ABSTRACT
PIWI-interacting RNAs (piRNAs), a subset of small non-coding RNAs enriched in animal gonads, repress transposons by assembling with PIWI proteins to form potent gene-silencing RNP complexes, piRISCs. Accumulating evidence suggests that piRNAs are produced through three interdependent pathways; the de novo primary pathway, the ping-pong pathway, and the phased primary pathway. The de novo primary pathway in Drosophila ovaries produces primary piRNAs for two PIWI members, Piwi and Aub. Aub then initiates the ping-pong pathway to produce secondary piRNAs for AGO3. AGO3-slicer dependent cleavage subsequently produces secondary piRNAs for Aub. Trailer products of AGO3-slicer activity are consumed by the phased primary pathway to increase the Piwi-bound piRNA population. All these pathways are regulated by a number of piRNA factors in a highly coordinated fashion. Recent studies show that two Tudor-domain containing piRNA factors, Krimper (Krimp) and Qin/Kumo, play crucial roles in making Aub-AGO3 heterotypic ping-pong robust. This maintains the levels of piRNAs loaded onto Piwi and Aub to efficiently repress transposons at transcriptional and post-transcriptional levels, respectively.
Keywords: krimp, piRNA, PIWI, Qin, ping-pong, Tudor domain, Transposon
piRNAs are a germline specific class of 23–30 nucleotide (nt) small RNAs. Together with PIWI proteins, piRNAs repress the expression of transposons to prevent their invasion into the host genome and to ensure faithful transmission of the germline genome to the next generation.1-9 Most piRNAs are complementary to transposon transcripts enabling PIWI proteins to be guided to them.1-9 Drosophila has 3 PIWI proteins, Piwi, Aubergine (Aub) and AGO3. Aub and AGO3 accumulate at a perinuclear cytoplasmic region called the nuage (French for ‘cloud’), whereas Piwi localizes in the nucleus.6-10 PIWI proteins contain symmetrical dimethylarginine (sDMA) residues in their arginine-glycine (RG) and/or arginine-alanine (RA) rich regions, which are amino-terminally located.11-13 The sDMA modification is mediated by the arginine methyltransferase, Dart5 (also known as PRMT5 or Capsuléen), and is necessary for the production of piRNAs.11-13 A number of genes involved in piRNA pathways have been identified by genetic screenings in Drosophila.14-17 Many of them have Tud domains, which are well known to recognize methylated arginine residues, including sDMAs.18,19 Tud domain-containing piRNA factors interact with sDMA-modified PIWI proteins and play critical roles in piRNA pathways.18,19
Three piRNA biogenesis pathways in Drosophila germline cells
Drosophila germline cells produce piRNAs by at least 3 mechanisms; the de novo primary pathway, the ping-pong pathway and the phased primary pathway (Fig. 1).20-24 The production of de novo primary piRNAs is initiated by the processing of piRNA precursor transcripts that are transcribed from piRNA clusters, distinct genomic regions composed of a large number of truncated transposons.21,25 They are then loaded onto Piwi and Aub, but not AGO3, in the cytoplasm.26,27 Orientation of transposon fragments inserted into germline piRNA clusters does not depend on the polarity of transcription of the locus. In addition, transcription occurs at both strands of these loci; therefore, it is still not clear how the antisense bias observed for primary piRNAs is initially generated. The precursor transcripts from piRNA clusters are processed by an endonuclease, Zucchini (Zuc), to produce mature piRNAs.22,28-31 Piwi is then imported into the nucleus where it targets nascent transposon transcripts and represses transcription of their loci by inducing local heterochromatin formation.22,32-35 In contrast, both AGO3 and Aub exhibit endonuclease activity, termed slicer activity,20,36 to cleave transposon transcripts in the cytoplasm. The slicer activity of Aub directs cleavage of transposon mRNAs between nucleotides 10 and 11, measured from the 5′ end of its bound piRNA. This initiates the ping-pong pathway.20,21,26,27,36 The cleavage products are then transferred onto AGO3 with the help of a DEAD-box helicase, Vasa.37,38 Thus, most AGO3-bound piRNAs are sense orientated, and the slicer activity of AGO3 directs cleavage of antisense transposon transcripts.20,21,26,27 The cleavage products produced by AGO3 are transferred onto Aub, which may require another RNA helicase, yet to be identified,38 and are further processed into mature antisense secondary piRNAs.20,21,26,27 Aub can initiate another round of cleavage of sense transcripts to produce AGO3-bound secondary piRNAs.20,21,26,27 Therefore, the ping-pong pathway is a slicer-mediated chain reaction to consume transposon transcripts and produce piRNAs. Mutual amplification of sense and antisense piRNAs by Aub-AGO3 heterotypic ping-pong defines the 5′ end of complementary piRNAs that overlap by 10 nt from their 5′ ends.20,21
Figure 1.
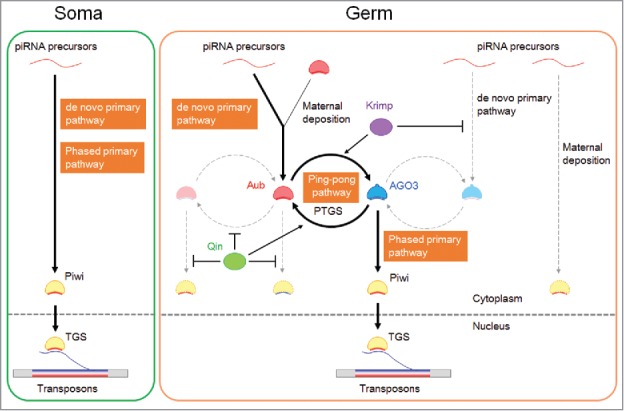
piRNA pathways in the Drosophila ovary. In germline cells, antisense piRNAs are produced and loaded onto Aub by the de novo primary pathway or through maternal deposition. Aub then triggers the ping-pong pathway to produce sense piRNAs bound to AGO3 and own antisense piRNAs by consuming transposon transcripts (post-transcriptional gene silencing; PTGS). AGO3 initiates the phased primary pathway to produce antisense piRNAs bound to Piwi. Very few Piwi-bound piRNAs are maternally deposited. Piwi translocates to the nucleus, where it provokes transcriptional gene silencing (TGS). In soma, only Piwi is expressed, and the de novo primary and phased primary pathways operate to produce antisense piRNAs bound to Piwi. Krimp promotes the Aub-AGO3 heterotypic ping-pong pathway and blocks de novo primary piRNA loading onto AGO3. Qin inhibits the Aub-Aub homotypic ping-pong pathway and the production of Piwi-bound phased primary piRNAs triggered by Aub. Sense and antisense piRNAs are indicated in blue and red, respectively.
The trailer sequences of AGO3 cleavage products from the ping-pong cycle are further processed by Zuc and then loaded onto Piwi (Fig. 1).23,24,39,40 Zuc defines both the 5′ and 3′ ends of the phased primary piRNAs.23,24 Piwi-bound phased piRNAs are antisense orientated.23,24,39,40 In addition, most Piwi-bound primary piRNAs are produced via the phased primary pathway, and very few piRNAs are produced via the de novo primary piRNA pathway in Drosophila ovarian germ cells, unlike in ovarian somatic cells where only the primary pathway operates because these cells lack both AGO3 and Aub.7-9,39,40 Phased primary piRNAs are also produced even in the absence of Aub or AGO3-slicer activity, via an alternative pathway, which produces pseudo-secondary piRNAs.39 Once phased primary piRNAs are loaded, Piwi translocates into the nucleus to repress transposons at the transcriptional level.22,32-35,39,40 Thus, AGO3 silences transposons indirectly by not only yielding Aub-bound antisense secondary piRNAs via the ping-pong pathway, but also by triggering Zuc-dependent phased piRNA production. Without AGO3, the de novo primary pathway is not affected, but neither the ping-pong pathway coupled with Aub nor the phased primary pathway is initiated, leading to a decrease in the number of antisense piRNAs.26,27 Loss of Aub function also shuts down the ping-pong pathway coupled with AGO3, and results in further impairment of Piwi-bound phased primary piRNA production.26,27 In the absence of Piwi, de novo primary piRNAs are loaded onto Aub, and initiate the ping-pong pathway with AGO3 to produce secondary piRNAs, but the phased primary piRNAs are not accumulated.23,24,39,40
Krimp
Krimper (Krimp) is a Tud domain-containing piRNA factor in Drosophila, and is localized in the nuage.10,41 krimp mutant ovaries show the typical phenotype of germline piRNA factor mutants; piRNA levels are decreased and transposons are derepressed.10,26 In krimp mutant ovaries, the cellular localization patterns of Piwi and Aub are not affected, but AGO3 is not localized in the nuage, where it is believed that the ping-pong pathway operates.10,26,41 These observations suggest that Krimp is involved in the ping-pong pathway by affecting the activity of AGO3.
Recently we showed that Krimp interacts with AGO3 through its Tud domain (Krimp-Tud).42 Surprisingly, Krimp-bound AGO3 does not contain sDMA, unlike other Tud containing proteins, most of which preferably associate with their substrates upon mono-methyl and/or di-methyl modification.42 We also found that Krimp-bound AGO3 is free from piRNAs.42 Simultaneously, Aravin's group reported that Krimp interacts not only with AGO3 but also with Aub through its Tud domain, but in contrast to the AGO3 interaction, that between Krimp and Aub is sDMA-dependent.43 Additionally, Aravin's group found that Krimp can form a homodimer through its amino-terminal region called the Krimp domain, which contains a coiled-coil domain.42,43 These results suggest a possible model for a molecular function of Krimp in the ping-pong pathway; Krimp binds unmodified AGO3 in the cytosol and then brings it to the nuage where AGO3 is sDMA-modified by Dart5. Krimp then further assembles ping-pong piRNA processing complexes through interaction with sDMA-modified Aub (sDMA-Aub with Krimp-Krimp-AGO3) to promote the sDMA modification of AGO3 and sense secondary piRNA loading onto AGO3 (Fig. 2).42,43
Figure 2.
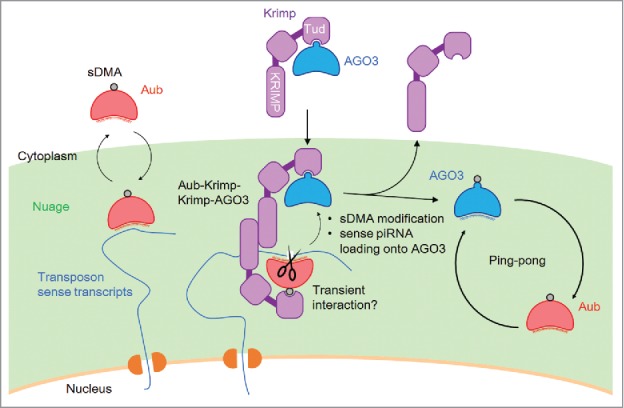
Model for Krimp functions in the ping-pong pathway. Krimp interacts with unmethylated and piRNA-free AGO3 to recruit it to the nuage, where Krimp also interacts transiently with Aub to promote sDMA modification and sense secondary piRNA loading onto AGO3. Aub shuttles between the cytoplasm and nuage. The recognition of target transcripts by the piRNA guide stably recruits Aub to nuage. sDMA-modified AGO3 is released from Krimp, and then initiates the ping-pong cycle with Aub. Sense and antisense piRNAs are indicated in blue and red, respectively.
A loss-of-function mutation in krimp caused complete loss of AGO3-bound sense piRNAs.42 As a consequence, there was a two-fold reduction in the level of Aub-bound antisense piRNAs.42 In sharp contrast, Aub-bound sense piRNAs were increased in krimp mutant ovaries. This suggests the presence of an aberrant Aub-Aub homotypic ping-pong reaction. However, fewer secondary piRNAs were detected in the mutant ovaries, and so transposons were only weakly repressed in the mutant ovaries. It seems that homotypic ping-pong is so inefficient in the production of secondary piRNAs that Drosophila ovaries have equipped Krimp to dominate Aub-AGO3 heterotypic ping-pong over homotypic ping-pong.42
Krimp is also expressed in ovarian soma, where it forms unique cytoplasmic granules named Krimp bodies.42,44 Unlike in the germline ping-pong pathway, Krimp is dispensable in the somatic primary piRNA pathway. However, AGO3 ectopically expressed in the ovarian somatic OSC cell line is sequestered to Krimp bodies; therefore, Krimp still has the ability to bind AGO3 in ovarian somatic cells.42 Under these conditions AGO3 is devoid of primary piRNAs.42 However, AGO3 starts to be associated with primary piRNAs when Krimp is depleted by RNAi, suggesting that Krimp acts as a regulator for AGO3, preventing it from being loaded with primary piRNAs.42 This function of Krimp is not unique in ovarian somas because Krimp sequesters endogenous AGO3 to Krimp bodies in aub and other germline piRNA factor mutant nurse cells.42,44 These findings suggest a model for dual functions of Krimp in the pong-pong pathway: Krimp promotes the Aub-AGO3 heterotypic ping-pong pathway and blocks de novo primary piRNA loading onto AGO3 (Fig. 1).
Qin/Kumo
Qin, also known as Kumo (Japanese for ‘cloud’), is also a Tud-domain protein involved in Drosophila germline piRNA production. It has a RING domain, two B-Box domains, and five Tud domains.45,46 Qin physically interacts with Aub through its Tud domains, and the interaction depends on Aub-sDMA modification.46 Qin localizes to the nuage, where it facilitates Aub-AGO3 interaction.45 Although qin loss-of-function in the ovaries does not change the total amount of piRNAs, the ratio of sense to antisense piRNAs increases; this is because of an increase in sense piRNAs in Aub-bound piRNA populations. Further analyses indicated that, in qin ovaries, secondary piRNA production seemed to switch from ‘productive’ Aub-AGO3 heterotypic ping-pong to ‘unproductive’ Aub-Aub homotypic ping-pong.45,47 Thus, it is concluded that Qin functions in blocking Aub-Aub homotypic ping-pong. This is likely to result in a substantial supply of antisense secondary piRNAs in nurse cells for efficiently destroying transposon mRNAs.45,47
In nurse cells of Drosophila ovaries, AGO3 initiates the production of the phased primary piRNAs that are loaded onto Piwi.39 In ago3 and qin double mutant ovaries, the levels of Aub-Aub homotypic ping-pong pairs and Piwi-bound piRNAs were higher than those in ago3 single mutant ovaries.39 Piwi in the double mutants associated preferably with sense piRNAs, as in ago3 mutant ovaries.39 Under circumstances where Aub-Aub homotypic ping-pong is somewhat dominant, cleavage products of AGO3 decrease, resulting in fewer phased piRNAs loaded onto Piwi. This would certainly increase the expression levels of transposons. Thus, to prevent this, Qin plays a role in suppressing Aub-Aub homotypic ping-pong. In this way, the amount of phased piRNAs is maintained and transposons are effectively repressed in Drosophila ovarian germ cells (Fig. 1).39
Conclusions
Each PIWI protein can harbor any piRNA via the de novo primary pathway, the ping-pong pathway, and probably others; however each pathway has precisely defined piRNAs and partner PIWI proteins.7-9,39,42 The cleavage of long antisense transcripts by AGO3 with sense piRNAs can initiate the production of antisense secondary piRNAs bound to Aub and further drives the production of antisense phased primary piRNAs bound to Piwi.23,24,39 Aub and Piwi, but not AGO3, can accumulate at the pole plasm in developing oocytes,21,48 implying the deposition of maternal Aub- and Piwi-bound piRNAs in progeny germline cells. In fact, Aub bound to antisense maternal piRNAs is indispensable to initiate the ping-pong pathway, and further antisense maternal piRNAs bound to Piwi are thought to be an important mark of source piRNA clusters on progeny genomes.49 Aub, which mainly binds to antisense piRNAs either derived from maternal deposition or the de novo primary pathway, not only directly cleaves transposon transcripts but is also indirectly involved in the production of Piwi-bound phased primary piRNAs by amplifying AGO3-bound sense piRNAs.21-26,39 Most Piwi-bound piRNAs appear to be produced via the phased primary pathway, and not the de novo primary pathway in Drosophila ovarian germline cells.39 In addition, extremely few Piwi-bound piRNAs are maternally deposited.39 Thus, the Aub-AGO3 heterotypic ping-pong pathway is essentially the only way to amplify the Piwi-bound antisense piRNAs in Drosophila ovarian germline cells. Two Tud domain piRNA factors, Krimp and Qin/Kumo, play respective roles to regulate the ping-pong pathway, and these proteins can synergistically make the Aub-AGO3 heterotypic ping-pong robust, thereby enforcing an antisense bias on germline piRNA pools.39,42 In addition to Krimp and Qin/Kumo, other piRNA factors and Tud domain proteins also collaborate to define the piRNA pathways. The molecular functions of these additional piRNA factors are unclear and need to be determined.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the Ministry of Education, Culture, Sports, Science and Technology (MEXT) and Grants-in-Aid for Scientific Research to K.S., Y.W.I., H.S. and M.C.S.
References
- 1.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet 2009; 10:94-108; PMID:19148191; http://dx.doi.org/ 10.1038/nrg2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 2009; 10:126-39; PMID:19165215; http://dx.doi.org/ 10.1038/nrm2632 [DOI] [PubMed] [Google Scholar]
- 3.Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell 2009; 136:656-68; PMID:19239887; http://dx.doi.org/ 10.1016/j.cell.2009.01.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siomi H, Siomi MC. On the road to reading the RNA-interference code. Nature 2009; 457:396-404; PMID:19158785; http://dx.doi.org/ 10.1038/nature07754 [DOI] [PubMed] [Google Scholar]
- 5.Thomson T, Lin H. The biogenesis and function of PIWI proteins and piRNAs: progress and prospect. Annu Rev Cell Dev Biol 2009; 25:355-76; PMID:19575643; http://dx.doi.org/ 10.1146/annurev.cellbio.24.110707.175327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Senti KA, Brennecke J. The piRNA pathway: a fly's perspective on the guardian of the genome. Trends Genet 2010; 26:499-509; PMID:20934772; http://dx.doi.org/: 10.1016/j.tig.2010.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol 2011; 12:246-58; PMID:21427766; http://dx.doi.org/ 10.1038/nrm3089 [DOI] [PubMed] [Google Scholar]
- 8.Iwasaki YW, Siomi MC, Siomi H. PIWI-Interacting RNA: Its Biogenesis and Functions. Annu Rev Biochem 2015; 84:405-33; PMID:25747396; http://dx.doi.org/ 10.1146/annurev-biochem-060614-034258 [DOI] [PubMed] [Google Scholar]
- 9.Guzzardo PM, Muerdter F, Hannon GJ. The piRNA pathway in flies: highlights and future directions. Curr Opin Genet Dev 2013; 23:44-52; PMID:23317515; http://dx.doi.org/ 10.1016/j.gde.2012.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lim AK, Kai T. Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc Natl Acad Sci USA 2007; 104:6714-9; PMID:17428915;http://dx.doi.org/ 10.1073/pnas.0701920104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kirino Y, Kim N, de Planell-Saguer M, Khandros E, Chiorean S, Klein PS, Rigoutsos I, Jongens TA, Mourelatos Z. Arginine methylation of Piwi proteins catalysed by dPRMT5 is required for Ago3 and Aub stability. Nat Cell Biol 2009; 11:652-8; PMID:19377467; http://dx.doi.org/ 10.1038/ncb1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nishida KM, Okada TN, Kawamura T, Mituyama T, Kawamura Y, Inagaki S, Huang H, Chen D, Kodama T, Siomi H, et al.. Functional involvement of Tudor and dPRMT5 in the piRNA processing pathway in Drosophila germlines. EMBO J 2009; 28:3820-31; PMID:19959991; http://dx.doi.org/ 10.1038/emboj.2009.365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirino Y, Vourekas A, Sayed N, de Lima Alves F, Thomson T, Lasko P, Rappsilber J, Jongens TA, Mourelatos Z. Arginine methylation of Aubergine mediates Tudor binding and germ plasm localization. RNA 2010; 16:70-8; PMID:19926723;http://dx.doi.org/ 10.1261/rna.1869710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishizu H, Siomi H, Siomi MC. Biology of PIWI-interacting RNAs: new insights into biogenesis and function inside and outside of germlines. Genes Dev 2012; 26:2361-73; PMID:23124062;http://dx.doi.org/ 10.1101/gad.203786.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Handler D, Meixner K, Pizka M, Lauss K, Schmied C, Gruber FS, Brennecke J. The genetic makeup of the Drosophila piRNA pathway. Mol Cell 2013; 50:762-77; PMID:23665231;http://dx.doi.org/ 10.1016/j.molcel.2013.04.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Czech B, Preall JB, McGinn J, Hannon GJ. A transcriptome-wide RNAi screen in the Drosophila ovary reveals factors of the germline piRNA pathway. Mol Cell 2013; 50:749-61; PMID:23665227; http://dx.doi.org/ 10.1016/j.molcel.2013.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muerdter F, Guzzardo PM, Gillis J, Luo Y, Yu Y, Chen C, Fekete R, Hannon GJ. A genome-wide RNAi screen draws a genetic framework for transposon control and primary piRNA biogenesis in Drosophila. Mol Cell 2013; 50:736-48;PMID:23665228;http://dx.doi.org/ 10.1016/j.molcel.2013.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siomi MC, Mannen T, Siomi H. How does the royal family of Tudor rule the PIWI-interacting RNA pathway? Genes Dev 2010; 24:636-46; PMID:20360382; http://dx.doi.org/ 10.1101/gad.1899210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen C, Nott TJ, Jin J, Pawson T. Deciphering arginine methylation: Tudor tells the tale. Nat Rev Mol Cell Biol 2011; 12:629-42; PMID:21915143; http://dx.doi.org/ 10.1038/nrm3185 [DOI] [PubMed] [Google Scholar]
- 20.Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 2007; 315:1587-90; PMID:17322028;http://dx.doi.org/ 10.1126/science.1140494 [DOI] [PubMed] [Google Scholar]
- 21.Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 2007; 128:1089-103; PMID:17346786; http://dx.doi.org/ 10.1016/j.cell.2007.01.043 [DOI] [PubMed] [Google Scholar]
- 22.Saito K, Inagaki S, Mituyama T, Kawamura Y, Ono Y, Sakota E, Kotani H, Asai K, Siomi H, Siomi MC. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature 2009; 461:1296-9; PMID:19812547; http://dx.doi.org/ 10.1038/nature08501 [DOI] [PubMed] [Google Scholar]
- 23.Mohn F, Handler D, Brennecke J. piRNA-guided slicing specifies transcripts for Zucchini-dependent, phased piRNA biogenesis. Science 2015; 348:812-7; PMID:25977553;http://dx.doi.org/ 10.1126/science.aaa1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han BW, Wang W, Li C, Weng Z, Zamore PD. piRNA-guided transposon cleavage initiates Zucchini-dependent, phased piRNA production. Science 2015; 348:817-21. PMID:25977554;http://dx.doi.org/ 10.1126/science.aaa1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamanaka S, Siomi MC, Siomi H. piRNA clusters and open chromatin structure. Mob DNA 2014; 5:22; PMID:25126116;http://dx.doi.org/ 10.1186/1759-8753-5-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, Hannon GJ. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 2009; 137:522-35; PMID:19395010; http://dx.doi.org/ 10.1016/j.cell.2009.03.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li C, Vagin VV, Lee S, Xu J, Ma S, Xi H, Seitz H, Horwich MD, Syrzycka M, Honda BM, et al.. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell 2009; 137:509-21; PMID:19395009; http://dx.doi.org/ 10.1016/j.cell.2009.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pane A, Wehr K, Schüpbach T. zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev Cell 2007; 12:851-62; PMID:17543859; http://dx.doi.org/: 10.1016/j.devcel.2007.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishimasu H, Ishizu H, Saito K, Fukuhara S, Kamatani MK, Bonnefond L, Matsumoto N, Nishizawa T, Nakanaga K, Aoki J, et al.. Structure and function of Zucchini endoribonuclease in piRNA biogenesis. Nature 2012; 491:284-7; PMID:23064230; http://dx.doi.org/ 10.1038/nature11509 [DOI] [PubMed] [Google Scholar]
- 30.Ipsaro JJ, Haase AD, Knott SR, Joshua-Tor L, Hannon GJ. The structural biochemistry of Zucchini implicates it as a nuclease in piRNA biogenesis. Nature 2012; 491:279-83; PMID:23064227; http://dx.doi.org/ 10.1038/nature11502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Voigt F, Reuter M, Kasaruho A, Schulz EC, Pillai RS, Barabas O. Crystal structure of the primary piRNA biogenesis factor Zucchini reveals similarity to the bacterial PLD endonuclease Nuc. RNA 2012; 18:2128-34; PMID:23086923;http://dx.doi.org/ 10.1261/rna.034967.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klenov MS, Sokolova OA, Yakushev EY, Stolyarenko AD, Mikhaleva EA, Lavrov SA, Gvozdev VA. Separation of stem cell maintenance and transposon silencing functions of Piwi protein. Proc Natl Acad Sci USA 2011; 108:18760-5; PMID:22065765;http://dx.doi.org/ 10.1073/pnas.1106676108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sienski G, Dönertas D, Brennecke J. Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell. 2012; 151:964-80; PMID:23159368; http://dx.doi.org/ 10.1016/j.cell.2012.10.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rozhkov NV, Hammell M, Hannon GJ. Multiple roles for Piwi in silencing Drosophila transposons. Genes Dev 2013; 27:400-12; PMID:23392609; http://dx.doi.org/ 10.1101/gad.209767.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Le Thomas A, Rogers AK, Webster A, Marinov GK, Liao SE, Perkins EM, Hur JK, Aravin AA, Tóth KF. Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes Dev 2013; 27:390-9; PMID:23392610; http://dx.doi.org/ 10.1101/gad.209841.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nishida KM, Saito K, Mori T, Kawamura Y, Nagami-Okada T, Inagaki S, Siomi H, Siomi MC. Gene silencing mechanisms mediated by Aubergine piRNA complexes in Drosophila male gonad. RNA 2007; 13:1911-22; PMID:17872506; http://dx.doi.org/ 10.1261/rna.744307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiol J, Spinelli P, Laussmann MA, Homolka D, Yang Z, Cora E, Couté Y, Conn S, Kadlec J, Sachidanandam R, et al.. RNA clamping by Vasa assembles a piRNA amplifier complex on transposon transcripts. Cell 2014; 157:1698-711; PMID:24910301; http://dx.doi.org/ 10.1016/j.cell.2014.05.018 [DOI] [PubMed] [Google Scholar]
- 38.Nishida KM, Iwasaki YW, Murota Y, Nagao A, Mannen T, Kato Y, Siomi H, Siomi MC. Respective functions of two distinct Siwi complexes assembled during PIWI-interacting RNA biogenesis in Bombyx germ cells. Cell Rep 2015; 10:193-203; PMID:25558067; http://dx.doi.org/ 10.1016/j.celrep.2014.12.013 [DOI] [PubMed] [Google Scholar]
- 39.Wang W, Han BW, Tipping C, Ge DT, Zhang Z, Weng Z, Zamore PD. Slicing and Binding by Ago3 or Aub Trigger Piwi-Bound piRNA Production by Distinct Mechanisms. Mol Cell 2015; 59:819-30; PMID:26340424; http://dx.doi.org/ 10.1016/j.molcel.2015.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Senti KA, Jurczak D, Sachidanandam R, Brennecke J. piRNA-guided slicing of transposon transcripts enforces their transcriptional silencing via specifying the nuclear piRNA repertoire. Genes Dev 2015; 29:1747-62; PMID:26302790;http://dx.doi.org/ 10.1101/gad.267252.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nagao A, Sato K, Nishida KM, Siomi H, Siomi MC. Gender-Specific Hierarchy in Nuage Localization of PIWI-Interacting RNA Factors in Drosophila. Front Genet 2011; 2:55; PMID:22303351; http://dx.doi.org/ 10.3389/fgene.2011.00055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sato K, Iwasaki YW, Shibuya A, Carninci P, Tsuchizawa Y, Ishizu H, Siomi MC, Siomi H. Krimper Enforces an Antisense Bias on piRNA Pools by Binding AGO3 in the Drosophila Germline. Mol Cell 2015; 59:553-63; PMID:26212455; http://dx.doi.org/ 10.1016/j.molcel.2015.06.024 [DOI] [PubMed] [Google Scholar]
- 43.Webster A, Li S, Hur JK, Wachsmuth M, Bois JS, Perkins EM, Patel DJ, Aravin AA. Aub and Ago3 Are Recruited to Nuage through Two Mechanisms to Form a Ping-Pong Complex Assembled by Krimper. Mol Cell 2015; 59:564-75; PMID:26295961; http://dx.doi.org/ 10.1016/j.molcel.2015.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Olivieri D, Senti KA, Subramanian S, Sachidanandam R, Brennecke J. The cochaperone shutdown defines a group of biogenesis factors essential for all piRNA populations in Drosophila. Mol Cell 2012; 47:954-69; PMID:22902557; http://dx.doi.org/ 10.1016/j.molcel.2012.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Z, Xu J, Koppetsch BS, Wang J, Tipping C, Ma S, Weng Z, Theurkauf WE, Zamore PD. Heterotypic piRNA Ping-Pong requires qin, a protein with both E3 ligase and Tudor domains. Mol Cell 2011; 44:572-84; PMID:22099305;http://dx.doi.org/ 10.1016/j.molcel.2011.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anand A, Kai T. The tudor domain protein kumo is required to assemble the nuage and to generate germline piRNAs in Drosophila. EMBO J 2012; 31:870-82; PMID:22157814;http://dx.doi.org/ 10.1038/emboj.2011.449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Z, Koppetsch BS, Wang J, Tipping C, Weng Z, Theurkauf WE, Zamore PD. Antisense piRNA amplification, but not piRNA production or nuage assembly, requires the Tudor-domain protein Qin. EMBO J 2014; 33:536-9; PMID:24652836; http://dx.doi.org/ 10.1002/embj.201384895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A, Hannon GJ. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science 2008; 322:1387-92; PMID:19039138; http://dx.doi.org/ 10.1126/science.1165171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Le Thomas A, Stuwe E, Li S, Du J, Marinov G, Rozhkov N, Chen YC, Luo Y, Sachidanandam R, Toth KF, et al.. Transgenerationally inherited piRNAs trigger piRNA biogenesis by changing the chromatin of piRNA clusters and inducing precursor processing. Genes Dev 2014; 28:1667-80; PMID:25085419; http://dx.doi.org/ 10.1101/gad.245514.114 [DOI] [PMC free article] [PubMed] [Google Scholar]


