore, when all participants were ranked according to the abundance of the discriminating bacterial taxa, a specific ordering was obtained, the so-called Index of Microbial Dysbiosis (IMD). This rank ordering reflected the patient's health status as it correlated with the increase of several immunological markers and intestinal complaints, where as no such associations were observed when subdividing the patients according to their clinical status. Interestingly, there were no significant correlations between the IMD and any of the psychological symptoms often associated with IBS. Our results corroborate the previous reports that the microbial composition could be used to further stratify IBS patients and thus complement the clinical subgroupings.21 In a recent study it was estimated that circa 33% of the IBS patients exhibit anxiety and 13% suffer from depression.22 In addition, it was found that somatisation was significantly increased in the IBS patients compared to controls with gastrointestinal symptoms but without IBS. Although the intestinal microbiota of these patients was not studied, in the light of the recent evidence it could be hypothesized that the etiology of the disease in the patients with psychological symptoms differs from those with microbial aberrations. Taken together, these results suggest that the discriminant microbial profile could be used as an objective measure of disturbed bowel functions and therefore a potential novel way of stratifying this heterogeneous patient material in future studies and in clinical practise.
To verify our findings in a larger patient material we combined the microbiota data form the British subjects18 with that of a Swedish PI-IBS cohort reported recently by Sundin and colleagues.23 Both studies were conducted with an identical analysis pipeline, including the well established mechanical lysis for extraction of faecal DNA,24,25 the HITChip phylogenetic microarray platform,19 and the normalization and analysis software suite.26 The combined data aimed to determine whether the PI-IBS patients from 2 countries share similarities in their microbiota compared to healthy controls. The Swedish cohort introduced new subjects to both healthy controls (n=16) and PI-IBS patients (n=13, Rome III) therefore doubling the sample size to a total of 51 subjects. Remarkably, the significant difference in the microbiota profiles between the healthy and PI-IBS patients was replicated (p < 0.05, Fig. 1). The strongest difference was observed with the significant increase in the Bacteroides ssp. and decrease of members of the Uncultured Clostridiales in the PI-IBS patients. Interestingly, both Bacteroides fragilis and Uncultured Clostridiales have been identified as bistable microbial groups and predicted to act as tipping points of the gut ecosystem in opposite ways.17 However, not all of the IMD taxa reported in the original study18 appeared as significantly different between the patients and controls, and there were new taxa that were significantly increased in the combined PI-IBS patient group. One of them was Dialister, which was recently identified as bistable i.e., being either abundant or nearly absent in contrast to most other bacteria that show gradual abundance distribution.17 Dialister has also previously been associated with IBS.21,27 This highlights the need of conducting the future research in large cohorts in order to verify the separating species between PI-IBS and healthy controls.
Figure 1.
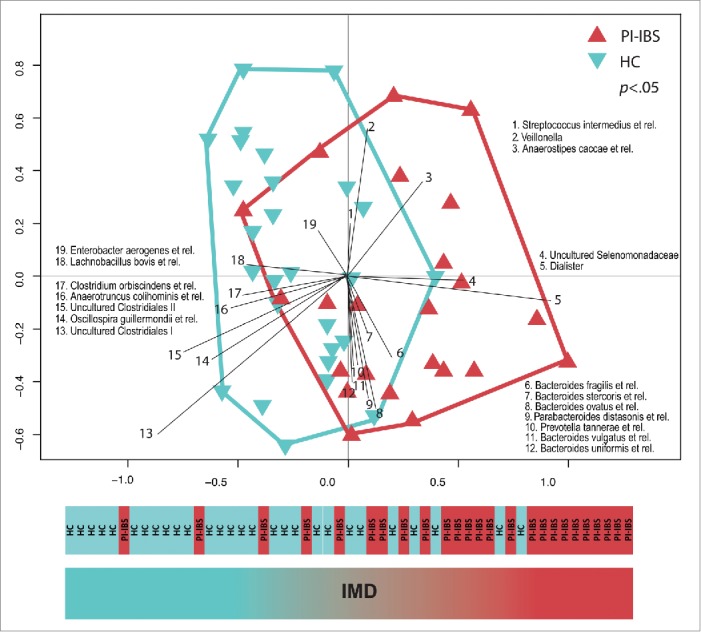
Intestinal microbiota of PI-IBS patients is significantly different form that of healthy controls (HC). The difference was measured with baggedRDA that was used to identify a microbial signature that separates HC from PI-IBS patients from the two study cohorts. The 19 taxa responsible for the separation are listed. Abundance of the discriminating taxa creates the so-called Index of Microbial Dysbiosis (IMD).
The role of Bacteroides spp. in the pathophysiology of IBS is controversial, probably reflecting the heterogeneity of the patients as well as the genus Bacteroides. Previous studies have reported opposing results regarding the relevance of Bacteroides genera in IBS, as there is evidence for both increased28-30 as well as decreased21,31 abundance in the patients. Interestingly, elevated levels of Bacteroides spp were among the microbiota changes that have been associated with susceptibility to enteric pathogens in mice.32 It has been suggested that susceptibility to infections could increase due to a imbalanced microbial composition, since complex interactions between the microbiota and host immune system might reduce the resistance to episodes of gastroenteritis. Recently, similar observations were made in humans by Dicksved and colleagues who followed abattoir workers who have a high risk of contracting C. jejuni infection.33 The participants donated faecal samples at the start of employment and were then regularly followed. The subjects who became C. jejuni infected had increased levels of Bacteroides species prior the infection, as opposed to those individuals who did not contract the disease during the follow-up period. The individuals who remained healthy had significantly higher levels of Uncultured Clostridiales.33 Similar to these findings, it was recently shown that the subjects contracting a gastroenteritis had increased levels of Bacteroides spp. and decreased levels of Firmicutes immediately after the infection.34 Although the infectious agents in this study were diverse, the differences in the microbiota profile were similar to what we found. Altogether, these data from several studies allow to hypothesize that the subjects with a microbial profile resembling the IMD could be more susceptible for contracting a gastrointestinal infection and later developing PI-IBS. More studies on patients before and after gastroenteritis are required to verify this hypothesis. Moreover, the possibility remains that the microbiota composition is not causally related to IBS. Recent experiments in mice have indicated that innate35 and mucosal immune system36 play a deterministic role in shaping the gut microbiota. Hence, the observed IMD may be a reflection of the activity of the host's immune system. While this alternative explanation may offer new insight in the etiology of IBS, it confirms the significance and application potential of IMD in stratifying IBS subjects.
Associations Between Microbiota and Host Gene Expression
To further analyze the crosstalk between the host and the microbiota, we concentrated on the 27 bacterial taxa of IMD and separately studied the associations between bacterial abundance and the host gene expression in mucosal surface of rectum, the last part of large intestine. We mapped the microarray-derived gene expression data from rectal biopsies to known biological functions using Gene Set Analysis (GSA). There were numerous statistically significant associations between the abundance of the IBS-type microbiota and the level of expressed host genes that suggested an association between the discriminant microbiota and the physical barrier integrity of the host. The most prominent negative correlation was observed between 7 Bacteroides ssp and the expression of glycine, serine and threonine metabolism pathway. In general, glycine, serine and threonine are known to be important in the maintenance of gut integrity and the structure of the mucin layer.37 For example, the majority of the dietary threonine is utilized for synthesising the secretory mucin and its dietary restriction results into impaired gut barrier.38 Moreover, the mucus structure and the extent of its glycosylation shape the microbiota composition39 and similar compositional changes have also been previously associated with barrier function abnormalities. Fucosyltransferase-2 is involved in the formation of the ABO-blood groups and its activity also creates adhesion receptor for several microbes.40 Therefore, it is interesting that FUT2 deficient mice (Fut2−/−) have been shown to display similar differences in their microbiota composition to what was found in our study, i.e. increased amounts of several Bacteroides species and decreased levels of Clostridiales.41
In addition to alterations in the mucosal barrier genes, we identified several other pathways, including those that regulate cell junctions and inflammatory responses, to be associated with the IMD bacterial taxa. The results led us to hypothesize that both the physical and immunological gut barrier of subjects with an IBS-type microbiota has been compromised resulting in a low-grade inflammation, a feature of some IBS patients. Changes in the inflammatory response, barrier function and microbiota composition could therefore lead into changes in the other host parameters such as gut-brain axis receiving signals from these systems.42 Moreover, we observed that the IMD correlated with the IBS symptoms and clinical features whereas the scores of anxiety and depression did not correlate with the IMD, indicating that patients with a more psychological basis for the disease have a rather undisturbed microbiota. This is in line with previous findings21 and of great significance as it allows proposing a model in which some subjects with a relatively healthy microbiota report IBS symptoms while others with a microbiota resembling that of the IBS patients have no complaints as they have compensating factors that overcome the microbial component and hence are scored healthy (Fig. 2).
Figure 2.
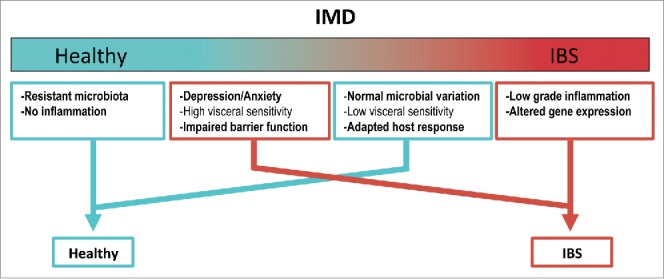
Proposed model on how microbial aberrations could explain the disease etiology in IBS. The PI-IBS patients can be stratified according to the IMD (or more generally the microbial composition) to those with IBS-like microbiota (red) and those with a composition resembling healthy individuals (turquoise). Objective measurements such as low-grade inflammation and altered gene expression correlated with the IMD. Psychological aspects, such as depression or anxiety do not correlate with the IMD and these patients, though diagnosed with IBS, show a healthy-like microbiota profile. The correlations between host response and microbiota aberration identified in our study are indicated in bold.
Conclusions
IBS has a heavy impact on the health care system43 and currently the patient diagnosis is based on subjective symptoms and exclusion of other gastrointestinal diseases. Results from our study18 as well as from others21 suggest that diagnosis based on symptoms alone is not optimal and should be accompanied with objective markers, such as the microbiota composition. We introduced an Index of Microbial Dysbiosis (IMD) that can potentially be used to objectively stratify patients into smaller groups where the patients with psychological basis of the syndrome could be separated from those with a peripheral gut abnormality. By using a relatively small but carefully phenotyped cohort we identified several associations between the IMD taxa, hosts' gene expression and clinical markers suggesting that impairment of intestinal barrier may underlie both immunological and microbiological deviations often associated with PI-IBS.18 Here we show that the microbial signatures that were observed in our British PI-IBS cohort were reproduced in an independent Swedish PI-IBS cohort (Fig. 1). This testifies for the robustness of these signatures in different cohorts and countries as well of the used microbial analysis pipeline. The main observed microbial aberration included an increased level of Bacteroides spp and decreased level of Uncultured Clostridiales, both recently identified as contrasting tipping point taxa that reflect alternative stable states of the gut ecosystem.17 Recently, similar bacterial signatures have been also detected in subjects prior to gastroenteritis,33,34 suggesting that perhaps the microbial composition already at the time of the initial infection predisposes certain subjects for PI-IBS. This is of significance as it allows not only to diagnose these subjects but also may provide basis for treatment. Dietary corrections represent the major approach in managing the IBS symptoms11 and it is expected that their efficacy depends on the individual's gut microbiota as microbial activity on food metabolism directly or indirectly contributes to many IBS symptoms.7 This hypothesis is supported by a pilot study among pediatric IBS patients in US.27,44 Of note, those children whose symptoms decreased during the test diet had lower abundance of Bacteroides and Dialister spp. that we found to typify the PI-IBS microbiota, further stressing the relevance of these organisms in IBS. We believe that the proposed model (Fig. 2) built on microbiota-based patient stratification will not only give insight into the observed microbiota heterogeneity among IBS patients as well as its partial overlap with healthy controls, but also represents a concrete step toward improved diagnostics and novel treatment options with improved efficacy.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We are grateful to our collaborators, notably Prof Robin Spiller MD and Prof Robert Jan Brummer MD, for helpful discussions.
Funding
This work was partly supported by the Finland Academy of Sciences (grants 137389, 141140 and 1272870) and the unrestricted Spinoza of the Netherlands Organization for Scientific Research.
References
- 1.Spiller R, Aziz Q, Creed F, Emmanuel A, Houghton L, Hungin P, Jones R, Kumar D, Rubin G, Trudgill N, et al.. Guidelines on the irritable bowel syndrome: mechanisms and practical management. Gut 2007; 56:1770-98; PMID:17488783; http://dx.doi.org/ 10.1136/gut.2007.119446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Longstreth GF, Thompson WG, Chey WD, Houghton LA, Mearin F, Spiller RC. Functional bowel disorders. Gastroenterology 2006; 130:1480-91; PMID:16678561; http://dx.doi.org/ 10.1053/j.gastro.2005.11.061 [DOI] [PubMed] [Google Scholar]
- 3.Camilleri M. Peripheral mechanisms in irritable bowel syndrome. N Engl J Med 2012; 367:1626-35; PMID:23094724; http://dx.doi.org/ 10.1056/NEJMra1207068 [DOI] [PubMed] [Google Scholar]
- 4.Saito YA, Mitra N, Mayer EA. Genetic approaches to functional gastrointestinal disorders. Gastroenterology 2010; 138:1276-85; PMID:20176021; http://dx.doi.org/ 10.1053/j.gastro.2010.02.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koloski NA, Jones M, Kalantar J, Weltman M, Zaguirre J, Talley NJ. The brain–gut pathway in functional gastrointestinal disorders is bidirectional: a 12-year prospective population-based study. Gut 2012; 61:1284-90; PMID:22234979; http://dx.doi.org/ 10.1136/gutjnl-2011-300474 [DOI] [PubMed] [Google Scholar]
- 6.Simren M, Barbara G, Flint HJ, Spiegel BMR, Spiller RC, Vanner S, Verdu EF, Whorwell PJ, Zoetendal EG. Intestinal microbiota in functional bowel disorders: a Rome foundation report. Gut 2013; 62:159-76; PMID:22730468; http://dx.doi.org/ 10.1136/gutjnl-2012-302167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rajilic-Stojanovic M, Jonkers DM, Salonen A, Hanevik K, Raes J, Jalanka J, de Vos WM, Manichanh C, Golic N, Enck P, et al.. Intestinal Microbiota And Diet in IBS: Causes, Consequences, or Epiphenomena? Am J Gastroenterol 2015; 110:278-87; PMID:25623659; http://dx.doi.org/ 10.1038/ajg.2014.427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salonen A, de Vos WM, Palva A. Gastrointestinal microbiota in irritable bowel syndrome: present state and perspectives. Microbiology 2010; 156:3205-15; PMID:20705664; http://dx.doi.org/ 10.1099/mic.0.043257-0 [DOI] [PubMed] [Google Scholar]
- 9.Whelan K. Probiotics and prebiotics in the management of irritable bowel syndrome: a review of recent clinical trials and systematic reviews. Curr Opin Clin Nutr Metab Care 2011; 14:581-7; PMID:21892075; http://dx.doi.org/ 10.1097/MCO.0b013e32834b8082 [DOI] [PubMed] [Google Scholar]
- 10.Menees SB, Maneerattannaporn M, Kim HM, Chey WD. The efficacy and safety of rifaximin for the irritable bowel syndrome: a systematic review and meta-analysis. Am J Gastroenterol 2012; 107:28-35; quiz 6; PMID:22045120; http://dx.doi.org/ 10.1038/ajg.2011.355 [DOI] [PubMed] [Google Scholar]
- 11.Staudacher HM, Irving PM, Lomer MCE, Whelan K. Mechanisms and efficacy of dietary FODMAP restriction in IBS. Nat Rev Gastro Hepat 2014; 11:256-66; http://dx.doi.org/ 10.1038/nrgastro.2013.259 [DOI] [PubMed] [Google Scholar]
- 12.Rodriguez LA, Ruigomez A. Increased risk of irritable bowel syndrome after bacterial gastroenteritis: cohort study. BMJ 1999; 318:565-6; PMID:10037630; http://dx.doi.org/ 10.1136/bmj.318.7183.565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chaudhary NA, Truelove SC. The irritable colon syndrome. A study of the clinical features, predisposing causes, and prognosis in 130 cases. The Quarterly journal of medicine 1962; 31:307-22; PMID:13878459 [PubMed] [Google Scholar]
- 14.Spiller R, Garsed K. Postinfectious irritable bowel syndrome. Gastroenterology 2009; 136:1979-88; PMID:19457422; http://dx.doi.org/ 10.1053/j.gastro.2009.02.074 [DOI] [PubMed] [Google Scholar]
- 15.Zoetendal EG, de Vos WM. Effect of diet on the intestinal microbiota and its activity. Curr Opin Gastroenterol 2014; 30:189-95; PMID:24457346; http://dx.doi.org/ 10.1097/MOG.0000000000000048 [DOI] [PubMed] [Google Scholar]
- 16.de Vos WM, Nieuwdorp M. Genomics: A gut prediction. Nature 2013; 498:48-9; PMID:23719383; http://dx.doi.org/ 10.1038/nature12251 [DOI] [PubMed] [Google Scholar]
- 17.Lahti L, Salojärvi J, Salonen A, Scheffer M, de Vos WM. Tipping elements in the human intestinal ecosystem. Nature communications 2014; 5:4344; PMID:25003530; http://dx.doi.org/ 10.1038/ncomms5344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jalanka-Tuovinen J, Salojärvi J, Salonen A, Immonen O, Garsed K, Kelly FM, Zaitoun A, Palva A, Spiller RC, de Vos WM. Faecal microbiota composition and host-microbe cross-talk following gastroenteritis and in postinfectious irritable bowel syndrome. Gut 2014; 63:1737-45; PMID:24310267; http://dx.doi.org/ 10.1136/gutjnl-2013-305994 [DOI] [PubMed] [Google Scholar]
- 19.Rajilic-Stojanovic M, Heilig HG, Molenaar D, Kajander K, Surakka A, Smidt H, de Vos WM. Development and application of the human intestinal tract chip, a phylogenetic microarray: analysis of universally conserved phylotypes in the abundant microbiota of young and elderly adults. Environ Microbiol 2009; 11:1736-51; PMID:19508560; http://dx.doi.org/ 10.1111/j.1462-2920.2009.01900.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Claesson MJ, O'Sullivan O, Wang Q, Nikkilä J, Marchesi JR, Smidt H, de Vos WM, Ross RP, O'Toole PW. Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine. PLoS ONE 2009; 4:e6669; PMID:19693277; http://dx.doi.org/ 10.1371/journal.pone.0006669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeffery IB, O'Toole PW, Ohman L, Claesson MJ, Deane J, Quigley EM, Simren M. An irritable bowel syndrome subtype defined by species-specific alterations in faecal microbiota. Gut 2012; 61:997-1006; PMID:22180058; http://dx.doi.org/ 10.1136/gutjnl-2011-301501 [DOI] [PubMed] [Google Scholar]
- 22.Patel P, Bercik P, Morgan DG, Bolino C, Pintos-Sanchez MI, Moayyedi P, Ford AC. Irritable bowel syndrome is significantly associated with somatisation in 840 patients, which may drive bloating. Aliment Pharmacol Ther 2015; 41:449-58; PMID:25586008; http://dx.doi.org/ 10.1111/apt.13074 [DOI] [PubMed] [Google Scholar]
- 23.Sundin J, Rangel I, Fuentes S, Heikamp-de Jong I, Hultgren-Hornquist E, de Vos WM, Brummer RJ. Altered faecal and mucosal microbial composition in post-infectious irritable bowel syndrome patients correlates with mucosal lymphocyte phenotypes and psychological distress. Aliment Pharmacol Ther 2015; 41:342-51; PMID:25521822; http://dx.doi.org/ 10.1111/apt.13055 [DOI] [PubMed] [Google Scholar]
- 24.Salonen A, Nikkilä J, Jalanka-Tuovinen J, Immonen O, Rajilic-Stojanovic M, Kekkonen RA, Palva A, de Vos WM. Comparative analysis of fecal DNA extraction methods with phylogenetic microarray: effective recovery of bacterial and archaeal DNA using mechanical cell lysis. J Microbiol Methods 2010; 81:127-34; PMID:20171997; http://dx.doi.org/ 10.1016/j.mimet.2010.02.007 [DOI] [PubMed] [Google Scholar]
- 25.Dore J, Ehrlich SD, Levenez F, Pelletier E, Alberti A, Bertrand L, Bork P, Costea PI, Sunagawa S, Guarner F, et al.. and IHMS Consortium IHMS_SOP 06 V1: Standard operating procedure for fecal samples DNA extraction, Protocol Q. International Human Microbiome Standards 2015. [Google Scholar]
- 26.Salojärvi J, Lahti L. Microbiome R package (v. 0.99.31). 2014. [Google Scholar]
- 27.Chumpitazi BP, Hollister EB, Oezguen N, Tsai CM, McMeans AR, Luna RA, Savidge TC, Versalovic J, Shulman RJ. Gut microbiota influences low fermentable substrate diet efficacy in children with irritable bowel syndrome. Gut microbes 2014; 5:165-75; PMID:24637601; http://dx.doi.org/ 10.4161/gmic.27923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kassinen A, Krogius-Kurikka L, Makivuokko H, Rinttilä T, Paulin L, Corander J, Malinen E, Apajalahti J, Palva A. The fecal microbiota of irritable bowel syndrome patients differs significantly from that of healthy subjects. Gastroenterology 2007; 133:24-33; PMID:17631127; http://dx.doi.org/ 10.1053/j.gastro.2007.04.005 [DOI] [PubMed] [Google Scholar]
- 29.Parkes GC, Rayment NB, Hudspith BN, Petrovska L, Lomer MC, Brostoff J, Whelan K, Sanderson JD. Distinct microbial populations exist in the mucosa-associated microbiota of sub-groups of irritable bowel syndrome. Neurogastroenterol Motil 2012; 24:31-9; PMID:22070725; http://dx.doi.org/ 10.1111/j.1365-2982.2011.01803.x [DOI] [PubMed] [Google Scholar]
- 30.Ponnusamy K, Choi JN, Kim J, Lee SY, Lee CH. Microbial community and metabolomic comparison of irritable bowel syndrome faeces. J Med Microbiol 2011; 60:817-27; PMID:21330412; http://dx.doi.org/ 10.1099/jmm.0.028126-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rajilic-Stojanovic M, Biagi E, Heilig HG, Kajander K, Kekkonen RA, Tims S, de Vos WM. Global and deep molecular analysis of microbiota signatures in fecal samples from patients with irritable bowel syndrome. Gastroenterology 2011; 141:1792-801; PMID:21820992; http://dx.doi.org/ 10.1053/j.gastro.2011.07.043 [DOI] [PubMed] [Google Scholar]
- 32.Haag L-M, Fischer A, Otto B, Plickert R, Kühl AA, Göbel UB, et al. Intestinal Microbiota Shifts towards Elevated Commensal Escherichia coli Loads Abrogate Colonization Resistance against Campylobacter jejuni in Mice. Plos One 2012; 7(5):e35988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dicksved J, Ellström P, Engstrand L, Rautelin H. Susceptibility to Campylobacter Infection Is Associated with the Species Composition of the Human Fecal Microbiota. mBio 2014; 5(5):e0212-14; PMID:25227462; http://dx.doi.org/ 10.1128/mBio.01212-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Youmans BP, Ajami NJ, Jiang ZD, Campbell F, Wadsworth WD, Petrosino JF, DuPont HL, Highlander SK. Characterization of the human gut microbiome during travelers' diarrhea. Gut Microbes 2015; 6(2):110-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chassaing B, Ley RE, Gewirtz AT. Intestinal epithelial cell toll-like receptor 5 regulates the intestinal microbiota to prevent low-grade inflammation and metabolic syndrome in mice. Gastroenterology 2014; 147:1363-77 e17; PMID:25172014; http://dx.doi.org/ 10.1053/j.gastro.2014.08.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salzman NH, Hung K, Haribhai D, Chu H, Karlsson-Sjoberg J, Amir E, Teggatz P, Barman M, Hayward M, Eastwood D, et al.. Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol 2010; 11:76-83; PMID:19855381; http://dx.doi.org/ 10.1038/ni.1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hansson GC. Role of mucus layers in gut infection and inflammation. Curr Opin Microbiol 2012; 15:57-62; PMID:22177113; http://dx.doi.org/ 10.1016/j.mib.2011.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Faure M, Moennoz D, Montigon F, Mettraux C, Breuille D, Ballevre O. Dietary threonine restriction specifically reduces intestinal mucin synthesis in rats. J Nutr 2005; 135:486-91; PMID:15735082 [DOI] [PubMed] [Google Scholar]
- 39.Wacklin P, Tuimala J, Nikkilä J, Tims S, Mäkivuokko H, Alakulppi N, et al.. Faecal Microbiota Composition in Adults Is Associated with the FUT2 Gene Determining the Secretor Status. Plos One 2014; 9(4): e94863; http://dx.doi.org/ 10.1371/journal.pone.0094863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moulds JM, Nowicki S, Moulds JJ, Nowicki BJ. Human blood groups: incidental receptors for viruses and bacteria. Transfusion 1996; 36:362-74; PMID:8623141; http://dx.doi.org/ 10.1046/j.1537-2995.1996.36496226154.x [DOI] [PubMed] [Google Scholar]
- 41.Kashyap PC, Marcobal A, Ursell LK, Smits SA, Sonnenburg ED, Costello EK, Higginbottom SK, Domino SE, Holmes SP, Relman DA, et al.. Genetically dictated change in host mucus carbohydrate landscape exerts a diet-dependent effect on the gut microbiota. Proc Natl Acad Sci U S A 2013; 110:17059-64; PMID:24062455; http://dx.doi.org/ 10.1073/pnas.1306070110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mayer EA, Tillisch K, Gupta A. Gut/brain axis and the microbiota. J Clin Invest 2015; 125:926-38; PMID:25689247; http://dx.doi.org/ 10.1172/JCI76304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hillilä MT, Färkkilä NJ, Färkkilä MA. Societal costs for irritable bowel syndrome - a population based study. Scandinavian Journal of Gastroenterology 2010; 45:582-91; PMID:20166844; http://dx.doi.org/ 10.3109/00365521003637211 [DOI] [PubMed] [Google Scholar]
- 44.Chumpitazi BP, Cope JL, Hollister EB, Tsai CM, McMeans AR, Luna RA, Versalovic J, Shulman RJ. Randomised clinical trial: gut microbiome biomarkers are associated with clinical response to a low FODMAP diet in children with the irritable bowel syndrome. Aliment Pharmacol Ther 2015; 42:418-27; PMID:26104013; http://dx.doi.org/ 10.1111/apt.13286 [DOI] [PMC free article] [PubMed] [Google Scholar]


