Cryptococcus neoformans is a fungal pathogen that causes disease in immunocompromised individuals, especially in HIV+/AIDS patients, and organ transplant recipients. Patients who present with cryptococcosis primarily die from chronic meningoencephalitis (CME), and disease may follow acute infection or reactivation of latent infection.1 Despite the introduction of Combination Antiretroviral Therapy (cART) and anti-fungal therapy, over 600,000 deaths per year have been reported worldwide.1 This is in large part due to the ability of the fungus to successfully infect a diverse range of hosts with different body temperatures.2 In addition, current treatment regimens are expensive and difficult to provide in resource limited settings because they are very toxic and require careful monitoring. Consequently, efforts are constantly undertaken to identify new antifungal drug regimens with fewer side effects. To date, laboratories have relied on cell, tissue, and animal based models to investigate new drug targets. However, it remains unclear to what extent, the current infection models fully represent this complex host-pathogen interaction.
Several models currently exist to study C. neoformans infection. In vitro models include the use of cell lines; primarily, monocytes,3 and macrophages,4 and also dendritic cells, neutrophils, eosinophils, and lymphocytes.5 In addition, in vivo models have been established and include vertebrate models of mouse, rat, and rabbit.6,7 Because cryptococcal virulence has been suggested to evolve in more primitive organisms as C. neoformans was first an environmental fungus that became an accidental pathogen,8 other host models have been used to investigate its interaction. These models include invertebrates, such as Caenorhabditis elegans,9 Acantamoeba castellanii,10 Dictyostelium discoideum,11 and most recently the lepidoptera, Galleria mellonella.12 Galleria is the greater wax moth and is used during its larval stage to screen for virulence with a diverse array of pathogens. This model affords several advantages, most important of which is that it can withstand mammalian body temperature, but also that its larvae are easy to inoculate, inexpensive, and have host hemocytes that are able to phagocytose C. neoformans. Therefore, this model has been used to test cryptococcal virulence, and antifungal efficacy.12 Since 2005, when this model was introduced for C. neoformans, there have been 25 papers published using the Galleria model with C. neoformans. Because we observed some inconsistencies in select experiments with this model when compared with murine infection models, we undertook a systematic large scale study to compare the virulence of 20 different clinical C. neoformans strains with cellular, murine, and Galleria infection models. Our results demonstrate that while the model is beneficial to study some virulence parameters with standard passaged laboratory strains, virulence does not consistently correlate with murine infection models. We propose that Galleria infection does not conclusively model the host-pathogen interaction of C. neoformans, and most importantly cannot substitute for mammalian models of C. neoformans infections.
First, clinical C. neoformans strains (n = 20) were characterized with respect to known virulence traits. Strains were randomly selected from our collection, and included both serotype A (n = 15) and serotype D (n = 5) strains from different parts in the world (United States of America and India). Also included was the standard strain, H99, which was first isolated from a patient at Duke University in 1978,13 and is the predominant strain used in published C. neoformans literature. All strains were found to exhibit variable doubling times, cell, and capsule sizes in rich glucose media, and laccase activity in L-DOPA media (Table 1). Doubling times ranged from 2.7 h (strain H99) to 5.2 h (strain M8A), and laccase activity ranged from 0.886 U (strain I65) to 1.038 U (strain JEC21) normalized to 1.000 U of strain H99. Next, phagocytosis indices, killing rates, and intracellular replication rates (IPR) by co-cultivation in the murine macrophage cell line J774.16 were determined for all strains. Phagocytosis ranged from 8 - 55%, and accordingly intracellular killing and IPR were also found to be variable (Table 1). In summary, these randomly selected clinical C. neoformans strains exhibited marked variability with respect to virulence traits that were consistent with the previously reported variability of replicative life span (RLS) in these strains, which ranged from 8 generations (strain M8A) to 99 generations (strain M12A).14
Table 1.
In vitro characterization of clinical C. neoformans strains
| Strain # | Strain serotype | Strain name | Doubling times (h) | Cell size (μm) | Capsule size (μm) | Laccase activity (U) | Phagocytosis indices (%) | Killing post phagocytosis (%) |
|---|---|---|---|---|---|---|---|---|
| 1 | A | H99 | 2.7 + 0.19 | 6.867 + 1.00 | 0.8566 + 0.35 | 1.000 | 48.38 | 35.92 |
| 2 | D | I114 | 3.3 + 0.25 | 6.961 + 0.98 | 0.6491 + 0.43 | 0.922 | 32.96 | - |
| 3 | A | I47 | 3.4 + 0.29 | 7.920 + 1.59 | 0.8348 + 0.57 | 0.968 | 33.46 | 20.55 |
| 4 | A | I55 | 2.8 + 0.48 | 6.797 + 0.61 | 0.7312 + 0.24 | 0.955 | 36.75 | 25.62 |
| 5 | A | I58 | 3.1 + 0.17 | 8.072 + 1.03 | 0.8870 + 0.28 | 1.020 | 44.00 | 19.51 |
| 6 | A | I65 | 3.6 + 0.24 | 7.540 + 1.09 | 0.7031 + 0.51 | 0.886 | 17.79 | 27.63 |
| 7 | A | ISG12 | 3.7 + 0.41 | 9.041 + 1.24 | 1.2440 + 0.43 | 0.894 | 18.06 | 12.11 |
| 8 | D | J22 | 2.9 + 0.50 | 7.341 + 1.12 | 1.0770 + 0.47 | 0.889 | 24.35 | 23.01 |
| 9 | D | J9 | 3.1 + 0.37 | 7.956 + 1.16 | 0.6764 + 0.42 | 0.914 | 8.31 | 46.37 |
| 10 | D | JEC21 | 3.7 + 0.41 | 7.629 + 1.06 | 0.9114 + 0.50 | 1.038 | 34.25 | 30.29 |
| 11 | A | M12A | 2.9 + 0.17 | 6.286 + 0.81 | 0.6905 + 0.43 | 0.906 | 39.33 | 30.51 |
| 12 | A | M12B | 3.9 + 0.17 | 6.751 + 0.90 | 0.6162 + 0.48 | 0.937 | 55.71 | 17.43 |
| 13 | A | M511B | 3.2 + 0.19 | 8.157 + 0.94 | 1.1220 + 0.28 | 0.965 | 38.83 | 21.90 |
| 14 | A | M7A | 3.6 + 0.32 | 6.528 + 0.83 | 0.6809 + 0.43 | 0.896 | 20.42 | 43.06 |
| 15 | A | M7E | 4.3 + 0.10 | 7.467 + 1.30 | 1.0130 + 0.43 | 0.924 | 18.75 | 57.56 |
| 16 | A | M8A | 5.2 + 0.25 | 6.285 + 0.92 | 0.5851 + 0.37 | 0.943 | 16.10 | 56.79 |
| 17 | A | M9A | 4.6 + 0.27 | 6.938 + 1.14 | 0.6149 + 0.55 | 0.909 | 13.04 | 42.01 |
| 18 | D | RC-2 | 3.3 + 0.46 | 11.100 + 1.51 | 2.8650 + 0.35 | 0.900 | 17.94 | 25.34 |
| 19 | A | SB4 | 2.8 + 0.46 | 8.572 + 0.89 | 0.9422 + 0.51 | 0.917 | 24.36 | 34.96 |
| 20 | A | W911A | 3.6 + 0.28 | 8.884 + 1.57 | 1.3070 + 0.55 | 0.960 | 29.17 | 44.86 |
Next, virulence of these strains was compared in the insect model, Galleria mellonella, as well as a murine intravenous (i.v.) infection model. Galleria (n = 20) were infected with 3 different doses [2 × 104, 105, or 106 colony forming units (CFU)] of individual strains in 3 independent experiments. BALB/c mice (n = 8) were infected i.v. with 106 CFU. Survival was observed daily and mortality was recorded (Fig. 1A). Virulence varied for clinical strains in both models. In the i.v. model, survival ranged from 8 days to 45 days (mice sacrificed at 45 days). Premorbid mice exhibited hunched backs, and decreased movement consistent with signs of secondary cryptococcal meningoencephalitis. Survival of Galleria correlated in the majority of the strains (70%) between the 3 doses (p < 0.05) (Fig. 1B). However, most importantly, a relationship in survival between both animal models, Galleria and mice, was also not found (Fig. 1C).
Figure 1.
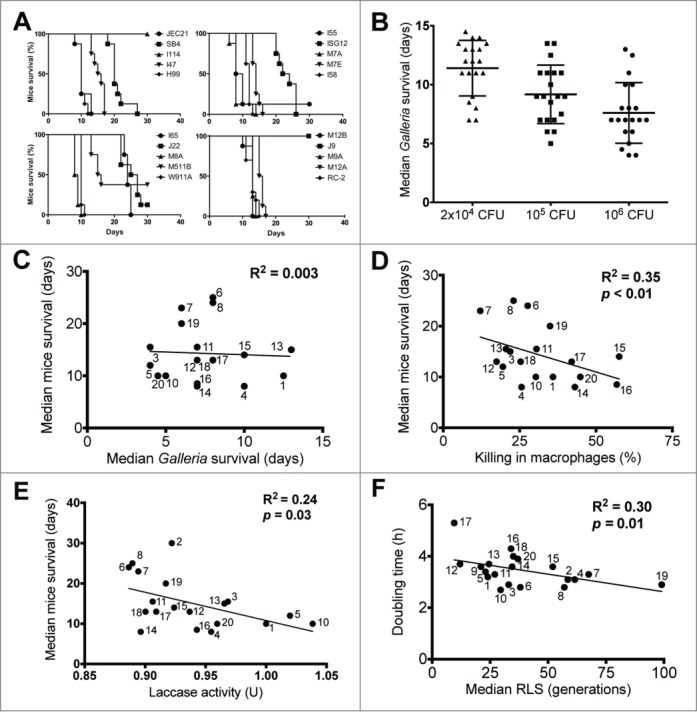
Relationships between different strain-specific characteristics of C. neoformans. (A) Kaplan-Meier survival curves of mice infected with clinical C. neoformans strains divided into 4 arbitrary groups (top left: JEC21, SB4, I114, I47, H99; top right: I55, ISG12, M7A, M7E, I58; bottom left: I65, J22, M8A, M511B, W911A; bottom right: M12B, J9, M9A, M12A, RC-2). (B) Scatter plot of Galleria infected with variable doses of clinical C. neoformans strains (symbols represent the infection dose: triangle = 2×104 CFU, square = 105 CFU, circle = 106). (C) No significant relationship was found between mice and Galleria survival. Symbol legends represent the assigned strain numbers listed in Table 1. Note that strains #2 and #9 were omitted from this comparison because infected mice were sacrificed at day 45 to terminate the study. Strains #12 and #18 are represented by the same symbol because they induced similar survival in mice and Galleria. (D) An inverse correlative relationship was found between mice survival and intracellular killing of C. neoformans strains in macrophages. Strains #2 and #9 were omitted from this comparison. (E) An inverse correlative relationship was found between mice survival and laccase activity of C. neoformans strains. (F) An inverse correlative relationship was found between the doubling time and the median replicative life span of C. neoformans strains. Strains #2 and #9 were omitted from this comparison. Correlation considered when Pearson's correlation coefficient had p < 0.05.
Given the lack of correlation between the survival of Galleria and the survival of mice, we explored correlation with other commonly assessed virulence traits. These experiments established that intracellular phagocytosis or killing by murine macrophages, as well as laccase activity does not correlate with virulence in Galleria. Instead, killing by murine macrophages, and laccase activity were found to moderately correlate with murine survival (p < 0.05) (Fig. 1D, E). Lastly, correlation with RLS, a variable, but also reproducible trait of yeast strains was examined. Previous studies have shown that older cryptococcal cells are more virulent;14 however, the relevance of the RLS of a strain to its virulence had not been examined. These data now demonstrate that although a strong correlation between doubling time and RLS in clinical strains was established (Fig. 1F), no correlation between the RLS of a strain and its virulence, either in Galleria, or in BALB/c mice was found.
The observed variability in the outcome of cryptococcal meningoencephalitis is attributed to differences in host immunity, as well as differences in potency of the virulence associated with individual Cryptococcus strains.1,15 Consequently, many studies on mutants and potential drug targets seek to screen and compare the virulence of individual Cryptococcus strains. Phenotypic characterization of human infection with C. neoformans can be mimicked in different infection models.5-7,9,12,16 Although murine infection models have been well established and have the advantage of close correlation with human pathology, strain virulence can also be assessed in more primitive infection models, such as Galleria mellonella, which are considerably cheaper.12
We compared Galleria and murine infection models, and also correlated outcomes with in vitro laccase activity, IPR, phagocytosis, killing, and growth times. Specifically, easily phagocytosed strains, namely “high-uptake” strains have been described as hypocapsular, have enhanced laccase activity, and importantly have been associated with central nervous system fungal burden and patient death.15 In our study, we found such high-uptake strains to have significantly enhanced laccase activity, and high virulence in mice, but not in Galleria. These data reaffirm that the virulence of clinical C. neoformans strains varies greatly, highlight some important differences between the various infection models, and ultimately suggest that the Galleria infection model cannot reliably predict virulence in the murine model. Notably, our high-uptake strains did not have a significantly smaller capsule size, nor a significantly smaller cell size. Capsular enlargement has been shown to be statistically similar among strains and not predictive of virulence.17 One study has also concluded that the virulence for C. neoformans may be an emergent property, rather than deterministic in the Galleria model of infection.18
High-throughput screening of mutants and of antifungal agents seeks to work with the most cost efficient model. Virulence of C. neoformans is mediated by many different virulence traits, which interact with the host in a complex manner. The most important virulence traits are the presence of a polysaccharide capsule, as well the ability to grow at 37°C;1,8 however, multiple other genes contribute to virulence, and either their up- or down-regulation can alter virulence as was recently described even with C. gattii strains used to infect Galleria.17 Several important findings were documented in this study. First, we established that internalization of the fungus did not depend on the doubling time of different C. neoformans strains; however, strains with a long doubling time were killed intracellularly faster than strains with a short doubling time. Virulence in Galleria was dependent on the different doses of C. neoformans inoculated for most of the strains. Despite this finding, we were not able to find a significant correlation between the virulence in Galleria, and in the most widely used animal model, the mouse. In the latter host, killing by macrophages and host survival were found to strongly correlate, but the same could not be established in Galleria. This suggests that variables usually reported to measure the virulence of a strain, i.e. phagocytosis and intracellular killing, are not being replicated in all the strains infecting the insect host, Galleria. These findings suggest that C. neoformans virulence data obtained in the Galleria model should be cautiously applied to the virulence of a pathogen in mammalian hosts. It should be acknowledged that for the 2 highly passaged laboratory strains, H99 and RC2, some correlation between Galleria and murine virulence was observed. It has been described that laboratory passage selects for fast growing, high passaged strains with phenotypes different from low passaged strains.19 These findings also highlight the importance of relying on multiple models of infection, particularly with respect to high-throughput screening. One group20 has found a way to reliably screen deletion mutants in a single strain using a multi-host approach that involves first, a C. elegans host, and second, a Galleria host, and this approach was successfully validated in a murine model of systemic cryptococcosis. This multi-host approach should greatly aid high-throughput screening of C. neoformans mutants and antifungal agents when limited to a single strain and not to compare virulence among different strains.
Lastly, we analyzed the replicative life span of the different clinical isolates. This was done since the short-living mucoid variant of strain RC2.14 is known to be hypervirulent in mice.21 Also, for a pathogen, the ability to live long and also divide fast could potentially aid in colonization; or alternatively, the ability to live short, but divide slower could aid in persistence. We found no correlation between RLS and virulence; however, interestingly an inverse correlation between doubling time and RLS was observed. This finding is not consistent with previously published data in S. cerevisiae that enhanced longevity often comes with a fitness cost.22 However, more recent data in this yeast23 highlights that genetic variability also contributes to variability in longevity. For facultative intracellular pathogens, such as C. neoformans, genetic variants could potentially be selected through the host response.
All animal experiments were carried out with the approval of the Albert Einstein College of Medicine Institute for Animal Studies under protocol number 20091015 as approved by the Institutional Animal Care and Use Committee at Einstein. C. neoformans strains used in this study are listed in Supplemental Methods (Table S2). Standard yeast culture media as outlined (Table S3) were employed. C. neoformans cells (n = 100) were imaged at 100X magnification on an Olympus AX70 microscope. Pictures were taken with a Qimaging Retiga 1300 digital camera using the Qcapture Suite V2.46 software (Qimaging, British Columbia, Canada) and size was measured with Adobe Photoshop CS5 for Macintosh. Log phase C. neoformans cells in 2% YPD were diluted to an OD of 0.01 and grown at 37 °C with agitation for in vitro studies. Growth curves were determined in a Bioscreen-C Automated Growth Curve Analysis System (Growth Curves USA). Standard in vitro phagocytosis indices, and macrophage killing assays were done in J774.16 cells as previously described.24 IPR and laccase activity was determined as previously described.15 For infection with Galleria mellonella, a 10 μl suspension of 2×104, 105, or 106 C. neoformans cells was used to infect larvae (n = 20) (Vanderhorst Wholesale, Inc., OH) in the last proleg as described previously.16 For infection with mice, 106 C. neoformans cells were used to infect 6-8 week old female BALB/c mice (n = 8) (National Cancer Institute, Bethesda, MD) intravenously as previously described.25,26 Standard statistical analysis and non-parametric tests, such as Student's t-test, Log-rank, Pearson correlation, and Wilcoxon rank sum tests were performed using Prism version 5 (Graphpad) or Microsoft Excel 2011 for Macintosh. Differences were considered significant if p < 0.05.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
Funding
B.C.F. is supported by NIH awards R01 AI059681 and R21 AI114259. For a portion of this work, T.B. was supported by the Institutional AIDS Training Grant, “HIV, AIDS and Opportunistic Infections” (T32 AI007501).
References
- 1.Perfect JR, Casadevall A. Cryptococcosis. Infect Dis Clin North Am 2002; 16:837-74, v-vi; PMID:12512184; http://dx.doi.org/ 10.1016/S0891-5520(02)00036-3 [DOI] [PubMed] [Google Scholar]
- 2.Mandal P, Banerjee U, Casadevall A, Nosanchuk JD. Dual infections with pigmented and albino strains of Cryptococcus neoformans in patients with or without human immunodeficiency virus infection in India. J Clin Microbiol 2005; 43:4766-72; PMID:16145139; http://dx.doi.org/ 10.1128/JCM.43.9.4766-4772.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alvarez M, Burn T, Luo Y, Pirofski LA, Casadevall A. The outcome of Cryptococcus neoformans intracellular pathogenesis in human monocytes. BMC Microbiol 2009; 9:51; PMID:19265539; http://dx.doi.org/ 10.1186/1471-2180-9-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gilbert AS, Wheeler RT, May RC. Fungal Pathogens: Survival and Replication within Macrophages. Cold Spring Harb Perspect Med 2014; 5:a019661; PMID:25384769; http://dx.doi.org/ 10.1101/cshperspect.a019661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sabiiti W, May RC, Pursall ER. Experimental models of cryptococcosis. International J Microbiol 2012; 2012:626745; http://dx.doi.org/ 10.1155/2012/626745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carroll SF, Guillot L, Qureshi ST. Mammalian model hosts of cryptococcal infection. Comparative Med 2007; 57:9-17 [PubMed] [Google Scholar]
- 7.Goldman DL, Casadevall A, Cho Y, Lee SC. Cryptococcus neoformans meningitis in the rat. Lab Invest 1996; 75:759-70; PMID:8973471 [PubMed] [Google Scholar]
- 8.Casadevall A, Steenbergen JN, Nosanchuk JD. 'Ready made' virulence and ‘dual use’ virulence factors in pathogenic environmental fungi–the Cryptococcus neoformans paradigm. Curr Opinion Microbiol 2003; 6:332-7; http://dx.doi.org/ 10.1016/S1369-5274(03)00082-1 [DOI] [PubMed] [Google Scholar]
- 9.Mylonakis E, Ausubel FM, Perfect JR, Heitman J, Calderwood SB. Killing of Caenorhabditis elegans by Cryptococcus neoformans as a model of yeast pathogenesis. Proc Natl Acad Sci USA 2002; 99:15675-80; PMID:12438649; http://dx.doi.org/ 10.1073/pnas.232568599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Steenbergen JN, Shuman HA, Casadevall A. Cryptococcus neoformans interactions with amoebae suggest an explanation for its virulence and intracellular pathogenic strategy in macrophages. Proc Natl Acad Sci USA 2001; 98:15245-50; PMID:11742090; http://dx.doi.org/ 10.1073/pnas.261418798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steenbergen JN, Nosanchuk JD, Malliaris SD, Casadevall A. Cryptococcus neoformans virulence is enhanced after growth in the genetically malleable host Dictyostelium discoideum. Infect Immun 2003; 71:4862-72; PMID:12933827; http://dx.doi.org/ 10.1128/IAI.71.9.4862-4872.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mylonakis E, Moreno R, El Khoury JB, Idnurm A, Heitman J, Calderwood SB, Ausubel FM, Diener A. Galleria mellonella as a model system to study Cryptococcus neoformans pathogenesis. Infect Immun 2005; 73:3842-50; PMID:15972469; http://dx.doi.org/ 10.1128/IAI.73.7.3842-3850.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perfect JR, Casadevall A. The history of Cryptococcus and Cryptococcosis Cryptococcus: from human pathogen to model yeast Washington, DC: ASM Press; pp 2011:17-26 [Google Scholar]
- 14.Bouklas T, Pechuan X, Goldman DL, Edelman B, Bergman A, Fries BC. Old Cryptococcus neoformans cells contribute to virulence in chronic cryptococcosis. MBio 2013; 4:e00455-13; PMID:23943761; http://dx.doi.org/ 10.1128/mBio.00455-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sabiiti W, Robertson E, Beale MA, Johnston SA, Brouwer AE, Loyse A, Jarvis JN, Gilbert AS, Fisher MC, Harrison TS, et al.. Efficient phagocytosis and laccase activity affect the outcome of HIV-associated cryptococcosis. J Clin Invest 2014; 124:2000-8; PMID:24743149; http://dx.doi.org/ 10.1172/JCI72950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cotter G, Doyle S, Kavanagh K. Development of an insect model for the in vivo pathogenicity testing of yeasts. FEMS Immunol Med Microbiol 2000; 27:163-9; PMID:10640612; http://dx.doi.org/ 10.1111/j.1574-695X.2000.tb01427.x [DOI] [PubMed] [Google Scholar]
- 17.Firacative C, Duan S, Meyer W. Galleria mellonella model identifies highly virulent strains among all major molecular types of Cryptococcus gattii. PloS One 2014; 9:e105076; PMID:25133687; http://dx.doi.org/ 10.1371/journal.pone.0105076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia-Solache MA, Izquierdo-Garcia D, Smith C, Bergman A, Casadevall A. Fungal virulence in a lepidopteran model is an emergent property with deterministic features. MBio 2013; 4:e00100-13; PMID:23631914; http://dx.doi.org/ 10.1128/mBio.00100-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ronald J, Tang H, Brem RB. Genomewide evolutionary rates in laboratory and wild yeast. Genetics 2006; 174:541-4; PMID:16816417; http://dx.doi.org/ 10.1534/genetics.106.060863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Desalermos A, Tan X, Rajamuthiah R, Arvanitis M, Wang Y, Li D, Kourkoumpetis TK, Fuchs BB, Mylonakis E. A multi-host approach for the systematic analysis of virulence factors in Cryptococcus neoformans. J Infect Dis 2015; 211:298-305; PMID:25114160; http://dx.doi.org/ 10.1093/infdis/jiu441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jain N, Li L, Hsueh YP, Guerrero A, Heitman J, Goldman DL, Fries BC. Loss of allergen 1 confers a hypervirulent phenotype that resembles mucoid switch variants of Cryptococcus neoformans. Infect Immun 2009; 77:128-40; PMID:18955480; http://dx.doi.org/ 10.1128/IAI.01079-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delaney JR, Murakami CJ, Olsen B, Kennedy BK, Kaeberlein M. Quantitative evidence for early life fitness defects from 32 longevity-associated alleles in yeast. Cell Cycle 2011; 10:156-65; PMID:21191185; http://dx.doi.org/ 10.4161/cc.10.1.14457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stumpferl SW, Brand SE, Jiang JC, Korona B, Tiwari A, Dai J, Seo JG, Jazwinski SM. Natural genetic variation in yeast longevity. Genome Res 2012; 22:1963-73; PMID:22955140; http://dx.doi.org/ 10.1101/gr.136549.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jain N, Cook E, Xess I, Hasan F, Fries D, Fries BC. Isolation and characterization of senescent Cryptococcus neoformans and implications for phenotypic switching and pathogenesis in chronic cryptococcosis. Eukaryotic cell 2009; 8:858-66; PMID:19411622; http://dx.doi.org/ 10.1128/EC.00017-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huffnagle GB, Yates JL, Lipscomb MF. Immunity to a pulmonary Cryptococcus neoformans infection requires both CD4+ and CD8+ T cells. J Exp Med 1991; 173:793-800; PMID:1672543; http://dx.doi.org/ 10.1084/jem.173.4.793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mukherjee S, Lee S, Mukherjee J, Scharff MD, Casadevall A. Monoclonal antibodies to Cryptococcus neoformans capsular polysaccharide modify the course of intravenous infection in mice. Infect Immun 1994; 62:1079-88; PMID:8112842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jain N, Wickes BL, Keller SM, Fu J, Casadevall A, Jain P, Ragan MA, Banerjee U, Fries BC. Molecular epidemiology of clinical Cryptococcus neoformans strains from India. J Clin Microbiol 2005; 43:5733-42; PMID:16272511; http://dx.doi.org/ 10.1128/JCM.43.11.5733-5742.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Casadevall A, Spitzer ED, Webb D, Rinaldi MG. Susceptibilities of serial Cryptococcus neoformans isolates from patients with recurrent cryptococcal meningitis to amphotericin B and fluconazole. Antimicrob Agents Chemother 1993; 37:1383-6; PMID:8328793; http://dx.doi.org/ 10.1128/AAC.37.6.1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kwon-Chung KJ, Edman JC, Wickes BL. Genetic association of mating types and virulence in Cryptococcus neoformans. Infect Immun 1992; 60:602-5; PMID:1730495 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


