Abstract
Intestinal immunity is subject to complex and fine-tuned regulation dictated by interactions of the resident microbial community and their gene products with host innate cells. Deterioration of this delicate process may result in devastating autoinflammatory diseases, including inflammatory bowel disease (IBD), which primarily comprises Crohn's disease (CD) and ulcerative colitis (UC). Efficacious interventions to regulate proinflammatory signals, which play critical roles in IBD, require further scientific investigation. We recently demonstrated that rebalancing intestinal immunity via the surface layer protein A (SlpA) from Lactobacillus acidophilus NCFM potentially represents a feasible therapeutic approach to restore intestinal homeostasis. To expand on these findings, we established a new method of purifying bacterial SlpA, a new SlpA-specific monoclonal antibody, and found no SlpA-associated toxicity in mice. Thus, these data may assist in our efforts to determine the immune regulatory efficacy of SlpA in humans.
Keywords: bacterial protein isolation, colonic inflammation, gut microbiota, intestinal immune regulation, surface layer protein A
Introduction
It is estimated that over 1 million individuals in the US suffer from inflammatory bowel disease (IBD), which is characterized by relapsing chronic inflammation of the intestinal tract, especially the colon.1-3 Moreover, it has consistently been noted that patients suffering from IBD have an altered or imbalanced gut microbiota, specifically reduced bacterial diversity, referred to as dysbiosis.3 In fact, many studies indicate that dysfunctional immune responses are likely elicited by this imbalanced microflora.4 Functional gut homeostasis between the gut epithelium, mucosal immune cells, and the resident gut microbiota is established by regulatory immune mechanisms elicited by the trillions of microbes and their interactions with numerous pathogen recognition receptors (PRRs), including C-type lectin receptors (CLRs) expressed in the gastrointestinal (GI) tract.5,6 Deterioration or alternation of these tightly regulated mechanisms by detrimental signals (microbial gene products, metabolites, altered cytokine milieu, etc.) can have devastating consequences, resulting in autoinflammatory diseases, including IBD. These uncontrolled immune signals (e.g., IL-1β) are generated by highly activated innate cells, including dendritic cells (DCs), during microbial dysbiosis4 and create an unbalanced microenvironment, wherein released soluble mediators activate intestine-infiltrating pathogenic T lymphocyte subsets (e.g., Th1, Th17), and even proinflammatory regulatory T cells (Tregs),7-9 resulting in tissue damage and gut barrier dysfunction.22 The cellular and molecular mechanisms induced by gut microbes, their gene products, and their metabolites, as well as how these relevant factors potentially interact to determine the activation and differentiation of innate cells and T cell subsets at distal sites, require further scientific investigation.
In our recent publications, we demonstrated that transient colonization of the colon with a strain of Lactobacillus acidophilus deficient in lipoteichoic acid (LTA), NCK2025, significantly mitigated chemically-induced and T cell-mediated colitis.10-12 Analyzed mechanisms suggested that the induction of regulatory IL-10+ DCs and functional Tregs, activation of pErk1/2, and the downregulation of critical downstream signals (Akt1, p38)13 are key elements involved in the amelioration of murine colitis in our models.10-14 Furthermore, this L. acidophilus strain lacking LTA significantly diminished inflammation-promoting colonic polyposis in the Apclox468 x TS4-Cre mouse model via the regulation of proinflammatory innate and T cell subsets.9 Thus, we theorized that the cross-talk between a specific surface molecule expressed by the LTA-deficient L. acidophilus, called SlpA, with intestinal innate cells (e.g., DCs) not only suppresses pathogenic inflammation, but also potentially modulates the expression of epigenetically-regulated, colorectal cancer (CRC)-associated genes,15 and restores gut homeostasis to ablate colonic polyps.9,16,17 These data galvanized our laboratory to more thoroughly define this protective role of bacterial SlpA in a murine colitis model. Accordingly, we hypothesized that the SlpA molecule interacts with colonic innate cells (e.g., DCs) via its cognate receptor, SIGNR3, to regulate proinflammatory signals (e.g., IL-1β) in order to reshape the functional balance of intestinal homeostasis, resulting in significant mitigation of T cell-induced colitis. The rationale for this hypothesis being that 3 L. acidophilus surface layer proteins (SlpA, SlpB, and SlpX) have been observed to interact with PRRs,18,19 which activate intestinal innate cells; however, information about the functions of these Slps, and in particular, SlpA, is relatively limited.20,21,22 To specifically determine the effects of SlpA and its binding to SIGNR3 on intestinal cells, and the consequences thereafter, the upp counter-selective knockout strategy10 was used to generate a new strain of L. acidophilus, called NCK2187, which expresses only SlpA on its surface.23 This new bacterial strain was critical to definitively elucidate the role of SlpA in controlling pathogenic inflammation, as oral treatment with purified SlpA or the bacteria expressing only SlpA on their surfaces resulted in significant clinical improvement of murine colitis. Furthermore, our data showed that SlpA plays a critical role in controlling immune responses upon its interaction with SIGNR3, resulting in the amelioration of induced colitis, protection of intestinal barrier integrity, and maintenance of the gut bacterial composition.23 To build upon these observations, we have optimized the purification of SlpA to investigate its physiological effects when orally administrated to mice, and have evaluated whether this protein is able to resist the harsh conditions of the GI microenvironment, both important factors that may affect the feasibility of its use in potential clinical trials.
Isolation and Detection of L. acidophilus Surface Layer Protein A
S-layers are paracrystalline (glyco) protein arrays that are present in abundance on the cell surface of a subset of eubacteria and archaea. Among the functional roles that have been attributed to S-layers,24 their binding to PRRs,18 including CLRs, has been found to be critical to their potential immunogenic capacity.19,25 Consistent with our goals to further clarify the regulatory role of SlpA in controlling downstream signals during the interaction with its cognate receptor, SIGNR3, and to make this technology suitable for clinical trials, we first sought to improve the process of SlpA isolation and purification. For this purpose, we used sodium chloride (NaCl) (5 M), instead of lithium chloride (LiCl), which is more commonly used for SlpA purification (Fig. 1A).26 The rationale being that SlpA purified by LiCl may potentially induce toxicity when orally administrated to experimental animals, resulting in the induction of low-grade inflammation and potential intestinal tissue damage. To avoid non-SlpA protein contamination in our isolation technique, we employed the LTA-, SlpB-, and SlpX-deficient L. acidophilus NCK2187 strain. Visualization of the isolated protein by SDS-PAGE showed a single protein band of the expected size for SlpA (46 kDa, Fig. 1B). An automated mass spectrometry microbial identification system that uses Matrix Assisted Laser Desorption Ionization Time-of-Flight technology (MALDI-TOF) indicated 97 unique spectra and 55 unique peptides generated post-trypsinization of the protein isolate, which identified 2 possible proteins [gi|58336516 (SlpA) and gi|362076610 (SlpB)] (Fig. 1C). MALDI-TOF data were then analyzed on Scaffold1.27 Further evaluation revealed that the peptides generated cover 54% of SlpA and 18% of SlpB (highlighted, Fig. 1D). However, NCK2187 bacteria do not express SlpB, and the peptides generated, one of which was recognized as a potential component of SlpB, were generated from the C-terminal region of SlpA, which is conserved between SlpA and SlpB. (red box, Fig. 1D). Thus, it was concluded that no single unique peptide from SlpB was identified. Therefore, mass spectrometry and SDS-PAGE analyses demonstrated that the identity of the purified SlpA protein was retained whether purified by NaCl or by LiCl (Fig. 1).
Figure 1.
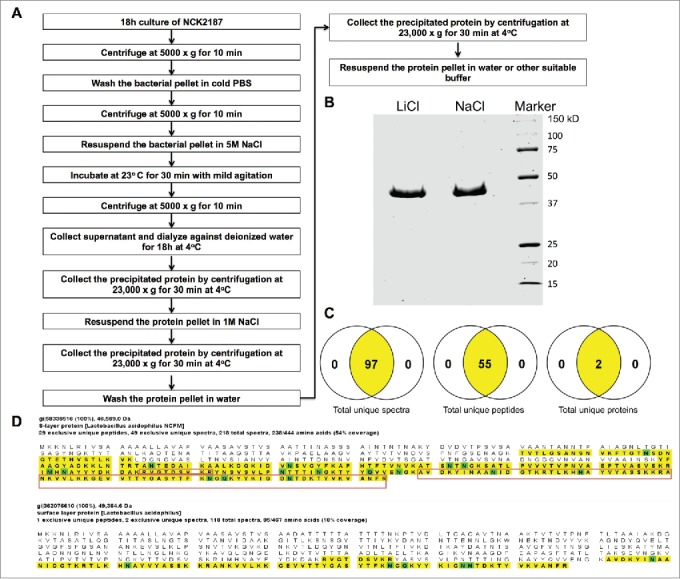
L. acidophilus-SlpA isolation by NaCl. L. acidophilus-SlpA was isolated and purified, as described previously,23 with substitution of LiCl with NaCl. (A) SDS-PAGE containing 2.5 μg of LiCl− and NaCl−isolated SlpA stained with Coomassie blue to visualize the purified protein. (B) Mass spectrometry data analyzed on the Scaffold27 platform showed 97 unique spectra with 55 unique peptides with the possibility of 2 proteins (C). The predicted protein gi|58336516 (SlpA) shows 54% coverage whereas gi|362076610 (SlpB) reveals only 18% of coverage (highlighted portion, D). The regions of SlpB matching the generated peptides are common between SlpA and SlpB (shown in the red box, D), and no single unique peptide from SlpB was identified.
To assess any potential toxicity of the isolated SlpA, groups of C57BL/6 mice were orally gavaged with purified SlpA (0, 150, 300, 600 μg/100 μL per mouse) every other day for a total of 4 treatments. Subsequently, the blood chemistry profiles of these animals were analyzed. Obtained data demonstrated that oral treatment of the mice with varying doses of SlpA did not significantly alter whole blood biochemical values that are typically assessed in a routine metabolic panel in these animals (Fig. 2). Changes in enzyme activity or concentration of other analytes in the blood were used as metrics of tissue damage or physiologic stress. Function of the urinary system (Fig. 2B) and of the hepatocellular and biliary systems (Figs. 2C–D & F) was evaluated and found to be unaffected by treatment. The electrolytes, sodium, potassium, calcium, and phosphorus, were also measured to gauge any changes in hydration status, excretional activity, or global cellular damage within the treated mice (Fig. 2E). No statistical differences were found in any of the parameters when comparing the controls and those mice receiving varying doses of SlpA administration, indicating no overt evidence of toxicity with oral treatment using SlpA in these animals.
Figure 2.
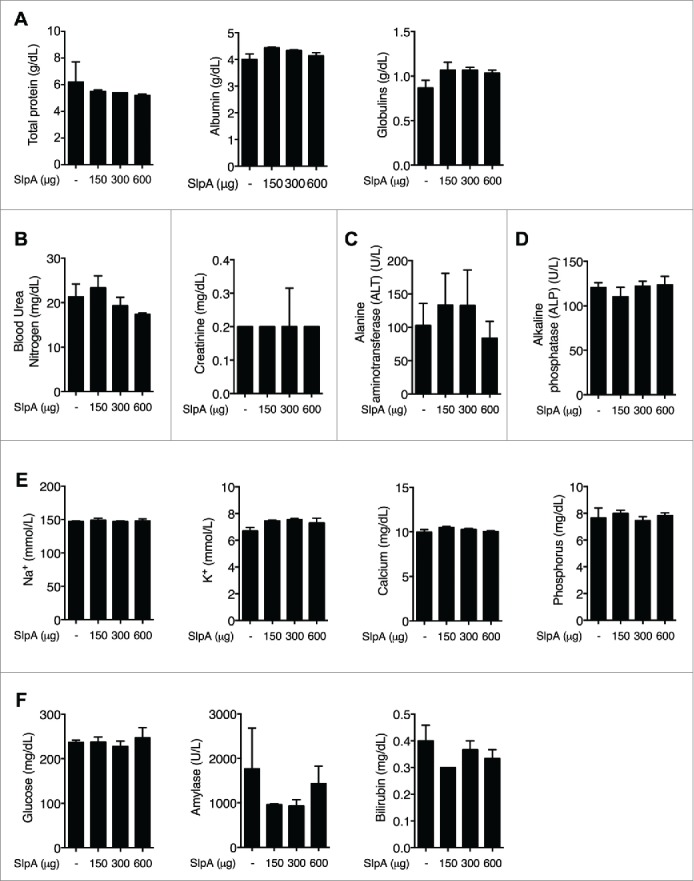
NaCl−purified SlpA is not overtly toxic to mice. (A–F) C57BL/6 mice were treated orally every other day with SlpA (0, 150, 300, 600 μg/100μL per mouse), for a total of 4 times. One week later, mice were sacrificed and a whole blood chemistry profile was generated for each mouse with a comprehensive metabolic chemistry panel, using a VetScan V2S analyzer. All animal experiments were performed under the guidelines of the Animal Welfare Act and the Public Health Policy on Humane Care, and with approval by the Institutional Animal Care and Use Committee (IACUC protocol 201406559) at the University of Florida. Data represent observations from 4 independent experiments (n = 5) and are shown as mean ± standard error of the mean.
We then sought to generate a specific monoclonal antibody against purified SlpA.30,31 To this end, groups of C57BL/6 mice were immunized with purified SlpA with killed L. gasseri as an adjuvant for 3 months (every week/100 μg of SlpA). Subsequently, spleen cells were derived to generate hybridoma cells producing a monoclonal antibody (mAb) recognizing SlpA. As seen in Fig. 3, the antibody derived from one of our hybridoma cell clones, BM1, recognized SlpA by Western blot (Fig. 3A). Furthermore, this mAb also recognized SlpA on the surface of SlpA-coated beads, and on SlpA-pulsed RAW 264.7 macrophages (Figs. 3B–C), respectively. We have reported immunomodulatory effects by the purified SlpA in murine colons, suggesting SlpA dissolved in PBS resists the hostile acidic milieu of the upper GI tract and enzymatic degradation within the intestinal lumen to reach the colon. To verify this, we established an ELISA using the mAb, BM1, that can detect SlpA (Fig. 3D). To evaluate the sensitivity of the BM1 to recognize SlpA, we coated ELISA plates with serial dilutions of purified SlpA (1 μg/mL down to 16 ng/mL). We observed a dose-dependent decrease in SlpA detection; the smallest concentration of SlpA that could be detected was 32 ng/mL (Fig. 3E). The sensitivity of the BM1 antibody may be able to be further enhanced after its purification. Data clearly showed that using this developed ELISA, SlpA can be detected in the fecal samples from mono-associated germ-free C57B/6 mice (Fig. 3F), indicating, as mentioned above, that SlpA can likely resist the harsh conditions of the GI system. These data may be useful for initiating Phase I clinical trials using NaCl−purified SlpA to demonstrate its ability to potentially downregulate induced colonic inflammation in human patients.
Figure 3.
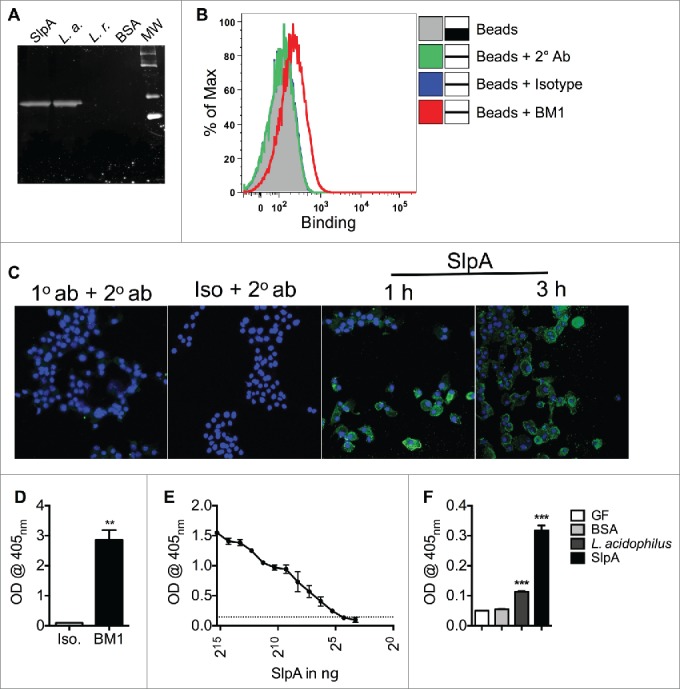
Generated mAb BM1 recognizes L. acidophilus-SlpA. C57BL/6 mice were immunized once a week for 3 months with 100 μg of SlpA, and 300 μg of heat-killed Lactobacillus gasseri as adjuvant. Polyclonal sera were tested for recognition of isolated SlpA by Western Blot (WB), and splenic cells from SlpA-reactive mice were fused with Sp2/0 myeloma cells at a ratio of 7:1. Hybridomas were seeded on a semi-solid medium for clone selection and screening. Subsequently, clones were screened by ELISA for SlpA reactivity. Reactive clones were isotyped and all IgM secretors removed. Clone BM1 (IgG) was selected for its ability to recognize SlpA by WB (A), flow cytometry (B), confocal microscopy (C), and ELISA (D, E, F). (A). L. acidophilus-SlpA detection by WB with BM1. 100 ng of purified SlpA, 108 CFU L. acidophilus (L. a.), 108 CFU L. reuteri (L. r.), or 100 ng of BSA proteins were separated by SDS-PAGE, transferred onto a PVDF membrane, and detected by BM1. (B). L. acidophilus-SlpA detection with BM1 by flow cytometry. Carboxylated Dynabeads were coated with purified SlpA and the reactivity of the BM1 mAb confirmed by Canto II flow cytometry. Data were analyzed by FlowJo. Experiments were performed at least 3 times with similar trends. (C). L. acidophilus-SlpA detection with BM1 by confocal microscopy. RAW 264.7 cells were pulsed for 1 or 3 hrs with NaCl purified SlpA (10 μg/mL). Subsequently, cells were fixed and stained with BM1 mAb for detection by confocal microscopy. Cells were incubated with BM1 mAb overnight. Cells were washed and subsequently incubated with a secondary antibody (Alexa Fluor 488 anti-mouse IgG1, 1:100) for 4 hrs. Nuclei were stained with DAPI (15 min) and visualized by a Zeiss confocal microscope. (D). L. acidophilus-SlpA detection with BM1 by ELISA. ELISA plates were coated with 500 ng of purified SlpA overnight, and binding by BM1 was tested thereafter. (E). Two-fold serial dilutions of SlpA were coated on ELISA plate overnight, and binding of BM1 was tested. (F). Germ-free (GF) mice were orally treated with 109 CFU L. acidophilus, 150 μg of SlpA, or left untreated. Fecal pellets from these mice were used to coat ELISA plates; BSA was used as a negative control. BM1 mAb only bound to plates coated with feces derived from treated mice. All animal experiments were performed under the guidelines of the Animal Welfare Act and the Public Health Policy on Humane Care, and with approval by the Institutional Animal Care and Use Committee (IACUC protocol 201406559) at the University of Florida. Data represent observations from 4 independent experiments (n = 4) and are shown as mean ± standard error of the mean. ** denotes statistical significance p < 0.01, ***p < 0.001.
Concluding Remarks
To gain further insights into the physiological effects of SlpA, studies have been performed to elucidate the feasibility of Phase I clinical trials using this protein. It appears that SlpA, using the newly employed purification method, does not elicit potential toxicity when administered orally to animals, and that the structural epitope(s) of this bacterial protein can still be recognized by the mAb generated in our laboratory, even after it is excreted in the feces. The same antibody may potentially be used for the detection of SlpA in colonic tissues, to evaluate its processing by immune cells. We are currently working on optimizing the isolation and the labeling of this antibody for its use in tissue staining. Nonetheless, further mechanistic studies, such as those focusing on the local and peripheral effects, and evaluation of targeted and untargeted metabolomics in treated animals, are required to demonstrate the role of SlpA on host physiology. These studies will also shed light on the effects of SlpA on other intestinal immune cells, including epithelial cells, colonic B cells, which mount critical humoral immune responses (e.g., IgA), and innate lymphoid cells (ILCs) in steady state and with colonic disease.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgment
We thank Dr. Todd Klaenhammer for fruitful discussion.
Funding
This work was supported by NIH R01 AI093370, the Department of Defense CA111002, the NIH/NCRR Clinical & Translational Science Award, a grant from GatoradeTrust Funds distributed by the University of Florida, Department of Medicine, and the Florida Breast Cancer foundation to the University of Florida (UL1 RR029890).
References
- 1.Kappelman MD, Rifas-Shiman SL, Kleinman K, Ollendorf D, Bousvaros A, Grand RJ, Finkelstein JA. The prevalence and geographic distribution of Crohn's disease and ulcerative colitis in the United States. Clin Gastroenterol Hepatol 2007; 5:1424-9; PMID:17904915; http://dx.doi.org/ 10.1016/j.cgh.2007.07.012 [DOI] [PubMed] [Google Scholar]
- 2.Kappelman MD, Moore KR, Allen JK, Cook SF. Recent trends in the prevalence of Crohn's disease and ulcerative colitis in a commercially insured US population. Dig Dis Sci 2013; 58:519-25; PMID:22926499; http://dx.doi.org/ 10.1007/s10620-012-2371-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walsh CJ, Guinane CM, O'Toole PW, Cotter PD. Beneficial modulation of the gut microbiota. FEBS Lett 2014; 588:4120-30; PMID:24681100; http://dx.doi.org/ 10.1016/j.febslet.2014.03.035 [DOI] [PubMed] [Google Scholar]
- 4.Major G, Spiller R. Irritable bowel syndrome, inflammatory bowel disease and the microbiome. Curr Opin Endocrinol Diabetes Obes 2014; 21:15-21; PMID:24296462; http://dx.doi.org/ 10.1097/MED.0000000000000032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee YK, Mazmanian SK. Has the microbiota played a critical role in the evolution of the adaptive immune system? Science 2010; 330:1768-73; PMID:21205662; http://dx.doi.org/ 10.1126/science.1195568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell 2014; 157:121-41; PMID:24679531; http://dx.doi.org/ 10.1016/j.cell.2014.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neurath MF. Cytokines in inflammatory bowel disease. Nat Rev Immunol 2014; 14:329-42; PMID:24751956; http://dx.doi.org/ 10.1038/nri3661 [DOI] [PubMed] [Google Scholar]
- 8.Geremia A, Biancheri P, Allan P, Corazza GR, Di Sabatino A. Innate and adaptive immunity in inflammatory bowel disease. Autoimmun Rev 2014; 13:3-10; PMID:23774107; http://dx.doi.org/ 10.1016/j.autrev.2013.06.004 [DOI] [PubMed] [Google Scholar]
- 9.Khazaie K, Zadeh M, Khan MW, Bere P, Gounari F, Dennis K, Blatner NR, Owen JL, Klaenhammer TR, Mohamadzadeh M. Abating colon cancer polyposis by Lactobacillus acidophilus deficient in lipoteichoic acid. Proc Natl Acad Sci U S A 2012; 109:10462-7; PMID:22689992; http://dx.doi.org/ 10.1073/pnas.1207230109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mohamadzadeh M, Pfeiler EA, Brown JB, Zadeh M, Gramarossa M, Managlia E, Bere P, Sarraj B, Khan MW, Pakanati KC, et al.. Regulation of induced colonic inflammation by Lactobacillus acidophilus deficient in lipoteichoic acid. Proc Natl Acad Sci U S A 2011; 108 Suppl 1:4623-30; PMID:21282652; http://dx.doi.org/ 10.1073/pnas.1005066107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khan MW, Zadeh M, Bere P, Gounaris E, Owen J, Klaenhammer T, Mohamadzadeh M. Modulating intestinal immune responses by lipoteichoic acid-deficient Lactobacillus acidophilus. Immunotherapy 2012; 4:151-61; PMID:22339459; http://dx.doi.org/ 10.2217/imt.11.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zadeh M, Khan MW, Goh YJ, Selle K, Owen JL, Klaenhammer T, Mohamadzadeh M. Induction of intestinal pro-inflammatory immune responses by lipoteichoic acid. J Inflamm (Lond) 2012; 9:7; PMID:22423982; http://dx.doi.org/ 10.1186/1476-9255-9-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saber R, Zadeh M, Pakanati KC, Bere P, Klaenhammer T, Mohamadzadeh M. Lipoteichoic acid-deficient Lactobacillus acidophilus regulates downstream signals. Immunotherapy 2011; 3:337-47; PMID:21395377; http://dx.doi.org/ 10.2217/imt.10.119 [DOI] [PubMed] [Google Scholar]
- 14.Mohamadzadeh M, Owen JL. Reprogramming intestinal immunity is the answer to induced pathogenic inflammation. Immunotherapy 2011; 3:1415-7; PMID:22091675; http://dx.doi.org/ 10.2217/imt.11.140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Varley KE, Mitra RD. Bisulfite Patch PCR enables multiplexed sequencing of promoter methylation across cancer samples. Genome Res 2010; 20:1279-87; PMID:20627893; http://dx.doi.org/ 10.1101/gr.101212.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lightfoot YL, Yang T, Sahay B, Mohamadzadeh M. Targeting aberrant colon cancer-specific DNA methylation with lipoteichoic acid-deficient Lactobacillus acidophilus. Gut Microbes 2012; 4:84-8; PMID:23137966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang T, Owen JL, Lightfoot YL, Kladde MP, Mohamadzadeh M. Microbiota impact on the epigenetic regulation of colorectal cancer. Trends Mol Med 2013; 19:714-25; PMID:24051204; http://dx.doi.org/ 10.1016/j.molmed.2013.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Underhill DM, Ozinsky A. Toll-like receptors: key mediators of microbe detection. Curr Opin Immunol 2002; 14:103-10; PMID:11790539; http://dx.doi.org/ 10.1016/S0952-7915(01)00304-1 [DOI] [PubMed] [Google Scholar]
- 19.Weis WI, Taylor ME, Drickamer K. The C-type lectin superfamily in the immune system. Immunol Rev 1998; 163:19-34; PMID:9700499; http://dx.doi.org/ 10.1111/j.1600-065X.1998.tb01185.x [DOI] [PubMed] [Google Scholar]
- 20.Avall-Jaaskelainen S, Palva A. Lactobacillus surface layers and their applications. FEMS Microbiol Rev 2005; 29:511-29; PMID:15935509; http://dx.doi.org/ 10.1016/j.fmrre.2005.04.003 [DOI] [PubMed] [Google Scholar]
- 21.Sara M, Sleytr UB. S-Layer proteins. J Bacteriol 2000; 182:859-68; PMID:10648507; http://dx.doi.org/ 10.1128/JB.182.4.859-868.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Avall-Jaaskelainen S, Lindholm A, Palva A. Surface display of the receptor-binding region of the Lactobacillus brevis S-layer protein in Lactococcus lactis provides nonadhesive lactococci with the ability to adhere to intestinal epithelial cells. Appl Environ Microbiol 2003; 69:2230-6; PMID:12676705; http://dx.doi.org/ 10.1128/AEM.69.4.2230-2236.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lightfoot YL, Selle K, Yang T, Goh YJ, Sahay B, Zadeh M, Owen JL, Colliou N, Li E, Johannssen T, et al.. SIGNR3-dependent immune regulation by Lactobacillus acidophilus surface layer protein A in colitis. Embo J 2015; 34(7):881-95; PMID:25666591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lightfoot YL, Mohamadzadeh M. Tailoring gut immune responses with lipoteichoic acid-deficient Lactobacillus acidophilus. Front Immunol 2013; 4:25; PMID:23390423; http://dx.doi.org/ 10.3389/fimmu.2013.00025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Konstantinov SR, Smidt H, de Vos WM, Bruijns SC, Singh SK, Valence F, Molle D, Lortal S, Altermann E, Klaenhammer TR, et al.. S layer protein A of Lactobacillus acidophilus NCFM regulates immature dendritic cell and T cell functions. Proc Natl Acad Sci U S A 2008; 105:19474-9; PMID:19047644; http://dx.doi.org/ 10.1073/pnas.0810305105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Konstantinov SR, Smidt H, Akkermans AD, Casini L, Trevisi P, Mazzoni M, De Filippi S, Bosi P, de Vos WM. Feeding of Lactobacillus sobrius reduces Escherichia coli F4 levels in the gut and promotes growth of infected piglets. FEMS Microbiol Ecol 2008; 66:599-607; PMID:18537838; http://dx.doi.org/ 10.1111/j.1574-6941.2008.00517.x [DOI] [PubMed] [Google Scholar]
- 27.Searle BC. Scaffold: a bioinformatic tool for validating MS/MS-based proteomic studies. Proteomics 2010; 10:1265-9; PMID:20077414; http://dx.doi.org/ 10.1002/pmic.200900437 [DOI] [PubMed] [Google Scholar]
- 28.Center SA. Interpretation of liver enzymes. Vet Clin North Am Small Anim Pract 2007; 37:297-333, vii; PMID:17336677; http://dx.doi.org/ 10.1016/j.cvsm.2006.11.009 [DOI] [PubMed] [Google Scholar]
- 29.Thrall MA, Weiser G, Allison RW, Campbell TW, eds. Veterinary Hematology and Clinical Chemistry. Ames, Iowa: Wiley-Blackwell, 2012. [Google Scholar]
- 30.Bergeron RJ, Bharti N, Singh S, McManis JS, Wiegand J, Green LG. Vibriobactin antibodies: a vaccine strategy. J Med Chem 2009; 52:3801-13; PMID:19492834; http://dx.doi.org/ 10.1021/jm900119q [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simrell CR, Klein PA. Antibody responses of tumor-bearing mice to their own tumors captured and perpetuated as hybridomas. J Immunol 1979; 123:2386-94; PMID:226630 [PubMed] [Google Scholar]


