Abstract
In metazoans, the Wnt signaling pathway plays a key role in the regulation of binary decisions during development. During this process different sets of target genes are activated in cells where the Wnt pathway is active (classic target genes) versus cells where the pathway is inactive (opposite target genes). While the mechanism of transcriptional activation is well understood for classic target genes, how opposite target genes are activated in the absence of Wnt remains poorly characterized. Here we discuss how the key transcriptional mediator of the Wnt pathway, the TCF family member POP-1, regulates opposite target genes during C. elegans development. We examine recent findings suggesting that the direction of the transcriptional output (activation or repression) can be determined by the way TCF is recruited and physically interacts with its target gene.
Keywords: C. elegans, POP-1, REF-2, SYS-1, transcription, TCF, Wnt signaling, Zic, β-catenin
Introduction
The Wnt/β-catenin pathway (or canonical Wnt pathway) is an essential signaling cascade that plays key roles during the development of many animals and its misregulation is involved in several human diseases.1,2 Activation of this pathway leads to stabilization of the transcriptional co-activator β-catenin, which enters the nucleus and associates with transcription factors of the TCF family. When the pathway is inactive TCF is not associated with β-catenin, which is degraded. Most direct transcriptional targets of this pathway follow a “classic” type of regulation: they contain TCF binding sites in their cis-regulatory region and are activated in presence of Wnt by the TCF:β-catenin complex and repressed in the absence of Wnt by TCF. However, in several organisms, a few direct target genes have been observed to follow an “opposite” type of regulation: they are repressed in the presence of Wnt by the TCF:β-catenin complex and activated in the absence of Wnt by TCF.3,4 How TCF mediates this atypical regulation is poorly understood.
In C. elegans, a variant of the Wnt/β-catenin pathway called the Wnt/β-catenin asymmetry pathway regulates many asymmetric divisions along the antero-posterior axis of the embryo and the larva.5-9 Activation of this pathway induces stabilization of the β-catenin protein SYS-1 and partial nuclear export of the TCF factor POP-1. In the posterior daughter, where the pathway is active, there is a high nuclear concentration of SYS-1/β-catenin and a low nuclear concentration of POP-1/TCF leading to the formation of a POP-1:SYS-1 complex. In the anterior daughter the pathway is inactive, there is a low nuclear concentration of SYS-1, a high nuclear concentration of POP-1, and nuclear POP-1 is mostly free of SYS-1 (Fig. 1).8 The mechanism by which target genes are activated by this pathway in posterior daughters is well characterized and follows the “classic target gene” logic of the Wnt/β-catenin pathway targets. Examples include the activation of the GATA transcription factor gene end-1 in the EMS lineage,10,11 the Nkx transcription factor gene ceh-22 in the distal tip cell precursor,12 the Meis transcription factor gene psa-3 in the T lineage,13 the homeodomain transcription factor gene ceh-10 in the AIY lineage14 or the GATA transcription factor gene egl-18 in seam cell lineages.15 These targets contain TCF binding sites in their cis-regulatory elements and are directly activated by the POP-1:SYS-1 complex in the posterior daughter and directly repressed by POP-1 in the anterior daughter (Fig. 1, classic target genes). On the contrary, how target genes are activated in anterior daughters remains poorly characterized. Here we will first examine evidence that some anterior genes are indirectly regulated by POP-1 via a posterior repressor. We will then discuss recent data suggesting another scenario where other anterior genes are directly activated by POP-1 via an atypical mechanism.
Figure 1.
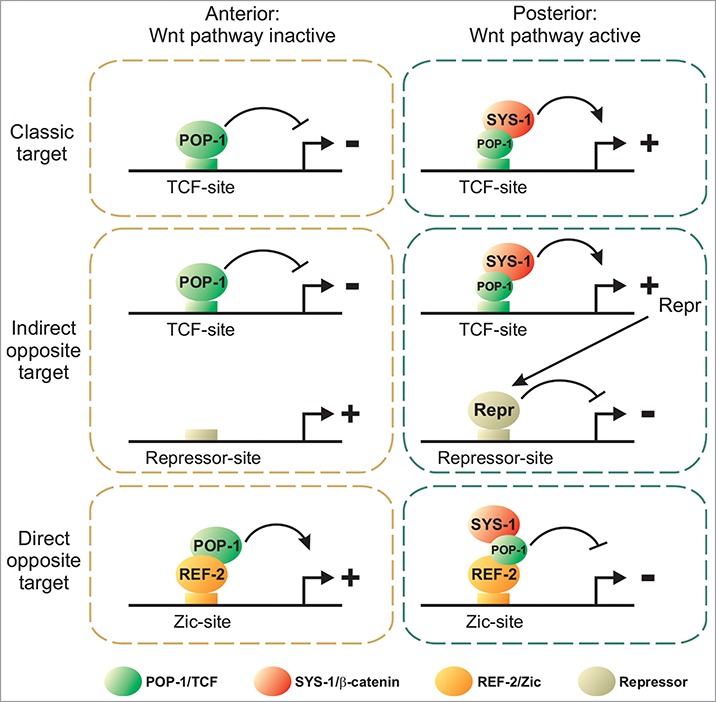
Regulation of target genes by the Wnt/β-catenin asymmetry pathway. Classic targets are directly activated in the posterior daughter by the POP-1:SYS-1 complex. Indirect opposite targets are repressed in the posterior daughter via a posterior repressor (Repr). Direct opposite targets are activated in the anterior daughter by a POP-1:REF-2 complex. Smaller POP-1 circles in the posterior daughter represent lower nuclear concentration.
Indirect Regulation of Opposite Targets via a Repressor
One simple model to explain the asymmetric activation of target genes in anterior daughters is an indirect regulation by a posterior repressor (Fig. 1, indirect opposite target). In this scenario, the POP-1:SYS-1 complex directly activates the expression of a transcriptional repressor in the posterior daughter via TCF binding sites present in its cis-regulatory elements. This repressor then directly represses the expression of anterior targets via repressor binding sites in their cis-regulatory regions. In the anterior daughter, the repressor is not expressed allowing the expression of anterior targets. While this mechanism has not been fully demonstrated, there is evidence that some anterior target genes are indirectly repressed in posterior daughters.
The best characterized example is probably the EMS lineage of the embryo. In the early embryo, the EMS blastomere divides asymmetrically to generate the anterior MS blastomere (precursor of mesoderm) and the posterior E blastomere (precursor of endoderm). The NKX transcription factor gene ceh-51 is expressed in MS while the GATA transcription factor genes end-1 and end-3 are expressed in E. The POP-1:SYS-1 complex represses the expression of ceh-51 in E, therefore restricting its expression to MS.16,17 While the mechanism by which the POP-1:SYS-1 complex represses ceh-51 in E remains to be fully characterized, it is at least in part indirect via the activation of end-1 and end-3 expression.
There is also evidence of indirect repression in other lineages, for example in the SMDD/AIY neuronal lineage. In the embryo the SMDD/AIY mother divides asymmetrically to generate the anterior SMDD motoneuron and the posterior AIY interneuron. Following asymmetric division, the POP-1:SYS-1 complex directly activates the expression of the homeodomain transcription factor gene ceh-10 in AIY via TCF binding sites.14 In addition, in ceh-10 null mutants AIY markers are completely lost in AIY, and SMDD markers become partially derepressed in AIY. This suggests that CEH-10 could play in part the role of the repressor in the SMDD/AIY lineage. However the mechanism through which CEH-10 represses SMDD markers is unknown.
This indirect regulation of anterior target genes involves an additional transcriptional step (transcriptional activation/repression of the repressor gene), which could result in a temporal shift between the activation of posterior targets (direct) and anterior targets (indirect) and thus a delay in the fate specification program of anterior daughters in comparison to posterior daughters. In addition, as time between successive cell divisions in the C. elegans embryo is very short (around 30 min at 20°C)18 the time delay imposed by a transcriptional intermediate step could be an important constraint for the embryo. Interestingly, recent data suggest that C. elegans has also developed a more direct way to activate the expression of anterior target genes.
Direct Activation of Opposite Targets by POP-1/TCF via an Atypical Mechanism
We have recently characterized how the LIM homeodomain transcription factor gene ttx-3 is activated in the anterior daughter of an embryonic neuronal lineage.19 Following asymmetric cell division, ttx-3 expression is activated in the anterior daughter (the SMDD/AIY neuroblast which generates the SMDD motoneuron and AIY interneuron), but not the posterior daughter (the SIAD/SIBV neuroblast which generates the SIAD and SIBV motoneurons). This activation is mediated by a Zic transcription factor (REF-2) via a Zic binding site present in the ttx-3 cis-regulatory region. In the anterior daughter, the POP-1 protein directly binds the REF-2 protein and the REF-2:POP-1 complex activates ttx-3 expression via the Zic binding site (Fig. 1, direct opposite target). In the posterior daughter, SYS-1 blocks this activation by binding to POP-1. These results suggest that POP-1 can directly activate the transcription of an anterior target without an intermediate step of transcription. This is an atypical mode of action for a TCF transcription factor, and whether other anterior target genes in C. elegans are regulated by a similar mechanism remains to be determined. This atypical mechanism could be conserved in other animals. Indeed, we were able to reconstitute this system in mammalian cell cultures.19 In addition, it has been observed in vertebrates that the Zic2 protein directly binds TCF4 and that the Zic2:TCF4:β-catenin complex cannot activate transcription via TCF binding sites.20
One important question that remains is why does the interaction with REF-2 invert the transcriptional activity of POP-1, converting POP-1 into an activator in the absence of SYS-1 and a repressor in the presence of SYS-1. Studies conducted on 2 opposite target genes in Drosophila (Ugt36Bc and Tiggrin) can provide an insight into the mechanism.21,22 TCF activates these targets in the absence of β-catenin and represses them in the presence of β-catenin via an atypical TCF binding site present in their cis-regulatory regions. This atypical TCF binding site differs in sequence from the classic TCF binding sites observed in classic target genes. Interestingly, TCF adopts distinct conformations when bound to classic vs. atypical binding sites (Fig. 2A, B).22 This suggests that TCF can exist in 2 different conformations. In the first conformation, TCF acts as a repressor in the absence of β-catenin and as an activator when bound to β-catenin. In the second conformation, TCF acts as an activator in the absence of β-catenin and as a repressor when bound to β-catenin. We speculate that REF-2, by binding to POP-1, could induce a conformational change in POP-1 similar to the one observed when TCF interacts with atypical binding sites in Drosophila, inverting the transcriptional output (Fig. 2C). In the end, the difference of transcriptional output between classic and opposite direct targets may be determined by the way TCF is recruited to its target.
Figure 2.
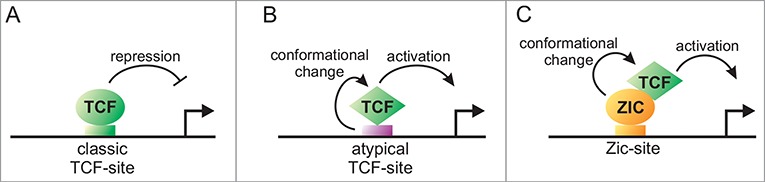
Conversion of TCF from a repressor to an activator via a conformational change. TCF acts as a repressor on classic TCF-sites (A). In Drosophila, binding to an atypical TCF-site induces a conformational change in TCF that converts TCF into an activator (B). We speculate that the Zic transcription factor REF-2 may induce a similar conformational change in the TCF transcription factor POP-1 converting it into an activator (C).
More work is required to understand the biochemical basis of this intriguing shift in TCF transcriptional activity and the analysis of additional direct opposite targets is needed to characterize the generality and diversity of the mechanisms at play. Some important misregulated genes in Wnt-related cancers are direct opposite target genes (such as p16 or E-cadherin),3,23,24 therefore the characterization of this atypical mode of regulation will certainly have important implications for the understanding of human diseases.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Ute Rothbächer and members of our lab for comments on the manuscript.
Funding
Work in the V.B. lab is funded by an ATIP/Avenir startup grant from CNRS/INSERM, grants from Sanofi-Aventis, from the Fédération pour la Recherche sur le Cerveau and from the Agence Nationale de la Recherche (ANR-14-CE11–0001 and ANR-11-LABX-0054).
References
- 1.MacDonald BT, Tamai K, He X. Wnt/β-catenin signaling: components, mechanisms, and diseases. Dev Cell 2009; 17:9-26; PMID:19619488; http://dx.doi.org/ 10.1016/j.devcel.2009.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clevers H, Nusse R. Wnt/β-catenin signaling and disease. Cell 2012; 149:1192-205; PMID:22682243; http://dx.doi.org/ 10.1016/j.cell.2012.05.012 [DOI] [PubMed] [Google Scholar]
- 3.Hoverter NP, Waterman ML. A Wnt-fall for gene regulation: repression. Sci Signaling 2008; 1:pe43; PMID:18827220; http://dx.doi.org/ 10.1126/scisignal.139pe43 [DOI] [PubMed] [Google Scholar]
- 4.Cadigan KM. TCFs and Wnt/β-catenin signaling: more than one way to throw the switch. Curr Top Dev Biol 2012; 98:1-34; PMID:22305157; http://dx.doi.org/ 10.1016/B978-0-12-386499-4.00001-X [DOI] [PubMed] [Google Scholar]
- 5.Kaletta T, Schnabel H, Schnabel R. Binary specification of the embryonic lineage in Caenorhabditis elegans. Nature 1997; 390:294-8; PMID:9384382; http://dx.doi.org/ 10.1038/36869 [DOI] [PubMed] [Google Scholar]
- 6.Lin R, Hill RJ, Priess JR. POP-1 and anterior-posterior fate decisions in C. elegans embryos. Cell 1998; 92:229-39; PMID:9458047; http://dx.doi.org/ 10.1016/S0092-8674(00)80917-4 [DOI] [PubMed] [Google Scholar]
- 7.Mizumoto K, Sawa H. Two betas or not two betas: regulation of asymmetric division by β-catenin. Trends Cell Biol 2007; 17:465-73; PMID:17919911; http://dx.doi.org/ 10.1016/j.tcb.2007.08.004 [DOI] [PubMed] [Google Scholar]
- 8.Phillips BT, Kimble J. A new look at TCF and β-catenin through the lens of a divergent C. elegans Wnt pathway. Dev Cell 2009; 17:27-34; PMID:19619489; http://dx.doi.org/ 10.1016/j.devcel.2009.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bertrand V, Hobert O. Lineage programming: navigating through transient regulatory states via binary decisions. Curr Opin Genet Dev 2010; 20:362-8; PMID:20537527; http://dx.doi.org/ 10.1016/j.gde.2010.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maduro MF, Kasmir JJ, Zhu J, Rothman JH. The Wnt effector POP-1 and the PAL-1/Caudal homeoprotein collaborate with SKN-1 to activate C. elegans endoderm development. Dev Biol 2005; 285:510-23; PMID:16084508; http://dx.doi.org/ 10.1016/j.ydbio.2005.06.022 [DOI] [PubMed] [Google Scholar]
- 11.Shetty P, Lo MC, Robertson SM, Lin R. C. elegans TCF protein, POP-1, converts from repressor to activator as a result of Wnt-induced lowering of nuclear levels. Dev Biol 2005; 285:584-92; PMID:16112103; http://dx.doi.org/ 10.1016/j.ydbio.2005.07.008 [DOI] [PubMed] [Google Scholar]
- 12.Lam N, Chesney MA, Kimble J. Wnt signaling and CEH-22/tinman/Nkx2.5 specify a stem cell niche in C. elegans. Curr Biol 2006; 16:287-95; PMID:16461282; http://dx.doi.org/ 10.1016/j.cub.2005.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arata Y, Kouike H, Zhang Y, Herman MA, Okano H, Sawa H. Wnt signaling and a Hox protein cooperatively regulate psa-3/Meis to determine daughter cell fate after asymmetric cell division in C. elegans. Dev Cell 2006; 11:105-15; PMID:16824957; http://dx.doi.org/ 10.1016/j.devcel.2006.04.020 [DOI] [PubMed] [Google Scholar]
- 14.Bertrand V, Hobert O. Linking asymmetric cell division to the terminal differentiation program of postmitotic neurons in C. elegans. Dev Cell 2009; 16:563-75; PMID:19386265; http://dx.doi.org/ 10.1016/j.devcel.2009.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorrepati L, Thompson KW, Eisenmann DM. C. elegans GATA factors EGL-18 and ELT-6 function downstream of Wnt signaling to maintain the progenitor fate during larval asymmetric divisions of the seam cells. Development 2013; 140:2093-102; PMID:23633508; http://dx.doi.org/ 10.1242/dev.091124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Broitman-Maduro G, Owraghi M, Hung WW, Kuntz S, Sternberg PW, Maduro MF. The NK-2 class homeodomain factor CEH-51 and the T-box factor TBX-35 have overlapping function in C. elegans mesoderm development. Development 2009; 136:2735-46; PMID:19605496; http://dx.doi.org/ 10.1242/dev.038307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Owraghi M, Broitman-Maduro G, Luu T, Roberson H, Maduro MF. Roles of the Wnt effector POP-1/TCF in the C. elegans endomesoderm specification gene network. Dev Biol 2010; 340:209-21; PMID:19818340; http://dx.doi.org/ 10.1016/j.ydbio.2009.09.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol 1983; 100:64-119; PMID:6684600; http://dx.doi.org/ 10.1016/0012-1606(83)90201-4 [DOI] [PubMed] [Google Scholar]
- 19.Murgan S, Kari W, Rothbacher U, Iche-Torres M, Melenec P, Hobert O, Bertrand V. Atypical Transcriptional Activation by TCF via a Zic Transcription Factor in C. elegans Neuronal Precursors. Dev Cell 2015; 33:737-45; PMID:26073017; http://dx.doi.org/ 10.1016/j.devcel.2015.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pourebrahim R, Houtmeyers R, Ghogomu S, Janssens S, Thelie A, Tran HT, Langenberg T, Vleminckx K, Bellefroid E, Cassiman JJ, et al.. Transcription factor Zic2 inhibits Wnt/β-catenin protein signaling. J Biol Chem 2011; 286:37732-40; PMID:21908606; http://dx.doi.org/ 10.1074/jbc.M111.242826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blauwkamp TA, Chang MV, Cadigan KM. Novel TCF-binding sites specify transcriptional repression by Wnt signalling. EMBO J 2008; 27:1436-46; PMID:18418383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang CU, Blauwkamp TA, Burby PE, Cadigan KM. Wnt-mediated repression via bipartite DNA recognition by TCF in the Drosophila hematopoietic system. PLoS genetics 2014; 10:e1004509; PMID:25144371; http://dx.doi.org/ 10.1371/journal.pgen.1004509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Delmas V, Beermann F, Martinozzi S, Carreira S, Ackermann J, Kumasaka M, Denat L, Goodall J, Luciani F, Viros A, et al.. Beta-catenin induces immortalization of melanocytes by suppressing p16INK4a expression and cooperates with N-Ras in melanoma development. Genes Dev 2007; 21:2923-35; PMID:18006687; http://dx.doi.org/ 10.1101/gad.450107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jamora C, DasGupta R, Kocieniewski P, Fuchs E. Links between signal transduction, transcription and adhesion in epithelial bud development. Nature 2003; 422:317-22; PMID:12646922; http://dx.doi.org/ 10.1038/nature01458 [DOI] [PMC free article] [PubMed] [Google Scholar]


