Abstract
The intestinal mucus is a pivotal part of our intestinal protection. It provides slow diffusion of protective molecules, trapping of luminal material as bacteria and smooth transport in the small intestine. In colon it restricts bacterial access to the epithelium limiting the responses to the enormous bacterial load present at this location. The development of these systems depends on the microbiota composition as seen in our recent study comparing the mucus phenotype in 2 colonies kept in different husbandries within the same SPF animal facility. One colony had impenetrable colonic mucus while the other colony had more penetrable mucus. The mucus phenotypes were transmitted via the microbiota and clear differences in its composition could be detected. Candidates associated with the different colonies were identified but the observed mucus difference could not be assigned to a specific bacterium.
Keywords: colon, commensal bacteria, intestine, ileum, mucus, Muc2, penetrability, proteomics
Abbreviations
- FISH
fluorescent in situ hybridization
- SPF
specific pathogen free.
The Microbiota Composition is Important to Form a Functional Protective Intestinal Mucus Layer
The mutualistic relationship with our microbiota has recently attracted a lot of attention, and understanding how we can manage this close contact in a balanced way is fascinating. The main site of interaction is the secreted mucus overlaying the epithelial surfaces. The mucus is a material with special properties that provides important functions for the tissue. It enables smooth transport through the gut, keeps a hydrated environment at the epithelium and traps material limiting hazardous exposure. The mucus has adapted to the requirements of different locations of the intestine serving more as a barrier that decreases diffusion in the small intestine and as a denser and more impenetrable barrier in the colon.1 Penetrability experiments in small intestinal mucus reveal that it can allow 2 µm beads to get through, but still bacteria are not frequently found in contact with the epithelium and are kept at a distance from the crypt openings.2 This is likely mediated by a number of mechanisms, with secreted antimicrobial proteins or peptides contributing considerably.3 The local environment close to the epithelium is microaerobic and contains reactive oxygen which also restricts bacteria from residing at the epithelial surface.4 Components of the immune system, with secreted IgA among others, are also involved in creating a balance in host-microbiota interactions.5 In colon, especially in the more distal region, the secreted mucus forms a different structure with a stacked laminated appearance. This mucus is impenetrable to beads 2 µm in size and is made up by newly secreted mucus from the goblet cells.6 The compact mucus barrier confines most bacteria to its luminal side, limiting direct bacterial-host interaction. Proteolytic and possibly other processes act on the mucus to change its structure when reaching further into the lumen, making the mucus more penetrable and giving access to bacteria within this outer layer. The altered properties at different intestinal locations occur although the mucus components are largely the same, built around the large, multimeric and extensively glycosylated structural component Muc2.7,8
The colonic impenetrable mucus layer is not well developed in germ-free mice, indicating that the formation of the protective mucus layer appears in response to the presence of bacteria.6 Bacterial components are also able to stimulate increased mucus release supporting this idea.9 This would argue for a general effect of bacteria, but our recent study revealed that the composition of the intestinal microbiota is important for the development of an impenetrable mucus layer.10 We have in that study compared 2 different colonies of C57BL/6 mice bred in 2 separate rooms of the same SPF facility.10 The microbiota of these mouse colonies (referred to as Room 1 and Room 2) was analyzed by 16S rDNA sequencing and was found to be divergent from each other, as we clearly demonstrated by principal coordinates analysis (PCoA). Both colonies appeared overall healthy and only a minor difference in body weight could be detected, with leaner mice in Room 2. The mucus in the small intestine of the 2 colonies had a similar phenotype, with penetrable and removable mucus of comparable thickness. In colon the mucus thickness was alike but the properties revealed unexpected differences between the rooms, with an impenetrable inner mucus layer in mice from Room 1 but with increased penetrability of the mucus in mice from Room 2. These latter mice, with a more penetrable inner mucus layer, also had more bacteria closer to the epithelium but still without excessive bacterial contact (Fig. 1). The arising question thus became: what is a normally developed mucus system? In non-inflamed human controls we have previously observed a thick mucus layer in colon that is impenetrable to 2 µm beads, which would argue for this phenotype to be most common in subjects with a natural variation in the microbiota composition. If this was also the prevailing phenotype in rodents was investigated using wild mice caught in their natural habitat. Many of these animals had a more complex gut community, including worms, and many of them displayed a thick, impenetrable mucus barrier. Development of a mucus layer that can separate most microbes from the epithelium could thus be considered to be the phenotype of choice to best maintain homeostasis. Although healthy, the animals in Room 2 displayed subtle changes in the epithelium with a slight increase in the number of proliferating cells, which could indicate an elevated inflammatory tone.
Figure 1.
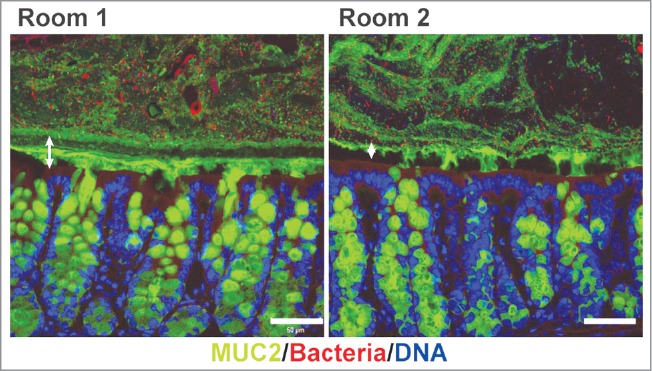
Bacteria penetration in colonic mucus of mice from the 2 rooms. Immunostaining of colon sections of mice from Room 1 and Room 2 detecting mucus through its main component, the Muc2 mucin (green), bacteria with FISH using a general 16S rDNA probe (red) and counterstaining of DNA with Hoechst (blue). The inner mucus layer in colon of mice from Room 1 restricts bacterial contact with the epithelium, while bacteria are clearly less separated from the epithelium by mucus in mice from Room 2. The dark area between the mucus and the epithelium is due to shrinkage during fixation. The inner mucus layers are marked by double arrows, scale bars are 50 µm.
That the changes observed in the mucus system were dependent on the microbiota was concluded by conventionalization experiments. The microbiota from the 2 colonies was transferred to germ-free mice, with undeveloped colonic mucus. The mucus phenotypes in these mice evolved to resemble the mucus in the microbiota donors: the microbiota from donors in Room 1 transmitted an impenetrable mucus layer, while microbiota from donors in Room 2 rendered the colonized mice with a more penetrable mucus layer. The microbiota was thus found to be a key factor in stimulating the development of an impenetrable mucus layer, but not due to the presence of bacteria in general. Triggering or impairing the system could be caused by specific bacteria or specific groups, something that is not well understood yet.
Bacteria with potential to affect the mucus properties
Microbiota sequencing was performed on samples from lumen and rinsed tissue containing attached mucus, both at distal small intestine and distal colon. Immunostainings did not reveal bacteria in the crypts or epithelium, indicating that bacteria identified in the mucus-containing samples most likely were mucus associated. Transformation to looser outer mucus allows penetrance of bacteria and some species find their niche here. One such candidate previously described is Akkermansia muciniphila that is common in the human intestine and have a niche in the mucus where it digests mucin carbohydrates. 11 Several beneficial effects have been associated with this bacterium. 12,13 In our study we did not detect A. muciniphila, but instead Mucispirillum was the main candidate having a mucus associated niche in the distal colon. This is a Gram-negative species within the phylum Deferribacteres.14 The importance of a mucus niche for this bacterium can be seen as its abundance is increased during mucus overproduction in helminth infections.15,16 Mucispirillum has been suggested to be associated with a healthy gut.17
Comparison of the microbiota composition in the 2 colonies revealed differences but only a few candidates possibly contributing to the formation of an impenetrable mucus layer were identified. Allobaculum was found to have an increased relative abundance in both small intestine and colon in animals from Room 1, but was also present in mice from Room 2. This species showed decreased abundance in animals fed a high-fat diet indicating association with health, but was in our study increased in mice that also displayed a more penetrable mucus phenotype when fed another diet. This would make it less unlikely to be a bacterium that promotes development of the mucus system. Another species almost exclusively found in mice from Room 1 with higher abundance in colon was Anaerostipes. This is an acetate-utilizing, butyrate-producing bacterium.18,19 Butyrate is a preferred energy source for colonic epithelial cells and the majority of the absorbed butyrate is metabolized by the epithelium.20 Local production by bacteria residing in the mucus with a relatively close proximity to the epithelium would contribute to utilization of this energy in the distal colon. Impaired butyrate absorption and capacity of the intestinal mucosa to oxidize butyrate have been associated with both intestinal inflammation and carcinogenesis.20
Bacteria with increased abundance associated with penetrable mucus were Desulfovibrio, Bacteroides, Parabacteroides, Prevotella and bacteria from the Helicobacteriaceae family and TM7 phyla. Several of these have been linked to intestinal inflammation.21-26 Of these, Prevotella, Desulfovibrio and an unknown genus in the Helicobacteriaceae family were almost only found in Room 2. Prevotella has the ability to degrade the sulfated mucin glycans, a modification that is more abundant in distal colon, which could affect the mucus layer.27,28 Desulfovibrio derive most of its metabolic energy from sulfate reduction, cooperating in bacterial community metabolism and generating hydrogen sulfide,29 which is a highly toxic compound that can damage the epithelium and impact health.30 Helicobacter species as H. hepaticus are known to trigger inflammation in susceptible hosts 31 and can possibly get through the mucus and access the epithelium by constitutive or induced systems as shown for H. pylori.32,33
There could be positive or negative effects of specific bacteria on the mucus, but more likely such a complex system is based on combinations of both detrimental and cooperative relationships between the host and the different members of the microbiota. In Room 1 some candidates that could have beneficial effects on mucus development have been identified in our study.10 In addition, a number of species that in contrast were more exclusive in Room 2 and therefore could have detrimental outcomes on the mucus were also suggested. These bacterial effects could lead to various consequences in the mucus, such as to trigger excessive responses that hamper the mucus development, affect the extracellular environment with impact on mucus layer formation or promote increased mucus degradation allowing more bacteria penetration.
Mucus proteome alterations in response to bacteria
The effect of bacteria on the mucus was addressed by performing proteomics on isolated mucus from ileal and colonic explants. The protein profile from Room 1 and Room 2 was compared and was found to be overall similar.10 Analyzing the data with a focus on proteins only present in one of the rooms identified some room-specific components (Fig. 2). These proteins could be of importance for the mucus phenotype but could also reflect the detection threshold of the method. In ileal mucus of mice from Room 1 we found 26 proteins that were not detected in Room 2, with 9 additional proteins with shared expression in distal colon of Room 1. Among the Room 1-ileum exclusive proteins we could find Trefoil factor 3, Angiogenin 4, Alpha-defensin 20, Murinoglobin 1 and Protein-glutamine gamma-glutamyltransferase 2, which could all have an impact in mucus formation or bacterial control. In ileal mucus from Room 2 we identified 20 unique proteins but these were mostly known to be intracellular and an effect on mucus structure was less obvious. When performing a pathway analysis the KEGG term “bacterial invasion of epithelial cells” was significant in the ileum of mice in Room 1 (p = 7.579 × 10e3) but not in Room 2. This term could indicate a response to the presence of bacteria leading to adaptation of the tissue at colonization. This could give the tissue a better barrier toward the microbiota to manage the colonization.
Figure 2.
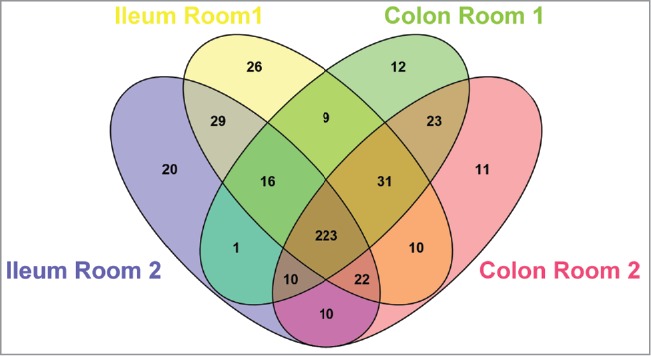
Protein composition of mucus from mice of the 2 colonies. Venn diagram of the proteins identified in mucus of ileum and colon in mice from Room 1 and Room 2, showing the number of shared and unique components.
In colonic mucus, 12 proteins were specific for Room 1 at this location and 11 for Room 2. Only a couple of possibly interesting candidates, Leukotriene A4 hydrolase and Serine protease inhibitor kazal-type 4, associated with protease activity, were found specifically in Room 1. We could thus not find specific proteins that would account for the variation seen in the penetrability phenotypes of ileum and colon in both rooms. However, some of the protein differences found could relate to a different status quo of the immune system in the animals from Room 1, indirectly pointing to the effects of the microbiota in the formation of an impenetrable mucus barrier, as part of the defense mechanisms to keep the commensals at a safe distance from the epithelium.
In conclusion, our data display a close relationship between the microbiota composition and mucus properties. Some bacteria that might stimulate mucus layer formation have been suggested but this needs to be further studied in detail. Besides, some bacteria that have been previously associated with inflammation were observed in mice in Room 2, which had a penetrable mucus layer resulting in a modestly increased bacterial contact with the epithelium. In these mice a slightly elevated inflammatory tone was seen and could be a result of enhanced responses to the microbiota or specific bacteria. Increased bacterial exposure of the epithelium and mucus defects have been observed in several mouse models of colitis and in ulcerative colitis patients, demonstrating a link between mucus protection and intestinal health.34 Our results suggest that the interaction at the intestinal surface is not as simple as “a single species provides all benefits” or “a single species is all detrimental.” The complexity of the microbiota and the host responses occurring at all different levels must be accounted for to fully understand this intricate relationship.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgment
We acknowledge the CCI unit at the University of Gothenburg for assistance with confocal microscopy.
Funding
The original work was supported by the Swedish Research Council (no. 7461, 21027, 22220-01-5) The Swedish Cancer Foundation, The Knut and Alice Wallenberg Foundation, IngaBritt and Arne Lundberg Foundation, Sahlgren’s University Hospital (LUA-ALF), Wilhelm and Martina Lundgren’s Foundation, Assar Gabrielsson’s foundation, Clas Groschinskys foundation, Torsten och Ragnar Söderbergs Stiftelser, The Sahlgrenska Academy, National Institute of Allergy and Infectious Diseases (U01AI095473, U01AI095776-03:9006862), the content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH, and The Swedish Foundation for Strategic Research The Mucus-Bacteria-Colitis Center (MBC) of the Innate Immunity Program.
References
- 1.Johansson ME, Sjovall H, Hansson GC. The gastrointestinal mucus system in health and disease. Nat Rev Gastroenterol Hepatol 2013; 10:352-61; PMID:23478383; http://dx.doi.org/ 10.1038/nrgastro.2013.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ermund A, Schutte A, Johansson ME, Gustafsson JK, Hansson GC. Studies of mucus in mouse stomach, small intestine, and colon. I. Gastrointestinal mucus layers have different properties depending on location as well as over the Peyer's patches. Am J Physiol Gastrointest Liver Physiol 2013; 305:G341-7; PMID:23832518; http://dx.doi.org/ 10.1152/ajpgi.00046.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mukherjee S, Hooper LV. Antimicrobial defense of the intestine. Immunity 2015; 42:28-39; PMID:25607457; http://dx.doi.org/ 10.1016/j.immuni.2014.12.028 [DOI] [PubMed] [Google Scholar]
- 4.Espey MG. Role of oxygen gradients in shaping redox relationships between the human intestine and its microbiota. Free Radic Biol Med 2013; 55:130-140; PMID:23127782; http://dx.doi.org/ 10.1016/j.freeradbiomed.2012.10.554 [DOI] [PubMed] [Google Scholar]
- 5.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science 2012; 336:1268-73; PMID:22674334; http://dx.doi.org/ 10.1126/science.1223490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johansson ME, Phillipson M, Petersson J, Velcich A, Holm L, Hansson GC. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci U S A 2008; 105:15064-9; PMID:18806221; http://dx.doi.org/ 10.1073/pnas.0803124105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ambort D, Johansson ME, Gustafsson JK, Ermund A, Hansson GC. Perspectives on mucus properties and formation-lessons from the biochemical world. Cold Spring Harb Perspect Med 2012; 2; pii: a014159; PMID:23125206; http://dx.doi.org/ 10.1101/cshperspect.a014159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodriguez-Pineiro AM, Bergstrom JH, Ermund A, Gustafsson JK, Schutte A, Johansson ME, Hansson GC. Studies of mucus in mouse stomach, small intestine, and colon. II. Gastrointestinal mucus proteome reveals Muc2 and Muc5ac accompanied by a set of core proteins. Am J Physiol Gastrointest Liver Physiol 2013; 305:G348-56; PMID:23832517; http://dx.doi.org/ 10.1152/ajpgi.00047.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petersson J, Schreiber O, Hansson GC, Gendler SJ, Velcich A, Lundberg JO, Roos S, Holm L, Phillipson M. Importance and regulation of the colonic mucus barrier in a mouse model of colitis. Am J Physiol Gastrointest Liver Physiol 2011; 300:G327-33; PMID:21109593; http://dx.doi.org/ 10.1152/ajpgi.00422.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jakobsson HE, Rodriguez-Pineiro AM, Schutte A, Ermund A, Boysen P, Bemark M, Sommer F, Backhed F, Hansson GC, Johansson ME. The composition of the gut microbiota shapes the colon mucus barrier. EMBO Rep 2015; 16:164-77; PMID:25525071; http://dx.doi.org/ 10.15252/embr.201439263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Derrien M, Collado MC, Ben-Amor K, Salminen S, de Vos WM. The Mucin degrader Akkermansia muciniphila is an abundant resident of the human intestinal tract. Appl Environ Microbiol 2008; 74:1646-8; PMID:18083887; http://dx.doi.org/ 10.1128/AEM.01226-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM, de Vos WM, Cani PD. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci U S A 2013; 110:9066-71; PMID:23671105; http://dx.doi.org/ 10.1073/pnas.1219451110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kang CS, Ban M, Choi EJ, Moon HG, Jeon JS, Kim DK, Park SK, Jeon SG, Roh TY, Myung SJ, Gho YS, Kim JG, Kim YK. Extracellular vesicles derived from gut microbiota, especially Akkermansia muciniphila, protect the progression of dextran sulfate sodium-induced colitis. PLoS ONE 2013; 8:e76520; PMID:24204633; http://dx.doi.org/ 10.1371/journal.pone.0076520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robertson BR, O'Rourke JL, Neilan BA, Vandamme P, On SL, Fox JG, Lee A. Mucispirillum schaedleri gen. nov., sp. nov., a spiral-shaped bacterium colonizing the mucus layer of the gastrointestinal tract of laboratory rodents. Int J Syst Evol Microbiol 2005; 55:1199-204; PMID:15879255; http://dx.doi.org/ 10.1099/ijs.0.63472-0 [DOI] [PubMed] [Google Scholar]
- 15.Li RW, Wu S, Li W, Navarro K, Couch RD, Hill D, Urban JF Jr. Alterations in the porcine colon microbiota induced by the gastrointestinal nematode Trichuris suis. Infect Immun 2012; 80:2150-7; PMID:22493085; http://dx.doi.org/ 10.1128/IAI.00141-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Houlden A, Hayes KS, Bancroft AJ, Worthington JJ, Wang P, Grencis RK, Roberts IS. Chronic Trichuris muris Infection in C57BL/6 Mice Causes Significant Changes in Host Microbiota and Metabolome: Effects Reversed by Pathogen Clearance. PLoS ONE 2015; 10:e0125945; PMID:25938477; http://dx.doi.org/ 10.1371/journal.pone.0125945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Belzer C, Gerber GK, Roeselers G, Delaney M, DuBois A, Liu Q, Belavusava V, Yeliseyev V, Houseman A, Onderdonk A, Cavanaugh C, Bry L. Dynamics of the microbiota in response to host infection. PLoS ONE 2014; 9:e95534; PMID:25014551; http://dx.doi.org/ 10.1371/journal.pone.0095534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwiertz A, Hold GL, Duncan SH, Gruhl B, Collins MD, Lawson PA, Flint HJ, Blaut M. Anaerostipes caccae gen. nov., sp. nov., a new saccharolytic, acetate-utilising, butyrate-producing bacterium from human faeces. Syst Appl Microbiol 2002; 25:46-51; PMID:12086188; http://dx.doi.org/ 10.1078/0723-2020-00096 [DOI] [PubMed] [Google Scholar]
- 19.Eeckhaut V, Van IF, Croubels S, De BS, Haesebrouck F, Ducatelle R, Louis P, Vandamme P. Butyrate production in phylogenetically diverse Firmicutes isolated from the chicken caecum. Microb Biotechnol 2011; 4:503-12; PMID:21375722; http://dx.doi.org/ 10.1111/j.1751-7915.2010.00244.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, Brummer RJ. Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther 2008; 27:104-19; PMID:17973645; http://dx.doi.org/ 10.1111/j.1365-2036.2007.03562.x [DOI] [PubMed] [Google Scholar]
- 21.Garrett WS, Gallini CA, Yatsunenko T, Michaud M, DuBois A, Delaney ML, Punit S, Karlsson M, Bry L, Glickman JN, Gordon JI, Onderdonk AB, Glimcher LH. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe 2010; 8:292-300; PMID:20833380; http://dx.doi.org/ 10.1016/j.chom.2010.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gophna U, Sommerfeld K, Gophna S, Doolittle WF, Veldhuyzen van Zanten SJ. Differences between tissue-associated intestinal microfloras of patients with Crohn's disease and ulcerative colitis. J Clin Microbiol 2006; 44:4136-41; PMID:16988016; http://dx.doi.org/ 10.1128/JCM.01004-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verma R, Verma AK, Ahuja V, Paul J. Real-time analysis of mucosal flora in patients with inflammatory bowel disease in India. J Clin Microbiol 2010; 48:4279-82; PMID:20861337; http://dx.doi.org/ 10.1128/JCM.01360-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Devkota S, Wang Y, Musch MW, Leone V, Fehlner-Peach H, Nadimpalli A, Antonopoulos DA, Jabri B, Chang EB. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10-/-mice. Nature 2012; 487:104-8; PMID:22722865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Noor SO, Ridgway K, Scovell L, Kemsley EK, Lund EK, Jamieson C, Johnson IT, Narbad A. Ulcerative colitis and irritable bowel patients exhibit distinct abnormalities of the gut microbiota. BMC Gastroenterol 2010; 10:134; PMID:21073731; http://dx.doi.org/ 10.1186/1471-230X-10-134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuehbacher T, Rehman A, Lepage P, Hellmig S, Folsch UR, Schreiber S, Ott SJ. Intestinal TM7 bacterial phylogenies in active inflammatory bowel disease. J Med Microbiol 2008; 57:1569-76; PMID:19018031; http://dx.doi.org/ 10.1099/jmm.0.47719-0 [DOI] [PubMed] [Google Scholar]
- 27.Wright DP, Rosendale DI, Robertson AM. Prevotella enzymes involved in mucin oligosaccharide degradation and evidence for a small operon of genes expressed during growth on mucin. FEMS Microbiol Lett 2000; 190:73-9; PMID:10981693; http://dx.doi.org/ 10.1111/j.1574-6968.2000.tb09265.x [DOI] [PubMed] [Google Scholar]
- 28.Rho JH, Wright DP, Christie DL, Clinch K, Furneaux RH, Roberton AM. A novel mechanism for desulfation of mucin: identification and cloning of a mucin-desulfating glycosidase (sulfoglycosidase) from Prevotella strain RS2. J Bacteriol 2005; 187:1543-51; PMID:15716424; http://dx.doi.org/ 10.1128/JB.187.5.1543-1551.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carbonero F, Benefiel AC, Alizadeh-Ghamsari AH, Gaskins HR. Microbial pathways in colonic sulfur metabolism and links with health and disease. Front Physiol 2012; 3:448; PMID:23226130; http://dx.doi.org/ 10.3389/fphys.2012.00448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carbonero F, Benefiel AC, Gaskins HR. Contributions of the microbial hydrogen economy to colonic homeostasis. Nat Rev Gastroenterol Hepatol 2012; 9:504-18; PMID:22585131; http://dx.doi.org/ 10.1038/nrgastro.2012.85 [DOI] [PubMed] [Google Scholar]
- 31.Fox JG, Ge Z, Whary MT, Erdman SE, Horwitz BH. Helicobacter hepaticus infection in mice: models for understanding lower bowel inflammation and cancer. Mucosal Immunol 2011; 4:22-30; PMID:20944559; http://dx.doi.org/ 10.1038/mi.2010.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sterzenbach T, Bartonickova L, Behrens W, Brenneke B, Schulze J, Kops F, Chin EY, Katzowitsch E, Schauer DB, Fox JG, Suerbaum S, Josenhans C. Role of the Helicobacter hepaticus flagellar sigma factor FliA in gene regulation and murine colonization. J Bacteriol 2008; 190:6398-408; PMID:18689480; http://dx.doi.org/ 10.1128/JB.00626-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Celli JP, Turner BS, Afdhal NH, Keates S, Ghiran I, Kelly CP, Ewoldt RH, McKinley GH, So P, Erramilli S, Bansil R. Helicobacter pylori moves through mucus by reducing mucin viscoelasticity. Proc Natl Acad Sci U S A 2009; 106:14321-6; PMID:19706518; http://dx.doi.org/ 10.1073/pnas.0903438106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johansson ME, Gustafsson JK, Holmen-Larsson J, Jabbar KS, Xia L, Xu H, Ghishan FK, Carvalho FA, Gewirtz AT, Sjovall H, Hansson GC. Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut 2014; 63:281-91; PMID:23426893 [DOI] [PMC free article] [PubMed] [Google Scholar]


