Abstract
X-chromosome inactivation, which was discovered by Mary Lyon in 1961 results in random silencing of one X chromosome in female mammals. This review is dedicated to Mary Lyon, who passed away last year. She predicted many of the features of X inactivation, for e.g., the existence of an X inactivation center, the role of L1 elements in spreading of silencing and the existence of genes that escape X inactivation. Starting from her published work here we summarize advances in the field.
Keywords: X chromosome, X inactivation, escape from X inactivation
Introduction
This review is in honour of Mary Lyon who passed away last year. She has inspired my work and that of the members of my laboratory who have studied the regulation of the mammalian X chromosome. I initially met Mary Lyon in Boston when I was a post-doctoral fellow in Samuel Latt’s laboratory at Harvard. Her visit was a highlight of my training. Throughout my career I continued to enjoy meeting her at the International Mammalian Genome Society conferences and the X inactivation meetings.
Lyon’s law
‘It is here suggested that this mosaic phenotype is due to the inactivation of one or other X chromosome early in embryonic development.’
‘Thus, the general picture concerning heterozygotes for sex-linked genes in man is one of variable expression, which accords with the predicted result of random inactivation of one or the other X chromosome.’
Mary Lyon formulated her X-chromosome inactivation (XCI) hypothesis in 1961 based on her observations in female mice heterozygous for a mutation in an X-linked gene that controls coat colour (for e.g. tabby), and based on known facts at the time that X0 mice are viable (Welshons and Russell 1959) and that female cells contain a heteropyknotic X chromosome (Ohno and Hauschka 1960). She interpreted the variegated coat colour as due to clonal growth of cells with random silencing of one X chromosome (Lyon 1961). Similar variegation in coat colour is seen in female mice with the Cattanach X; autosome insertion (figure 1a). Mary Lyon predicted that her XCI hypothesis, now deemed a law, would be applicable to humans (Lyon 1962). A main consequence of XCI is to equalize the dosage of X-linked gene expression (dosage compensation) between male and female mammals (see Gartler, same issue). A second type of dosage compensation balances expression between X-linked genes and autosomal genes by upregulation of genes on the active X chromosome (Disteche 2012; Deng et al. 2014). Thus, XCI also prevents overexpression of X-linked genes in female cells with two X chromosomes (Lin et al. 2011). Early studies in female mouse preimplantation embryos demonstrated evidence of halving X-linked gene expression, thus pinpointing the timing of random XCI (Epstein et al. 1978; Kratzer and Gartler 1978). Silencing of one allele of X-linked genes is then clonally inherited in female somatic cells. Mary Lyon favoured the existence of three major steps for XCI: initiation, spreading and maintenance (Lyon 1988).
Figure 1.
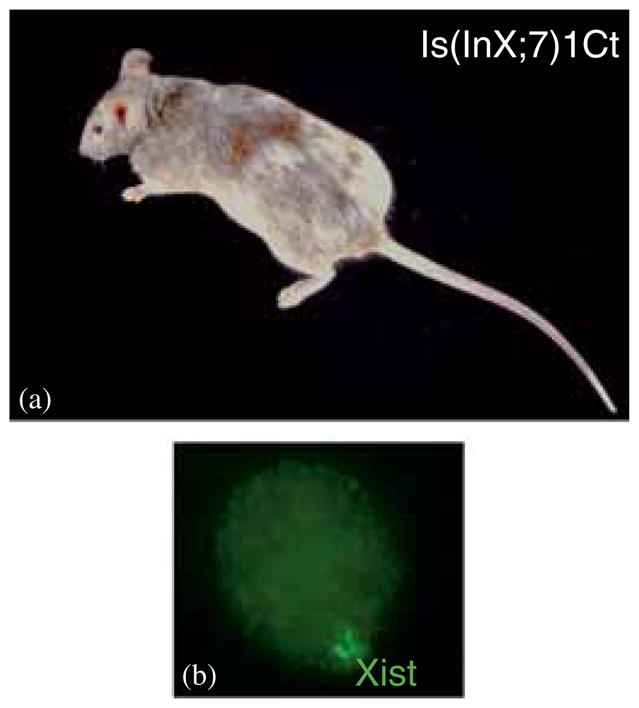
(a) Female mouse with the Cattanach insertion [Is(In7;X)1Ct] shows variegation of her coat colour due to spreading of XCI in the inserted portion of chromosome 7, which silences coat colour markers. (b) Nucleus from a female mouse fibroblast after Xist RNA-FISH (green). The inactive X is coated with Xist RNA.
X inactivation centre and XCI initiation
‘One would therefore not expect all points on the X to act independently with regards to inactivation. There might be some center or centres from which the inactivation spreads.’
Based on analyses of X; autosome translocations in which silencing can spread into the attached autosome, both Lyon and Russell suggested the possibility of an X-inactivation centre (XIC) required for the onset of XCI (Lyon et al. 1964; Russell 1964). Location of the XIC on the mouse and human X chromosomes was further defined by examining patterns of XCI in multiple additional cases of X; autosome translocations or other types of X rearrangements (Brockdorff et al. 1991; Leppig et al. 1993). The search was on to find the key molecular element(s) within the XIC essential for initiation.
The main key element turned out to be a gene that encodes a long noncoding RNA (lncRNA) called X inactive-specific transcript (Xist). XIST was initially discovered in humans by Carolyn Brown and Hunt Willard who singled out this lncRNA as a critical factor for XCI based on its location within the XIC and its unique expression pattern that is completely female-specific in adult somatic cells (Brown et al. 1991; Brown et al. 1992). The homologous Xist gene was then identified in mice where expression was detected at a critical stage of embryo development (Brockdorff et al. 1992; Kay et al. 1993). XIST/Xist RNA coats the inactive X chromosome in cis and thus becomes detectable as a cloud within the nucleus of somatic cells using RNA-FISH (Clemson et al. 1996) (figure 1b). Subsequent studies showed that insertion of the XIC including Xist on autosomes induces silencing at great distances, and that deletions/mutations of Xist perturbs XCI (Penny et al. 1996; Lee and Jaenisch 1997). The role of Xist and of all elements of the XIC which include several other lncRNAs and controlling elements is still under study (Payer and Lee 2008; Gendrel and Heard 2014). In mice, where there are two waves of silencing in early development imprinted XCI is initiated on the paternal X chromosome at day four after fertilization (Okamoto et al. 2005). Imprinted paternal XCI persists in extraembryonic tissues (Takagi and Sasaki 1975; West et al. 1977), while it is followed by X reactivation in the inner cell mass and random XCI at the blastocyst stage (Mak et al. 2004). XCI depends on levels of Xist expression, which are controlled by its antisense Tsix and a series of lncRNAs located at the XIC (Galupa and Heard 2015). The XIC also contains the protein-coding gene Rnf12, whose product activates Xist expression based on its dosage (Gontan et al. 2012). Pluripotency factors such as OCT4 and NANOG control Xist and Tsix expression, preventing XCI in pluripotent cells such as embryonic stem (ES) cells (Navarro et al. 2008). Induced differentiation of cultured ES cells triggers the onset of random XCI, which has greatly facilitated experimentation.
Surprisingly, in human and rabbit, XCI onset is delayed and the paternal or maternal X chromosome is randomly silenced in some cells at early stages of development (Okamoto et al. 2011), thus skipping the imprinted XCI observed in mouse. The organization and function of the XIC also differ between human and mouse. Whereas the antisense Tsix RNA and/or its transcription are critical for regulation of Xist in mouse, this is not the case in human (Chang and Brown 2010). Important questions remain, for e.g., about mechanisms that ensures that only one active X per diploid set of autosomes persists. Tsix may protect the active X from silencing (Gayen et al. 2015). The choice of which X chromosome becomes silenced is also under study; one important element is the X controlling element (Xce) locus (Cattanach 1975) whose molecular identity remains elusive (Morey and Avner 2010; Thorvaldsen et al. 2012). One possibility is that structural oscillations in topological domains at the XIC may influence choice in individual cells (Giorgetti et al. 2014). It has also been proposed that initiation of XCI may be stochastic, with subsequent selection of cells with the appropriate number of X chromosomes expressed, i.e. a single X chromosome per diploid cell (Monkhorst et al. 2008).
XCI spreading
‘It is suggested that interspersed repetitive elements of the LINE type, in which the X chromosome is particularly rich, act as booster elements to promote the spread of XIST mRNA.’
Gartler and Riggs originally proposed that there would be way-stations that help spreading of silencing along the inactive X chromosome (Gartler and Riggs 1983). However, how exactly Xist RNA spreads along the X chromosome is still controversial (Engreitz et al. 2013; Simon et al. 2013). One possibility is that Xist RNA binds to preferred sites in a saltatory way and spreads from a limited number of recruitment sites (Pinter et al. 2012). High resolution microscopy of single cells show fewer Xist RNA molecules over the inactive X chromosome than expected from previous studies on bulk cells, suggesting a hit-and-run model (Sunwoo et al. 2015). Importantly, new studies have identified proteins recruited by Xist RNA, which directly or indirectly facilitate gene silencing (Chu et al. 2015; McHugh et al. 2015; Minajigi et al. 2015). For example, SHARP (also called SPEN) interacts with Xist RNA and recruit SMRT that activates the histone deacetylase HDAC3 for silencing (McHugh et al. 2015). Among the most enriched proteins recruited by Xist RNA is the nuclear matrix protein HnrnpK, which is involved in recruitment of the PRC1 and PRC2 complexes for deposition of the repressive marks H2AK119ub and H3K27me3 (Hasegawa et al. 2010; Chu et al. 2015; McHugh et al. 2015; Minajigi et al. 2015). Histone modifications including deacetylation of histones, methylation of H3K27 and ubiquitination of H3K119 are early events in establishing silencing (Jeppesen and Turner 1993; Boggs et al. 2002; Plath et al. 2002; Heard and Disteche 2006; Marks et al. 2009). The A-repeat within the Xist gene is essential for gene silencing (Wutz et al. 2002) and is a key element for the binding interactors (Chu et al. 2015). Thus, important new elements are being discovered that connect Xist RNA to the deposition of specific epigenetic modifications put in place to implement stable and heritable gene silencing (see maintenance) (Gendrel and Heard 2014).
‘... and that there will be a similar effect when autosomal genes are translocated to the X-chromosome’
Silencing via XIST/Xist RNA spreading in cis can also recruit silencing factors along autosomal segments attached to the inactive X following a translocation or insertion (figure 1a). Silencing is less efficient along autosomal regions (Sharp et al. 2002), suggesting that specific elements enriched on the X help spreading and maintenance. Mary Lyon proposed that such elements might be LINE1 repeats, which are particularly abundant on the X chromosome (Lyon 1998). Indeed, the core of the condensed inactive X chromosome is enriched in L1 elements (Chow et al. 2010; Deng et al. 2015). Accordingly, inefficient discontinuous spreading is observed along autosomal segments with few L1 elements (Tang et al. 2010). However, cell selection plays an important role in the observed patterns of inactivation in X;autosome translocations, which should be interpreted with caution (Disteche et al. 1979). An interesting application of the power of XIST in inducing cis autosomal silencing is the correction of trisomy 21 by insertion of a highly expressed XIST transgene on one human chromosome 21 in trisomic cells (Jiang et al. 2013). This restores normal gene expression and cellular phenotypes, thus offering hope for helping individuals with Down’s syndrome or other autosomal trisomies, at least in cells accessible to treatment such as bone marrow.
XCI maintenance
‘Thus, it is at present considered that methylation is part of the mechanism for stabilizing inactivation, after spreading has occurred’
Early studies identified a key molecular feature that locks silencing, i.e. DNA methylation at CpG islands of X-linked genes (Riggs 1975; Gartler and Riggs 1983). Particularly telling were experiments in which a methylated DNA plasmid containing the X-linked gene HPRT was shown to remain silent after transfection into HPRT-deficient cells, but became competent after removal of DNA methylation by 5-azacytidine (Liskay and Evans 1980; Venolia et al. 1982; Venolia and Gartler 1983). Maintenance of XCI is also ensured by the histone modifications that are progressively added throughout early development (see spreading). Later events include replacement of histone H2A by macrohistone H2A (Costanzi and Pehrson 1998), and DNA methylation of CpG islands implemented by the methylases Dnmt3a/b and maintained by Dnmt1 (Norris et al. 1991). Different genes become silent at different times in concordance with epigenetic changes (Gendrel et al. 2012). Maintenance of XCI requires synergy of Xist RNA, histone modifications and DNA methylation (Csankovszki et al. 2001). However, loss of any one of the element does not necessarily affect silencing. For example, EED, a component of the PRC2 complex that mediate H3K27me3 is dispensable for initiation and maintenance of XCI in embryos (Kalantry et al. 2006).
X chromosome 3D structure
‘The cytology evidence was provided by Ohno and Hauschka (1960), who showed that in cells of various tissues of female mice one chromosome was heteropycnotic.’
The inactive X chromosome forms the Barr body (Barr and Bertram 1949) visible as a condensed heteropycnotic structure in interphase nuclei of female cells (Ohno and Hauschka 1960). The modalities of condensation of the inactive X are only beginning to be deciphered using genome-wide analyses of chromatin structure by chromatin conformation studies including Hi-C. Both in human and mouse, the inactive X chromosome forms a bipartite structure of two super-domains separated by a boundary (Rao et al. 2014; Deng et al. 2015; Minajigi et al. 2015). The superdomains differ between human and mouse but the boundary between domains is partially conserved and contains the macrosatellite locus Dxz4 (Deng et al. 2015), which binds CTCF specifically on the inactive X (Chadwick 2008; Horakova et al. 2012a, b). CTCF is a zinc finger protein widely known to help organize chromatin in topologically associated domains (TADs) (Dixon et al. 2012). In mouse, the boundary between superdomains on the inactive X appears to represent a nucleolus-associated domain (NAD) (Deng et al. 2015).
‘Knowledge of the fine structure of the embryo at this stage may provide some clue whether or not attachment of the X chromosome to a site is a likey mechanism’
Mary Lyon suggested that the inactive X may occupy a preferred site in the nucleus (Lyon 1971). Such preferred locations for the inactive X are proximity to either the nuclear membrane (Barr and Bertram 1949) or the nucleolus (Zhang et al. 2007). These preferred locations are in agreement with findings in other systems, suggesting that the lamina and/or the nucleolus represent ‘Velcro’ elements for heterochromatin (Padeken and Heun 2014). Interestingly, XIST interactors include proteins that help anchor chromosomes to the nuclear membrane such as the lamin B receptor (LBR) (Chu et al. 2015; McHugh et al. 2015; Minajigi et al. 2015). Our own data suggest that proximity of the inactive X to the nucleolus may be facilitated by specific elements such as the lncRNA genes, Firre and Dxz4, which bind CTCF specifically on the inactive X. Knockdown of Firre causes loss of the repressive mark H3K27me3 on the X chromosome, suggesting a role in maintenance of heterochromatin potentially related to positioning (Yang et al. 2015).
X reactivation
‘These observations provide the first evidence with a true X-linked gene (Oct) for an age-related decrease in the stability of the X-inactivation mechanism.’
X chromosome regulation in females represents a cycle of inactivation and reactivation (Gartler and Riggs 1983; Gartler et al. 1992). In precursor female germ cells both X chromosomes become active by a process of reactivation that progresses along the X, genes closest to Xist being reactivated last (Sugimoto and Abe 2007). This reactivation ensures that each haploid female germ cell contains an active X chromosome. Interestingly, haploid cells derived from female germ cells have a high X:autosome expression ratio due to upregulation of the active X (Leeb and Wutz 2011). Immediately after fertilization, there is reactivation of the paternal X which is largely silenced in sperm (Okamoto et al. 2004). A second wave of reactivation occurs in the inner cell mass at blastocyst stage prior to the onset of random XCI (Mak et al. 2004). In cells where reactivation occurs XIST becomes silent and repressive histone marks are lost (Ohhata and Wutz 2012).
X reactivation can also occur in somatic cells in relation to ageing as Mary Lyon first described (Wareham et al. 1987). Aberrant reactivation is also observed in congenital or acquired diseases (see below). For example, abnormal X-linked gene expression is seen in ICF syndrome, which is due to a mutation in the methylase Dnmt3b (Hansen et al. 2000). Persistence of XIST/Xist in somatic cells is not necessarily required for stable silencing (Brown and Willard 1994). However, an induced Xist deletion caused X reactivation and cancer in mice after a long period of time (Yildirim et al. 2013). Reactivation can also be induced in iPS cells following dedifferentiation of somatic cells (Lessing and Lee 2013). While this can easily be induced in mouse by adding pluripotent factors this is not always the case in human cells where variable patterns are observed (Lessing and Lee 2013). The presence of two active X chromosomes is rarely observed in undifferentiated human ES cell lines unless they are in a ‘naive’ state (Ware et al. 2014). Interestingly, X reactivation in human pluripotent stem cells is accompanied by coating with the lncRNA XACT prior to loss of XIST RNA (Vallot et al. 2015).
Escape from XCI
‘… it is still possible that inactivation of one X does not take place in man, or that it differs in some way from the process in the mouse... ’
‘The other possible explanation is that the X chromosome of man has a short pairing segment, that is not normally inactivated, and that it is duplication or deficiency of this region which gives rise to the abnormal phenotypes observed.’
Mary Lyon hypothesized that some genes, probably located in the pseudoautosomal region (PAR) of pairing between the X and Y chromosomes, would escape XCI (Lyon 1962). She puzzled about differences in phenotypes between X0 mice that can reproduce and 45,X women who have abnormal phenotypes and are infertile. Subsequent studies have shown that the mouse and human PARs contain very different sets of genes (Disteche et al. 1992). In addition, genes outside the PAR can also escape XCI with significant differences in the list of escape genes between mouse and human (Berletch et al. 2010). In human, about 15% of X-linked genes escape XCI compared to 3–7% in mouse (Carrel et al. 1999; Yang et al. 2010). Some of the escape genes that reside outside of the PAR have retained a Y-linked paralogue (Lahn and Page 1997). In fact, a subset of these X/Y genes are conserved on the sex chromosomes in multiple mammalian species, possibly because they encode for critical proteins and are highly dosage-sensitive (Bellott et al. 2014; Cortez et al. 2014).
Escape from XCI can vary between tissues and individuals. We recently completed a study of XCI and escape in multiple mouse tissues using RNA-seq to test allele-specific expression in F1 mice with skewed XCI based on frequent SNPs (figure 2) (Berletch et al. 2015). While, a subset of escape genes were common between tissues, others were tissue-specific. Interestingly, many genes found to escape XCI in adult mouse tissues differ from those reported in trophoblastic cells derived from placenta in which XCI is paternally imprinted, suggesting significant differences between imprinted and random XCI (Calabrese et al. 2012; Corbel et al. 2013; Finn et al. 2014). In addition, levels of expression from the inactive X can vary for a given gene, suggesting tissue-specific dosage effects of escape. Thus, escape from XCI may be a source of tissue-specific sex differences. In human, SNP analyses have also shown tissue and individual variability (Cotton et al. 2013). Differential methylation levels at X-linked CpG islands and gene bodies has also helped identify escape genes in many human tissue types (Lister et al. 2013; Cotton et al. 2015; Schultz et al. 2015). Indeed, escape genes are often depleted in repressive marks associated with XCI and enriched in marks associated with active gene transcription (Berletch et al. 2011). In addition, escape genes are located at the periphery of the silent domain of the inactive X chromosome where they apparently interact with each other (Splinter et al. 2011; Deng et al. 2015).
Figure 2.

Differences and commonalities in the distribution of genes (pink) that escape X inactivation on the mouse X chromosome (centromere to the left) between tissues in vivo: ovary (top), spleen (middle), and brain (bottom). Pink bars indicate the position of escape genes on schematics of the mouse X chromosome (centromere at left) (see Berletch et al. 2015).
Escape genes that lack a functionally equivalent Y paralogue are a potential source of sex-specific differences in gene expression and thus, candidates for sex-specific phenotypes (Berletch et al. 2011). One example is the histone demethylase KDM6A encoded by a gene that escapes XCI in multiple species. Kdm6a is more highly expressed in female cells and regulates a set of reproduction-related homeobox genes (Rhox6 and Rhox9) in a female-specific manner (Berletch et al. 2013). Whether other escape genes contribute to sex-specific differences is still under study.
XCI, X aneuploidy and disease
‘Facts that remain unexplained are that an X0 female and an XXY male show any abnormality, and that an X0 female in man differs in phenotype from that in mouse’
Escape gene dosage is dependent on the number of X chromosomes present, thus escape genes are candidates for phenotypes associated with X aneuploidy. Indeed, aberrant copy number of escape genes (PAR or non-PAR) is thought to be associated with abnormal phenotypes in Turner and Klinefelter syndromes (Zinn et al. 2007; Tartaglia et al. 2010a, b). For example, loss of one copy of the SHOX gene located in the PAR explains the short stature in Tuner syndrome, whereas three copies of this gene in XXY individuals explain the tall stature in Klinefelter syndrome (Blaschke and Rappold 2006). Specific escape genes have been implicated in mental impairment; for example, KDM5C and IQSEC2 deletions or mutations cause X-linked intellectual disability both in males and females, consistent with dosage sensitivity (Santos-Reboucas et al. 2011; Simensen et al. 2013; Fieremans et al. 2015). Further, cognitive deficiencies have been reported in individuals carrying microduplications of KDM5C and IQSEC2 associated with abnormally high expression (Moey et al. 2015). Similarly, mutations and deletions in KDM6A have been discovered in patients with Kabuki syndrome characterized by intellectual disability, growth retardation, skeletal abnormalities, and visceral malformations (Lederer et al. 2012; Miyake et al. 2012). Female carrier of mutations also show abnormalities, consistent with dosage anomalies (Lindgren et al. 2013). Some of these patients show symptoms overlapping with Turner syndrome, termed Turner–Kabuki syndrome, suggesting a potential link to other genes that escape XCI.
X-linked mutations cause diseases with widely different consequences in males and females. Males are often affected because they have only one X chromosome, so that recessive mutations cause abnormal phenotypes. Females can compensate by having patches of cells that express the normal allele, or by strong selection (skewing) for cells that express the normal allele (Deng et al. 2014). Skewing of XCI can be very extensive or only affect the tissue in which proper expression is critical (Migeon 2014). Random distribution of patches of cells with one X active can be extensive as shown by a recent study of female mice with a different X-linked fluorescent reporter on each allele, in which in situ visualization of XCI distribution revealed surprisingly extensive skewing of XCI (Wu et al. 2014). For example, one mouse had half of her brain with silencing of the maternal X and the other with silencing of the paternal X.
Mutations in escape genes and abnormal dosage have been linked to noncongenital disease as well. For example, KDM6A mutations have been observed in renal carcinoma as well as other cancer types (van Haaften et al. 2009; Dalgliesh et al. 2010). Interestingly, KDM6A seems to function as a gender-specific tumour suppressor in T-cell acute lymphoblastic leukaemia, where only males with the disease had inactivating mutations in the demethylase (Van der Meulen et al. 2015). Additionally, aberrant hypomethylation of the X chromosome along with loss of part or one entire X can occur in breast cancer cells (Sun et al. 2015). Extensive reactivation of the X chromosome has been documented in breast cancer (Chaligne et al. 2015).
Summary
In summary, the X chromosome inactivation law proposed by Mary Lyon has helped us to understand not only basic principles of gene silencing, heterochromatin structure and nuclear organization, but has also led to discoveries of new master switches such as the lncRNA Xist and to a better understanding of X-linked diseases and of sex-specific differences.
Acknowledgments
This work was supported by grants GM046883 and GM113943 (CMD) from the National Institutes of Health. JB is also supported by grant MH105768 from the National Institutes of Health. We thank X. Deng for critical reading of the manuscript.
References
- Barr ML, Bertram EG. A morphological distinction between neurones of the male and female, and the behaviour of the nucleolar satellite during accelerated nucleoprotein synthesis. Nature. 1949;163:676. doi: 10.1038/163676a0. [DOI] [PubMed] [Google Scholar]
- Bellott DW, Hughes JF, Skaletsky H, Brown LG, Pyntikova T, Cho TJ, et al. Mammalian Y chromosomes retain widely expressed dosage-sensitive regulators. Nature. 2014;508:494–499. doi: 10.1038/nature13206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berletch JB, Yang F, Disteche CM. Escape from X inactivation in mice and humans. Genome Biol. 2010;11:213. doi: 10.1186/gb-2010-11-6-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berletch JB, Yang F, Xu J, Carrel L, Disteche CM. Genes that escape from X inactivation. Hum Genet. 2011;130:237–245. doi: 10.1007/s00439-011-1011-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berletch JB, Deng X, Nguyen DK, Disteche CM. Female bias in Rhox6 and 9 regulation by the histone demethylase KDM6A. PLoS Genet. 2013;9:e1003489. doi: 10.1371/journal.pgen.1003489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berletch JB, Ma W, Yang F, Shendure J, Noble WS, Disteche CM, Deng X. Escape from X inactivation varies in mouse tissues. PLoS Genet. 2015;11:e1005079. doi: 10.1371/journal.pgen.1005079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaschke RJ, Rappold G. The pseudoautosomal regions, SHOX and disease. Curr Opin Genet Dev. 2006;16:233–239. doi: 10.1016/j.gde.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Boggs BA, Cheung P, Heard E, Spector DL, Chinault AC, Allis CD. Differentially methylated forms of histone H3 show unique association patterns with inactive human X chromosomes. Nat Genet. 2002;30:73–76. doi: 10.1038/ng787. [DOI] [PubMed] [Google Scholar]
- Brockdorff N, Kay G, Smith S, Keer JT, Hamvas RM, Brown SD, Rastan S. High-density molecular map of the central span of the mouse X chromosome. Genomics. 1991;10:17–22. doi: 10.1016/0888-7543(91)90478-w. [DOI] [PubMed] [Google Scholar]
- Brockdorff N, Ashworth A, Kay GF, McCabe VM, Norris DP, Cooper PJ, et al. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71:515–526. doi: 10.1016/0092-8674(92)90519-i. [DOI] [PubMed] [Google Scholar]
- Brown CJ, Willard HF. The human X-inactivation center is not required for maintenance of X-chromosome inactivation. Nature. 1994;368:154–156. doi: 10.1038/368154a0. [DOI] [PubMed] [Google Scholar]
- Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, Willard HF. A gene from the region of the human X inactivation center is expressed exclusively from the inactive X chromosome. Nature. 1991;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- Brown CJ, Hendrich BD, Rupert JL, Lafreniere RG, Xing Y, Lawrence J, Willard HF. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- Calabrese JM, Sun W, Song L, Mugford JW, Williams L, Yee D, et al. Site-specific silencing of regulatory elements as a mechanism of X inactivation. Cell. 2012;151:951–963. doi: 10.1016/j.cell.2012.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrel L, Cottle AA, Goglin KC, Willard HF. A first-generation X-inactivation profile of the human X chromosome. Proc Natl Acad Sci USA. 1999;96:14440–14444. doi: 10.1073/pnas.96.25.14440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattanach BM. Control of chromosome inactivation. Annu Rev Genet. 1975;9:1–18. doi: 10.1146/annurev.ge.09.120175.000245. [DOI] [PubMed] [Google Scholar]
- Chadwick BP. DXZ4 chromatin adopts an opposing conformation to that of the surrounding chromosome and acquires a novel inactive X-specific role involving CTCF and antisense transcripts. Genome Res. 2008;18:1259–1269. doi: 10.1101/gr.075713.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaligne R, Popova T, Mendoza-Parra MA, Saleem MA, Gentien D, Ban K, et al. The inactive X chromosome is epigenetically unstable and transcriptionally labile in breast cancer. Genome Res. 2015;25:488–503. doi: 10.1101/gr.185926.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang SC, Brown CJ. Identification of regulatory elements flanking human XIST reveals species differences. BMC Mol Biol. 2010;11:20. doi: 10.1186/1471-2199-11-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow JC, Ciaudo C, Fazzari MJ, Mise N, Servant N, Glass JL, et al. LINE-1 activity in facultative heterochromatin formation during X chromosome inactivation. Cell. 2010;141:956–969. doi: 10.1016/j.cell.2010.04.042. [DOI] [PubMed] [Google Scholar]
- Chu C, Zhang QC, da Rocha ST, Flynn RA, Bharadwaj M, Calabrese JM, et al. Systematic discovery of Xist RNA binding proteins. Cell. 2015;161:404–416. doi: 10.1016/j.cell.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, McNeil JA, Willard HF, Lawrence JB. XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. J Cell Biol. 1996;132:259–275. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbel C, Diabangouaya P, Gendrel AV, Chow JC, Heard E. Unusual chromatin status and organization of the inactive X chromosome in murine trophoblast giant cells. Development. 2013;140:861–872. doi: 10.1242/dev.087429. [DOI] [PubMed] [Google Scholar]
- Cortez D, Marin R, Toledo-Flores D, Froidevaux L, Liechti A, Waters PD, et al. Origins and functional evolution of Y chromosomes across mammals. Nature. 2014;508:488–493. doi: 10.1038/nature13151. [DOI] [PubMed] [Google Scholar]
- Costanzi C, Pehrson JR. Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals. Nature. 1998;393:599–601. doi: 10.1038/31275. [DOI] [PubMed] [Google Scholar]
- Cotton AM, Ge B, Light N, Adoue V, Pastinen T, Brown CJ. Analysis of expressed SNPs identifies variable extents of expression from the human inactive X chromosome. Genome Biol. 2013;14:R122. doi: 10.1186/gb-2013-14-11-r122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotton AM, Price EM, Jones MJ, Balaton BP, Kobor MS, Brown CJ. Landscape of DNA methylation on the X chromosome reflects CpG density, functional chromatin state and X-chromosome inactivation. Hum Mol Genet. 2015;24:1528–1539. doi: 10.1093/hmg/ddu564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csankovszki G, Nagy A, Jaenisch R. Synergism of Xist RNA, DNA methylation, and histone hypoacetylation in maintaining X chromosome inactivation. J Cell Biol. 2001;153:773–784. doi: 10.1083/jcb.153.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalgliesh GL, Furge K, Greenman C, Chen L, Bignell G, Butler A, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463:360–363. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Berletch JB, Nguyen DK, Disteche CM. X chromosome regulation: diverse patterns in development, tissues and disease. Nat Rev Genet. 2014;15:367–378. doi: 10.1038/nrg3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Ma W, Ramani V, Hill A, Yang F, Ay F, et al. Bipartite structure of the inactive mouse X chromosome. Genome Biol. 2015;16:152. doi: 10.1186/s13059-015-0728-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disteche CM. Dosage compensation of the sex chromosomes. Annu Rev Genet. 2012;46:537–560. doi: 10.1146/annurev-genet-110711-155454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disteche CM, Brannan CI, Larsen A, Adler DA, Schorderet DF, Gearing D, et al. The human pseudoautosomal GM-CSF receptor alpha subunit gene is autosomal in mouse. Nat Genet. 1992;1:333–336. doi: 10.1038/ng0892-333. [DOI] [PubMed] [Google Scholar]
- Disteche CM, Eicher EM, Latt SA. Late replication in an X-autosome translocation in the mouse: correlation with genetic inactivation and evidence for selective effects during embryogenesis. Proc Natl Acad Sci USA. 1979;76:5234–5238. doi: 10.1073/pnas.76.10.5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engreitz JM, Pandya-Jones A, McDonel P, Shishkin A, Sirokman K, Surka C, et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. 2013;341:1237973. doi: 10.1126/science.1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein CJ, Smith S, Travis B, Tucker G. Both X chromosomes function before visible X-chromosome inactivation in female mouse embryos. Nature. 1978;274:500–503. doi: 10.1038/274500a0. [DOI] [PubMed] [Google Scholar]
- Fieremans N, Van Esch H, de Ravel T, Van Driessche J, Belet S, Bauters M, Froyen G. Microdeletion of the escape genes KDM5C and IQSEC2 in a girl with severe intellectual disability and autistic features. Eur J Med Genet. 2015;58:324–327. doi: 10.1016/j.ejmg.2015.03.003. [DOI] [PubMed] [Google Scholar]
- Finn EH, Smith CL, Rodriguez J, Sidow A, Baker JC. Maternal bias and escape from X chromosome imprinting in the midgestation mouse placenta. Dev Biol. 2014;390:80–92. doi: 10.1016/j.ydbio.2014.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galupa R, Heard E. X-chromosome inactivation: new insights into cis and trans regulation. Curr Opin Genet Dev. 2015;31:57–66. doi: 10.1016/j.gde.2015.04.002. [DOI] [PubMed] [Google Scholar]
- Gartler SM, Dyer KA, Goldman MA. Mammalian X chromosome inactivation. Mol Genet Med. 1992;2:121–160. doi: 10.1016/b978-0-12-462002-5.50010-8. [DOI] [PubMed] [Google Scholar]
- Gartler SM, Riggs AD. Mammalian X-chromosome inactivation. Annu Rev Genet. 1983;17:155–190. doi: 10.1146/annurev.ge.17.120183.001103. [DOI] [PubMed] [Google Scholar]
- Gayen S, Maclary E, Buttigieg E, Hinten M, Kalantry S. A primary role for the Tsix lncRNA in maintaining random X-chromosome inactivation. Cell Rep. 2015;11:1251–1265. doi: 10.1016/j.celrep.2015.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel AV, Heard E. Noncoding RNAs and epigenetic mechanisms during X-chromosome inactivation. Annu Rev Cell Dev Biol. 2014;30:561–580. doi: 10.1146/annurev-cellbio-101512-122415. [DOI] [PubMed] [Google Scholar]
- Gendrel AV, Apedaile A, Coker H, Termanis A, Zvetkova I, Godwin J, et al. Smchd1-dependent and -independent pathways determine developmental dynamics of CpG island methylation on the inactive x chromosome. Dev Cell. 2012;23:265–279. doi: 10.1016/j.devcel.2012.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgetti L, Galupa R, Nora EP, Piolot T, Lam F, Dekker J, et al. Predictive polymer modeling reveals coupled fluctuations in chromosome conformation and transcription. Cell. 2014;157:950–963. doi: 10.1016/j.cell.2014.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gontan C, Achame EM, Demmers J, Barakat TS, Rentmeester E, van IW, et al. RNF12 initiates X-chromosome inactivation by targeting REX1 for degradation. Nature. 2012;485:386–390. doi: 10.1038/nature11070. [DOI] [PubMed] [Google Scholar]
- Hansen RS, Stoger R, Wijmenga C, Stanek AM, Canfield TK, Luo P, et al. Escape from gene silencing in ICF syndrome: evidence for advanced replication time as a major determinant. Hum Mol Genet. 2000;9:2575–2587. doi: 10.1093/hmg/9.18.2575. [DOI] [PubMed] [Google Scholar]
- Hasegawa Y, Brockdorff N, Kawano S, Tsutui K, Tsutui K, Nakagawa S. The matrix protein hnRNP U is required for chromosomal localization of Xist RNA. Dev Cell. 2010;19:469–476. doi: 10.1016/j.devcel.2010.08.006. [DOI] [PubMed] [Google Scholar]
- Heard E, Disteche CM. Dosage compensation in mammals: fine-tuning the expression of the X chromosome. Genes Dev. 2006;20:1848–1867. doi: 10.1101/gad.1422906. [DOI] [PubMed] [Google Scholar]
- Horakova AH, Calabrese JM, McLaughlin CR, Tremblay DC, Magnuson T, Chadwick BP. The mouse DXZ4 homolog retains Ctcf binding and proximity to Pls3 despite substantial organizational differences compared to the primate macrosatellite. Genome Biol. 2012a;13:R70. doi: 10.1186/gb-2012-13-8-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horakova AH, Moseley SC, McLaughlin CR, Tremblay DC, Chadwick BP. The macrosatellite DXZ4 mediates CTCF-dependent long-range intrachromosomal interactions on the human inactive X chromosome. Hum Mol Genet. 2012b;21:4367–4377. doi: 10.1093/hmg/dds270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppesen P, Turner BM. The inactive X chromosome in female mammals is distinguished by a lack of histone H4 acetylation, a cytogenetic marker for gene expression. Cell. 1993;74:281–289. doi: 10.1016/0092-8674(93)90419-q. [DOI] [PubMed] [Google Scholar]
- Jiang J, Jing Y, Cost GJ, Chiang JC, Kolpa HJ, Cotton AM, et al. Translating dosage compensation to trisomy 21. Nature. 2013;500:296–300. doi: 10.1038/nature12394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalantry S, Mills KC, Yee D, Otte AP, Panning B, Magnuson T. The Polycomb group protein Eed protects the inactive X-chromosome from differentiation-induced reactivation. Nat Cell Biol. 2006;8:195–202. doi: 10.1038/ncb1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay GF, Penny GD, Patel D, Ashworth A, Brockdorff N, Rastan S. Expression of Xist during mouse development suggests a role in the initiation of X chromosome inactivation. Cell. 1993;72:171–182. doi: 10.1016/0092-8674(93)90658-d. [DOI] [PubMed] [Google Scholar]
- Kratzer PG, Gartler SM. HGPRT activity changes in preimplantation mouse embryos. Nature. 1978;274:503–504. doi: 10.1038/274503a0. [DOI] [PubMed] [Google Scholar]
- Lahn BT, Page DC. Functional coherence of the human Y chromosome. Science. 1997;278:675–680. doi: 10.1126/science.278.5338.675. [DOI] [PubMed] [Google Scholar]
- Lederer D, Grisart B, Digilio MC, Benoit V, Crespin M, Ghariani SC, et al. Deletion of KDM6A, a histone demethylase interacting with MLL2, in three patients with Kabuki syndrome. Am J Hum Genet. 2012;90:119–124. doi: 10.1016/j.ajhg.2011.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT, Jaenisch R. Long-range cis effects of ectopic X-inactivation centers on a mouse autosome. Nature. 1997;386:275–279. doi: 10.1038/386275a0. [DOI] [PubMed] [Google Scholar]
- Leeb M, Wutz A. Derivation of haploid embryonic stem cells from mouse embryos. Nature. 2011;479:131–134. doi: 10.1038/nature10448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppig KA, Brown CJ, Bressler SL, Gustashaw K, Pagon RA, Willard HF, Disteche CM. Mapping of the distal boundary of the X-inactivation center in a rearranged X chromosome from a female expressing XIST. Hum Mol Genet. 1993;2:883–887. doi: 10.1093/hmg/2.7.883. [DOI] [PubMed] [Google Scholar]
- Lessing D, Lee JT. X chromosome inactivation and epigenetic responses to cellular reprogramming. Annu Rev Genomics Hum Genet. 2013;14:85–110. doi: 10.1146/annurev-genom-091212-153530. [DOI] [PubMed] [Google Scholar]
- Lin H, Halsall JA, Antczak P, O’Neill LP, Falciani F, Turner BM. Relative overexpression of X-linked genes in mouse embryonic stem cells is consistent with Ohno’s hypothesis. Nat Genet. 2011;43:1169–1170. doi: 10.1038/ng.992. author reply 1171–1172. [DOI] [PubMed] [Google Scholar]
- Lindgren AM, Hoyos T, Talkowski ME, Hanscom C, Blumenthal I, Chiang C, et al. Haploinsufficiency of KDM6A is associated with severe psychomotor retardation, global growth restriction, seizures and cleft palate. Hum Genet. 2013;132:537–552. doi: 10.1007/s00439-013-1263-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liskay RM, Evans RJ. Inactive X chromosome DNA does not function in DNA-mediated cell transformation for the hypoxanthine phosphoribosyltransferase gene. Proc Natl Acad Sci USA. 1980;77:4895–4898. doi: 10.1073/pnas.77.8.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, et al. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341:1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon M. Gene action in the X-chromosome of the mouse (Mus musculus L) Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- Lyon MF. Sex chromatin and gene action in the mammalian X-chromosome. Am J Hum Genet. 1962;14:135–148. [PMC free article] [PubMed] [Google Scholar]
- Lyon MF. Possible mechanisms of X chromosome inactivation. Nat New Biol. 1971;232:229–232. doi: 10.1038/newbio232229a0. [DOI] [PubMed] [Google Scholar]
- Lyon MF. The William Allan memorial award address: X-chromosome inactivation and the location and expression of X-linked genes. Am J Hum Genet. 1988;42:8–16. [PMC free article] [PubMed] [Google Scholar]
- Lyon MF. Some milestones in the history of X-chromosome inactivation. Annu Rev Genet. 1992;26:16–28. doi: 10.1146/annurev.ge.26.120192.000313. [DOI] [PubMed] [Google Scholar]
- Lyon MF. X-chromosome inactivation: a repeat hypothesis. Cytogenet Cell Genet. 1998;80:133–137. doi: 10.1159/000014969. [DOI] [PubMed] [Google Scholar]
- Lyon MF, Searle AG, Ford CE, Ohno S. A mouse translocation suppressing sex-linked variegation. Cytogenetics. 1964;3:306–323. doi: 10.1159/000129820. [DOI] [PubMed] [Google Scholar]
- Mak W, Nesterova TB, de Napoles M, Appanah R, Yamanaka S, Otte AP, Brockdorff N. Reactivation of the paternal X chromosome in early mouse embryos. Science. 2004;303:666–669. doi: 10.1126/science.1092674. [DOI] [PubMed] [Google Scholar]
- Marks H, Chow JC, Denissov S, Francoijs KJ, Brockdorff N, Heard E, Stunnenberg HG. High-resolution analysis of epigenetic changes associated with X inactivation. Genome Res. 2009;19:1361–1373. doi: 10.1101/gr.092643.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh CA, Chen CK, Chow A, Surka CF, Tran C, McDonel P, et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 2015;521:232–236. doi: 10.1038/nature14443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migeon BR. Females are mosaic: X inactivation and sex differences in disease. Oxford University Press; Oxford: 2014. [Google Scholar]
- Minajigi A, Froberg JE, Wei C, Sunwoo H, Kesner B, Colognori D, et al. A comprehensive Xist interactome reveals cohesin repulsion and an RNA-directed chromosome conformation. Science. 2015 doi: 10.1126/science.aab2276. pii, aab2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake N, Mizuno S, Okamoto N, Ohashi H, Shiina M, Ogata K, et al. KDM6A point mutations cause Kabuki syndrome. Hum Mutat. 2012;34:108–110. doi: 10.1002/humu.22229. [DOI] [PubMed] [Google Scholar]
- Moey C, Hinze SJ, Brueton L, Morton J, McMullan DJ, Kamien B, et al. Xp11.2 microduplications including IQSEC2, TSPYL2 and KDM5C genes in patients with neurodevelopmental disorders. Eur J Hum Genet. 2015 doi: 10.1038/ejhg.2015.123. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monkhorst K, Jonkers I, Rentmeester E, Grosveld F, Gribnau J. X inactivation counting and choice is a stochastic process: evidence for involvement of an X-linked activator. Cell. 2008;132:410–421. doi: 10.1016/j.cell.2007.12.036. [DOI] [PubMed] [Google Scholar]
- Morey C, Avner P. Genetics and epigenetics of the X chromosome. Ann N Y Acad Sci. 2010;1214:E18–E33. doi: 10.1111/j.1749-6632.2010.05943.x. [DOI] [PubMed] [Google Scholar]
- Navarro P, Chambers I, Karwacki-Neisius V, Chureau C, Morey C, Rougeulle C, Avner P. Molecular coupling of Xist regulation and pluripotency. Science. 2008;321:1693–1695. doi: 10.1126/science.1160952. [DOI] [PubMed] [Google Scholar]
- Norris DP, Brockdorff N, Rastan S. Methylation status of CpG-rich islands on active and inactive mouse X chromosomes. Mamm Genome. 1991;1:78–83. doi: 10.1007/BF02443782. [DOI] [PubMed] [Google Scholar]
- Ohhata T, Wutz A. Reactivation of the inactive X chromosome in development and reprogramming. Cell Mol Life Sci. 2012;70:2443–2461. doi: 10.1007/s00018-012-1174-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S, Hauschka TS. Allocycly of the X-chromosome in tumors and normal tissues. Cancer Res. 1960;20:541–545. [PubMed] [Google Scholar]
- Okamoto I, Otte AP, Allis CD, Reinberg D, Heard E. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science. 2004;303:644–649. doi: 10.1126/science.1092727. [DOI] [PubMed] [Google Scholar]
- Okamoto I, Arnaud D, Le Baccon P, Otte AP, Disteche CM, Avner P, Heard E. Evidence for de novo imprinted X-chromosome inactivation independent of meiotic inactivation in mice. Nature. 2005;438:369–373. doi: 10.1038/nature04155. [DOI] [PubMed] [Google Scholar]
- Okamoto I, Patrat C, Thepot D, Peynot N, Fauque P, Daniel N, et al. Eutherian mammals use diverse strategies to initiate X-chromosome inactivation during development. Nature. 2011;472:370–374. doi: 10.1038/nature09872. [DOI] [PubMed] [Google Scholar]
- Padeken J, Heun P. Nucleolus and nuclear periphery: velcro for heterochromatin. Curr Opin Cell Biol. 2014;28:54–60. doi: 10.1016/j.ceb.2014.03.001. [DOI] [PubMed] [Google Scholar]
- Payer B, Lee JT. X chromosome dosage compensation: how mammals keep the balance. Annu Rev Genet. 2008;42:733–772. doi: 10.1146/annurev.genet.42.110807.091711. [DOI] [PubMed] [Google Scholar]
- Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- Pinter SF, Sadreyev RI, Yildirim E, Jeon Y, Ohsumi TK, Borowsky M, Lee JT. Spreading of X chromosome inactivation via a hierarchy of defined Polycomb stations. Genome Res. 2012;22:1864–1876. doi: 10.1101/gr.133751.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plath K, Mlynarczyk-Evans S, Nusinow DA, Panning B. Xist RNA and the mechanism of x chromosome inactivation. Annu Rev Genet. 2002;36:233–278. doi: 10.1146/annurev.genet.36.042902.092433. [DOI] [PubMed] [Google Scholar]
- Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs AD. X inactivation, differentiation, and DNA methylation. Cytogenet Cell Genet. 1975;14:9–25. doi: 10.1159/000130315. [DOI] [PubMed] [Google Scholar]
- Russell LB. Another Look at the Single-Active-X Hypothesis. Trans N Y Acad Sci. 1964;26:726–736. doi: 10.1111/j.2164-0947.1964.tb01939.x. [DOI] [PubMed] [Google Scholar]
- Santos-Reboucas CB, Fintelman-Rodrigues N, Jensen LR, Kuss AW, Ribeiro MG, Campos M, et al. A novel nonsense mutation in KDM5C/JARID1C gene causing intellectual disability, short stature and speech delay. Neurosci Lett. 2011;498:67–71. doi: 10.1016/j.neulet.2011.04.065. [DOI] [PubMed] [Google Scholar]
- Schultz MD, He Y, Whitaker JW, Hariharan M, Mukamel EA, Leung D, et al. Human body epigenome maps reveal noncanonical DNA methylation variation. Nature. 2015;523:212–216. doi: 10.1038/nature14465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp AJ, Spotswood HT, Robinson DO, Turner BM, Jacobs PA. Molecular and cytogenetic analysis of the spreading of X inactivation in X;autosome translocations. Hum Mol Genet. 2002;11:3145–3156. doi: 10.1093/hmg/11.25.3145. [DOI] [PubMed] [Google Scholar]
- Simensen RJ, Rogers RC, Collins JS, Abidi F, Schwartz CE, Stevenson RE. Short-term memory deficits in carrier females with KDM5C mutations. Genet Couns. 2013;23:31–40. [PubMed] [Google Scholar]
- Simon MD, Pinter SF, Fang R, Sarma K, Rutenberg-Schoenberg M, Bowman SK, et al. High-resolution Xist binding maps reveal two-step spreading during X-chromosome inactivation. Nature. 2013;504:465–469. doi: 10.1038/nature12719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splinter E, de Wit E, Nora EP, Klous P, van de Werken HJ, Zhu Y, et al. The inactive X chromosome adopts a unique three-dimensional conformation that is dependent on Xist RNA. Genes Dev. 2011;25:1371–1383. doi: 10.1101/gad.633311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto M, Abe K. X chromosome reactivation initiates in nascent primordial germ cells in mice. PLoS Genet. 2007;3:e116. doi: 10.1371/journal.pgen.0030116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Prodduturi N, Sun SY, Thompson EA, Kocher JA. Chromosome X genomic and epigenomic aberrations and clinical implications in breast cancer by base resolution profiling. Epigenomics. 2015:1–12. doi: 10.2217/epi.15.43. [DOI] [PubMed] [Google Scholar]
- Sunwoo H, Wu JY, Lee JT. The Xist RNA-PRC2 complex at 20-nm resolution reveals a low Xist stoichiometry and suggests a hit-and-run mechanism in mouse cells. Proc Natl Acad Sci USA. 2015;112:E4216–E4225. doi: 10.1073/pnas.1503690112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi N, Sasaki M. Preferential inactivation of the paternally derived X chromosome in the extraembryonic membranes of the mouse. Nature. 1975;256:640–642. doi: 10.1038/256640a0. [DOI] [PubMed] [Google Scholar]
- Tang YA, Huntley D, Montana G, Cerase A, Nesterova TB, Brockdorff N. Efficiency of Xist-mediated silencing on autosomes is linked to chromosomal domain organisation. Epigenet Chromatin. 2010;3:10. doi: 10.1186/1756-8935-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartaglia N, Cordeiro L, Howell S, Wilson R, Janusz J. The spectrum of the behavioral phenotype in boys and adolescents 47, XXY (Klinefelter syndrome) Pediatr Endocrinol Rev. 2010a;8(suppl 1):151–159. [PMC free article] [PubMed] [Google Scholar]
- Tartaglia NR, Howell S, Sutherland A, Wilson R, Wilson L. A review of trisomy X (47,XXX) Orphanet J Rare Dis. 2010b;5:8. doi: 10.1186/1750-1172-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsen JL, Krapp C, Willard HF, Bartolomei MS. Nonrandom X chromosome inactivation is influenced by multiple regions on the murine X chromosome. Genetics. 2012;192:1095–1107. doi: 10.1534/genetics.112.144477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallot C, Ouimette JF, Makhlouf M, Feraud O, Pontis J, Come J, et al. Erosion of X chromosome inactivation in human pluripotent cells initiates with XACT coating and depends on a specific heterochromatin landscape. Cell Stem Cell. 2015;16:533–546. doi: 10.1016/j.stem.2015.03.016. [DOI] [PubMed] [Google Scholar]
- Van der Meulen J, Sanghvi V, Mavrakis K, Durinck K, Fang F, Matthijssens F, et al. The H3K27me3 demethylase UTX is a gender-specific tumor suppressor in T-cell acute lymphoblastic leukemia. Blood. 2015;125:13–21. doi: 10.1182/blood-2014-05-577270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Haaften G, Dalgliesh GL, Davies H, Chen L, Bignell G, Greenman C, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet. 2009;41:521–523. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venolia L, Gartler SM. Comparison of transformation efficiency of human active and inactive X-chromosomal DNA. Nature. 1983;302:82–83. doi: 10.1038/302082a0. [DOI] [PubMed] [Google Scholar]
- Venolia L, Gartler SM, Wassman ER, Yen P, Mohandas T, Shapiro LJ. Transformation with DNA from 5-azacytidine-reactivated X chromosomes. Proc Natl Acad Sci USA. 1982;79:2352–2354. doi: 10.1073/pnas.79.7.2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware CB, Nelson AM, Mecham B, Hesson J, Zhou W, Jonlin EC, et al. Derivation of naive human embryonic stem cells. Proc Natl Acad Sci USA. 2014;111:4484–4489. doi: 10.1073/pnas.1319738111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wareham KA, Lyon MF, Glenister PH, Williams ED. Age related reactivation of an X-linked gene. Nature. 1987;327:725–727. doi: 10.1038/327725a0. [DOI] [PubMed] [Google Scholar]
- Welshons WJ, Russell LB. The Y-chromosome as the bearer of male determining factors in the mouse. Proc Natl Acad Sci USA. 1959;45:560–566. doi: 10.1073/pnas.45.4.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West JD, Frels WI, Chapman VM, Papaioannou VE. Preferential expression of the maternally derived X chromosome in the mouse yolk sac. Cell. 1977;12:873–882. doi: 10.1016/0092-8674(77)90151-9. [DOI] [PubMed] [Google Scholar]
- Wu H, Luo J, Yu H, Rattner A, Mo A, Wang Y, et al. Cellular resolution maps of X chromosome inactivation: implications for neural development, function, and disease. Neuron. 2014;81:103–119. doi: 10.1016/j.neuron.2013.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wutz A, Rasmussen TP, Jaenisch R. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat Genet. 2002;30:167–174. doi: 10.1038/ng820. [DOI] [PubMed] [Google Scholar]
- Yang F, Babak T, Shendure J, Disteche CM. Global survey of escape from X inactivation by RNA-sequencing in mouse. Genome Res. 2010;20:614–622. doi: 10.1101/gr.103200.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Deng X, Ma W, Berletch JB, Rabaia N, Wei G, et al. The lncRNA Firre anchors the inactive X chromosome to the nucleolus by binding CTCF and maintains H3K27me3 methylation. Genome Biol. 2015:1–17. doi: 10.1186/s13059-015-0618-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildirim E, Kirby JE, Brown DE, Mercier FE, Sadreyev RI, Scadden DT, Lee JT. Xist RNA is a potent suppressor of hematologic cancer in mice. Cell. 2013;152:727–742. doi: 10.1016/j.cell.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang LF, Huynh KD, Lee JT. Perinucleolar targeting of the inactive X during S phase: evidence for a role in the maintenance of silencing. Cell. 2007;129:693–706. doi: 10.1016/j.cell.2007.03.036. [DOI] [PubMed] [Google Scholar]
- Zinn AR, Roeltgen D, Stefanatos G, Ramos P, Elder FF, Kushner H, et al. A Turner syndrome neurocognitive phenotype maps to Xp22.3. Behav Brain Funct. 2007;3:24. doi: 10.1186/1744-9081-3-24. [DOI] [PMC free article] [PubMed] [Google Scholar]


