Abstract
Using a DNA polymerase-coupled assay and FRET-based helicase assays, in this work we show that a monomer of S. cerevisiae Pif1 can unwind double-stranded DNA. The helicase activity of a Pif1 monomer is modulated by the nature of the 3′-ssDNA tail of the substrate and its effect on a Pif1-dependent re-winding activity that is coupled to the opening of dsDNA. We propose that in addition to the ssDNA site on the protein that interacts with the translocating strand, Pif1 has a second site that binds the 3′-ssDNA of the substrate. Interaction of DNA with this site modulates the degree to which re-winding counteracts unwinding. Depending on the nature of the 3′-tail and the length of the duplex DNA to be unwound this activity is sufficiently strong to mask the helicase activity of a monomer. In excess Pif1 over the DNA the Pif1-dependent re-winding of the opened DNA strongly limits unwinding, independent of the 3′-tail. We propose that in this case binding of DNA to the second site is precluded and modulation of the Pif1-dependent re-winding activity is largely lost.
Graphical Abstract
INTRODUCTION
S. cerevisiae Pif1 is a 5′ to 3′ helicase that belongs to the SF1B family of helicases 1–4 and it participates in a wide range of DNA processing steps, both in the nucleus and in mitochondria 1,4–9. In mitochondria Pif1 plays a role in mitochondrial DNA repair and maintenance 3,4,10,11. The nuclear form of Pif1 has both telomeric and non-telomeric functions. Pif1 has been proposed to participate in Okazaki fragment processing, where in conjunction with the Dna2 helicase/nuclease it helps processing long flaps that cannot be cleaved by the FEN1 nuclease 7,12–14. Growing experimental evidence also points to a role of Pif1 in preventing genomic instability via destabilization of G4-DNA structures (i.e. G-quadruplex)15–17. Pif1 binds tightly to G4-DNAs and it unwinds them 15,18. Genome wide analysis showed that Pif1 associates with G4 motifs in vivo and resolution of these structures by Pif1 is proposed to be a means to facilitate progression of replication fork, thereby preventing replication fork stalling and DNA damage 15,16. In contrast to this, it has also been shown that in the absence of Pif1 replication fork progression across either G4- or hairpin-forming DNA sequences is unaffected, rather it has been suggested that Pif1 might promotes replication fork movement across a DNA protein barrier 19. Recently, it has been shown that Pif1 has a role in promoting DNA synthesis during break-induced replication 20 and in crossover recombination 20–22. In vitro Pif1 stimulates extension by DNA polymerase δ of a Rad51-generated D-loop 20.
Pif1 acts also as a negative regulator of telomerase, inhibiting telomere elongation and de novo telomere addition 6,9,23–25. In the absence of Pif1 telomeres are longer 9,23 and this was shown to originate from a direct effect of Pif1 on the telomerase, due to its displacement from the telomeric end 2,6,26. In addition to its effect on telomere length, deletion of PIF1 leads to an even more striking effect on the de novo telomere addition pathway to double stranded DNA breaks 23–25. Deletion of PIF1 results in a 200- to 1000-fold increase in gross chromosomal rearrangements that in turn lead to chromosome instability 24,25. The activity of Pif1 on de novo telomere addition is via a direct effect on the telomerase 23 and it requires phosphorylation of Thr763 and Ser766 in the TLSS motif at the C-terminus of the protein 27. Phosphorylation of this motif has also been shown to be important for the activity of Pif1 in BIR 22.
While Pif1 is a monomer in solution, we showed that Pif1 dimerizes upon binding to DNA 28. DNA-induced dimerization of Pif1 is observed on ssDNA as well as with model tailed- and forked-dsDNA substrates. A dimer of Pif1 also forms on DNA unwinding substrates in the presence of saturating concentrations of non-hydrolyzable ATP analogs 28. Based on these observations we suggested that a dimeric form of the enzyme might constitute the pre-initiation complex required for unwinding activity. We also showed that a monomer of Pif1 is a 5′-3′ ssDNA translocase that can displace a protein tightly bound to ssDNA 29. Moreover, while in excess enzyme Pif1 has unwinding activity 26,30,31, our initial data showed that a Pif1 monomer retains limited unwinding activity even on a forked-dsDNA substrate with the ssDNA regions comprised of homo-oligodeoxythymine 29. Single-molecule work from the Ha group showed that indeed a Pif1 monomer is a translocase and it cannot unwind a dsDNA region that does not contain a 3′-ssDNA end 18. However, the single-molecule work also showed that a monomer unwinds RNA-DNA and a G4 DNA structure 18, indicating that a Pif1 monomer has intrinsic unwinding activity that is strongly influenced by the nature of the substrate. Moreover, recent work from the Raney group showed that Pif1 catalyzes annealing of complementary ssDNA 32. Depending on the nature of the substrate, this activity of Pif1 could counteract unwinding, leading to an apparently inefficient helicase.
Starting from the premise that a Pif1 monomer cannot unwind dsDNA 18 or, if it does so, unwinding is limited to just a few base-pairs 29, in this work we designed a simple experimental strategy to determine whether a Pif1 monomer has any dsDNA unwinding activity. The assay is based on coupling the activity of Pif1, or lack thereof, to the primer extension activity of a DNA polymerase that does not catalyze efficient strand displacement DNA synthesis. Using phage T7 DNA polymerase we show that a monomer of Pif1 is sufficient to stimulate primer extension activity of the polymerase. FRET-based unwinding experiments performed under multiple- and single-turnover conditions show that stimulation of primer synthesis originates from the ability of a Pif1 monomer to unwind dsDNA. Surprisingly, we find that dsDNA unwinding by a Pif1 monomer is strongly influenced by the nature of the 3′-end of the substrate. We provide evidence strongly suggesting that interaction of the 3′-end region of the substrate with a Pif1 monomer modulates its re-winding activity. We also show that when Pif1 is in excess relative to the substrate its re-winding activity strongly limits unwinding, independent of the nature of the 3′-tail. The data show that the re-winding activity does not reside in the N-terminus region of the protein and that the C-terminus region appears to have a role in modulating both unwinding and re-winding of dsDNA.
RESULTS
Monomer Pif1 stimulates the primer extension activity of phage T7 DNA polymerase
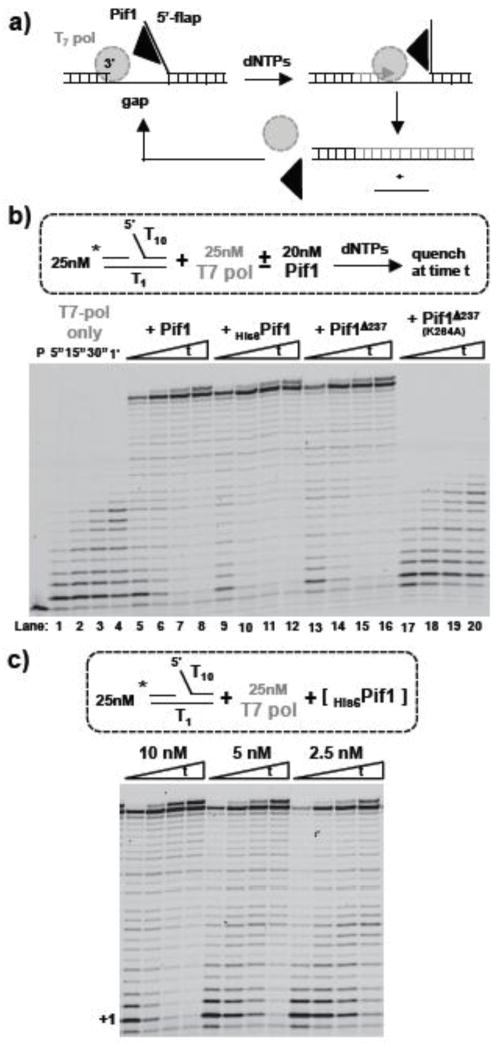
In order to test whether a monomer of Pif1 has any dsDNA unwinding activity, we implemented an assay that couples unwinding activity to the primer extension activity of a DNA polymerase. The fact that phage T7 DNA polymerase (T7-Pol) does not catalyze efficient strand displacement DNA synthesis 33 provides a simple means to monitor the opening a dsDNA downstream by an added helicase as stimulation of primer extension. This is schematically shown in Figure 1a; provided a 5′-ssDNA flap as an entry point for Pif1, if the downstream duplex is unwound, the reaction can be monitored by the extension of a labeled primer by the polymerase.
Figure 1. Monitoring Pif1 activity via a T7 DNA polymerase primer extension assay. a).
Schematic of the assay. b) Extension of a Cy3-labeled primer (P) after forming a complex of 25 nM DNA and 25 nM T7-Pol in the absence or presence of 20 nM of different Pif1 constructs and starting the reaction with 100 μM dNTPs. c) Same reactions as in b) but at lower Pif1 concentrations.
Consistent with the reported inability of T7-Pol to catalyze significant strand displacement 33, control experiments indicate that, independent of the flap length (not shown), the polymerase catalyzes limited strand displacement DNA synthesis into a 23 bp dsDNA placed downstream of the primer (Figure 1b, lanes 1–4). When the reactions are performed at a concentration of Pif1 that favors a monomer bound to the DNA, stimulation of primer extension activity of T7-Pol is observed within the first few seconds. Stimulation of primer extension occurs with both untagged and N-terminus His6-tagged full-length Pif1 (Figure 1b, lanes 5–12). It has been shown that the non-conserved N-terminus of Pif1 is not necessary for helicase activity under conditions of excess Pif1 over the DNA concentration 34. Thus, we generated a Pif1 construct missing the first 237 amino acids (Figure S1a). We note that although at saturating concentration of ATP deletion of the N-terminus has little effect on the DNA dependent ATPase activity (Figure S1b), the KATP of Pif1238–859 is ~ 4-fold higher than for full-length Pif1, indicating that removal of the N-terminus affects one or more steps in the binding/hydrolysis cycle (Figure S1c). In the T7-Pol coupled assay Pif1238–859 stimulates primer extension to the same extent as full-length Pif1 (Figure 1b, lanes 13–16). However, Pif1238–859/K264A that lacks ATPase activity does not stimulate primer extension by T7-Pol (Figure 1b, lanes 17–20). This indicates that at the low Pif1 concentration used, binding of the enzyme to the 5′-flap is not sufficient for stimulation of T7-Pol strand displacement activity (e.g. like a single-stranded DNA binding protein 35). Moreover, an active ATPase is required, indicating that either unwinding or translocation by Pif1 are involved (see below for this point). We note that the reactions were performed in the presence of only dNTPs, thus hydrolysis by Pif1 of dNTPs (dATP only, not shown) is sufficient for stimulation of primer extension.
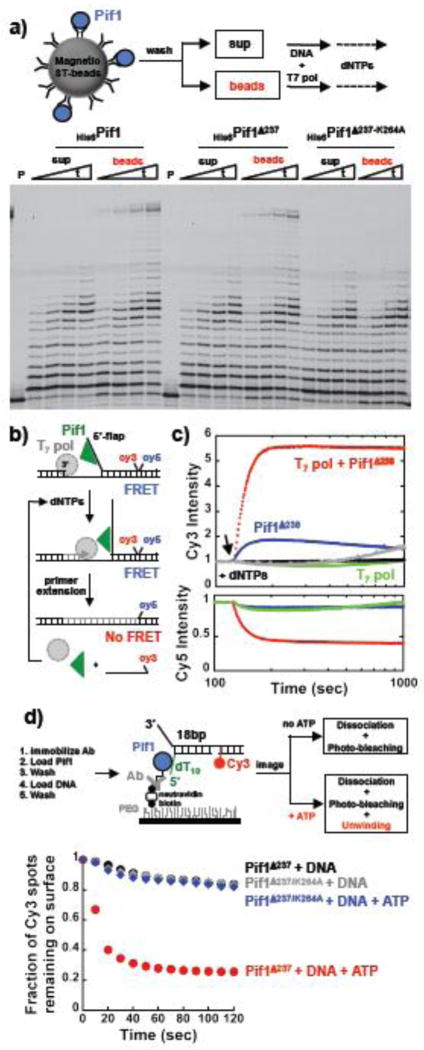
Stimulation of the primer extension activity of T7-Pol was observed also with Pif1 concentrations that are well below the concentration of the DNA (Figure 1c). At a concentration 10-fold lower than the DNA concentration, Pif1 is still able to stimulate primer extension. Under conditions of excess DNA over the enzyme we showed that a monomer of Pif1 binds to the DNA 28,29, thus the activity of this species is sufficient for stimulation of primer extension. In order to provide further evidence that a monomer of Pif1 is indeed sufficient for this reaction, we used a strategy similar to the one recently employed at the single molecule level to study unwinding by a monomer of Pif118. Pif1 in solution is a monomer and therefore coupling to a surface will report on the activities of this species. The scheme of the reaction is shown in Figure 2a. Pif1 was first bound via its N-terminus His6-tag to biotinylated penta-His antibody followed by coupling to magnetic streptavidin beads. After several washing steps to remove unbound Pif1, the resulting supernatant and bead fractions were tested in a T7-Pol primer extension reaction. Stimulation of primer extension activity was observed only with the bead fraction containing the bound full-length Pif1 or its shorter version missing the first 237 amino acids. Also, no stimulation of primer extension was observed with the Pif1 version that has no ATPase activity (K264A), consistent with the data in solution. These data provide support to the conclusion that the activity of a monomer of Pif1 is sufficient to stimulate the primer extension activity of T7-Pol.
Figure 2. A monomer of Pif1 is sufficient to stimulate T7-DNA polymerase activity by unwinding the downstream dsDNA. a).
Primer extension reactions performed with Pif1 immobilized to streptavidin bead via a biotinylated penta-His antibody. b) Schematic of the FRET-based unwinding assays. c) FRET-based assays using 20 nM DNA and monitoring the strand displacement DNA synthesis/unwinding by 20 nM T7-Pol (green) or 20 nM T7-Pol and 15 nM Pif1238–859 (red) or 15 nM Pif1238–859 (blue). The same experiments for the ATPase inactive Pif1 are shown in black and gray. The arrow indicates the time of addition of 100 μM dNTPs. d) Schematic of single molecule approach to detect helicase activity of Pif1 monomers immobilized on a surface. The graph shows the fraction of Cy3 spots remaining on the surface after addition of either image buffer or image buffer + ATP. Pif1238–859 or its ATPase inactive variant K264A were used in these experiments.
Stimulation of T7-Pol primer extension is due to unwinding of the downstream duplex by Pif1
The data in the previous sections show that Pif1 stimulates T7-Pol activity and this requires an active ATPase. Moreover, a monomer of Pif1 is sufficient for stimulation. However, we showed that a monomer of Pif1 is a 5′-3′ translocase that retains limited unwinding activity 29, thus the strand displacement activity of the polymerase could be coupled to Pif1 ssDNA translocation in the 5′-3′ direction and trapping of the newly displaced strand. Alternatively, stimulation of primer extension activity of T7-Pol could originate because of unwinding of the duplex downstream by Pif1. In this case either Pif1 alone can unwind the entire duplex region or Pif1 could unwind just a few base-pairs, with the polymerase synthesizing behind and trapping the newly opened DNA. In this latter case and in the case where translocation of Pif1 were to be coupled to the strand displacement activity of T7-Pol, no unwinding should be detected in the absence of the polymerase.
In order to examine the effect of the polymerase and Pif1 independent of each other, we employed a fluorescence-based assay that uses a substrate that contains a Cy3-Cy5 FRET pair at a nick at the end of a 23 bp duplex region to be unwound/strand displaced and 5′-dT5 as entry point for Pif1 (Figure 2b). Addition of T7-Pol and dNTPs shows no detectable change in either Cy3 or Cy5 fluorescence, i.e. no FRET change (Figure 2c). This is consistent with the primer extension assays showing that T7-Pol catalyzes strand displacement of only ~ 9–10 bp into the downstream duplex, not sufficient to release ssDNA and generate a FRET change. Addition of Pif1 to the reaction leads to a large increase in Cy3 fluorescence accompanied by a concomitant decrease in Cy5 fluorescence, i.e. loss of FRET. Consistent with the gel-based assays, these data show that Pif1 stimulates primer extension by the polymerase. To our surprise a smaller but detectable increase in Cy3 signal is also observed when Pif1 alone and dNTPs are added to the substrate. The same is not true when the ATPase deficient Pif1 is used in the assays. Similar experiments were performed also with a stopped-flow apparatus using substrates with different lengths of the dsDNA to be strand displaced/unwound and a 5′-dT10 as entry point for Pif1 (Figure S2). These data and the ones in the previous sections strongly suggest that the unwinding activity of monomer Pif1 rather than its translocation activity leads to opening of the duplex and under multiple turnovers in the presence of the polymerase, primer extension leads to completion of the reaction.
Next, we used a single molecule approach similar to the one used by the Ha group to test the helicase activity of a monomer of Pif118. The experimental strategy used is schematically shown in Figure 2d and the assay is based on the disappearance of the Cy3 signal upon unwinding and release of the substrate by Pif1. For these experiments we used substrates that contain a 5′-dT10 as entry point for Pif1, a 22 nt 3′-ssDNA and a 18 bp duplex region to be unwound followed by a 18 bp dsDNA with a Cy3 at the 5′-end of a nick. After immobilization of Pif1238–859 (or the K264A variant) on the surface and binding of the Cy3-labeled substrate, the Cy3 fluorescence of individual spots on the surface was monitored over time in the absence and presence of added ATP. The fraction of Cy3 spots remaining on the surface as a function of time, determined from single photo-bleaching/dissociation events, is shown in Figure 2d. In the absence of ATP very little dissociation/photo-bleaching is observed; however, upon addition of ATP the Cy3 signal quickly disappears. Moreover, when the ATPase inactive Pif1 is used in the experiment, little change in the Cy3 signal over time is observed after addition of ATP. These data, together with the ones in Figure 1c and Figure 2a, support the conclusion that a monomer of Pif1 can unwind dsDNA.
A monomer of Pif1 unwinds fork-DNA substrates in multiple turnovers
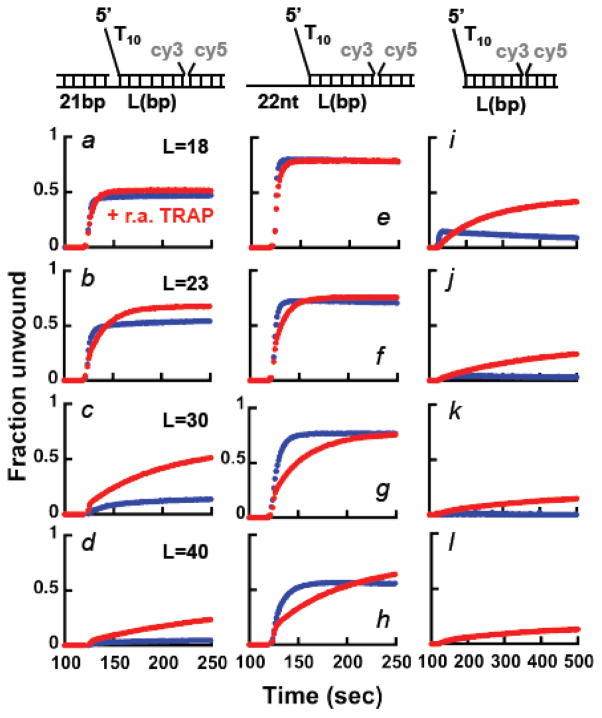
The findings in the previous are surprising because we showed that under monomer conditions Pif1 has limited helicase activity 29 and single-molecule work showed that a monomer of Pif1 could not unwind dsDNA 18. In order to examine this point further, we used FRET-based assays and performed multiple turnover unwinding experiments under condition that favor a Pif1 monomer bound to DNA and tested whether different regions of the substrate would affect unwinding. After forming a Pif1-DNA complex (15 nM protein and 20 nM DNA), the reactions were initiated either by addition of 0.5 mM ATP or 0.5 mM ATP and a 3.5-fold excess of an unlabeled trap (same as the Cy3-labeled strand) to prevent re-annealing of the unwound dsDNA (r.a. TRAP). The fraction of ssDNA formed was calculated using the signal of ssDNA in the presence of Pif1 as a reference for the unwound state (not shown).
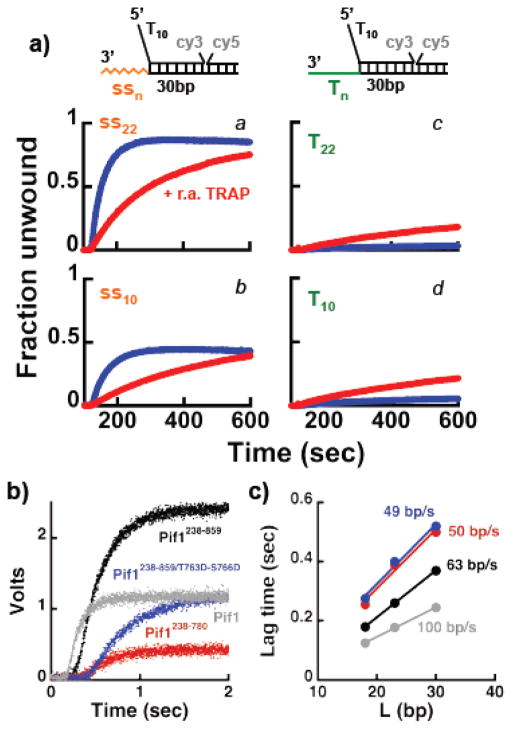
When a primer is annealed to the 3′-tail of the substrate (Figure 3, panels a–d), in the presence of ATP unwinding is observed for the shorter dsDNA lengths. However, the fraction of DNA unwound decreases significantly as the duplex length increases, consistent with the observations in Figure S2, where unwinding was driven by dNTPs. Moreover, for the longer duplex lengths addition of the trap to prevent re-annealing together with ATP stimulates the reaction. Surprisingly, with a substrate containing a 22 nt 3′-ssDNA tail (rather than a dsDNA) unwinding by Pif1238–859 is observed for all duplex lengths, with ~50% ssDNA formed even with a 40 bp duplex (Figure 3, panels e–h). Under these conditions, inclusion with ATP of the trap to prevent re-annealing does not stimulate the reaction further. Rather, some Pif1 appears to be trapped by the addition of ssDNA at a concentration 3.5-fold higher than the substrate. The same results were also obtained with full-length Pif1 (Figure S3a), showing that the observed behavior does not originate from having deleted the N-terminus region of the protein. These data differ from the expected behavior if the annealing activity of Pif1 were limiting unwinding 32. Addition of a trap to prevent re-annealing should stimulate the fraction of DNA being unwound; this is indeed what is observed for substrates containing a dsDNA region rather than a ssDNA 3′-tail or no 3′ssDNA tail or when Pif1 is in excess relative to the DNA (see below). We take this as an indication that for a monomer Pif1 bound to substrates containing a 3′-ssDNA tail of mixed sequence composition, the unwinding activity of Pif1 is not limited by its annealing activity 32.
Figure 3. A monomer of Pif1 has helicase activity under multiple turnover conditions.
FRET-based unwinding experiments monitoring the increase in Cy3 fluorescence using 20 nM of the indicated substrates in complex with 15 nM Pif1238–859. The reactions were started by addition of either 0.5 mM ATP (blue) or 0.5 mM ATP and a trap to prevent re-annealing of the unwound dsDNA in 3.5-fold excess (3.5x) relative to the DNA (r.a. TRAP, red).
Finally, we performed the same experiments using substrates that do not contain a 3′-ssDNA tail (Figure 3, panels i–l). In this case a small amount of unwinding is detected for the shorter 18 bp duplex and again unwinding of the longer duplexes is stimulated by the addition of the trap to prevent re-annealing. These data are consistent with the reported lower efficiency of unwinding by Pif1 of substrates that do not contain a 3′-ssDNA tail 26,30. The observation that a trap to prevent re-annealing of the unwound strand stimulates the unwinding of substrates that either lack the 3′-ssDNA end or contain a 3′-dsDNA suggests that the reported annealing activity of Pif1 32 may have a role in limiting unwinding even for a Pif1 monomer. Yet, with neither of these substrates the trap stimulates unwinding to the extent observed with substrates containing a 3′-ssDNA tail of mixed sequence composition.
A Pif1 monomer unwinds dsDNA in a single turnover
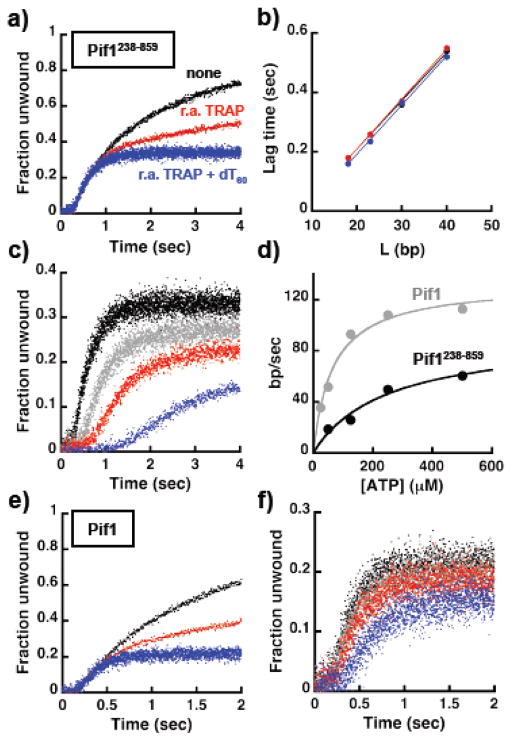
The data presented so far argue that a monomer Pif1 has unwinding activity. Next, we performed stopped-flow experiments, following the large increase in Cy3 fluorescence due to loss of FRET upon unwind, to determine the rate of unwinding of a Pif1 monomer. Experiments were performed at low DNA concentration (20 nM) with 15 nM pre-bound Pif1238–859, a condition where a monomer binds to the DNA. The experiments were started either by addition of ATP or “ATP + re-annealing trap” or “ATP + re-annealing and protein traps”. Control experiments show that 100–1000 nM of dT60 is a good protein trap for Pif1 (Figure S3b) 30,31. The fraction unwound was calculated using the maximum fluorescence change observed in experiments with T7-DNA polymerase and using substrates with a primer annealed to the 22 nt 3′-tail (as in Figure S2). In absence of any trap, unwinding of a substrate containing a 22 nt 3′-ssDNA tail and a 23 bp duplex region is characterized by an initial lag followed by a large increase reporting on unwinding under multiple turnovers (Figure 4a). Consistent with the data presented in Figure 3, addition of the trap to prevent re-annealing (r.a. TRAP) does not increase the extent of unwinding but rather it partly works as a trap for Pif1 (Figure 4a, red), as indicated by its effect on the multiple turnover region of the progress curve (Figure S3c). Single turnover conditions are achieved when dT60 is included with ATP (Figure 4a, blue) and even under these condition a Pif1 monomer can unwind ~ 30% of the substrate. The lag times for experiments performed using different lengths of the dsDNA to be unwound are shown in Figure 4b and they follow a linear dependence, as it would be expected for a helicase/translocase 36–39. Moreover, the values of the lag time is affected little by the presence of the traps, suggesting that for short time scales multiple turnovers of Pif1 do not affect the observed lag time. Thus, the rates of unwinding determined from the slopes in Figure 4b were averaged yielding a value of 60.4 ± 0.8 bp/sec. Unwinding experiments were also performed at different ATP concentrations in the presence of either the re-annealing trap (not shown) or re-annealing and protein traps (Figure 4c). As expected for unwinding driven by the ATPase activity of the helicase, as the ATP concentration decreases the lag time increases. The unwinding rate as function of ATP determined from data performed at the different duplex lengths is shown in Figure 4d and the calculated apparent KATP is 246 ± 104 μM, consistent with the value determined for the ssDNA-dependent ATPase activity (Figure S1c).
Figure 4. A monomer of Pif1 can unwind dsDNA in a single turnover. a).
Stopped-flow FRET-based unwinding by 15 nM Pif1238–859 of a substrate (20 nM) with a 23 bp dsDNA, a 5′-dT10 and a 22 nt 3′-tail of mixed sequence composition. The reactions were started by addition of either 0.5 mM ATP (black) or 0.5 mM ATP + 3.5x of the r.a. TRAP (red) or 0.5 mM ATP + 3.5x of the r.a. TRAP + 250 nM dT60 as protein trap (blue). b) Lag times as function of dsDNA length for experiments performed in the absence (black) or the presence of the r.a. TRAP (red) or the r.a TRAP + dT60 (blue). c) ATP concentration dependence (500 μM, black; 250 μM, gray; 125 μM, red; 50 μM, blue) of unwinding by Pif1238–859 using the same substrate as in a). d) Rate of unwinding as a function of ATP concentration determined using different lengths of the dsDNA for Pif1238–859 (black) and full-length Pif1 (gray). e,f) Same experiments as in a) and c) using full-length Pif1.
The same experiments were also performed with full-length Pif1 (Figure 4e,f and S3d). The rate of unwinding as a function of ATP is shown in Figure 4d. For full-length Pif1 the calculated apparent KATP is 66 ± 12 μM, ~ four times lower than for Pif1 missing the first 237 amino acids, as observed for the ssDNA-dependent ATPase (Figure S1c). This strongly suggests that during unwinding one or more steps in the ATPase cycle must be different between the two proteins. Also, at saturating ATP concentration full-length Pif1 has an unwinding rate faster than Pif1 missing the first 237 amino acids, suggesting that albeit not necessary for unwinding the N-terminus region contributes to it. In addition, under single turnover conditions the ATP concentration dependence of the signal amplitude is different for full-length Pif1 and its shorter variant (Figure 4f vs 4c). At saturating ATP concentration the amplitude of reaction is lower for full-length Pif1 compared to Pif1238–859. This is not simply due to a difference in the fraction of bound enzyme at the beginning of the reaction. The same behavior is observed also when the experiments are performed at higher DNA concentration (see Figure 6b). This suggests that full-length Pif1 has lower processivity.
Figure 6. Deletion of the C-terminus of Pif1 affects the unwinding activity of the monomer. a).
FRET-based unwinding experiments under multiple turnover conditions using 15 nM Pif1238–780 and 20 nM of the indicated substrates, starting the reaction by addition of either 0.5 mM ATP (blue) or 0.5 mM ATP and 3.5x of the r.a. TRAP. b) Stopped-flow experiment comparing the unwinding activity of a monomer of different Pif1 constructs (75 nM) using a substrate (100 nM) with a 23 bp dsDNA, a 5′-dT10 and a 22 nt 3′-tail of mixed sequence composition. c) Lag times as a function of dsDNA length obtained at the higher concentrations used in b). The calculated rate of unwinding of the different Pif1 constructs is shown, with the colors matching the ones in b).
The nature of the 3′-tail regulates the ability of a monomer of Pif1 to unwind
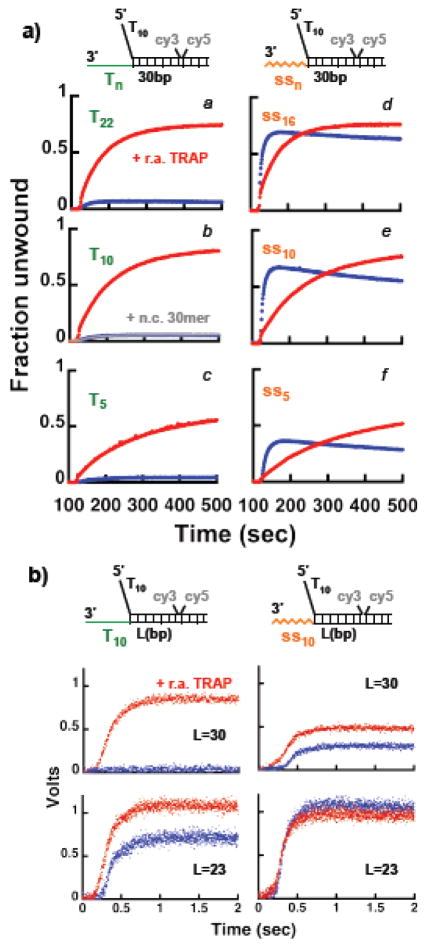
The finding that a Pif1 monomer efficiently unwinds substrates with the 22 nt 3′-ssDNA tail is unexpected. Our previous work examining the translocation and unwinding property of monomer Pif1 showed that although a 3′-ssDNA tail comprised of a homo-oligomer dT8 stimulates unwinding 29, it appears to do so to a lesser extent than what we observe with these new substrates. In order to clarify this possible inconsistency in our own data, we performed FRET-based unwinding experiments with a substrate containing a 30 bp duplex and 3′-ssDNA tails containing a different number of thymines. Under identical conditions where Pif1238–859 unwinds the substrate with the 22 nt 3′-tail of mixed sequence, unwinding of a substrate with a dT22 3′-tail is limited (Figure 5a, panel a). The inability to unwind this substrate is not due to the lack of Pif1 binding, as inclusion with ATP of the trap to prevent re-annealing clearly stimulates unwinding activity. Moreover, unwinding of substrates that contain shorter dT10 and dT5 3′-tails is limited as well (Figure 5a, panels b and c), indicating that the lack of unwinding activity does not originate from an inhibitory effect of the length of the 3′-tail. The observation that on these substrates unwinding by Pif1 is stimulated by the addition of the trap to prevent re-annealing suggests that a 3′-dTn tail does not limit the DNA re-winding activity of Pif1 (see below). This is further corroborated by the observation that if a 30 nt ssDNA that does not have complementary to the dsDNA unwound is used as a trap, the unwinding of the substrate with a 3′-dT10 tail is not stimulated (Figure 5a, panel b).
Figure 5. The helicase activity of a monomer of Pif1 depends on the nature of the 3′ssDNA tail. a).
FRET-based unwinding experiments under multiple turnover conditions using 15 nM Pif1238–859 and 20 nM of a substrate with a 30 bp dsDNA and different lengths of the 3′-tail comprised of either an oligo-dT (panels a–c) or a mixed sequence (panels d–f). The reactions were started by addition of either 0.5 mM ATP (blue) or 0.5 mM ATP and 3.5x of the r.a. TRAP. b) Stopped-flow experiments using Pif1238–859 (40 nM) and substrates (50 nM) containing different lengths of the dsDNA and either a 3′-dT10 (left panels) or 10 nt of mixed sequence (right panels). The reactions were started by addition of either 0.5 mM ATP + 0.5μM dT60 (blue) or 0.5 mM ATP + 0.5 μM dT60 + 3.5x of the r.a. TRAP (red).
Next, we tested whether the peculiar strong unwinding activity of the substrates with the 22 nt 3′-ssDNA tail was due to our arbitrary choice of the sequence of this ssDNA. FRET-based unwinding experiments were performed with substrates containing a 30 bp duplex and shorter 3′-tails, where nucleotides were progressively removed from the 3′-end of the original 22-mer (Figure 5, panels d–f). For any length of these new 3′-ssDNA tails the unwinding activity of Pif1 is higher than with the oligo-dT 3′-tails. Also, a substrate with a 22 nt 3′-tail with a different mixed sequence composition is unwound to a similar extent as the original 3′-tail we chose (data not shown). Finally, full-length Pif1 shows a similar sensitivity to the nature of the 3′-ssDNA tail (Figure S4a), indicating that removal of the N-terminus region in Pif1238–859 is not the reason for the lack of unwinding observed with the oligo-dT 3′-tail.
One possible explanation for the behavior observed with the dTn 3′-tails could be that the formed Pif1-DNA complex is either weaker or not as competent for unwinding as the one formed with a 3′-tail of mixed composition. If this were the case, in a single-turnover Pif1 should unwind a substrate with a dTn 3′-tail to a lesser extent than one with a 3′-tail of mixed composition, even in the presence of a trap to prevent re-annealing. Figures 5b shows single-turnover stopped-flow unwinding experiments of dsDNA that contain either a 3′-tail with dT10 or 10 nt of mixed sequence, comparing the effect of adding with ATP the trap to prevent re-annealing. Pif1238–859 unwinds the 30 bp dsDNA with the dT10 3′-tail in a single-turnover only in the presence of a trap for re-annealing. This shows that in a single turnover Pif1238–859 is competent for unwinding a substrate with the dT10 3′-tail, even more so than the one with the 3′-tail of mixed composition. For this latter substrate the presence of the re-annealing trap is not required to observe unwinding, albeit it stimulates the reaction and some Pif1 dependent re-rewinding may still occur even with a 3′-tail of mixed sequence composition. This last point is further supported by the observation that in the absence of the trap to prevent re-annealing the lag time is slightly longer, as it would be expected if re-winding were to counteract unwinding. Nonetheless, the data strongly suggest that 3′-tails of mixed sequence composition interact differently with Pif1 and this interaction limits the re-winding activity of the monomer; oligo-dT 3′-tails do not appear to limit this Pif1-dependent activity, thus leading to an apparent lack of unwinding unless the trap to prevent re-annealing is added with ATP. However, the effect of the oligo-dT 3′-tail is not absolute and Pif1238–859 can unwind in part a shorter 23 bp dsDNA even in the absence of a trap to prevent re-annealing (Figure 5b). Similar results were obtained also with full-length Pif1 (Figure S4b). We note that as the dsDNA is shortened, it is possible that few base-pairs at the end of the duplex are not unwound catalytically by the helicase40,41 and this would result in an apparent higher unwinding activity when shorter dsDNAs are used. In order to determine how many base-pairs Pif1 is able to unwind into a 30 bp dsDNA substrate when the 3′-ssDNA is comprised of oligo-dT, we placed a single 2-aminopurine (2-AP) at different positions in the dsDNA along the Pif1 translocating strand and monitored the increase in 2-AP fluorescence due to dsDNA opening (Figure S5). When the 3′-tail is comprised of 10 nt of mixed sequence, in a single turnover Pif1 appears to open the 22nd base-pair, independent of the presence of a trap to prevent re-annealing. This is consistent with the observation in Figure 5b that with this substrate Pif1 unwinds 30 bp dsDNA even in the absence of the trap to prevent re-annealing. However, with a 3′-dT10 tail and in the absence of a trap to prevent re-annealing Pif1 appears to open the 16th but not the 22nd base-pair; the 22nd base-pair is opened only when the trap to prevent re-annealing is included. These data confirm that the effect of the oligo-dT 3′-tail is not absolute and Pif1 is able to unwind 17–21bp into a 30 bp duplex without generating fully unwound product, as confirmed by the experiments in Figure 5b.
During unwinding the C-terminus region of Pif1 stabilizes the enzyme on the substrate
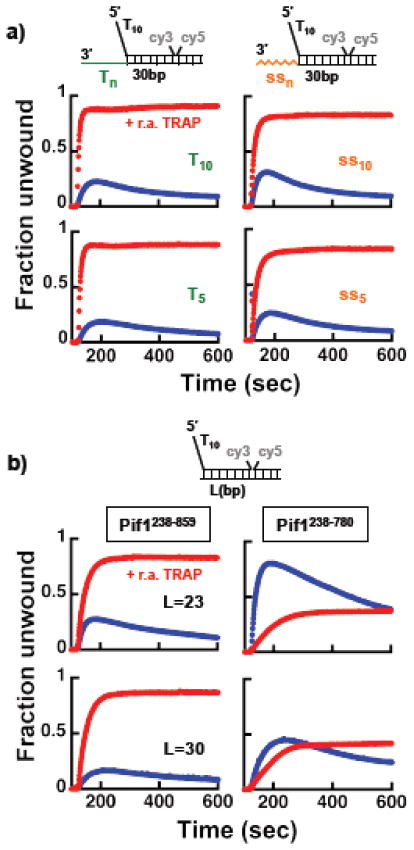
The data in the previous section show that the ability of a Pif1 monomer to unwind dsDNA is modulated by the nature of the 3′-ssDNA tail and how it interacts with Pif1. Moreover, the observation that both full-length Pif1 and the construct missing the N-terminus have a similar behavior argues that the interaction of the 3′-tail occurs with either the helicase core or the non-conserved C-terminus region or both. In order to test whether the C-terminus region has any role in modulating the activities of Pif1 we generated a Pif1 construct (Pif1238–780) missing the last 79 amino acids. Deletion of these amino acids does not have major effects on the DNA-dependent ATPase activity and the KATP (Figure S1). Similar to Pif1238–859, under multiple turnover condition a Pif1238–780 monomer unwinds a 30 bp dsDNA that contains 3′-ssDNA tails of mixed composition (Figure 6a). However, at difference with Pif1238–859, Pif1238–780 does not unwind substrates with a dTn 3′-tail even when the trap to prevent re-annealing is included with ATP. This suggests that removal of the last 79 amino acids affects the response of Pif1 to the different nature of the 3′-ssDNA tail.
The unwinding activity of Pif1238–780 was examined also under single-turnover conditions using higher DNA concentrations to minimize possible effects due to differences in affinities for the substrate. Figure 6b shows FRET-based unwinding experiments using a 23 bp dsDNA with a 3′-tail of 22 nt mixed composition. Similar to what observed at the lower DNA concentration in Figure 4, full-length Pif1 unwinds at a faster rate than Pif1238–859 (100 vs 63 bp/sec) (Figure 6c); however, the amplitude of unwinding is lower, again suggesting that full-length Pif1 has lower processivity despite having a faster rate of unwinding. Pif1238–780 unwinds these substrates with a rate similar to Pif1238–859 (50 bp/sec vs. 63 bp/sec), yet for Pif1238–780 the amplitude of unwinding is much lower for any length examined. Experiments performed at higher Pif1 concentrations show the same behavior (not shown), indicating that the lower amplitude of unwinding for Pif1238–780 does not originate from a lower affinity for the DNA substrate. These data strongly suggests that removal of the last 79 amino acids decreases processivity (i.e. Pif1238–780 has a higher dissociation rate). Also, we generated Pif1238–859 with phosphomimetic changes at two positions within the TLSS motif at the C-terminus (Figure S1), shown to be important for the activity of Pif1 in DSB repair and BIR 22,27. Pif1238–859/T763D–S766D unwinds with a similar rate (49 bp/sec) but to a lower extent than Pif1238–859, suggesting that introducing two negative charges in this region is sufficient to affect the processivity of Pif1.
Under conditions of excess enzyme the Pif1-dependent re-winding activity is not limited by the nature of the 3′-ssDNA tail
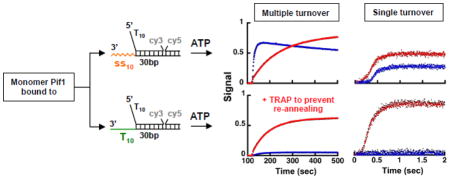
The data in the previous sections show that for a monomer of Pif1 the 3′-ssDNA tail of the substrate modulates its unwinding, with Pif1-dependent re-winding playing a limited role for 3′-tails of mixed sequence. We note that the ability of Pif1 to catalyze annealing of complementary ssDNA has been recently reported for the full-length protein under conditions of excess enzyme over the DNA 32. Multiple turnover FRET-based unwinding experiments performed with a 10-fold excess of full-length Pif1 or Pif1238–859 show that for any length of the duplex the fraction unwound reaches a maximum followed by a gradual decay in signal (Figure S6). The slow decaying phase is eliminated by the addition with ATP of the trap to prevent re-annealing. Over these long times ATP is being depleted by the ATPase activity of Pif1 and we ascribe this phase to the spontaneous re-annealing of the unwound ssDNA. However, for the longer duplex lengths addition of the trap to prevent re-annealing has a profound effect on the extent of unwinding, largely stimulating the reaction. Thus, consistent with the recent work from the Raney group 32, under conditions of excess enzyme the Pif1-dependent re-winding activity limits unwinding, so much so that for the longer dsDNA lengths this activity appears to dominate. Moreover, in the absence of the trap to prevent re-annealing the extent of unwinding is limited also for the Pif1 construct missing the first 237 amino acids, strongly arguing that the effect of the Pif1-dependent re-winding activity cannot originate from N-terminus region of the protein.
Next, we tested whether under conditions of excess Pif1 3′-ssDNA tails of mixed sequence composition or oligo-dT would affect differently the Pif1-dependent re-rewinding activity, as observed for a monomer. Multiple turnover unwinding experiments performed with a 10-fold excess of Pif1238–859 using a 30 bp dsDNA with different 3′-tails are shown in Figure 7a. Similar unwinding behavior is observed with 3′-tails of either mixed sequence or oligo-dT, strongly suggesting that in excess Pif1 the re-winding activity is insensitive to the nature of the 3′-tail. To this point, the left panels in Figure 7b show unwinding experiments performed with a 10-fold excess of Pif1238–859 using substrates that do not contain a 3′-tail. Addition of the trap to prevent re-annealing stimulates unwinding even on these substrates, indicating that in excess Pif1 re-winding coupled to unwind is independent of the 3′-tail altogether. Next, we tested whether the C-terminus region of Pif1 has any role in this. The right panels in Figure 7b show the same experiments performed with a 10-fold excess of Pif1238–780. To our surprise, in the absence of the trap for re-annealing Pif1238–780 unwinds these substrates to a larger extent than Pif1238–859. Moreover, for this Pif1238–780 construct addition of the trap to prevent re-annealing does not stimulate unwinding.
Figure 7. In excess Pif1 unwinding is limited by the Pif1-dependent annealing activity. a).
FRET-based unwinding experiments using 20 nM of the indicated substrates and 200 nM Pif1238–859, starting the reactions by addition of either 0.5 mM ATP (blue) or 0.5 mM ATP and 3.5x r.a. TRAP (red). b) Same experiments as in a) comparing the unwinding activity of Pif1238–859 and Pif1238–780 using substrates that do not contain a 3′-tail.
DISCUSSION
Ensemble assays to monitor helicase activity are in general based on all-or-none reactions where the formation of ssDNA product is detected either by the release of radiolabeled ssDNA or by changes in a fluorescence signal (i.e. FRET). Independent of the signal monitored, the dsDNA substrate must be fully unwound for the ssDNA product to be detected. However, at least two scenarios can be thought of where this would be a limitation to the ability to detect helicase activity. First, if the helicase is able to unwind only a few base-pairs, the substrate would re-anneal upon dissociation of the enzyme and thus the reaction would be scored as negative. We recently proposed this possibility to explain the limited helicase activity of Pif1 under monomer conditions 29. Second, the unwinding activity of a helicase could be underestimated if the protein can catalyze re-annealing of the substrate that is being unwound. The ability of Pif1 and other helicases to catalyze annealing of complementary ssDNA has been documented 32,42–49. Therefore, we designed an assay that should bypass these two complications in determining the unwinding ability of a helicase. The assay is taken directly from a page of phage T7 biochemistry. It has been shown that T7 DNA polymerase (T7-Pol) does not catalyze efficient strand displacement DNA synthesis across a duplex DNA bound on the template downstream of the primer 33. Strand displacement activity by the T7-Pol is strongly stimulated either by binding of a single-stranded DNA binding protein to a 5′-flap in the duplex to be displaced or by the T7 DNA helicase initiating unwinding from the 5′-flap 35. The assay used in this work is based on the idea of coupling the unwinding activity of a heterologous helicase with the primer extension activity of the polymerase. In this work, we employed both a regular primer extension assay using denaturing polyacrylamide gel electrophoresis to monitor primer extension (i.e. unwinding) with single base-pair resolution and also FRET-based assays to be able to monitor the polymerase and helicase activities independent of each other. The T7 polymerase coupled assays and single molecule experiments showed that a monomer of Pif1 has dsDNA unwinding activity independent of the presence of the polymerase, an unexpected finding based on our previous observations and the ones from the Ha group 18,29.
The ability of a Pif1 monomer to unwind dsDNA is strongly modulated by the nature of the non-translocating strand (3′-ssDNA tail) available to form the initial Pif1-DNA complex. In absence of a 3′-tail unwinding of dsDNA by a Pif1 monomer is inefficient even in multiple turnovers. We believe that this is not exclusively due to a lower affinity of the Pif1-DNA complex; unwinding in a single turnover of dsDNA without a 3′-tail remains inefficient even at relatively high DNA concentrations (Figure S7), where formation of the pre-initiation complex on DNA should not be limited by the affinity of Pif1. However, consistent with the single-molecule observations by the Ha group 18, in the absence of a 3′-ssDNA tail a monomer of Pif1 is able to unwinding a RNA-DNA hybrid in a single turnover (Figure S7). The reason why Pif1 can unwind efficiently the RNA-DNA hybrid compared to a dsDNA is currently unknown. It is possible that a region of Pif1 interacts with RNA-DNA hetero-duplex making the enzyme competent for unwinding. For dsDNA, the data show that interaction with a 3′-ssDNA tail is needed to poise a monomer of Pif1 for unwinding. However, not all 3′-tails are made equal. The surprising finding in this work is that a Pif1 monomer unwinds dsDNA when the 3′-tail is comprised of ssDNA of mixed sequence composition. This effect does not originate from a secondary structure formed by the 3′-tail or a specific sequence; only five nucleotides are sufficient for stimulating unwinding. Even when this region of the substrates is in dsDNA form it allows for a Pif1 monomer to unwind at least a short dsDNA. However, a Pif1 monomer does not unwind a 30 bp dsDNA with the 3′-ssDNA tails comprised of homo-oligodeoxythymines (oligo-dT). This explains our previous observation of limited unwinding activity of a Pif1 monomer, where the experiments were performed in the presence of heparin to trap the protein but in the absence of a trap to prevent re-annealing 29. The question then is how a mix sequence or an oligo-dT 3′-tail modulate the unwinding activity of a Pif1 monomer.
Multiple turnover experiments where a trap to prevent re-annealing is added together with ATP show that for the substrates with an oligo-dT 3′-tail addition of the trap strongly stimulates unwinding, while for a 3′-tail of mixed composition the trap to prevent re-annealing functions like a partial protein trap. The ability of Pif1 to unwind a 30 bp dsDNA with an oligo-dT 3′-tail only in the presence of the trap to prevent re-annealing is true even in a single turnover, indicating that a Pif1 monomer is fully competent for unwinding but this is not accompanied by the net release of product. However, the effect of the trap is not absolute and Pif1 can open at least 17-21 bp. Recent work by the Raney group showed that under conditions of large excess of protein relative to the DNA Pif1 stimulates annealing of complementary ssDNA 32, while in defect protein a Pif1 monomer contributes little to this activity. However, our data argue that even for a Pif1 monomer annealing can strongly counteract unwinding. We propose that for longer dsDNA and with an oligo-dT 3′-tail Pif1 undergoes multiple cycles of unwinding and re-winding with the net result of no unwound product being generated. The presence of the trap to prevent re-annealing tips the system towards full product formation. With a mix sequence composition instead re-annealing does not counteract unwinding to the same extent and thus there is net product formation. We note that with the oligo-dT 3′-tail and in the presence of the trap to prevent re-annealing, unwound product is detected on the same time scale as with a 3′-tail of mixed sequence composition in the absence of the trap. Thus, the data suggest that with the oligo-dT 3′-tail and in the absence of the trap Pif1 unwinding and re-winding occur on the same time scale. We would like to point out that unwinding and re-winding by a helicase because a protein-dependent re-winding activity would lead to the same result as if the helicase were to partially unwind the dsDNA, switch DNA strand and translocate out of the unwound duplex thereby leading to re-zipping. Such a mechanism has been shown to occur for different helicases 50–54. Although at this stage we cannot differentiate these possibilities, we note that the duplex region to be unwound is identical in both substrates and strand switching followed by translocation on the opposite strand would have to depend on the nature of the 3′-tail.
We do not currently know the mechanism that allows for the 3′-tail to modulate the re-winding activity of Pif1. However, the data point to the presence on Pif1 of a second site that binds the 3′-ssDNA strand of the substrate. This second DNA binding site must be separate from the ssDNA site on the protein that interacts with the translocating strand. Interaction of DNA with this second site modulates the degree to which re-winding counteracts unwinding. Also, this second DNA binding site on Pif1 cannot be within the 237 amino acids at the N-terminus of the protein, as Pif1 missing this region shows sensitivity to the 3′-tail. We note that even though Pif1 lacking the first 237 amino acids behaves similar to the full-length protein, deletion of this region has an effect on the unwinding behavior under single turnover. The full-length protein unwinds at a faster rate although with an apparent lower processivity. Moreover, the KATP for unwinding is lower for the full-length protein than for the shorter version. These differences strongly suggest that albeit not necessary for unwinding, the N-terminus contributes to modulating one or more steps during the cycle of ATP hydrolysis coupled to unwinding. Also, deletion of the non-conserved C-terminus region strongly affects the ability of a monomer of Pif1 to unwind dsDNA. Removal of this region leads to lower processivity, possibly by increasing the dissociation of the helicase during unwinding. This is further confirmed by the observation that introducing two negative charges within the TLSS motif, that is phosphorylated in vivo 22,27, is sufficient to affect processivity. We note that the C-terminus of Pif1 is highly positively charged (15 Arg+Lys and 8 Asp+Glu) and it is possible that this region is in part responsible for interaction with the 3′-ssDNA tail. However, the data suggest that the C-terminus may not be the sole contributor to the second site on Pif1 that interacts with the 3′-ssDNA tail. If the second DNA site were to reside exclusively within the 79 amino acids at the C-terminus, its deletion should abrogate interaction with the 3′-tail. Thus, similar to what observed with dsDNA that does not have a 3′-tail, Pif1238–780 should not unwind substrates containing a 3′-tail. This is not the case. Albeit with lower processivity, Pif1238–780 unwinds dsDNA with a 22 nt 3′-tail of mixed composition even in a single turnover; however, Pif1238–780 has limited unwinding activity of dsDNA without a 3′-tail, even under multiple turnovers (not shown). This suggests that when the C-terminus is absent the 3′-tail is still able to interact with Pif1 and the second site has not been fully abrogated. At the same time, we would like to point out that at difference with Pif1238–859, in multiple turnover conditions Pif1238–780 does not unwind a dsDNA with a oligo-dT 3′-tails even when a trap to prevent re-annealing is added together with ATP. This suggests that although the C-terminus might not be the sole contributor to the second site interacting with the 3′-tail, it has a role in allowing Pif1 to discriminate the nature of this region of the substrate.
Consistent with the observation by the Raney group that under conditions of excess enzyme relative to DNA Pif1 accelerates annealing of complementary ssDNA 32, our data show that in excess Pif1 re-annealing strongly limits unwinding. At difference with a Pif1 monomer, under these conditions the Pif1-dependent re-winding activity is independent of whether the 3′-tail is an oligo-dT or a sequence of mixed composition. We showed that in excess Pif1 relative to the DNA formation of a dimer of Pif1 is favored on the DNA 28. Thus, we propose that if a dimer of Pif1 forms on the substrate it precludes binding of the 3′-ssDNA tail to the second site on Pif1 and re-annealing cannot be modulated by the 3′-tail. Potential functional roles of the Pif1-dependent annealing activity have been described 32. In the context of our observations, we propose a different and simpler view for the role of this activity. Binding of a Pif1 monomer or a DNA-induced Pif1 dimer regulates the potential of Pif1 as a dsDNA helicase by modulating the intrinsic Pif1-dependent annealing activity. We propose that under conditions of excess Pif1 the presence of a strong annealing activity would impose a requirement for a polymerase to catalyzed DNA synthesis on the non-translocating strand, thereby preventing re-winding of the opened DNA and thus allowing efficient unwinding. This could in part explain the recent observation that Pif1 allows DNA polymerase δ to synthetize kbps of DNA from a pre-formed D-loop 20. Also, we note that an annealing activity has been described for human Pif1 and it was ascribed to the N-terminus of the protein 45. Our data for ScPif1 show that the N-terminus region is not involved in its re-winding activity and argue that if annealing originates from a distinct domain, it must have moved within the protein from yeast to human. A potential candidate for an “annealing domain” in ScPif1 would be the non-conserved C-terminus that makes Pif1 unique among its family members. Interestingly, removal of the C-terminus of Pif1 has an effect on the annealing activity coupled to unwinding when the reactions are performed in excess enzyme. In these conditions, Pif1238–780 unwinds dsDNA to a higher extent when the trap to prevent re-annealing is not included with ATP. Taken at face value this observation would suggest that the C-terminus region is responsible for the annealing activity of Pif1. Although at this stage we cannot exclude that this is indeed the case in conditions of excess enzyme, the unwinding data in defect Pif1 relative to the DNA suggest the C-terminus might not be an “annealing domain” in its own right. With a 30 bp dsDNA containing an oligo-dT 3′-tail and in the absence of a trap to prevent re-annealing the data indicate that a monomer of Pif1238–859 undergoes multiple cycles of unwinding and re-winding. Thus, if the C-terminus were an annealing domain its removal should allow Pif1 to unwind these substrates in the absence of the trap to prevent re-annealing. This is not the case. Rather, in multiple turnovers Pif1238–780 does not unwind dsDNA with an oligo-dT 3′-tail even when the trap to prevent re-annealing is present.
In summary, we showed that a monomer of Pif1 is a dsDNA helicase and that this activity strongly depends on the nature of the DNA to be unwound and the initial complex Pif1 can form on it. The presence of a second DNA binding site on Pif1 explains most of our observations and it provides a working model where interaction of the 3′-ssDNA tail with this site modulates the Pif1-dependent rewinding activity of a monomer. Under conditions where a dimer is favored on DNA the second site becomes unavailable and re-annealing strongly counteracts unwinding. Neither the N-terminus nor the C-terminus of Pif1 appear to be essential for the unwinding activity of a monomer, although they clearly affect it; moreover, the C-terminus appears to have a complex set of interactions with the rest of the protein that affect both unwinding and annealing.
MATERIAL AND METHODS
Reagents and Buffers
All chemicals used were reagent grade. All solutions were prepared with distilled and deionized Milli-Q water. Oligonucleotides were purchased from Integrated DNA Technology (IDT, Coralville, IA). The sequences of the oligonucleotides used are listed in Table S1 and the substrates in Table S2.
Purification of Proteins
Full-length untagged Pif1 was purified as previously described 28. His6-tagged Pif1 constructs (full-length and the shorter variants comprising residues 238–859, 238–859K264A, 238–859T763D/S766D and 238–780) were cloned in pET28b and expressed in E. coli inducing with 0.7 mM IPTG and grown overnight at 16 °C. The cells were suspended in Buffer L (50 mM Tris-HCl pH 8.2, 400 mM NaCl, 1 mM EDTA, 0.5 mM DTT, 10% w/v sucrose, 1 mM PMSF) and following opening with sonication, polymin P was added to 0.3% v/v, the proteins recovered in the supernatant, Am2SO4 added at 0.35 g/ml and the resulting pellet re-suspended in Buffer T (50 mM Tris-HCl pH 7.8, 1 mM EDTA, 0.5 mM DTT, 20% v/v glycerol and 0.1 mM PMSF) + 300 mM NaCl. After extensive dialysis Pif1 was loaded on a High S column (BioRad) and eluted at 700mM NaCl, dialyzed in Buffer T + 200mM NaCl and loaded on a High Q column (BioRad) and collected in the flow-through. After dialysis in Buffer P (20 mM sodium phosphate pH 7.8, 400 mM NaCl, 10% v/v glycerol, 5 mM β-ME, 0.1 mM PMSF), Pif1 was loaded on HisPur Ni-NTA superflow agarose (Thermo Scientific), washed with 30 mM imidazole in Buffer P and eluted with 300 mM imidazole in Buffer P and dialyzed in storage buffer (50 mM Tris-HCl pH 8, 600 mM NaCl, 1 mM EDTA, 1 mM DTT, 40% v/v glycerol) and stored at −80 °C. Exonuclease deficient (gp55A7A) phage T7 DNA polymerase (T7-Pol = Pol + Thioredoxin) was a kind gift from Tom Ellenberger (Washington University School of Medicine, St. Louis).
Strand displacement assay
Primer extension reactions were carried out in Buffer SD (20 mM Hepes pH 7.4, 100 mM NaCl, 8 mM MgAc2, 1 mM DTT, 0.1 mg/mL BSA). The experiments were performed by pre-forming a DNA+T7 Pol complex (25 nM final concentration) or DNA+ T7 Pol + 20 nM Pif1, followed by addition of dNTP mix at a final concentration of 100 μM each. For primer extension assays with Pif1 bound to beads the reactions were performed in Buffer SD with 1 mg/mL of BSA. His6-Pif1 and PentaHis Biotin Conjugate (Qiagen) (4-fold excess) were incubated for 10 minutes followed by addition of excess MyOne Dynabead pre-equilibrated in Buffer SD. The beads were washed 3 times with Buffer SD and then re-suspended in equal volume of Buffer SD. Following separation, either the supernatant or the bead fractions were incubated with Cy3 labeled substrate and T7-Pol and the reaction started by addition of dNTPs, with a final concentration of the components of 25 nM DNA, 25 nM T7-Pol, 8 nM Pif1 on beads and 100 μM dNTPs. At the indicate times the reactions were stopped by the addition of 80 mM EDTA, 0.08% SDS. After addition of formamide (50% final), the samples were heated at 95 °C for 2 min and analyzed on a 12% denaturing polyacrylamide gel, pre-run for 2 hours in 0.5X TBE. The gels were scanned using a Typhoon 9400 Variable Mode Imager.
Unwinding experiments monitored by FRET
Manual addition experiments were performed with an L-format PC1 spectrofluorimeter (ISS, Champaign, IL). Cy3 fluorescence time courses were monitored with excitation at 520 nm and emission at 565 nm. The experiments were performed in Buffer S100 (20 mM Hepes pH 7.4, 8 mM MgAc2, 1 mM DTT, 100mM NaCl) at 20 °C by pre-forming a complex of DNA with a defect concentration of Pif1 (with or without T7-Pol) followed by the addition of either 1) dNTP mix, or 2) ATP, or 3) ATP + a trap to prevent re-annealing (same as the strand carrying the Cy3 label) in a 3.5-fold excess relative to the DNA concentration used (3.5x for short). Single turnover experiments were performed with a SX.20 Applied Photophysics stopped-flow by mixing a pre-formed DNA-Pif1 complex with either ATP or ATP with different combinations of traps. The Cy3 was excited at 510nm and fluorescence emission monitored with a 570 nm bandpass filter (Newport).
Unwinding experiments at the single-molecule level
The experiments were performed using an objective type TIRF microscope (Olympus IX71) equipped with a 532nm laser system as described previously55. The flow cells were assembled from a cover slip (VWR, 24×50 mm N.1) and a pre-drilled slide (VWR, 75×25×1 mm) by heat-curing a parafilm mask to generate a single flow channel. The channel was coated with a 1:20 mixture of biotin-PEG-SVA (average MW 5000) and mPEG-SVA (average MW 5000) (Laysan Bio). After binding of neutravidin (Thermo Scientific) to the surface, excess unbound neutravidin was washed out. Following binding of 50–60 nM PentaHis Biotin Conjugate (Qiagen) to the qsurface for 15 minutes, unbound antibody was washed out and 100nM His6-Pif1 added for 10 minutes and unbound enzyme washed out. 50–100 pM Cy3-labeled DNA was then added to the immobilized Pif1 and unbound DNA washed out. At this stage either image buffer (50mM Tris-HCl pH 8.3, 100mM NaCl, 8mM MgAc2, 0.1mM DTT, 0.1mg/mL BSA, 2mM Trolox, 0.8% w/v glucose, 165 U/mL glucose oxidase, 2170 U/mL catalase) or image buffer + 1mM ATP were added and fluorescence of the Pif1 - DNA-Cy3 spots on the surface monitored over time. Movies were collected at ~30 fps and analyzed using software packages provided by Taekjip Ha (University of Illinois, Urbana-Champaign).
Supplementary Material
Highlights.
A monomer of Pif1 unwinds double-stranded DNA.
The 3′-tail of the DNA substrate regulates the activity of a monomer of Pif1.
A second DNA site within the Pif1 monomer interacts with the 3′-tail.
The N- and C-terminus of Pif1 contribute to discriminating the 3′-tail and unwinding.
Acknowledgments
This work was support by the National Institutes of Health (GM098509 to R.G.). We thank Prof. Ellenberger for the T7 DNA polymerase and Prof. Lohman for suggestions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bessler JB, Torredagger JZ, Zakian VA. The Pif1p subfamily of helicases: region-specific DNA helicases? Trends Cell Biol. 2001;11:60–5. doi: 10.1016/s0962-8924(00)01877-8. [DOI] [PubMed] [Google Scholar]
- 2.Boule JB, Zakian VA. Roles of Pif1-like helicases in the maintenance of genomic stability. Nucleic Acids Res. 2006;34:4147–53. doi: 10.1093/nar/gkl561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lahaye A, Leterme S, Foury F. PIF1 DNA helicase from Saccharomyces cerevisiae. Biochemical characterization of the enzyme. J Biol Chem. 1993;268:26155–61. [PubMed] [Google Scholar]
- 4.Lahaye A, Stahl H, Thines-Sempoux D, Foury F. PIF1: a DNA helicase in yeast mitochondria. Embo J. 1991;10:997–1007. doi: 10.1002/j.1460-2075.1991.tb08034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bochman ML, Sabouri N, Zakian VA. Unwinding the functions of the Pif1 family helicases. DNA Repair (Amst) 9:237–49. doi: 10.1016/j.dnarep.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boule JB, Vega LR, Zakian VA. The yeast Pif1p helicase removes telomerase from telomeric DNA. Nature. 2005;438:57–61. doi: 10.1038/nature04091. [DOI] [PubMed] [Google Scholar]
- 7.Budd ME, Reis CC, Smith S, Myung K, Campbell JL. Evidence suggesting that Pif1 helicase functions in DNA replication with the Dna2 helicase/nuclease and DNA polymerase delta. Mol Cell Biol. 2006;26:2490–500. doi: 10.1128/MCB.26.7.2490-2500.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ivessa AS, Zhou JQ, Zakian VA. The Saccharomyces Pif1p DNA helicase and the highly related Rrm3p have opposite effects on replication fork progression in ribosomal DNA. Cell. 2000;100:479–89. doi: 10.1016/s0092-8674(00)80683-2. [DOI] [PubMed] [Google Scholar]
- 9.Zhou J, Monson EK, Teng SC, Schulz VP, Zakian VA. Pif1p helicase, a catalytic inhibitor of telomerase in yeast. Science. 2000;289:771–4. doi: 10.1126/science.289.5480.771. [DOI] [PubMed] [Google Scholar]
- 10.Doudican NA, Song B, Shadel GS, Doetsch PW. Oxidative DNA damage causes mitochondrial genomic instability in Saccharomyces cerevisiae. Mol Cell Biol. 2005;25:5196–204. doi: 10.1128/MCB.25.12.5196-5204.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Rourke TW, Doudican NA, Mackereth MD, Doetsch PW, Shadel GS. Mitochondrial dysfunction due to oxidative mitochondrial DNA damage is reduced through cooperative actions of diverse proteins. Mol Cell Biol. 2002;22:4086–93. doi: 10.1128/MCB.22.12.4086-4093.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stith CM, Sterling J, Resnick MA, Gordenin DA, Burgers PM. Flexibility of eukaryotic Okazaki fragment maturation through regulated strand displacement synthesis. J Biol Chem. 2008;283:34129–40. doi: 10.1074/jbc.M806668200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pike JE, Henry RA, Burgers PM, Campbell JL, Bambara RA. An alternative pathway for Okazaki fragment processing: resolution of fold-back flaps by Pif1 helicase. J Biol Chem. 2010;285:41712–23. doi: 10.1074/jbc.M110.146894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pike JE, Burgers PM, Campbell JL, Bambara RA. Pif1 helicase lengthens some Okazaki fragment flaps necessitating Dna2 nuclease/helicase action in the two-nuclease processing pathway. J Biol Chem. 2009;284:25170–80. doi: 10.1074/jbc.M109.023325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paeschke K, Bochman ML, Garcia PD, Cejka P, Friedman KL, Kowalczykowski SC, Zakian VA. Pif1 family helicases suppress genome instability at G-quadruplex motifs. Nature. 2013;497:458–62. doi: 10.1038/nature12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paeschke K, Capra JA, Zakian VA. DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell. 2011;145:678–91. doi: 10.1016/j.cell.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ribeyre C, Lopes J, Boule JB, Piazza A, Guedin A, Zakian VA, Mergny JL, Nicolas A. The yeast Pif1 helicase prevents genomic instability caused by G-quadruplex-forming CEB1 sequences in vivo. PLoS Genet. 2009;5:e1000475. doi: 10.1371/journal.pgen.1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou R, Zhang J, Bochman ML, Zakian VA, Ha T. Periodic DNA patrolling underlies diverse functions of Pif1 on R-loops and G-rich DNA. Elife. 2014;3:e02190. doi: 10.7554/eLife.02190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anand RP, Shah KA, Niu H, Sung P, Mirkin SM, Freudenreich CH. Overcoming natural replication barriers: differential helicase requirements. Nucleic Acids Res. 2012;40:1091–105. doi: 10.1093/nar/gkr836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilson MA, Kwon Y, Xu Y, Chung WH, Chi P, Niu H, Mayle R, Chen X, Malkova A, Sung P, Ira G. Pif1 helicase and Poldelta promote recombination-coupled DNA synthesis via bubble migration. Nature. 2013;502:393–6. doi: 10.1038/nature12585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saini N, Ramakrishnan S, Elango R, Ayyar S, Zhang Y, Deem A, Ira G, Haber JE, Lobachev KS, Malkova A. Migrating bubble during break-induced replication drives conservative DNA synthesis. Nature. 2013;502:389–92. doi: 10.1038/nature12584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vasianovich Y, Harrington LA, Makovets S. Break-induced replication requires DNA damage-induced phosphorylation of Pif1 and leads to telomere lengthening. PLoS Genet. 2014;10:e1004679. doi: 10.1371/journal.pgen.1004679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schulz VP, Zakian VA. The saccharomyces PIF1 DNA helicase inhibits telomere elongation and de novo telomere formation. Cell. 1994;76:145–55. doi: 10.1016/0092-8674(94)90179-1. [DOI] [PubMed] [Google Scholar]
- 24.Myung K, Chen C, Kolodner RD. Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature. 2001;411:1073–6. doi: 10.1038/35082608. [DOI] [PubMed] [Google Scholar]
- 25.Pennaneach V, Putnam CD, Kolodner RD. Chromosome healing by de novo telomere addition in Saccharomyces cerevisiae. Mol Microbiol. 2006;59:1357–68. doi: 10.1111/j.1365-2958.2006.05026.x. [DOI] [PubMed] [Google Scholar]
- 26.Boule JB, Zakian VA. The yeast Pif1p DNA helicase preferentially unwinds RNA DNA substrates. Nucleic Acids Res. 2007;35:5809–18. doi: 10.1093/nar/gkm613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Makovets S, Blackburn EH. DNA damage signalling prevents deleterious telomere addition at DNA breaks. Nat Cell Biol. 2009;11:1383–6. doi: 10.1038/ncb1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barranco-Medina S, Galletto R. DNA binding induces dimerization of Saccharomyces cerevisiae Pif1. Biochemistry. 2010;49:8445–54. doi: 10.1021/bi100984j. [DOI] [PubMed] [Google Scholar]
- 29.Galletto R, Tomko EJ. Translocation of Saccharomyces cerevisiae Pif1 helicase monomers on single-stranded DNA. Nucleic Acids Res. 2013;41:4613–27. doi: 10.1093/nar/gkt117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramanagoudr-Bhojappa R, Chib S, Byrd AK, Aarattuthodiyil S, Pandey M, Patel SS, Raney KD. Yeast Pif1 helicase exhibits a one-base-pair stepping mechanism for unwinding duplex DNA. J Biol Chem. 2013;288:16185–95. doi: 10.1074/jbc.M113.470013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duan XL, Liu NN, Yang YT, Li HH, Li M, Dou SX, Xi XG. G-quadruplexes significantly stimulate Pif1 helicase-catalyzed duplex DNA unwinding. J Biol Chem. 2015;290:7722–35. doi: 10.1074/jbc.M114.628008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramanagoudr-Bhojappa R, Byrd AK, Dahl C, Raney KD. Yeast Pif1 accelerates annealing of complementary DNA strands. Biochemistry. 2014;53:7659–69. doi: 10.1021/bi500746v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stano NM, Jeong YJ, Donmez I, Tummalapalli P, Levin MK, Patel SS. DNA synthesis provides the driving force to accelerate DNA unwinding by a helicase. Nature. 2005;435:370–3. doi: 10.1038/nature03615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramanagoudr-Bhojappa R, Blair LP, Tackett AJ, Raney KD. Physical and functional interaction between yeast Pif1 helicase and Rim1 single-stranded DNA binding protein. Nucleic Acids Res. 2013;41:1029–46. doi: 10.1093/nar/gks1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nandakumar D, Pandey M, Patel SS. Cooperative base pair melting by helicase and polymerase positioned one nucleotide from each other. Elife. 2015:4. doi: 10.7554/eLife.06562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fischer CJ, Lohman TM. ATP-dependent translocation of proteins along single-stranded DNA: models and methods of analysis of pre-steady state kinetics. J Mol Biol. 2004;344:1265–86. doi: 10.1016/j.jmb.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 37.Fischer CJ, Maluf NK, Lohman TM. Mechanism of ATP-dependent translocation of E. coli UvrD monomers along single-stranded DNA. J Mol Biol. 2004;344:1287–309. doi: 10.1016/j.jmb.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 38.Fischer CJ, Tomko EJ, Wu CG, Lohman TM. Fluorescence methods to study DNA translocation and unwinding kinetics by nucleic Acid motors. Methods Mol Biol. 2012;875:85–104. doi: 10.1007/978-1-61779-806-1_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fischer CJ, Wooten L, Tomko EJ, Lohman TM. Kinetics of motor protein translocation on single-stranded DNA. Methods Mol Biol. 2010;587:45–56. doi: 10.1007/978-1-60327-355-8_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Galletto R, Jezewska MJ, Bujalowski W. Unzipping mechanism of the double-stranded DNA unwinding by a hexameric helicase: quantitative analysis of the rate of the dsDNA unwinding, processivity and kinetic step-size of the Escherichia coli DnaB helicase using rapid quench-flow method. J Mol Biol. 2004;343:83–99. doi: 10.1016/j.jmb.2004.07.055. [DOI] [PubMed] [Google Scholar]
- 41.Jeong YJ, Levin MK, Patel SS. The DNA-unwinding mechanism of the ring helicase of bacteriophage T7. Proc Natl Acad Sci U S A. 2004;101:7264–9. doi: 10.1073/pnas.0400372101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen CF, Brill SJ. An essential DNA strand-exchange activity is conserved in the divergent N-termini of BLM orthologs. Embo J. 2010;29:1713–25. doi: 10.1038/emboj.2010.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheok CF, Wu L, Garcia PL, Janscak P, Hickson ID. The Bloom’s syndrome helicase promotes the annealing of complementary single-stranded DNA. Nucleic Acids Res. 2005;33:3932–41. doi: 10.1093/nar/gki712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garcia PL, Liu Y, Jiricny J, West SC, Janscak P. Human RECQ5beta, a protein with DNA helicase and strand-annealing activities in a single polypeptide. Embo J. 2004;23:2882–91. doi: 10.1038/sj.emboj.7600301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gu Y, Masuda Y, Kamiya K. Biochemical analysis of human PIF1 helicase and functions of its N-terminal domain. Nucleic Acids Res. 2008;36:6295–308. doi: 10.1093/nar/gkn609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Machwe A, Xiao L, Groden J, Matson SW, Orren DK. RecQ family members combine strand pairing and unwinding activities to catalyze strand exchange. J Biol Chem. 2005;280:23397–407. doi: 10.1074/jbc.M414130200. [DOI] [PubMed] [Google Scholar]
- 47.Masuda-Sasa T, Polaczek P, Campbell JL. Single strand annealing and ATP-independent strand exchange activities of yeast and human DNA2: possible role in Okazaki fragment maturation. J Biol Chem. 2006;281:38555–64. doi: 10.1074/jbc.M604925200. [DOI] [PubMed] [Google Scholar]
- 48.Muftuoglu M, Kulikowicz T, Beck G, Lee JW, Piotrowski J, Bohr VA. Intrinsic ssDNA annealing activity in the C-terminal region of WRN. Biochemistry. 2008;47:10247–54. doi: 10.1021/bi800807n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pike AC, Gomathinayagam S, Swuec P, Berti M, Zhang Y, Schnecke C, Marino F, von Delft F, Renault L, Costa A, Gileadi O, Vindigni A. Human RECQ1 helicase-driven DNA unwinding, annealing, and branch migration: insights from DNA complex structures. Proc Natl Acad Sci U S A. 2015;112:4286–91. doi: 10.1073/pnas.1417594112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dessinges MN, Lionnet T, Xi XG, Bensimon D, Croquette V. Single-molecule assay reveals strand switching and enhanced processivity of UvrD. Proc Natl Acad Sci U S A. 2004;101:6439–44. doi: 10.1073/pnas.0306713101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tomko EJ, Jia H, Park J, Maluf NK, Ha T, Lohman TM. 5′-Single-stranded/duplex DNA junctions are loading sites for E. coli UvrD translocase. Embo J. 2010;29:3826–39. doi: 10.1038/emboj.2010.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yodh JG, Stevens BC, Kanagaraj R, Janscak P, Ha T. BLM helicase measures DNA unwound before switching strands and hRPA promotes unwinding reinitiation. Embo J. 2009;28:405–16. doi: 10.1038/emboj.2008.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Comstock MJ, Whitley KD, Jia H, Sokoloski J, Lohman TM, Ha T, Chemla YR. Protein structure. Direct observation of structure-function relationship in a nucleic acid-processing enzyme. Science. 2015;348:352–4. doi: 10.1126/science.aaa0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Klaue D, Kobbe D, Kemmerich F, Kozikowska A, Puchta H, Seidel R. Fork sensing and strand switching control antagonistic activities of RecQ helicases. Nat Commun. 2013;4:2024. doi: 10.1038/ncomms3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nguyen B, Sokoloski J, Galletto R, Elson EL, Wold MS, Lohman TM. Diffusion of human replication protein A along single-stranded DNA. J Mol Biol. 2014;426:3246–61. doi: 10.1016/j.jmb.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.