Abstract
Cellular and molecular mechanisms that suppress small RNAs in oocytes while maintaining them in zygotes remain unknown. Signal-mediated regulation of small RNA biogenesis pathway is emerging as a theme for regulating small RNA production. We recently reported that ERK-mediated phosphorylation of Dicer, a central player in small RNA biogenesis, induced Dicer to move from the cytoplasm to the nucleus. Dicer phosphorylation inhibited its function, e.g., the production of 26G endo-siRNAs in the female germline. Moreover, our findings showed that the inhibition of Dicer function was necessary for normal progression of meiosis I and oogenesis, and that Dicer function had to be restored before fertilization for normal progression of embryogenesis. Thus, extracellular signal-dependent inhibition and then reactivation of Dicer is essential for oocyte-to-embryo transition. Strikingly, signal-induced Dicer translocation from the cytoplasm to nucleus is evolutionarily conserved from worm, flies, mice to humans thereby suggesting the ERK-mediated control of Dicer activity may be a generalized mechanism for regulating small RNA biogenesis.
Keywords: ERK Signaling, Dicer phosphorylation, oocyte-to-embryo transition
Introduction
Organismal development requires precise orchestration of multiple signaling networks and molecular pathways. A key molecular pathway that governs developmental transitions is the small non-coding RNA pathway such as the microRNA (miRNA) and the siRNA pathway.1,2 The miRNA and the siRNA pathways were uncovered as mediators of developmental transitions during C. elegans larval development, and regulate gene expression either through sequence specific degradation of mRNAs as a post-transcriptional gene silencing mechanism or through translational repression of mRNAs (eg., let-7 miRNA).3-5 Generation of the functional miRNA or siRNA is mediated by a series of processing steps; in particular the Dicer RNAse III enzyme cleaves the dsRNA or pre-miRNA into the final siRNA and miRNA forms, respectively.3,6,7 These small RNAs modulate gene expression under multiple developmental and tissue contexts, mediating diverse cellular responses like cell cycle regulation8 and differentiation.9 Molecular mechanisms that impose specificity on the small RNA biogenesis pathway remain to be fully elucidated. Argonaute proteins direct distinct species of small RNAs e.g., miRNA, piRNA, siRNAs into specific RNA Induced Silencing Complexes (RISC).10-12 Argonautes were thus thought to impose specificity in generation of distinct classes of small RNAs. However, since most Argonautes (e.g., Ago2) are ubiquitously expressed and often have the same phenotype as loss of Dicer,13,14 the question of how specificity is achieved in the pathway is still an intense area of investigation.
More recently, links between signaling pathways and small RNA biogenesis effectors have been uncovered suggesting that there may be context-dependent regulation of small RNA expression. For example, ERK phosphorylates TRBP2 protein which binds to Dicer and regulates its activity in vertebrates.15 Biochemically, phosphorylated TRBP2 stabilizes Dicer via direct binding. Functionally, however, binding of phosphorylated TRBP2 to Dicer can increase or decrease the abundance of miRNAs in cultured cells.15 An increase in miRNA abundance is predicted based on enhanced stability of Dicer; however, a decrease in miRNAs is unexpected and suggests that there may be additional levels of control that regulate or modulate Dicer activity, which in turn influences the abundance of miRNAs. Thus, post-translational control of catalytic enzymes is increasingly in focus as an important mechanism for context-dependent regulation of small RNAs. While multiple mechanisms are proposed in cultured cells, the in vivo context of such regulation, why such regulation would occur and what its biological consequences might be, are only just beginning to be elucidated.
The RAS-MPK-1 Pathway Coordinates C. elegans Germline Development
The C. elegans germline is organized in a distal to proximal polarity with respect to the uterus; the germline stem cells are at the distal end and mature oocytes at the proximal end16 (Fig. 1). Germ cells initiate meiosis in the transition zone (TZ) and progress through an extended meiotic prophase of pachytene. On the surface, the cells are packaged into a hexagonal symmetry via the plasma membrane (Fig. 1A). Due to incomplete cytokinesis, the distal mitotic region forms a syncytium whereby the cytoplasm is shared among the majority of the germ cells from distal mitotic region until the loop region (Fig. 1A). At the diplotene stage in the loop region the syncytial germ cells morph into individual oocytes. The oocytes then undergo growth and maturation during diakinesis in the proximal region.16 Genetic studies from many labs have shown that the RAS-MPK-1 (ERK) pathway is essential for oogenesis.17-19 For example, MPK-1 coordinately controls 7 distinct biological processes during oogenesis such as germ cell apoptosis and oocyte growth.18 The total MPK-1 protein is distributed homogeneously throughout the germline. However, the active form of MPK-1, visualized by an antibody specific to the diphosphorylated form of MPK-1 (dpMPK-1), displays a dynamic and spatially restricted localization pattern (Fig. 1B). MPK-1 is first activated and high during pachytene, low in the loop region, and then high again in proximal oocytes due to the sperm signal20 (Fig. 1B). Of note, dpMPK-1 is dramatically downregulated in the final oocyte right after it completes meiotic maturation and just before ovulation and fertilization21 (Fig. 1B).
Figure 1.
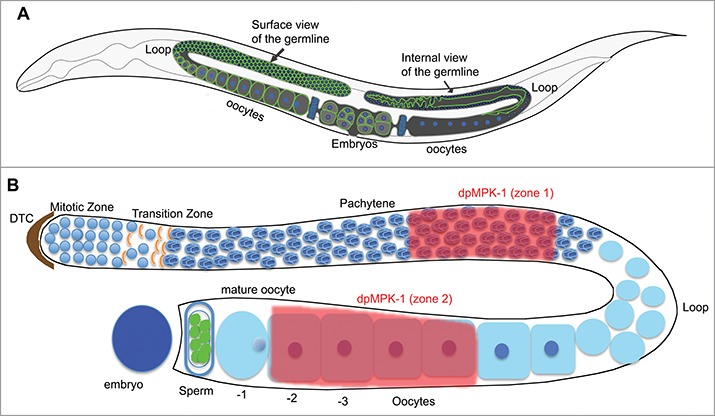
C. elegans germline organization. (A) C. elegans germline has an organized cell surface (left arm), with hexagonal plasma membranes. Due to incomplete cytokinesis in the distal region, the germline has a central rachis with shared cytoplasm (right arm). The two gonad arms end in common uterus. (B) Schematized view of one gonad arm. MPK-1 is active in mid-pachytene, downregulated in the loop, active proximal diakinetic oocytes and turned off in mature oocyte. Active ERK has not been demonstrated in early embryo.
Nuclear Translocation of Dicer is Dependent on ERK-Mediated Phosphorylation and is Conserved Evolutionarily
Expression of Dicer during oogenesis is conserved from C. elegans to mammals.1,21,22 In C. elegans, complete loss of Dicer (DCR-1 in worms) blocks oocyte meiotic maturation,23 whereas a reduction in Dicer function allows for oogenesis to proceed but results in early embryonic death.21 Spatially, Dicer is expressed throughout the germline.22 At the cellular level, Dicer is expressed in the cytoplasm and P granules.21 In a screen designed to identify targets of MPK-1 ERK in the C. elegans germline we identified, and biochemically confirmed, DCR-1 as an ERK target.17 Subsequently, we showed that human and mouse Dicer were also targets of ERK phosphorylation in vitro and in cell culture. Characterization of phosphorylated Dicer using phospho-specific antibodies during C. elegans germline development demonstrated that phospho-Dicer was nuclear from mid-pachytene onwards, overlapping with dpMPK-1 expression pattern. Thus, Dicer phosphorylation was dependent on the activation of MPK-1 and resulted in the translocation of phospho-Dicer from the cytoplasm to the nucleus, in vivo. Using the antibody generated against the C. elegans DCR-1, we showed that human and mouse Dicer both underwent cytoplasm-to-nucleus translocation in an ERK-dependent manner, highlighting the conserved nature of this regulation.21 Taken together, the evolutionary conservation of ERK-mediated Dicer phosphorylation and its subsequent translocation suggest that this phenomenon may have important functional consequences in different biological contexts.
Phosphorylation of Dicer is Necessary and Sufficient to Trigger its Nuclear Localization
In its canonical function, Dicer acts in the cytoplasm24 to generate miRNAs and siRNAs. However, Dicer can localize to the nucleus in many contexts.25 For example, in fission yeast, a C-terminally-tagged version of Dicer localizes to perinuclear foci.26 The mechanism by which Dicer translocates to the nucleus in yeast, however, is unclear as fusion of only the C-terminal end of Dicer to GFP is not sufficient to direct GFP to the nucleus. Thus, the C-terminal end of Dicer does not behave as a simple nuclear localization signal in yeast.27 Dicer can also localize to chromosomes during mitosis in vertebrate cultured cells.28 The localization of Dicer to chromosomes in this context was specific to rDNA loci, but did not correlate with processing of pre-rRNA or the methylation status of these loci, and the mechanism of this nuclear translocation remains at large.
In C. elegans, Dicer is phosphorylated by MPK-1 on the C-terminally located RNase IIIb and dsRNA binding domains, on Serine 1705 and Serine 1833 respectively, and this phosphorylation renders Dicer nuclear in the germline.21 Visualization of GFP tagged Dicer transgenes revealed that the wild-type Dicer transgene is nuclear in localization in the germline in regions with high active MPK-1, and cytoplasmic in regions with no active MPK-1. Phospho-mimetic Dicer transgenes (wherein Serine 1705 and / or 1833 were replaced with Glutamic acid) displayed a nuclear localization of Dicer protein throughout the germline. Lack of Dicer phosphorylation (wherein Serine 1705 and / or 1833 were replaced with Alanine) rendered the Dicer protein cytoplasmic and in P granules, demonstrating that phosphorylation is necessary and sufficient to support nuclear localization of Dicer.
However, it remains unclear how phosphorylated Dicer finds its way to the nucleus. It is highly likely that phosphorylation may recruit nuclear import proteins such as the RAN/Importin mediated system. But it is also likely, based on crystal structure predictions,29 that phosphorylation of the C-terminal tail of Dicer results in exposure of a nuclear localization signal in the very C-terminal tail, which results in its nuclear import. Current ongoing studies in multiple labs and future studies should help address these questions. Functionally, phosphorylation of Dicer has a large impact on its activity, and reveals a novel role for Dicer in regulating progression of meiosis I, and oocyte-to-embryo transition.
Impact of Dicer Phosphorylation on Small RNAs in C. elegans
Deep sequencing analysis of small RNAs (in the range between 20 and 30 nucleotide) from wild type, phospho-mimetic and unphosphorylatable Dicer transgenes revealed no significant differences in miRNA abundance between the dcr-1(0) and wild-type animals. Examination of specific miRNA species showed dramatically lowered expression of only the embryo-specific miR35-miR42 cluster in dcr-1(0) animals. However, lack of the miR35–42 cluster could be accounted for by the complete lack of embryo development in dcr-1(0) animals. It is likely that the perdurance of dcr-1(+) activity coming from maternal contribution of Dicer protein, when segregated from heterozygous mothers results in a persistence of miRNAs in the dcr-1(0) homozygous animals.
Thus, contrary to the dogma, we found no significant difference in miRNA abundance in animals that lack dcr-1. Therefore, 26G class endo-siRNAs, which are Dicer-dependent and abundant in the germline30,31 were assayed as an indicator for loss of Dicer activity. These endo-siRNAs are grouped into Class I or the spermatogenic class and Class II or the oogenic and the embryogenic class.30-33 Both Class I and Class II 26G RNAs are germline specific, and usually detected only in young adult or adult animals. Thus, we hypothesized that in the dcr-1(0) adult animals the 26G endo-siRNAs should not be impacted by maternal contribution of Dicer activity, which should be minimal by adulthood. The analysis of 26G RNAs demonstrated that (i), the 26G endo-siRNAs were depleted in dcr-1(0) animals, as predicted, (ii) wild-type Dicer transgene rescued the dcr-1(0) animal for 26G endo-siRNAs, and (iii) that both phospho-mimetic Dicer transgenes showed loss of the 26G endo-siRNAs suggesting that phosphorylation of Dicer inhibits its activity toward these small RNAs. Conversely, unphosphorylatable Dicer transgenes had different profiles. S1833A Dicer transgene lead to the generation of 26G endo-siRNAs, while S1705A did not. Thus the inability to phosphorylate Dicer at Serine 1833 results in a constitutively active protein whereas the S1705A mutation results in a protein that may have novel (but yet undetermined) activities. Cumulatively, we propose that differential phosphorylation of Dicer may regulate its ability to generate distinct populations of small RNAs.
Something Strange in the Neighborhood: A new Dicer-depletion-dependent Small RNA Species, Pseudo-miRNA
During the course of the above analysis, a very surprising observation was made. It was discovered that in dcr-1(0) animals some small RNAs were enriched 800–1000 fold when compared to wild-type animals. These small RNAs were 26–30-nucleotide in length (Fig. 2) and shared the core 21-nucleotide sequence with miRNAs. As Dicer generates miRNAs in the canonical pathway, we called these small RNA species “pseudo-miRNAs” since they were generated upon loss of Dicer. We propose that the pseudo-miRNAs are derived independent of dcr-1-processing of pre-miRNAs that accumulate under conditions of dcr-1 depletion. But is Drosha (DRSH-1 in C. elegans) required for pseudo-miRNA biogenesis? Drosha functions as part of the microprocessor complex to cleave the longer RNA precursor (pri-miRNA)34 and produce the pre-miRNA, which are cleaved by Dicer to generate miRNA. Assay of drsh-1(0) animals revealed that the pseudo-miRNAs are fully dependent on Drosha. This finding supports the hypothesis that pseudo-miRNAs are generated from the same pre-miRNA pool as their mature miRNA counterparts. Since the pseudo-miRNAs were only observed in conditions of dcr-1 depletion, we propose that abundance of pseudo-miRNAs can be used to track Dicer activity in vivo. For example, phospho-mimetic Dicer transgenes accumulate pseudo-miRNA 58 and 80 at 200–250 fold higher level compared to the wild-type transgene. In contrast, lack of Dicer phosphorylation resulted in either 10–fold15- (S1833A) or 80–120-fold increase (S1705A) in pseudo-miRNA 58 and 80 as compared to the wild-type transgene. These data suggest that phosphorylation of Dicer results in downregulation of Dicer activity (thus leading to production of the pseudo-miRNAs), while lack of phosphorylation at Serine 1833 renders the Dicer protein largely inactive. And S1705A Dicer protein may have as yet unidentified novel activities. The data also suggest that the inability to phosphorylate Serine 1705 or 1833 may confer on each Dicer variant a different level of activity, which may culminate in differences in pseudo-miRNA accumulation level.
Figure 2.
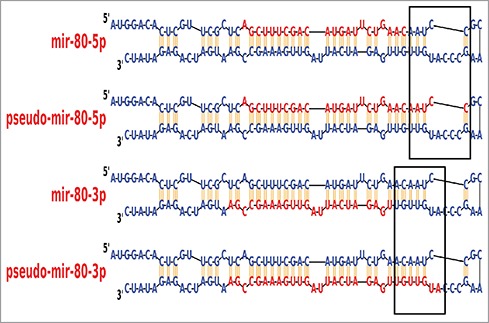
Psuedo-miRNAs. Small RNA species that are 26–30-nucleotide long, and share 21-nucleotide core sequence with miRNAs (box). These are generated in conjunction with the mature miRNA forms, but in the absence of Dicer activity.
These data demonstrate that Dicer phosphorylation modulates the normal small RNA repertoire and raises many exciting questions. What is the function of pseudo-miRNAs? Do pseudo-mioRNAs function similar to miRNAs and regulate gene expression? Are pseudo-miRNAs metabolic byproducts of small RNA pathway processing enzymes?
A role for ERK-mediated Regulation of Dicer During in oocyte-to-embryo Transition
In mice and C. elegans, loss of Dicer function in oocytes causes infertility.21,23 But, in both cases, oogenesis proceeds apparently normally at least until just before fertilization (or after oocyte maturation). In mouse oocytes that lack dicer function no detectable defects are observed in growing oocytes (during meiosis I) but rather after the oocytes mature.35 In C. elegans, genetic mosaic studies indicate that dcr-1 function is dispensable for oogenesis during meiosis I, but essential for embryogenesis: worms that lack dcr-1 function in the germline produce apparently normal oocytes, but embryos that die at varied stages of early development. Additionally, while systemic loss of dcr-1 function in worms results in endomitotic oocytes,23 genetic mosaic studies demonstrate that endomitotic phenotype arises from the loss of dcr-1 in somatic tissues, but not the germline.21 The somatic gonad in worms is known to initiate specific events during oocyte maturation and ovulation.36 Thus, even though Dicer is expressed at the RNA and protein level throughout the germline, it does not appear to function in the germline to promote oocyte development.
But, if Dicer is expressed in the female germline and has no apparent function in oocyte development, why is it there? Phospho-mimetic Dicer transgenic animals have normal oocyte development; however, the embryos die early and resemble animals that have lost Dicer specifically from the germline. This data suggests that phospho-mimetic Dicer behaves very much like loss of Dicer from the germline. Unphosphorylated Dicer transgenic animals, however, present with 2 phenotypes: increased MPK-1 activation (via S1833A), and defects in germline progression and oocyte development (via S1705A), suggesting that lack of phosphorylation of Dicer presented with new phenotypes, which were unlike systemic or tissue specific loss of Dicer. This suggests that Dicer needs to be phosphorylated for normal progression of oogenesis and oocyte development, but lack of phosphorylation results in aberrations in MPK-1 activation, meiotic progression and oocyte development. Conversely, presence of phosphorylated Dicer in the embryo is incompatible with embryonic development. Together with the genetic mosaic analysis, these data propose a model on how MPK-1 may regulate Dicer activity to mediate oocyte development and oocyte-to-embryo transition.
In the C. elegans germline MPK-1 is active in the mid-pachytene stage of oogenesis, downregulated in the loop region, reactivated in developing oocytes, and downregulated in the mature oocyte just before fertilization (Fig. 1). MPK-1 drives these oogenic processes at least in part through its phosphorylation and inhibition of Dicer function within developing oocytes. Thus, until the very end of oogenesis Dicer function is inhibited toward certain classes of small RNAs. Minute's prior to oocyte fertilization (upon maturation), a switch is flipped, and MPK-1 activity is downregulated, followed by the dephosphorylation of Dicer. Dephosphorylation of Dicer then renders it functional in anticipation of fertilization and embryogenesis. Thus, for oogenesis to occur normally, Dicer function must be inhibited or at least significantly altered, and for embryogenesis to occur normally, Dicer function must be reinitiated. The presence of active MPK-1 during oogenesis drives the former process, its absence allows for the latter process, and the precipitous switch from active to inactive MPK-1 that occurs in the terminal oocyte just before fertilization signals the exact timing of the oocyte-to-embryo transition and the likely reprogramming of small RNA production (Fig. 3).
Figure 3.
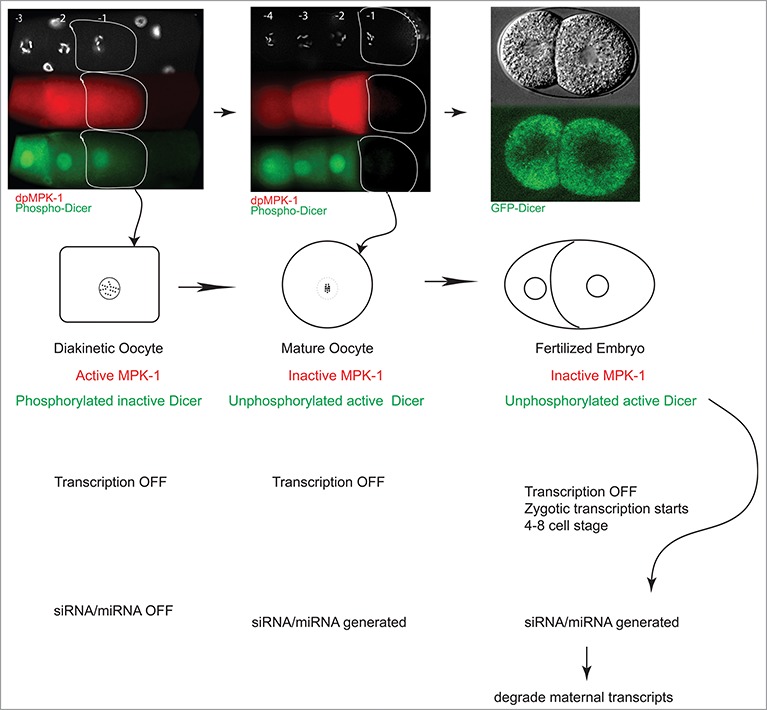
Model for why Dicer is so tightly regulated in the germline, and the embryo. During diakinesis (column 1), the oldest oocyte (−1) contains active MPK-1 and phosphorylated Dicer (green, inactive). As this oocyte exits diakinesis and matures (−1, column 2), MPK-1 is inactivated; Dicer is de-phosphorylated and presumably active. Active Dicer is then competent to generate small RNAs presumably in early embryos. These small RNAs may degrade maternal RNAs. Thus phosphorylation and dephosphorylation of Dicer appears to be necessary for the reprogramming of an oocyte to an embryo.
It is interesting to speculate (Fig. 3) that one reason for a tight control on Dicer activity during this critical phase of development is to protect certain maternal RNAs in the germline, and degrade them in a step-wise fashion upon embryo development, via generation of specific classes of endo-siRNAs (through unphosphorylated active Dicer). It would be very interesting in the future to determine whether lack of turning down maternal RNAs in the embryo results in embryonic lethality, and what the mechanisms might be that may govern this. Certainly, suppression of small RNAs in the maternal germline was demonstrated in mouse oocytes. Loss of the dgcr8 microprocessor gene,37,38 from mouse oocytes does not affect oocyte development, but results in embryonic death at E6.5, suggesting that miRNA activity is not necessary for oocyte development but essential for embryonic development. Interestingly much like in worms,21 loss of dicer in mouse oocytes also does not result in a clear decrease in miRNA levels, but rather endo-siRNA levels are dampened, suggesting that dicer may mediate its oocyte function primarily through siRNAs in mammals as well.39 While the precise profiles of small RNAs in the oocyte and early embryos remain to be clarified, our observations in aggregate support the model that a switch in the functions of the small RNA biogenesis pathway coincides with the oocyte-to-embryo transition, and open many exciting new directions.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work is supported by NIHGM98200 RO1, American Cancer Society ACS-RSG014–044-DDC, the Center for Genetics and Genomics UT MD Anderson Cancer Center and the Cancer Center Support Grant (CCSG-IRG).
References
- 1.Andl T, Murchison EP, Liu F, Zhang Y, Yunta-Gonzalez M, Tobias JW, Andl CD, Seykora JT, Hannon GJ, Millar SE. The miRNA-processing enzyme dicer is essential for the morphogenesis and maintenance of hair follicles. Curr Biol 2006; 16:1041-9; http://dx.doi.org/ 10.1016/j.cub.2006.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Svoboda P, Flemr M. The role of miRNAs and endogenous siRNAs in maternal-to-zygotic reprogramming and the establishment of pluripotency. EMBO Rep 2010; 11:590-7; PMID:20651740; http://dx.doi.org/ 10.1038/embor.2010.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 2001; 106:23-34; PMID:11461699; http://dx.doi.org/ 10.1016/S0092-8674(01)00431-7 [DOI] [PubMed] [Google Scholar]
- 4.Slack FJ, Basson M, Liu Z, Ambros V, Horvitz HR, Ruvkun G. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol Cell 2000; 5:659-69; PMID:10882102; http://dx.doi.org/ 10.1016/S1097-2765(00)80245-2 [DOI] [PubMed] [Google Scholar]
- 5.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000; 403:901-6; PMID:10706289; http://dx.doi.org/ 10.1038/35002607 [DOI] [PubMed] [Google Scholar]
- 6.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature 2004; 432:231-5; PMID:15531879; http://dx.doi.org/ 10.1038/nature03049 [DOI] [PubMed] [Google Scholar]
- 7.Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev 2001; 15:2654-9; PMID:11641272; http://dx.doi.org/ 10.1101/gad.927801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J, Shingara J, et al.. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res 2007; 67:7713-22; PMID:17699775; http://dx.doi.org/ 10.1158/0008-5472.CAN-07-1083 [DOI] [PubMed] [Google Scholar]
- 9.Ding XC, Slack FJ, Grosshans H. The let-7 microRNA interfaces extensively with the translation machinery to regulate cell differentiation. Cell Cycle 2008; 7:3083-90; PMID:18818519; http://dx.doi.org/ 10.4161/cc.7.19.6778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yigit E, Batista PJ, Bei Y, Pang KM, Chen CC, Tolia NH, Joshua-Tor L, Mitani S, Simard MJ, Mello CC. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell 2006; 127:747-57; PMID:17110334; http://dx.doi.org/ 10.1016/j.cell.2006.09.033 [DOI] [PubMed] [Google Scholar]
- 11.Gu W, Shirayama M, Conte D Jr, Vasale J, Batista PJ, Claycomb JM, Moresco JJ, Youngman EM, Keys J, Stoltz MJ, et al.. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol Cell 2009; 36:231-44; PMID:19800275; http://dx.doi.org/ 10.1016/j.molcel.2009.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wedeles CJ, Wu MZ, Claycomb JM. A multitasking Argonaute: exploring the many facets of C. elegans CSR-1. Chromosome Res 2013; 21:573-86; PMID:24178449; http://dx.doi.org/ 10.1007/s10577-013-9383-7 [DOI] [PubMed] [Google Scholar]
- 13.Schmitter D, Filkowski J, Sewer A, Pillai RS, Oakeley EJ, Zavolan M, Svoboda P, Filipowicz W. Effects of Dicer and Argonaute down-regulation on mRNA levels in human HEK293 cells. Nucleic Acids Res 2006; 34:4801-15; PMID:16971455; http://dx.doi.org/ 10.1093/nar/gkl646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tops BB, Plasterk RH, Ketting RF. The Caenorhabditis elegans Argonautes ALG-1 and ALG-2: almost identical yet different. Cold Spring Harb Symp Quant Biol 2006; 71:189-94; PMID:17381296; http://dx.doi.org/ 10.1101/sqb.2006.71.035 [DOI] [PubMed] [Google Scholar]
- 15.Paroo Z, Ye X, Chen S, Liu Q. Phosphorylation of the human microRNA-generating complex mediates MAPK/Erk signaling. Cell 2009; 139:112-22; PMID:19804757; http://dx.doi.org/ 10.1016/j.cell.2009.06.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hubbard EJ, Greenstein D. The Caenorhabditis elegans gonad: a test tube for cell and developmental biology. Dev Dyn 2000; 218, 2-22; PMID:10822256; http://dx.doi.org/ 10.1002/(SICI)1097-0177(200005)218:1%3c2::AID-DVDY2%3e3.0.CO;2-W [DOI] [PubMed] [Google Scholar]
- 17.Arur S, Ohmachi M, Nayak S, Hayes M, Miranda A, Hay A, Golden A, Schedl T. Multiple ERK substrates execute single biological processes in Caenorhabditis elegans germ-line development. Proc Natl Acad Sci U S A 2009; 106:4776-81; PMID:19264959; http://dx.doi.org/ 10.1073/pnas.0812285106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee MH, Ohmachi M, Arur S, Nayak S, Francis R, Church D, Lambie E, Schedl T. Multiple functions and dynamic activation of MPK-1 extracellular signal-regulated kinase signaling in Caenorhabditis elegans germline development. Genetics 2007; 177:2039-62; PMID:18073423; http://dx.doi.org/ 10.1534/genetics.107.081356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Church DL, Guan KL, Lambie EJ. Three genes of the MAP kinase cascade, mek-2, mpk-1/sur-1 and let-60 ras, are required for meiotic cell cycle progression in Caenorhabditis elegans. Development 1995; 121:2525-35; PMID:7671816 [DOI] [PubMed] [Google Scholar]
- 20.Miller MA, Nguyen VQ, Lee MH, Kosinski M, Schedl T, Caprioli RM, Greenstein D. A sperm cytoskeletal protein that signals oocyte meiotic maturation and ovulation. Science 2001; 291:2144-7; PMID:11251118; http://dx.doi.org/ 10.1126/science.1057586 [DOI] [PubMed] [Google Scholar]
- 21.Drake M, Furuta T, Suen KM, Gonzalez G, Liu B, Kalia A, Ladbury JE, Fire AZ, Skeath JB, Arur S. A requirement for ERK-dependent Dicer phosphorylation in coordinating oocyte-to-embryo transition in C. elegans. Dev Cell 2014; 31:614-28; PMID:25490268; http://dx.doi.org/ 10.1016/j.devcel.2014.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beshore EL, McEwen TJ, Jud MC, Marshall JK, Schisa JA, Bennett KL. C. elegans Dicer interacts with the P-granule component GLH-1 and both regulate germline RNPs. Dev Biol 2011; 350:370-81; PMID:21146518; http://dx.doi.org/ 10.1016/j.ydbio.2010.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knight SW, Bass BL. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science 2001; 293:2269-71; PMID:11486053; http://dx.doi.org/ 10.1126/science.1062039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tijsterman M, May RC, Simmer F, Okihara KL, Plasterk RH. Genes required for systemic RNA interference in Caenorhabditis elegans. Curr Biol 2004; 14:111-6; PMID:14738731; http://dx.doi.org/ 10.1016/j.cub.2003.12.029 [DOI] [PubMed] [Google Scholar]
- 25.Hu F, Lai EC, Okamura K. A signaling-induced switch in dicer localization and function. Dev Cell 2014; 31:523-4; PMID:25490263; http://dx.doi.org/ 10.1016/j.devcel.2014.11.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Emmerth S, Schober H, Gaidatzis D, Roloff T, Jacobeit K, Bühler M. Nuclear retention of fission yeast dicer is a prerequisite for RNAi-mediated heterochromatin assembly. Dev Cell 18:102-13; http://dx.doi.org/ 10.1016/j.devcel.2009.11.011 [DOI] [PubMed] [Google Scholar]
- 27.Barraud P, Emmerth S, Shimada Y, Hotz HR, Allain FH, Bühler M. An extended dsRBD with a novel zinc-binding motif mediates nuclear retention of fission yeast Dicer. EMBO J 2011; 30:4223-35; PMID:21847092; http://dx.doi.org/ 10.1038/emboj.2011.300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ando Y, Tomaru Y, Morinaga A, Burroughs AM, Kawaji H, Kubosaki A, Kimura R, Tagata M, Ino Y, Hirano H, et al.. Nuclear pore complex protein mediated nuclear localization of dicer protein in human cells. PLoS One 2011; 6, e23385; PMID:21858095; http://dx.doi.org/ 10.1371/journal.pone.0023385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacRae IJ, Zhou K, Doudna JA. Structural determinants of RNA recognition and cleavage by Dicer. Nat Struct Mol Biol 2007; 14:934-940; PMID:17873886; http://dx.doi.org/ 10.1038/nsmb1293 [DOI] [PubMed] [Google Scholar]
- 30.Han T, Manoharan AP, Harkins TT, Bouffard P, Fitzpatrick C, Chu DS, Thierry-Mieg D, Thierry-Mieg J, Kim JK. 26G endo-siRNAs regulate spermatogenic and zygotic gene expression in Caenorhabditis elegans. Proc Natl Acad Sci U S A 2009; 106:18674-9; PMID:19846761; http://dx.doi.org/ 10.1073/pnas.0906378106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang C, Montgomery TA, Gabel HW, Fischer SE, Phillips CM, Fahlgren N, Sullivan CM, Carrington JC, Ruvkun G. mut-16 and other mutator class genes modulate 22G and 26G siRNA pathways in Caenorhabditis elegans. Proc Natl Acad Sci U S A 2011; 108:1201-8; PMID:21245313; http://dx.doi.org/ 10.1073/pnas.1018695108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Conine CC, Batista PJ, Gu W, Claycomb JM, Chaves DA, Shirayama M, Mello CC. Argonautes ALG-3 and ALG-4 are required for spermatogenesis-specific 26G-RNAs and thermotolerant sperm in Caenorhabditis elegans. Proc Natl Acad Sci U S A 2010; 107:3588-93; PMID:20133686; http://dx.doi.org/ 10.1073/pnas.0911685107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kamminga LM, van Wolfswinkel JC, Luteijn MJ, Kaaij LJ, Bagijn MP, Sapetschnig A, Miska EA, Berezikov E, Ketting RF. Differential impact of the HEN1 homolog HENN-1 on 21U and 26G RNAs in the germline of Caenorhabditis elegans. PLoS Genet 2012; 8, e1002702; PMID:22829772; http://dx.doi.org/ 10.1371/journal.pgen.1002702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Warf MB, Johnson WE, Bass BL. Improved annotation of C. elegans microRNAs by deep sequencing reveals structures associated with processing by Drosha and Dicer. RNA 2011; 17:563-77; PMID:21307183; http://dx.doi.org/ 10.1261/rna.2432311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murchison EP, Stein P, Xuan Z, Pan H, Zhang MQ, Schultz RM, Hannon GJ. Critical roles for Dicer in the female germline. Genes Dev 2007; 21:682-93; PMID:17369401; http://dx.doi.org/ 10.1101/gad.1521307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCarter J, Bartlett B, Dang T, Schedl T. Soma-germ cell interactions in Caenorhabditis elegans: multiple events of hermaphrodite germline development require the somatic sheath and spermathecal lineages. Dev Biol 1997; 181:121-43; PMID:9013925; http://dx.doi.org/ 10.1006/dbio.1996.8429 [DOI] [PubMed] [Google Scholar]
- 37.Ma J, Flemr M, Stein P, Berninger P, Malik R, Zavolan M, Svoboda P, Schultz RM. MicroRNA activity is suppressed in mouse oocytes. Curr Biol 2010; 20:265-70; http://dx.doi.org/ 10.1016/j.cub.2009.12.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suh N, Baehner L, Moltzahn F, Melton C, Shenoy A, Chen J, Blelloch R. MicroRNA function is globally suppressed in mouse oocytes and early embryos. Curr Biol 2010; 20:271-7; http://dx.doi.org/ 10.1016/j.cub.2009.12.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Flemr M, Malik R, Franke V, Nejepinska J, Sedlacek R, Vlahovicek K, Svoboda P. A retrotransposon-driven dicer isoform directs endogenous small interfering RNA production in mouse oocytes. Cell 2013; 155:807-16; PMID:24209619; http://dx.doi.org/ 10.1016/j.cell.2013.10.001 [DOI] [PubMed] [Google Scholar]


