Abstract
The enteric nervous system (ENS) coordinates the major functions of the gastrointestinal tract. Its development takes place within a constantly changing environment which, after birth, culminates in the establishment of a complex gut microbiota. How such changes affect ENS development and its subsequent function throughout life is an emerging field of study that holds great interest but which is inadequately explored thus far. In this addendum, we discuss our recent findings showing that a component of the ENS, the enteric glial cell network that resides in the gut lamina propria, develops after birth and parallels the evolution of the gut microbiota. Importantly, this network was found to be malleable throughout life by incorporating new cells that arrive from the area of the gut wall in a process of directional movement which was controlled by the lumen gut microbiota. Finally, we postulate on the roles of the intestinal epithelium and the immune system as potential intermediaries between gut microbiota and ENS responses.
Keywords: enteric nervous system, enteric glial cells, gut microbiota, germ-free mice, gut immune system, inflammation, TLRs
Introduction
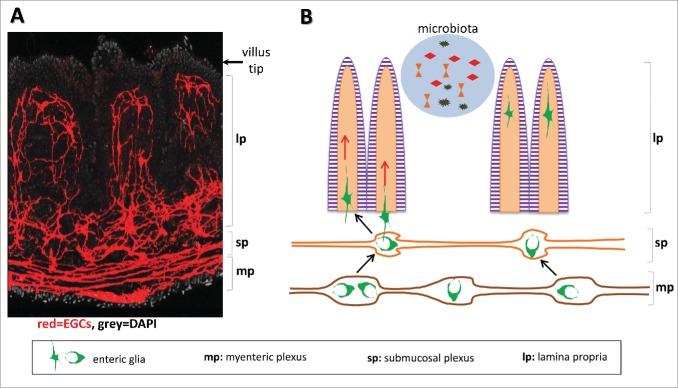
The enteric nervous system (ENS) is a vast and complex network of neurons and glial cells that extends throughout the length of the gastrointestinal tract and coordinates all essential functions of the organ. Neuronal cell bodies are exclusively located within enteric ganglia that are organized in 2 distinct plexi, the outer myenteric plexus sandwiched between the longitudinal and circular smooth muscles and the inner submucosal plexus located adjacent to the mucosa.1,2 Enteric glial cells (EGCs) closely associate with neurons within ganglia but are also distributed throughout the gut wall including the wider area of the mucosa where they form a network that extends to the tip of villi3,4 (Fig. 1A). In addition to their role in supporting neural circuits, EGCs have a role in strengthening the intestinal epithelial barrier5 and preventing overt gut inflammation6 as demonstrated in experimental systems. In humans, abnormalities in the structure of the ENS and increased cell death were detected in specimens from necrotising enterocolitis patients, a life-threatening inflammatory condition,7 and interestingly upregulation of disease markers was associated with EGC activation.8 These observations suggest multiple and diverse functions for these cells in gut homeostasis and warrant further investigation.
Figure 1.
The gut microbiota control the influx of EGCs in to the lamina propria. (A) The entire EGC network visualised following expression of the reporter protein in Sox10::CreERT2;Rosa26-tdTomato transgenic line. (B) Schematic depiction of the directional movement of EGCs from the gut wall located plexi toward the lamina propria.
Although the organization and connectivity of the ENS mainly forms during embryogenesis it persists postnatally for several weeks to attain a complete and fully functional neuroglial circuit.9,10 It was initially considered that embryonic development was proceeding in a sterile environment achieved by the barrier function of the placenta. However, this supposition has been challenged recently with data demonstrating the presence of a low-abundance, placenta-specific microbiome in both mice and humans the profile of which seems to be closer related to the oral microbiome.11-13 Furthermore, the meconium, the new-born's first intestinal discharge, harbours a variable microbiome which in some cases seems to be of intrauterine origin.14
Nonetheless, the gut lumen of mammals is further colonised by a seed of microorganisms acquired from the mother during parturition.15 Although comparative studies have shown significant variations in the composition of the early gut microbiome owing to factors such as mode of delivery15 and mother's diet,16 a 'core' of operational taxonomic units was identified in all cases pointing to a rudimentary microbial cluster that colonises the intestine immediately after birth.17 This initial seed expands and evolves gradually during the postnatal period to reach a mature adult state after weaning.18-20 Maturation and adaptation of the host immune system parallels that of gut microbiota, and there is strong experimental evidence demonstrating the extensive inter-dependence and cross-regulation of the 2 processes.21,22 Therefore, after birth the ENS is exposed to the growing complexity of the gut microbiota, its products and metabolites, as well as to a developing immune system. How these dramatic changes influence the postnatal stages of ENS development is unknown. Equally important is the question of whether and by what mechanism changes in gut microbiota as a result of aging,23 diet variations,24,25 or pharmacological interventions26 affect the function of the mature ENS.
Development of the EGC Network in the Mucosa Correlates with Maturation of the Gut Microbiota
Formation of the ENS plexi is complete by the time of birth although in the mouse a relatively small number of enteric neurons and (many more) glial cells are added during the first postnatal weeks.9 Nonetheless, by the time of birth, the mucosa has been invaded by neuronal processes with almost all the villi scoring positive for neuronal markers.27 In contrast, EGCs although present in the gut wall, are not observed in the lamina propria.27 To determine the time of appearance of mucosal EGCs, we examined ilea from animals at different postnatal stages and up to adulthood. This analysis revealed that full colonisation of the mucosa by EGCs occurs step-wise, initially during the suckling period and then after weaning to reach levels observed in the adult. Therefore, generation of the mucosal EGC network is a gradual process that parallels the maturation of the gut microbiota (and of the immune system) as it changes from the predominant species of the suckling period such as Lactobacilli, to species prominent in adults such as Bacteroidetes and Firmicutes.19
Mucosal EGCs are Replenished Throughout Life
In contrast to several constituent gut tissues (such as the epithelial cell layer, the villus vasculature, and the mucosal immune system) which are highly dynamic, the ENS has been portrayed as a rigid network that forms during development but remains unchanged thereafter. By employing Cre-LoxP-based genetic marking and fate mapping in adult animals, we have recently challenged this view and demonstrated that similar to other gut tissues the EGC network in the lamina propria does not remain unchanged but renews continuously. Adult mice carrying inducible Cre recombinase transgenes under the control of the glia-specific Sox10 or GFAP regulatory elements and the Cre-dependent Rosa26-Confetti reporter (Sox10::CreERT2;Rosa26-Confetti and GFAP::CreERT2;Rosa26-Confetti respectively) were injected with tamoxifen to activate Cre and induce expression of the reporter. Recombination of the Rosa26-Confetti cassette resulted in the stochastic expression of one out of 4 possible fluorescent proteins28 producing a mosaic of color -labeled EGCs. We noticed that shortly after induction (2–4 d post-induction) the majority of labeled EGCs were located in the area of the gut wall at the level of the plexi and very few colored cells were seen within the lamina propria. However, when animals were examined 2 weeks after induction there was a strong increase in the number of colored EGCs in the villi indicating that some cells that were initially labeled in the gut wall, or their progeny, migrated toward the lamina propria. Furthermore, the multicolour property of the Confetti reporter allowed us to demonstrate that the population of EGCs invading each villus was not clonal but instead originated from multiple independently labeled glial cells. Although the level of cell labeling differed between the 2 Cre drivers with Sox10::CreERT2 being considerably more efficient, both transgenic lines produced the same result establishing the directional movement of EGCs along the serosa-lumen axis which led us to investigate the role of indigenous lumen microbiota in this process (Fig. 1B). It should be noted that while expression of the GFAP locus was shown to be modulated by environmental cues such as tissue inflammation,29 to our knowledge, the same has not been reported for the Sox10 locus.
The Gut Microbiota Regulates the Influx of EGCs in to the Lamina Propria
To explore the potential role of luminal microbiota in driving the influx of EGCs in to the lamina propria, we compared adult mice maintained under sterile (germ-free) conditions to those raised in a specific pathogen-free (SPF; conventional) environment. The number of EGCs and the extent of the network they form were severely reduced in villi from germ-free mice. Importantly, when germ-free mice were conventionalised by the introduction of gut microbiota and examined 4 weeks later, a robust increase in the number and complexity of the mucosal EGC network was observed which was similar to that seen in conventionally raised animals. These findings highlight the plasticity and dynamic character of the mucosal EGC network in adult animals and reveal a key role for lumen microbiota in driving the influx of EGCs from the peripheral neuroglia plexi to the lamina propria.
To establish whether the continuous flow of EGCs is maintained by the presence of the microbiota, tamoxifen-induced Sox10::CreERT2;Rosa26-Confetti mice were treated with a cocktail of broad-spectrum antibiotics which results in a dramatic reduction in the diversity and volume of the microbiome, and the fate of labeled cells was compared to those from untreated littermates. In antibiotic-treated animals the flux of labeled EGCs in to villi was strongly diminished indicating that the continuous movement of EGCs along the serosa-lumen axis requires the steady supply of signals originating from the lumen microbiota.
Potential Mechanisms of Communication between Gut Microbiota and EGCs
The finding that the gut microbiota can regulate a component of the ENS at the fundamental level of structural organization and possibly connectivity opens a new avenue of investigation in gut physiology, and perhaps identifies the first step in understanding the mechanistic process that underpins the gut-brain axis.30 Within this context an important question emerging is the effective mechanism(s) via which gut microbiota stimulates the influx of EGCs in to the lamina propria. Here we provide some possible explanations and potential directions for future research, although at this point in time they represent only speculation (Fig. 2). The mechanisms discussed below are not mutually exclusive and could operate in concert.
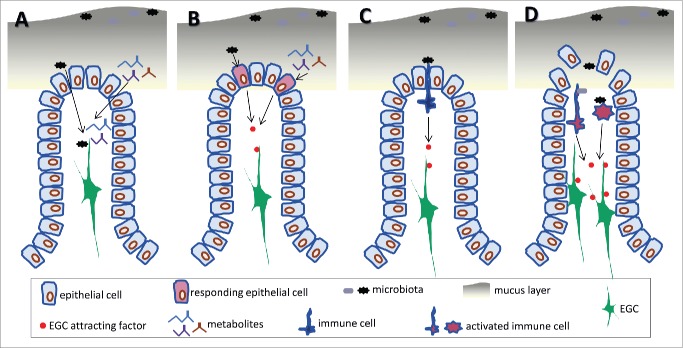
Figure 2.
Possible mechanisms underpinning the stimulation of EGCs by the gut microbiota. (A) Components or metabolites of the microbiota can gain access to the lamina propria and interact directly with EGCs. (B) Recognition of microbiota by the intestinal epithelium and production of soluble factors capable of attracting EGCs in to the villi. (C) Identification of microbiota by immune cells residing in the lamina propria and production of factors that attract EGCs. (D) Compromised integrity of the intestinal barrier leading to activation of immune cells and production of factors that act on EGCs.
One possibility is that EGCs are equipped with the required molecular apparatuses, for example expressing appropriate pattern recognition receptors such as TLRs, and thus capable of directly detecting and responding to the microbiota. Expression of certain members of the TLR family in the ENS has been documented31 and furthermore, at least in an experimental setting, EGCs could participate in immune regulation in vivo.32 Interestingly, TLR4 signaling was found to influence gastrointestinal motility,33 while TLR2 regulated intestinal inflammation in part by controlling the integrity of the ENS.34 In this scenario, elements of the microbiota will have to penetrate through the mucus layer and the epithelial barrier, and possibly in significant volume, in order to reach the mucosa if they are to be detected by the EGCs (Fig. 2A). Although this possibility cannot be excluded, it would seem more plausible in situations where there is a transient breach of the intestinal barrier such as during pathogenic infection or inflammation (Fig. 2D), or possibly in certain chronic conditions associated with reduced strength of the intestinal barrier (leaky gut syndrome), which however are only poorly defined at present.35 Alternatively, there could be an active transport by the intestinal epithelium of certain microbiota-derived components so that they are presented to host cells residing in the lamina propria including EGCs. Furthermore, EGCs might respond to products, such as short chain fatty acids (SCFAs), generated by the metabolic activity of the microbiota (Fig. 2A). SCFAs (primarily butyric acid) were recently shown to play key role in the development of colonic T regulatory cells36-38 and it is conceivable that might play an equally important role in the regulation of the ENS. Enteric neurons express SCFA receptors39 and exogenous supply of butyrate was shown to regulate enteric neuron activity and to control motility.40 Further exploration of this area could reveal important insights in to the mechanism of ENS response to the microbiome but also to changes in diet.
Another potential mechanism could involve the intestinal epithelium as the intervening cell type which by responding to the microbiota or its metabolic products produces mediators that act upon EGCs (Fig. 2B). The response of the intestinal epithelium to the gut microbiota has been documented and shown to involve TLR signaling.41,42 In addition, a recent report demonstrates the effect of butyric acid which via stabilization of the hypoxia-inducible transcription factor strengthens epithelial cell tight junctions and the intestinal barrier.43 Nonetheless, validation of this theory will have to await the identification of mediators produced by the intestinal epithelium in the presence of microbiota which can act upon EGCs and in particular mediators with chemotactic action.
A third possibility is the involvement of the immune system which has evolved to recognize and respond to foreign material and is abundant in the intestine owing to the presence of the luminal microflora.44 In this scenario, components of the gut microbiota are recognized by immune cells, predominantly in the area of the lamina propria which is adjacent to the lumen, resulting in the production of mediators that attract EGCs from the gut wall (Fig. 2C). Recent work identified a mechanism of cross-regulation between the ENS and the immune system where enteric neurons of the myenteric plexus produce CSF1 to attract macrophages which in turn produce BMP2 to support the operation of enteric neurons.45 Importantly, this cross-communication is regulated by the presence of the indigenous microbiota as in its absence the number of macrophages is severely reduced. Furthermore, Schwann cells, glial cells of the peripheral nervous system, have the capacity to respond to the activation of immune cells following traumatic injury by mobilising and readjusting their position to form tracks that allow neuronal process regeneration.46 Also, astrocytes respond to factors produced from activated immune cells following injury in the central nervous system.47 Collectively, these results make the case for a detailed study to delineate the molecular basis of the immune-ENS system cross-communication.
The mucosa is the part of the gut that is most amenable to microbiota insult especially when there is abrupt epithelial barrier breach (infection by pathogens, excessive immune response) or chronic barrier leakiness (leaky gut). It is conceivable that the EGC movement in to the mucosa might reflect a defense mechanism the purpose of which is to protect resident neuronal processes from damage and to contribute to the strengthening of the intestinal barrier preventing detrimental dissemination of microorganisms away from the intestinal canal. Consistent with this supposition are reports demonstrating the positive role of mucosal EGCs in strengthening the intestinal barrier48,49 and the fact that their ablation in an experimental setting results in the loss of intestinal homeostasis and severe inflammation in the small intestine.6 Nonetheless, EGCs might perform additional functions in the lamina propria which will be important to identify in future research.
In recent years, a number of laboratories have demonstrated the modulatory effects the gut microbiota has on the physiological function of host systems, including the central nervous system in a process termed "the gut-brain axis".50-52 Mechanistically, this process will most likely turn out to be complex and multidimensional. Nonetheless it is reasonable to assume that the ENS, owing to its physical proximity, is the nervous system best poised to respond to the gut microbiota and to relay important information. Recent work demonstrating the involvement of gut-brain neural circuits in mediating the beneficial effect of microbiota-produced metabolites,53 and on the other hand, the proposed link between gut dysbiosis and certain central nervous system diseases such as autism spectrum disorders,54,55 highlight the importance of fully understanding how the ENS communicates and responds to the gut microbiota and its products. This area of research promises to reveal important aspects of gut physiology and potentially novel therapeutic opportunities for certain diseases.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the Medical Research Council UK (grant-in-aid U117537087) and the Biotechnology and Biological Sciences Research Council (BBSRC, grant L022974/1) to V.P.
References
- 1.Furness JB, The enteric nervous system and neurogastroenterology. Nat Rev Gastroenterol Hepatol 2012; 9:286-94; PMID:22392290; http://dx.doi.org/ 10.1038/nrgastro.2012.32 [DOI] [PubMed] [Google Scholar]
- 2.Heanue TA, Pachnis V. Enteric nervous system development and Hirschsprung's disease: advances in genetic and stem cell studies. Nat Rev Neurosci 2007; 8:466-79; PMID:17514199; http://dx.doi.org/ 10.1038/nrn2137 [DOI] [PubMed] [Google Scholar]
- 3.Boesmans W, Lasrado R, Vanden Berghe P, Pachnis V. Heterogeneity and phenotypic plasticity of glial cells in the mammalian enteric nervous system. Glia 2015; 63:229-41; PMID:25161129; http://dx.doi.org/ 10.1002/glia.22746 [DOI] [PubMed] [Google Scholar]
- 4.Gulbransen BD, Sharkey KA. Novel functional roles for enteric glia in the gastrointestinal tract. Nat Rev Gastroenterol Hepatol 2012; 9:625-32; PMID:22890111; http://dx.doi.org/ 10.1038/nrgastro.2012.138 [DOI] [PubMed] [Google Scholar]
- 5.Neunlist M, Van Landeghem L, Mahe MM, Derkinderen P, des Varannes SB, Rolli-Derkinderen M. The digestive neuronal-glial-epithelial unit: a new actor in gut health and disease. Nat Rev Gastroenterol Hepatol 2013; 10:90-100; PMID:23165236; http://dx.doi.org/ 10.1038/nrgastro.2012.221 [DOI] [PubMed] [Google Scholar]
- 6.Bush TG, Savidge TC, Freeman TC, Cox HJ, Campbell EA, Mucke L, Johnson MH, Sofroniew MV. Fulminant jejuno-ileitis following ablation of enteric glia in adult transgenic mice. Cell 1998; 93:189-201; PMID:9568712; http://dx.doi.org/ 10.1016/S0092-8674(00)81571-8 [DOI] [PubMed] [Google Scholar]
- 7.Zhou Y, Yang J, Watkins DJ, Boomer LA, Matthews MA, Su Y, Besner GE. Enteric nervous system abnormalities are present in human necrotizing enterocolitis: potential neurotransplantation therapy. Stem Cell Res Ther 2013; 4:157; PMID:24423414; http://dx.doi.org/ 10.1186/scrt387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fagbemi AO, Torrente F, Puleston J, Lakhoo K, James S, Murch SH. Enteric neural disruption in necrotizing enterocolitis occurs in association with myenteric glial cell CCL20 expression. J Pediatr Gastroenterol Nutr 2013; 57:788-93; PMID:24280992; http://dx.doi.org/ 10.1097/MPG.0b013e3182a86fd4 [DOI] [PubMed] [Google Scholar]
- 9.Laranjeira C, Sandgren K, Kessaris N, Richardson W, Potocnik A, Vanden Berghe P, Pachnis V. Glial cells in the mouse enteric nervous system can undergo neurogenesis in response to injury. J Clin Invest 2011; 121:3412-24; PMID:21865647; http://dx.doi.org/ 10.1172/JCI58200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roberts RR, Murphy JF, Young HM, Bornstein JC. Development of colonic motility in the neonatal mouse-studies using spatiotemporal maps. Am J Physiol Gastrointest Liver Physiol 2007; 292:G930-8; PMID:17158255; http://dx.doi.org/ 10.1152/ajpgi.00444.2006 [DOI] [PubMed] [Google Scholar]
- 11.Aagaard K, Ma J, Antony KM, Ganu R, Petrosino J, Versalovic J. The placenta harbors a unique microbiome. Sci Transl Med 2014; 6:237ra65; PMID:24848255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fardini Y, Chung P, Dumm R, Joshi N, Han YW. Transmission of diverse oral bacteria to murine placenta: evidence for the oral microbiome as a potential source of intrauterine infection. Infect Immun 2010; 78:1789-96; PMID:20123706; http://dx.doi.org/ 10.1128/IAI.01395-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stout MJ, Conlon B, Landeau M, Lee I, Bower C, Zhao Q, Roehl KA, Nelson DM, Macones GA, Mysorekar IU. Identification of intracellular bacteria in the basal plate of the human placenta in term and preterm gestations. Am J Obstet Gynecol 2013; 208:226 e1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gosalbes MJ, Llop S, Valles Y, Moya A, Ballester F, Francino MP. Meconium microbiota types dominated by lactic acid or enteric bacteria are differentially associated with maternal eczema and respiratory problems in infants. Clin Exp Allergy 2013; 43:198-211; PMID:23331561; http://dx.doi.org/ 10.1111/cea.12063 [DOI] [PubMed] [Google Scholar]
- 15.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A 2010; 107:11971-5; PMID:20566857; http://dx.doi.org/ 10.1073/pnas.1002601107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma J, Prince AL, Bader D, Hu M, Ganu R, Baquero K, Blundell P, Alan Harris R, Frias AE, Grove KL, et al. . High-fat maternal diet during pregnancy persistently alters the offspring microbiome in a primate model. Nat Commun 2014; 5:3889; PMID:24846660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Del Chierico F, Vernocchi P, Petrucca A, Paci P, Fuentes S, Pratico G, Capuani G, Masotti A, Reddel S, Russo A, et al. . Phylogenetic and metabolic tracking of gut microbiota during perinatal development. PloS One 2015; 10:e0137347; PMID:26332837; http://dx.doi.org/ 10.1371/journal.pone.0137347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, Ley RE. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A 2011; 108 Suppl 1:4578-85; PMID:20668239; http://dx.doi.org/ 10.1073/pnas.1000081107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S. Host-gut microbiota metabolic interactions. Science 2012; 336:1262-7; PMID:22674330; http://dx.doi.org/ 10.1126/science.1223813 [DOI] [PubMed] [Google Scholar]
- 20.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol 2007; 5:e177; PMID:17594176; http://dx.doi.org/ 10.1371/journal.pbio.0050177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell 2014; 157:121-41; PMID:24679531; http://dx.doi.org/ 10.1016/j.cell.2014.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maynard CL, Elson CO, Hatton RD, Weaver CT. Reciprocal interactions of the intestinal microbiota and immune system. Nature 2012; 489:231-41; PMID:22972296; http://dx.doi.org/ 10.1038/nature11551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Claesson MJ, Cusack S, O'Sullivan O, Greene-Diniz R, de Weerd H, Flannery E, Marchesi JR, Falush D, Dinan T, Fitzgerald G, et al. . Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc Natl Acad Sci U S A 2011; 108 Suppl 1:4586-91; PMID:20571116; http://dx.doi.org/ 10.1073/pnas.1000097107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Turnbaugh PJ, Backhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 2008; 3:213-23; PMID:18407065; http://dx.doi.org/ 10.1016/j.chom.2008.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, et al. . Linking long-term dietary patterns with gut microbial enterotypes. Science 2011; 334:105-8; PMID:21885731; http://dx.doi.org/ 10.1126/science.1208344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol 2008; 6:e280; PMID:19018661; http://dx.doi.org/ 10.1371/journal.pbio.0060280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kabouridis PS, Lasrado R, McCallum S, Chng SH, Snippert HJ, Clevers H, Pettersson S, Pachnis V. Microbiota controls the homeostasis of glial cells in the gut lamina propria. Neuron 2015; 85:289-95; PMID:25578362; http://dx.doi.org/ 10.1016/j.neuron.2014.12.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Snippert HJ, van der Flier LG, Sato T, van Es JH, van den Born M, Kroon-Veenboer C, Barker N, Klein AM, van Rheenen J, Simons BD, et al. . Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 2010; 143:134-44; PMID:20887898; http://dx.doi.org/ 10.1016/j.cell.2010.09.016 [DOI] [PubMed] [Google Scholar]
- 29.von Boyen GB, Steinkamp M, Reinshagen M, Schafer KH, Adler G, Kirsch J. Proinflammatory cytokines increase glial fibrillary acidic protein expression in enteric glia. Gut 2004; 53:222-8; PMID:14724154; http://dx.doi.org/ 10.1136/gut.2003.012625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dinan TG, Cryan JF. The impact of gut microbiota on brain and behaviour: implications for psychiatry. Curr Opin Clin Nutr Metab Care 2015; 18(6):552-8; PMID:26372511 [DOI] [PubMed] [Google Scholar]
- 31.Ruhl A. Glial cells in the gut. Neurogastroenterol Motil 2005; 17:777-90; PMID:16336493; http://dx.doi.org/ 10.1111/j.1365-2982.2005.00687.x [DOI] [PubMed] [Google Scholar]
- 32.Cornet A, Savidge TC, Cabarrocas J, Deng WL, Colombel JF, Lassmann H, Desreumaux P, Liblau RS. Enterocolitis induced by autoimmune targeting of enteric glial cells: a possible mechanism in Crohn's disease? Proc Natl Acad Sci U S A 2001; 98:13306-11; PMID:11687633; http://dx.doi.org/ 10.1073/pnas.231474098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anitha M, Vijay-Kumar M, Sitaraman SV, Gewirtz AT, Srinivasan S. Gut microbial products regulate murine gastrointestinal motility via Toll-like receptor 4 signaling. Gastroenterology 2012; 143:1006-16 e4; PMID:22732731; http://dx.doi.org/ 10.1053/j.gastro.2012.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brun P, Giron MC, Qesari M, Porzionato A, Caputi V, Zoppellaro C, Banzato S, Grillo AR, Spagnol L, De Caro R, et al. . Toll-like receptor 2 regulates intestinal inflammation by controlling integrity of the enteric nervous system. Gastroenterology 2013; 145:1323-33; PMID:23994200; http://dx.doi.org/ 10.1053/j.gastro.2013.08.047 [DOI] [PubMed] [Google Scholar]
- 35.Barbara G, Zecchi L, Barbaro R, Cremon C, Bellacosa L, Marcellini M, De Giorgio R, Corinaldesi R, Stanghellini V. Mucosal permeability and immune activation as potential therapeutic targets of probiotics in irritable bowel syndrome. J Clin Gastroenterol 2012; 46 Suppl:S52-5; PMID:22955358; http://dx.doi.org/ 10.1097/MCG.0b013e318264e918 [DOI] [PubMed] [Google Scholar]
- 36.Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, Liu H, Cross JR, Pfeffer K, Coffer PJ, et al. . Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013; 504:451-5; PMID:24226773; http://dx.doi.org/ 10.1038/nature12726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, et al. . Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013; 504:446-50; PMID:24226770; http://dx.doi.org/ 10.1038/nature12721 [DOI] [PubMed] [Google Scholar]
- 38.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, Glickman JN, Garrett WS. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 2013; 341:569-73; PMID:23828891; http://dx.doi.org/ 10.1126/science.1241165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nohr MK, Pedersen MH, Gille A, Egerod KL, Engelstoft MS, Husted AS, Sichlau RM, Grunddal KV, Poulsen SS, Han S, et al. . GPR41/FFAR3 and GPR43/FFAR2 as cosensors for short-chain fatty acids in enteroendocrine cells vs FFAR3 in enteric neurons and FFAR2 in enteric leukocytes. Endocrinology 2013; 154:3552-64; PMID:23885020; http://dx.doi.org/ 10.1210/en.2013-1142 [DOI] [PubMed] [Google Scholar]
- 40.Soret R, Chevalier J, De Coppet P, Poupeau G, Derkinderen P, Segain JP, Neunlist M. Short-chain fatty acids regulate the enteric neurons and control gastrointestinal motility in rats. Gastroenterology 2010; 138:1772-82; PMID:20152836; http://dx.doi.org/ 10.1053/j.gastro.2010.01.053 [DOI] [PubMed] [Google Scholar]
- 41.Mukherji A, Kobiita A, Ye T, Chambon P. Homeostasis in intestinal epithelium is orchestrated by the circadian clock and microbiota cues transduced by TLRs. Cell 2013; 153:812-27; PMID:23663780; http://dx.doi.org/ 10.1016/j.cell.2013.04.020 [DOI] [PubMed] [Google Scholar]
- 42.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 2004; 118:229-41; PMID:15260992; http://dx.doi.org/ 10.1016/j.cell.2004.07.002 [DOI] [PubMed] [Google Scholar]
- 43.Kelly CJ, Zheng L, Campbell EL, Saeedi B, Scholz CC, Bayless AJ, Wilson KE, Glover LE, Kominsky DJ, Magnuson A, et al. . Crosstalk between Microbiota-Derived Short-Chain Fatty Acids and Intestinal Epithelial HIF Augments Tissue Barrier Function. Cell Host Microbe 2015; 17:662-71; PMID:25865369; http://dx.doi.org/ 10.1016/j.chom.2015.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science 2012; 336:1268-73; PMID:22674334; http://dx.doi.org/ 10.1126/science.1223490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muller PA, Koscso B, Rajani GM, Stevanovic K, Berres ML, Hashimoto D, Mortha A, Leboeuf M, Li XM, Mucida D, et al. . Crosstalk between muscularis macrophages and enteric neurons regulates gastrointestinal motility. Cell 2014; 158:300-13; PMID:25036630; http://dx.doi.org/ 10.1016/j.cell.2014.04.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arthur-Farraj PJ, Latouche M, Wilton DK, Quintes S, Chabrol E, Banerjee A, Woodhoo A, Jenkins B, Rahman M, Turmaine M, et al. . c-Jun reprograms Schwann cells of injured nerves to generate a repair cell essential for regeneration. Neuron 2012; 75:633-47; PMID:22920255; http://dx.doi.org/ 10.1016/j.neuron.2012.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bardehle S, Kruger M, Buggenthin F, Schwausch J, Ninkovic J, Clevers H, Snippert HJ, Theis FJ, Meyer-Luehmann M, Bechmann I, et al. . Live imaging of astrocyte responses to acute injury reveals selective juxtavascular proliferation. Nat Neurosci 2013; 16:580-6; PMID:23542688; http://dx.doi.org/ 10.1038/nn.3371 [DOI] [PubMed] [Google Scholar]
- 48.Savidge TC, Newman P, Pothoulakis C, Ruhl A, Neunlist M, Bourreille A, Hurst R, Sofroniew MV. Enteric glia regulate intestinal barrier function and inflammation via release of S-nitrosoglutathione. Gastroenterology 2007; 132:1344-58; PMID:17408650; http://dx.doi.org/ 10.1053/j.gastro.2007.01.051 [DOI] [PubMed] [Google Scholar]
- 49.Van Landeghem L, Chevalier J, Mahe MM, Wedel T, Urvil P, Derkinderen P, Savidge T, Neunlist M. Enteric glia promote intestinal mucosal healing via activation of focal adhesion kinase and release of proEGF. Am J Physiol Gastrointest Liver Physiol 2011; 300:G976-87; PMID:21350188; http://dx.doi.org/ 10.1152/ajpgi.00427.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Braniste V, Al-Asmakh M, Kowal C, Anuar F, Abbaspour A, Toth M, Korecka A, Bakocevic N, Ng LG, Kundu P, et al. . The gut microbiota influences blood-brain barrier permeability in mice. Sci Transl Med 2014; 6:263ra158; PMID:25411471; http://dx.doi.org/ 10.1126/scitranslmed.3009759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cryan JF, Dinan TG. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci 2012; 13:701-12; PMID:22968153; http://dx.doi.org/ 10.1038/nrn3346 [DOI] [PubMed] [Google Scholar]
- 52.Diaz Heijtz R, Wang S, Anuar F, Qian Y, Bjorkholm B, Samuelsson A, Hibberd ML, Forssberg H, Pettersson S. Normal gut microbiota modulates brain development and behavior. Proc Natl Acad Sci U S A 2011; 108:3047-52; PMID:21282636; http://dx.doi.org/ 10.1073/pnas.1010529108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.De Vadder F, Kovatcheva-Datchary P, Goncalves D, Vinera J, Zitoun C, Duchampt A, Bäckhed F, Mithieux G. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell 2014; 156:84-96; PMID:24412651; http://dx.doi.org/ 10.1016/j.cell.2013.12.016 [DOI] [PubMed] [Google Scholar]
- 54.Chaidez V, Hansen RL, Hertz-Picciotto I. Gastrointestinal problems in children with autism, developmental delays or typical development. J Autism Dev Disord 2014; 44:1117-27; PMID:24193577; http://dx.doi.org/ 10.1007/s10803-013-1973-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T, Codelli JA, Chow J, Reisman SE, Petrosino JF, et al. . Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell 2013; 155:1451-63; PMID:24315484; http://dx.doi.org/ 10.1016/j.cell.2013.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]