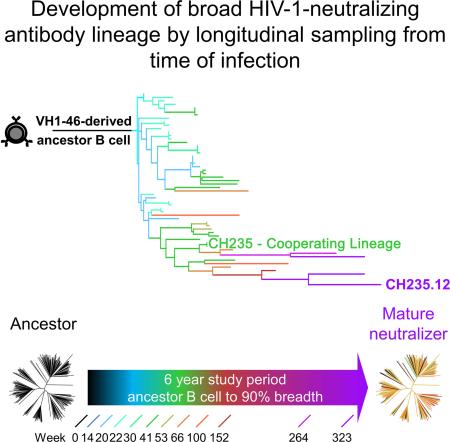
SUMMARY
Antibodies with ontogenies from VH1-2 or VH1-46-germline genes dominate the broadly neutralizing response against the CD4-binding site (CD4bs) on HIV-1. Here we define with longitudinal sampling from time-of-infection the development of a VH1-46-derived antibody lineage that matured to neutralize 90% of HIV-1 isolates. Structures of lineage antibodies CH235 (week 41 from time-of-infection, 18% breadth), CH235.9 (week 152, 77%) and CH235.12 (week 323, 90%) demonstrated the maturing epitope to focus on the conformationally invariant portion of the CD4bs. Similarities between CH235 lineage and five unrelated CD4bs lineages in epitope focusing, length-of-time to develop breadth, and extraordinary levels of somatic hypermutation suggested commonalities in maturation among all CD4bs antibodies. Fortunately, the required CH235-lineage hypermutation appeared substantially guided by the intrinsic mutability of the VH1-46 gene, which closely resembled VH1-2. We integrated our CH235-lineage findings with a second broadly neutralizing lineage and HIV-1 co-evolution to suggest a vaccination strategy for inducing both lineages.
Graphical Abstract
INTRODUCTION
Understanding the pathways and mechanisms of broadly neutralizing antibody (bnAb) induction is a critical goal of HIV-1 vaccine development (Bonsignori et al., 2012; Haynes, 2015; Haynes and Bradley, 2015; Haynes et al., 2012; Mascola and Haynes, 2013;). In chronic HIV-1 infections, breadth of plasma neutralization follows a uniform distribution and broad neutralization arises in ~50% of individuals after 5 years or more of infection (Hraber et al., 2014). The delayed appearance of bnAbs suggests roadblocks to their development, and one vaccine approach is to decipher these roadblocks and devise strategies to overcome them. It is possible that - because of the high diversity of antibodies resulting from recombination and somatic hypermutation (SHM) - different bnAb lineages may have different developmental pathways and roadblocks. However, for the CD4-binding site (CD4bs), a population-level analysis on 14 donors indicated only two general types of CD4bs bnAbs: VH-gene restricted and CDR H3-dominated (Zhou et al., 2015).
The VH-gene restricted classes arise from two highly similar VH -genes: VH1-2 and VH1-46 (Scheid et al., 2011; Wu et al., 2011). VH1-2*02 and VH1-46*01 share 93.4% (269/288) nucleotide sequence identity. Both classes give rise to antibodies that recognize the CD4bs via VH structural mimicry of the immunoglobulin-like N-terminal domain of CD4 (Zhou et al., 2010; Zhou et al., 2015). For the VH1-2 gene-derived antibodies, analysis of their ontogeny suggests two roadblocks based on: (i) a requirement for high levels of SHM (Klein et al., 2013; Scheid et al., 2009; Scheid et al., 2011; Wu et al., 2010), and (ii) weak binding of the inferred unmutated common ancestor (UCA) to gp120 (Jardine et al., 2013; McGuire et al., 2013; Scheid et al., 2011; Wu et al., 2011; Zhou et al., 2010; Zhou et al., 2015), although a definitive analysis from time-of-infection had not yet provided detail. In addition, several of the CD4bs bnAbs are autoreactive with ubiquitinase enzymes (Bonsignori et al., 2014; Liao et al., 2013; Liu et al., 2015).
Structure-based design of UCA-interacting immunogens has recently demonstrated a means to overcome this second roadblock, with priming of VH1-2 bnAb lineages in knock-in mice (Dosenovic et al., 2015; Jardine et al., 2015). However, the maturation of primed VH1-2 CD4bs B cell lineages to broad neutralization as well as the mechanism for the development of breadth remain unresolved.
For the VH1-46-derived antibodies, far less is known. Two chronically HIV-infected individuals, RU1 and RU8, have developed VH1-46-derived bnAbs, 1B2530 and 8ANC131 (Scheid et al., 2011). We recently described an African individual (donor CH505) who, over time, developed a CD4bs bnAb lineage (the CH103 lineage) that recognized the CD4 supersite through a CDR H3-dominated mode of interaction (Liao et al., 2013). Analysis of the co-evolution between virus and CH103 lineage demonstrated a second B cell lineage (the CH235 lineage) that cooperated by selection of escape mutants from the CH235 lineage that drove the CH103 bnAb lineage (Gao et al., 2014). Here we find that the CH235 lineage itself progressed to bnAb over 5 years of affinity maturation. We identify sequences of the CH235 lineage through longitudinal samples of 17 time points spanning weeks (wks) 6-323 post infection, assess neutralization breadth of sequential lineage members on a panel of ~200 diverse isolates, and determine Env-complexed crystal and EM structures for lineage members. We analyze the conformity (i.e. the level of shared mutation positions and identical sequence mutations) of CH235 lineage development relative to other VH gene-specific bnAb lineages in other donors, as well as the co-evolution of virus and CH235 lineage. Despite an early near-optimal binding orientation, the CH235 lineage required over 20% SHM to reach 90% neutralization breadth. Our results provide insight into the difficulties in focusing recognition to the conserved site of HIV-1 vulnerability, and suggest that CD4bs-directed antibodies, whether VH-gene restricted or CDR H3-dominated, face similar obstacles in development. For VH1-46- and VH1-2-derived CD4-mimic antibodies, the unique genetic mutability inherent in each of these two VH-germline genes helps to direct maturation, potentially providing an explanation for the prevalence of effective CD4bs antibodies derived from these two germline genes.
RESULTS
Sequencing of B cell antibody gene rearrangements in longitudinal samples
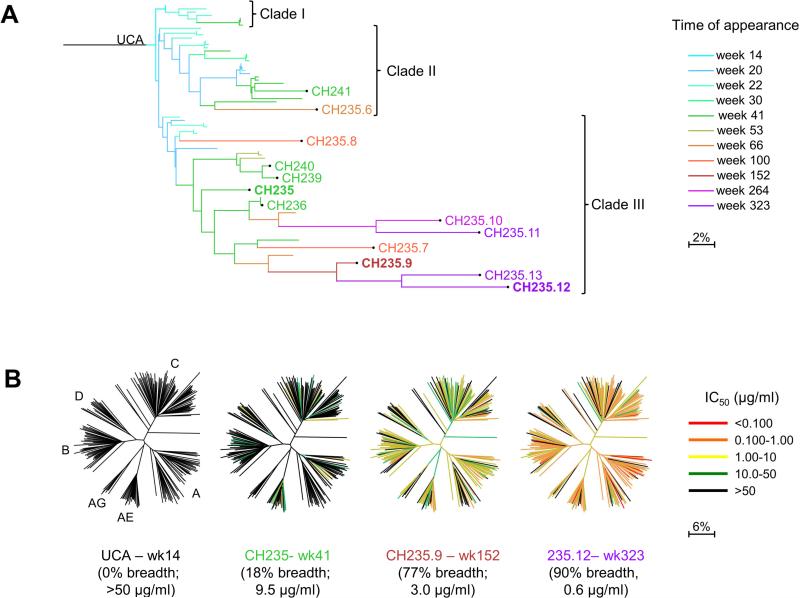
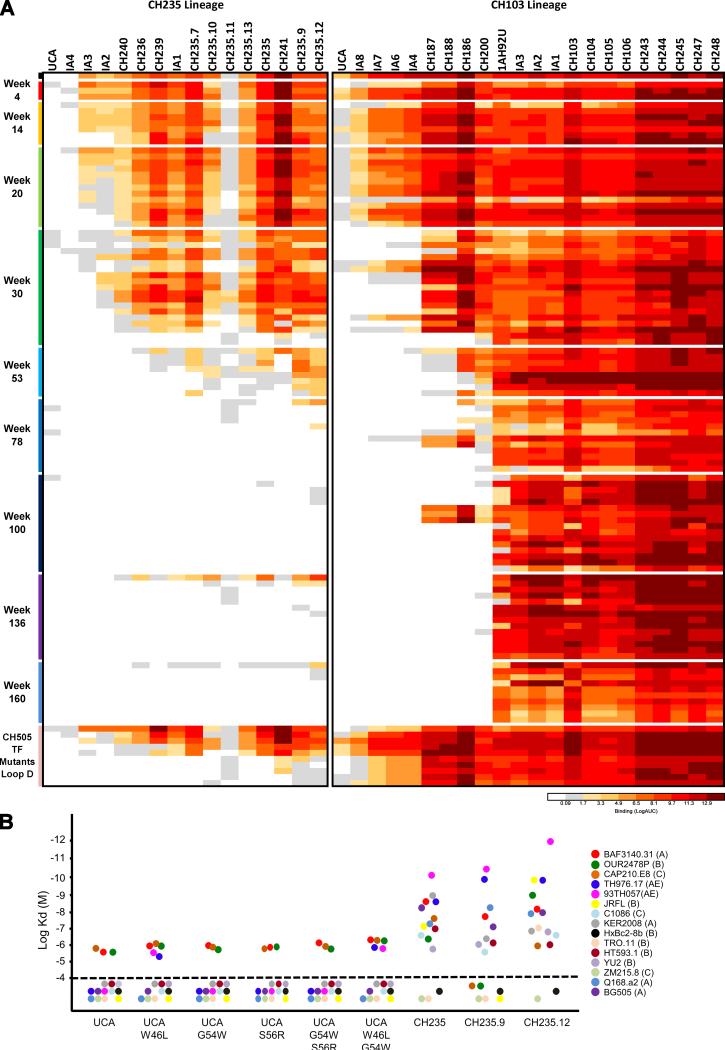
To understand the maturation of the cooperating CH235 lineage in donor CH505, we sought to identify sequences of lineage members at 17 time points, spanning wks 6 to 323 from time of infection. We first asked when we could detect members of the CH235 lineage. Next-generation sequencing (NGS) of antibody heavy chain gene rearrangements amplified from genomic DNA template of blood mononuclear cells from wk 6 to 152 (15 time points) identified a total of 479,028 unique, non-duplicated V-heavy sequences. The first V-heavy sequences belonging to the CH235 B cell lineage were found at wk 14, and additional CH235 lineage members were found at all subsequent time points. Only unique sequences in the CH235 lineage were further investigated and they were assigned to the earliest time-point (time-of-appearance) in which they were identified. Four V-heavy sequences were paired with the closest VL from isolated antibodies and produced as recombinant monoclonal antibodies (mAbs) (CH235.6 through CH235.9). From cultured memory B cells collected 41 wks post-transmission we had previously isolated five members of the CH235 lineage (CH235, CH236, CH239, CH240 and CH241) (Gao et al., 2014) and we have now isolated four additional members with natural VH and VL pairing from cultured memory B cells collected at wks 264 and 323 post-transmission: CH235.10 through CH235.13 (Figure 1A, Figure S1A and Table S1). CH235 lineage antibodies represented 0.018% of the total memory B cell repertoire and 0.5% of the CH505 TF gp120-specific memory B cell population.
Figure 1. CH235 Lineage, with Time of Appearance and Neutralization by Select Members.
(A) Phylogram of CH235 lineage. Phylogenetic tree is colored by first time (wk post-infection) from which sequences were obtained. Key members of the CH235 lineage are labeled. CH235.6, CH235.7, CH235.8 and CH235.9 VH were complemented with full heavy chain gene regions and paired with the VL from the closest natural antibody. (B) Neutralization dendrograms display single mAb neutralization of a genetically diverse panel of 199 HIV-1 isolates. Coloration is by IC50. See also Figure S1, Tables S1 and S2.
The CH235 lineage could be separated into three clades (clade I, II and III). Clade I showed a number of early lineage members, but no additional clade I sequences were observed after wk 30; clade II showed further development and included members CH241 (wk 41) and CH235.6 (wk 66), but no additional sequences were observed after wk 66; clade III developed through wk 323 and included antibodies CH235 (wk 41), CH235.9 (wk 152), and CH235.12 (wk 323) (Figure 1A).
CH235 lineage HIV-1 neutralization
To characterize the development of neutralization breadth in the CH235 lineage, we assessed antibodies in clade III for their ability to neutralize diverse HIV-1 isolates in a 199-isolate panel (Figure 1B and Table S2). No isolates were neutralized by the unmutated common ancestor (UCA), whereas 18% of the viruses were neutralized by CH235 at wk 41. By wk 152, CH235.9 neutralized 77% of viruses, although with a relatively weak potency of 3 μg/ml. By wk 323, however, CH235.12 was able to neutralize 90% of viruses, and the neutralization 50% inhibitory concentration (IC50) potency increased by 5-fold to 0.6 μg/ml.
We next analyzed the heterologous neutralization pattern of these antibodies to understand their development of broad neutralization (Figure S1B) (Georgiev et al., 2013). CH235 lineage members and previously identified HIV-1 bnAbs were clustered based on heterologous neutralization activity. CH235 neutralization activity was more similar to CD4bs bnAbs than to bnAbs with other epitope specificities. While the CH235 neutralization profile was the most divergent from other CD4bs bnAbs, CH235.9 and CH235.12 were much more similar to other CD4bs bnAbs and each other. Interestingly, despite VH1-46 usage, the CH235.9 and CH235.12 neutralizing profile was more similar to that of VH1-2-derived antibodies, such as VRC01, than VH1-46-derived antibodies, such as 8ANC131 (Figure S1B).
Crystal structures of CH235-lineage members with HIV-1 gp120
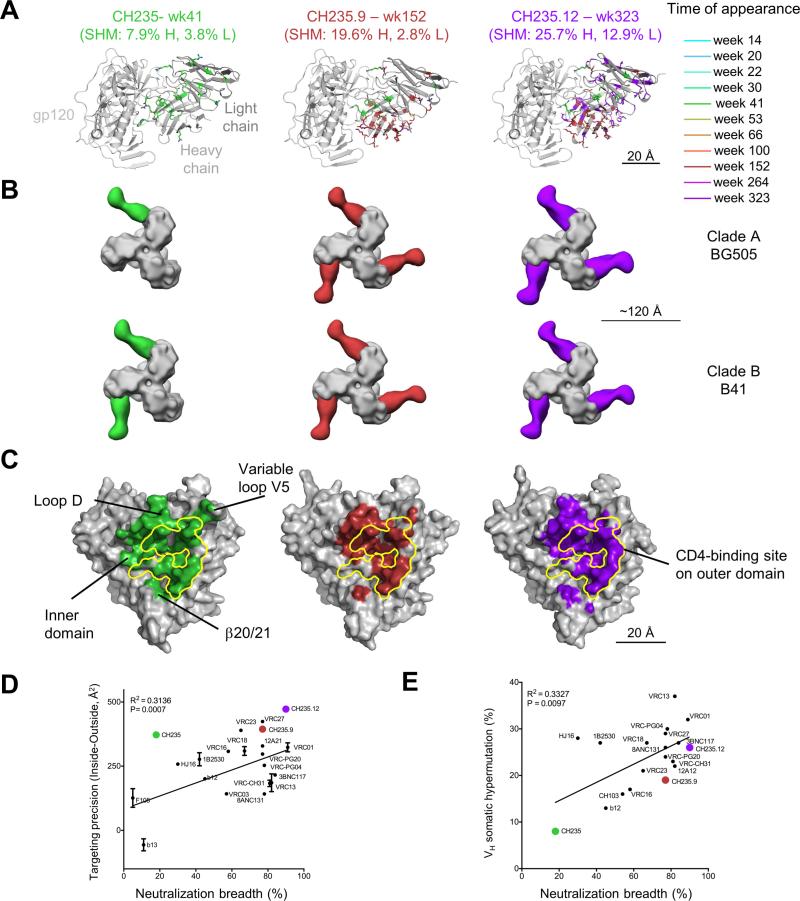
To provide structural insight into the recognition and maturation of the CH235 lineage, we prepared the antigen-binding fragments (Fabs) of antibodies CH235 (wk 41 from time of infection, 18% breadth), CH235.9 (wk 152, 77%) and CH235.12 (wk 323, 90%), and co-crystallized, solved and refined these in complex with the gp120 core of HIV-1 isolate strain (93TH057) (Figure 2, Table S3). We mapped the location of residues altered during SHM and observed changes throughout the variable domain (Figure 2A).
Figure 2. Structures of CH235-Lineage Members in Complex with HIV-1 Env.
(A) Co-crystal structures of the antigen-binding fragments (Fabs) of CH235-lineage members with core gp120. Structures are shown in ribbon diagram, with gp120 in gray and residues altered by SHM in stick representation colored by time-of-appearance. (B) Negative stain EM of Fabs of CH235-lineage members and trimeric HIV-1 Env from BG505 (top row) and B41 (bottom row). Structures in surface representation, with Env portions colored gray and Fabs by time-of-appearance. (C) Epitope displayed on the gp120 surface and colored by antibody time-of-appearance, with the vulnerable portion of the CD4bs highlighted in yellow and select regions labeled. (D) Targeting precision of CD4bs-directed antibodies vs neutralization breadth. (E) VH-gene SHM of CD4bs-directed antibodies vs neutralization breadth. See also Figure S2 and Table S3.
Comparison of the orientation of the VH of CH235 in Env binding with that of CD4, VRC01 and 8ANC131 (Scheid et al., 2011) showed that the CH235 VH domain mimicked CD4 in Env binding and was highly similar to the VH orientation and structure of the VRC01 and 8ANC131 VH chains: in particular, the VH1-46 of CH235 preserved key contacts mediated by the CDR H2 loop for the CD4 binding loop and for the gp120 D368 (Figure S2A,B).
Analysis of the angle of recognition for the CH235 lineage indicated little change during maturation, with CH235, CH235.9 and CH235.12 all clustering within the larger VRC01-class of antibodies. Interestingly, other VH1-46 antibodies clustered differently, with antibody 1B2530 from HIV-1-positive donor RU1 at a highly similar angle and 1.5 Å translated, and antibodies 8ANC131 and 8ANC134 from HIV-1-positive donor RU8 occupying a cluster about 55 degrees and 3.5 Å translated related to the CD4 (Figure S2C).
These results suggest that the gp120-antibody orientation was determined early in bnAb lineage ontogeny, with further maturation maintaining the same general orientation. Overall, the structures of CH235 lineage members with HIV-1 gp120 Env revealed CD4 mimicry. While the VH gene usage classifies the CH235 lineage within the VH1-46-derived 8ANC131 bnAb class, it is both functionally and structurally closer to the VRC01 class (Zhou et al., 2015).
Negative stain EM of CH235-lineage members with trimeric HIV-1 Env
To visualize the recognition of the CH235 lineage in the context of the HIV-1 Env trimer, we used negative stain EM to determine 3D-reconstructions of Fabs CH235, CH235.9 and CH235.12 bound to trimeric BG505 and B41 HIV-1 Env glycoproteins (Figure 2B)(Pugach et al., 2015; Sanders et al., 2013). Notably, the stoichiometry increased with antibody maturation, with CH235 (8% VH mutation) binding with a stoichiometry of 1:1 (BG505; Figure 2B, top, Figure S2D-F) or 2:1 (B41; Figure 2B, bottom, Figure S2D-F) Fabs per trimer and CH235.9 and CH235.12 (19% and 25% VH mutation, respectively) binding with a 3:1 Fab to trimer ratio (Figure 2B). We next compared the orientation and stoichiometry of CH235.12 Fab with that of CH103, a CDR H3-dependent CD4bs bnAb isolated from the same subject (Liao et al., 2013). EM analysis of either CH235.12 or CH103 Fab in complex with BG505 SOSIP.664 revealed structural differences between the CDR H3-dominated CH103 class bnAb and the 8ANC131-class CH235.12 bnAb and, in accordance with crystallographic results, the angle of approach of CH235 was similar to that of VRC01 and other CD4 mimicking bnAbs (Figure S2G).
Despite the CD4 mimicry by CH235, the trimer remained in a closed conformation when the CH235 lineage members were bound. However, the EM-derived model of CH103 in complex with BG505 revealed that CH103 either bound to or induced a more open version of the trimer. This conformation represents an intermediate state between the closed, compact trimer in complex with CH235 or VRC01, and the CD4-induced open model in complex with soluble CD4 or 17b Fab (Figure S2G). Similar to more mature CH235 lineage bnAb Fabs, bnAb CH103 bound to BG505 with a stoichiometry of 3 Fabs per trimer. (Figure S2G).
Maturation focuses CH235 lineage recognition to a conserved site of CD4 vulnerability
To gain insight into the structural consequences of maturation, we mapped the epitope of CH235 lineage members relative to the conformationally invariant CD4 supersite of vulnerability (Zhou et al., 2015). When we mapped the CH235 footprint on gp120, we observed portions of the CH235-binding surface on gp120 to be outside of the CD4 supersite of vulnerability (Figure 2C, left). This surface was reduced in CH235.9 and CH235.12 structures, especially on variable loop V5. Recognition by the CH235.12 antibody concentrated almost entirely on the CD4 supersite of vulnerability, with little interactions with the inner domain or variable loop V5; there was, however, a large remaining interaction with the conserved loop D region (Figure 2C, middle and right).
To quantify targeting precision, we computed the buried surface between antibodies and gp120 co-crystal complexes, for the region overlapping the CD4 supersite of vulnerability minus the region outside the vulnerable site. Overall targeting precision correlated with neutralization breadth (P=0.0007) (Figure 2D). The CH235-lineage antibodies all showed good targeting precision. We also analyzed the correlation of SHM versus neutralization breadth (P=0.0097) (Figure 2E): While the CH235 lineage generally trended towards lower SHM relative to neutralization breadth, all CD4bs bnAbs appeared to require a high degree of SHM, independent of whether the antibody derived from a specific VH-gene or used a CDR H3-dominated mode of recognition.
Overall, the results suggest that maturation requires a high degree of SHM to focus recognition onto the CD4 supersite of vulnerability and that this high degree of SHM is a general requirement of all CD4bs bnAb lineages, even those that begin with highly favorable orientations such as CH235.
Conformity of sequence evolution of CH235 lineage
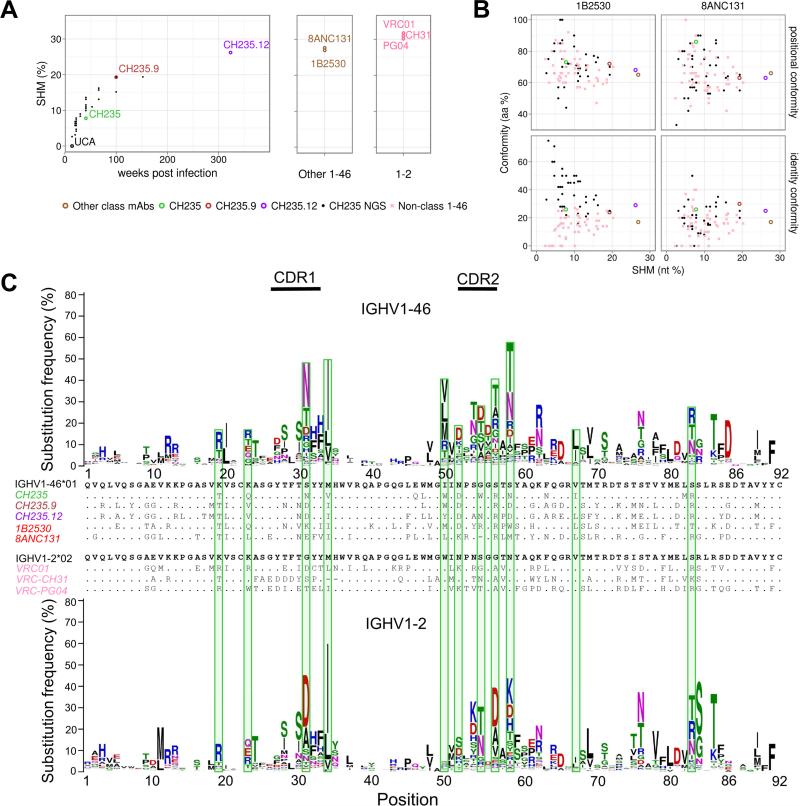
The mutation levels of CH235-lineage antibodies isolated 41 wks post infection from memory B cell cultures was markedly lower (range 7-11%;) than that of all previously reported VH1-46 and VH1-2 CD4bs bnAbs (>25%) (Scheid et al., 2011; Sui et al., 2010; Wu et al., 2010; Zhou et al., 2015) (Table S1). The mutation levels of CH235-lineage antibodies isolated up to 264 wks post infection increased to ~20%, but were still lower than those of most other bnAbs until 323 wks post infection (CH235.12 : 26% mutations) (Figure 3A).
Figure 3. Sequence Evolution of CH235 Lineage: SHM, Timing, and Conformity of CH235-Lineage Development from UCA to Antibody with 90% Breadth.
(A) Heavy chain SHM over time for the CH235 lineage (left panel). SHM levels of other VH1-46-derived CD4bs mAbs and selected VH1-2-derived VRC01-class mAbs are shown (middle and right panels, respectively); the time since infection is unknown for these mAbs. (B) Maturation conformity vs overall heavy chain SHM. Positional conformity (top row) is defined as the number of aa positions differing from the germline sequence in both the conforming and reference sequences, divided by the total number of aa changes in the conforming antibody. Identity conformity (bottom row) is defined as the number of such positions which are additionally mutated to the same residue, divided by the total number of mutations in the conforming antibody. Conformity to 1B2530 (left) and to 8ANC131 (right) is shown for both position and identity. (C) VH-gene mutability accounts for the majority of positional conformity of CH235 lineage. The mutability of the VH-gene for VH1-46 (top) and VH1-2 (bottom) is shown. Sequence logos are shown at each position; the height of each logo corresponds to the percent of mutated reads. Green bars are shown for SHM in antibody CH235, which are altered in over a quarter of VH1-46-derived antibodies. See also Figure S3 and Table S4.
To quantify the conformity of CH235-lineage antibodies to the two VH1-46-derived bnAbs (1B2530 from donor RU1 and 8ANC131 from donor RU8) (Scheid et al., 2011; Zhou et al., 2015), we analyzed the similarity of shared mutation positions (positional conformity) and shared identical mutations (identity conformity) of the VH genes (Figure 3B, Figure S3A). As a comparison, we also calculated the positional conformity and identity conformity of non-HIV-1 targeting antibodies isolated from 3 HIV-1 negative donors relative to template antibodies 1B2530 and 8ANC131. Positional conformity in SHM was spread over a large range (50-90%), and there did not seem to be much discrimination between VH1-46 in antibodies that effectively neutralized HIV-1 and those that did not (Figure 3B, top panels). Identity conformity in SHM was also spread over a large range (0-75%) (Figure 3B, bottom panels), and while little discrimination was observed between VH1-46 in antibodies that effectively neutralized HIV-1 and those that did not for antibody 8ANC131, there was discrimination among CD4bs antibodies when 1B2530 was used as a reference (Figure 3B, bottom left panel). The differences in CH235-lineage identify conformity to 1B2530 or to 8ANC131 may reflect the greater similarity of the recognition orientation of CH235-lineage members with 1B2530 (Figure S2C) and suggested that slight differences in recognition orientation can substantially alter factors associated with identity conformity.
Overall, these results indicated SHM in response to HIV-1 infection to proceed in a manner that depended less on functional selection and more on intrinsic properties of the VH1-46-gene, especially related to the position of residues that undergo SHM. To investigate further the contribution of the VH1-46 gene, we analyzed SHM observed in VH1-46 gene transcripts from three uninfected individuals (Figure 3C, top); notably, all 11 positions mutated in CH235, 1B2530 and 8ANC131 were also mutated among non-HIV-1 neutralizing antibodies with high frequency (≥20%). Moreover, the residue substitutions in CH235 were frequently found in the top three most commonly observed substitutions for that position in the VH1-46 gene. To quantify the impact of gene mutability, we compared the difference in probability distributions of positional and identity conformity for sequences simulated with and without taking into account the intrinsic VH1-46 gene mutability. The simulations showed that both positional and identity conformity shifted to a higher level of similarity when considering gene mutability (Figure S3B and S3C). Notably, a substantial shift in probability was observed for the positional conformity of CH235 (Figure S3B, Table S4A). Similar shifts in identity conformity were also observed for CH235 (Figure S3C, Table S4B). Thus, the intrinsic susceptibility at specific sites of the VH1-46 germline gene to mutation as well as to the frequency of specific mutations that existed at each of these sites appeared to be a dominant factor in the SHM alteration of the CH235 lineage. These results are in line with our previous finding that selection and mutability synergized during affinity maturation of an influenza HA-reactive clone from a non-HIV-1 infected person to hemagglutinin (HA) (Kepler et al., 2014): hence, the dominant role of intrinsic susceptibility at specific sites may be a more general biological phenomenon in dictating the course of SHM.
Because VH1-2 is genetically the most closely related germline gene to VH1-46, we also examined the mutability of the VH1-2 gene (Figure 3C, bottom). Consistent with VH1-46 antibodies, the mutated positions among VH1-2 derived bnAbs also showed high frequency of mutation among non-HIV-1 targeting VH1-2 antibodies, suggesting that gene mutability contributes to VH1-2 derived HIV-1 antibody evolution. Notably, the average mutability of the VH1-2 gene at positions where the CH235 antibody showed SHM was generally high: 9 of 15 positions mutated in CH235 antibody were also mutated in more than 15% of VH1-2-derived NGS reads. In 10 of these 15 positions, the mature VH1-2-derived bnAbs (VRC01, VRC-CH31 and VRC-PG04) also showed changes. When we analyzed mutability of other VH genes used by CD4bs bnAbs (VH1-69, VH3-23, VH3-30, and VH4-59) (Zhou et al., 2015) (Figure S3D), we observed gene mutability patterns different from that of VH1-46 while, in contrast, the mutability patterns of VH1-2 and VH1-46 were more similar (Table S4C). Despite the similarity between VH1-2 and VH1-46, we did observe that antibody sequences from CD4bs bnAbs of each gene segregated phylogenetically (Figure S3E), indicating differences in maturation pathway between bnAbs evolving from these two germline genes.
These data suggested that for both VH1-2 and VH1-46 germline genes-derived bnAbs, somatic mutations that lead to neutralization breadth appeared to be primarily determined by the intrinsic mutability of VH1-46 and VH1-2 germline genes. The differences in the intrinsic mutabilities of these VH genes may contribute to the high occurrence of CD4bs bnAbs that originate from either VH1-2 or VH1-46 (Zhou et al., 2015).
Interaction between CH235 and CH103 bnAb lineages
While gene mutability plays a role in determining the position where SHM occurs, binding between antibody and HIV-1 Env likely also plays a role in selecting or fixing a mutation. A hallmark of cooperating B cell lineages is that they interact at the same site as the bnAb lineage that is being driven (Gao et al., 2014).
To determine a mechanism whereby the initial interaction of the early CH235 and CH103 lineage members bind to the same or similar epitope and result in CH235 selection of escape mutants that stimulated the CH103 bnAb lineage (Gao et al., 2014), we evaluated cross-competition between early CH235 lineage antibodies and the CH103 lineage antibody CH106 in ELISA, as an example of early CH103 lineage development, and measured their association rate constant with surface plasmon resonance (SPR). Since both the CH235 and CH103 lineages bound to the loop D gp120 region, we asked if the early CH235 lineage antibodies could block the binding of the CH103 lineage mature antibody CH106, or block the binding of soluble (s)CD4 to CH505 TF gp120 Env. CH241 was the only antibody in the CH235 lineage that strongly blocked CH106 bnAb and sCD4 binding to CH505 gp120 (IC50 = 2.6 and 1.5 μg/ml, respectively) (Table S5A).
To confirm early dominance of the binding of CH103 lineage compared to the CH235 lineage to CH505 TF Env, we reversed the blocking assay and asked if bnAb CH106 could block the binding of biotinylated CH235, CH236, CH239, CH240 or CH241. CH106 strongly blocked the binding of all the CH235 mature antibodies with IC50s ranging from 2.3 μg/ml (for CH240) to 14.3 μg/ml (for CH241) (Table S5B). These data suggested that the earliest maturation intermediates of the CH235 lineage antibodies could not outcompete CH106 bnAb for binding to CH505 TF gp120 Env.
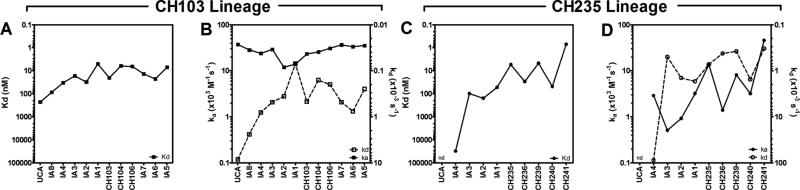
Affinity maturation in germinal centers is subjected to kinetic selection and involves improvement in dissociation rate constant (Kd) that is often driven by an improvement in the kinetic association rate (ka), which is a key variable in conferring a binding advantage for the cognate epitope to an antibody over other competing antibodies (Foote and Milstein, 1991; Kepler et al., 2014). We measured the ka and dissociation kinetic rate (kd) of the CH505 TF gp120 Env binding by CH235 and CH103 with SPR to identify differences that might explain the relative inability of the CH235 lineage to block the binding of the CH103 lineage bnAbs to autologous CH505 TF Env and found that the two lineages followed two distinct trajectories and modalities to increase their overall affinity.
The UCA of the CH103 lineage bound to CH505 TF Env with a Kd of 227 nM which increased one order of magnitude throughout affinity maturation (Figure 4A). The CH103 UCA displayed a fast association rate (ka = 37 × 103 M−1s−1) which was maintained across the intermediate and mature mAbs (ka = 11.9 - 37.3 ×103 M−1s−1), suggesting that maintaining the fast association rate was important for survival and maturation of the CH103 lineage (Figure 4B). In contrast, the CH235 lineage mAb Kd increased four orders of magnitude during affinity maturation (from 30.6 mM of IA4 - the earliest intermediate mAb in the CH235 lineage for which kinetic rates could be measured - to 0.7 nM of CH241) (Figure 4C). Such increase was predominantly facilitated by slower dissociation rates (kd) observed in later intermediates and mature mAbs, which decreased from 88.1 ×10−3 s−1 of IA4 to 0.33 ×10−3 s−1 of CH241 (Figure 4D). Conversely, CH235 lineage mAbs bound to CH505 TF gp120 Env with ka that started off an order of magnitude slower than CH103 UCA and its earlier intermediates (IA4 ka =2.9 ×103 M−1s−1) and only modestly improved - primarily between IA1 and CH235 mAbs - with the majority of the early CH235 mAbs having slower ka than CH103 mAbs (Figure 4D).
Figure 4. Binding Kinetics of CH103 and CH235 Lineage Antibodies.
Binding association (ka) and dissociation (kd) rates of the CH103 (A-B, squares) and CH235 (C-D, circles) lineage mAbs to CH505.TF gp120 Env were measured with SPR and used to calculate the dissociation rate constants (Kd). Kds are shown in A and C, ka (solid lines, plotted on the left y-axis) and kd (dashed lines, plotted on the right y-axis) are shown in B and D. See also Table S5.
Thus, the relative inability of wk 41 CH235 lineage antibodies to block early mature CH103 lineage mAbs could be explained by the observed differences in their association rates, and these data provide an explanation of how the CH235 antibody lineage exerted its cooperating function in driving autologous virus toward better neutralization by the CH103 antibody lineage without impeding concurrent development of the CH103 antibody lineage itself.
Late CH235 lineage broadly neutralizing antibodies neutralize autologous loop D escape viruses selected by early CH235 lineage members
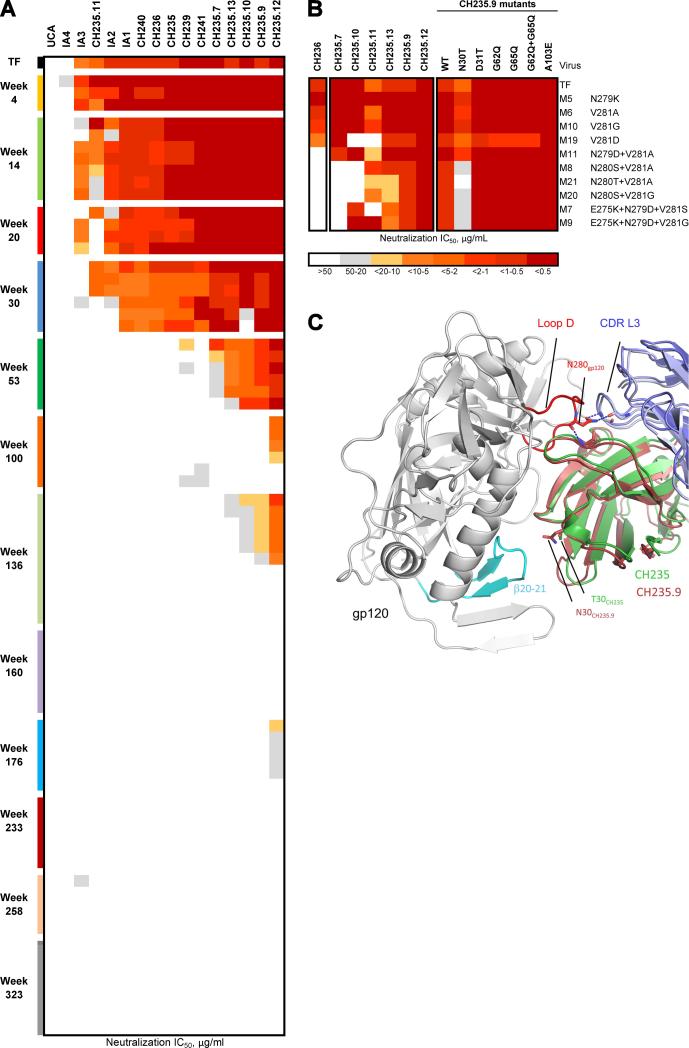
We have previously demonstrated that the CH235 lineage selected escape viruses with mutations in the loop D region of gp120 Env that rendered loop D mutant viruses more sensitive to the CH103 bnAb lineage and that autologous virus escaped from early CH235 lineage antibodies by wk 30 after infection (Gao et al., 2014). We have now isolated autologous viruses through wk 323 and determined the neutralization capacity of the late CH235 lineage bnAbs. Viruses partially sensitive to the later members of the CH235 lineage (particularly bnAbs CH235.9 and CH235.12) were found as late as wk 176 (Figure 5A, Table S6). These viruses still contained the loop D mutations that were selected by virus escape from early antibody members of the CH235 lineage (Gao et al., 2014). Hence, we tested the ability of the late CH235 lineage bnAbs to neutralize the panel of CH505 TF loop D mutants (Gao et al., 2014). Remarkably, CH235.9, CH235.12 and CH235.13 bnAbs acquired the ability to neutralize all loop D mutants that were resistant to the early members of the CH235 lineage (Figure 5B and Table S7). In particular, CH235.9, CH235.12 and CH235.13 neutralized CH505 TF gp120 M8, M20 and M21 (not neutralized by early lineage member CH236), which differed from CH505 TF gp120 M6 and M10 (neutralized by CH236) by a single mutation at position 280 (N280S for M8 and M20, and N280T for M21) (Figure 5B).
Figure 5. CH235 Lineage Antibodies Neutralization of Autologous Virus and CH505.TF Loop D Mutants.
(A) Heatmap analysis of neutralization of 76 pseudoviruses (row) by 16 CH235 lineage mAbs (column). Coloration is by IC50. This analysis extends previous observations on early CH235 lineage antibodies (Gao et al., 2014) by including late mAbs CH235.7, CH235.8, CH235.10, CH235.11, CH235.12 and CH235.13 and by adding pseudoviruses isolated from wk 136 to 323 post-transmission. (B) CH505 TF and loop D mutants M5, M6, M10, M19, M11, M7, M8, M9, M20 and M21 neutralization by CH236 mAb, late mAbs CH235.7, CH235.9 CH235.10, CH235.11, CH235.12, CH235.13 (left panel) and CH235.9 mAb mutants (right panel). Neutralization is expressed as IC50 μg/ml. CH505 TF sequence mutations are shown on the right. (C) The CDR H1 N30 (sticks, dark red) in CH235.9, which interacts with the β20- β21 loop in the bridging sheet of gp120 (cyan), is over 19Å away from the N280S mutation site in loop D (orange). See also Figure S4, Tables S6 and S7.
In the gp120-complexed structure, the side chains of N280 forms three hydrogen bonds with two residues in the CDR L3 and these hydrogen bonds are predicted to be disrupted by the N280S and N280T mutations (Figure S4A). Since the CH235.9 antibody had the VL of CH236, the direct implication was that mutations in the heavy chain were responsible for the ability of CH235.9 to neutralize loop D mutant viruses. Interestingly, CH235.7, which did not neutralize autologous viruses beyond wk 53, also had the VL of CH236 but, in contrast to CH235.9, failed to neutralize CH236-resistant loop D mutants M7, M8, M9, M20 and M21.
Therefore, we reverted the 5 amino acids (aa) in CH235.9 VH at gp120 contact positions that were different from those present in CH236 VH but not shared with CH235.7 VH: N30T and D31T in CDR H1, G62Q and G65Q in FR H3 and A103E in CDR H3 (Figure S4B). Five of the six CH235.9 mutants retained the ability to potently neutralize all the CH505 TF loop D mutant viruses. In contrast, the N30T mutation in CDR H1 reverted CH235.9 to the CH236 phenotype (CH236 has a threonine in position 30): M21 neutralization was abrogated, M20, M7 and M9 were near completely abrogated (CH235.9 N30T IC50 >44 μg/ml) and M8 IC50 increased 37-fold (CH235.9 IC50 = 0.66 μg/ml vs CH235.9 N30T IC50 = 24.31 μg/ml) (Table S7).
Thus, acquisition of extraordinary breadth in the CH235 bnAb lineage was associated with accumulation of somatic mutations in CDR H1 that enabled late CH235 lineage antibodies to neutralize autologous loop D mutant viruses that were escape mutants from early CH235 antibodies. CH235.9 bnAb residue N30 contacts R429 in the β20-β21 loop of the C4 region of gp120 Env, which is on the opposite face of the CD4bs from loop D (Figure 5C). In addition, CH505 TF has a glutamic acid in position 429 that is in close enough proximity to N30 to form a hydrogen bond.
These findings indicate a mechanism for acquiring the ability to neutralize loop D mutants via a compensatory mutation in the CH235 VHDJH which strengthens the binding to the gp120 C4 region by introducing hydrogen bonds that correct the loss of neutralization due to disruption of the hydrogen bonds between loop D and the CH235 mAb light chain.
CH235 and CH103 lineage antibody binding to CH505 gp120 Env
We tested the CH235 lineage antibodies for binding to 113 recombinant CH505 gp120 Env isolated from time of transmission to wk 160 post-transmission, including CH505 TF loop D mutant Envs (Figure 6A and Table S8). Of note, CH235.9 and CH235.12 bound to 4/22 and 8/22 Envs isolated from wk 136 and 160 post-transmission, respectively, including Envs from viruses that were also neutralized. We have previously reported Env binding to the initial members of the CH103 lineage (Hraber et al., 2015), and have now performed the same Env binding analysis of the CH103 lineage with 10 additional matured bnAb members of the CH103 lineage (Figure 6A and Table S8). We have used these data to select CH505 gp120 Env quasi-species that bound to mature and precursor bnAbs of both lineages, defining a series of CH505 Env immunogens now optimized and predicted to induce both bnAb lineages (Figure S5A).
Figure 6. Binding of CH235 and CH103 Lineage mAbs to Autologous CH505 (A) and CH235 UCA Binding to Heterologous HIV-1 Env Glycoproteins (B).
(A) Heatmap analysis of UCA, intermediate (IA) and mature CH235 and CH103 lineage mAbs binding to 113 CH505 autologous Env isolated from time of infection (TF) to 160 wks post-infection and to the CH505.TF mutants (Gao et al., Cell 2014). Mabs were tested in ELISA at concentrations ranging from 100 μg/ml to 0.6 ng/ml . Binding is expressed as a LogAUC. (B) Affinity of CH235 UCA, CH235 wild-type and select SHM variants to a panel of 15 heterologous gp120 Envs. See also Figure S5 and Table S8.
We had previously reported that CH235 UCA weakly reacted with CH505 TF gp120 at ~10 μM as determined by SPR (Gao et al., 2014). Here we show stronger binding of the CH235 UCA to 8/113 autologous CH505 gp120 Envs measured in ELISA (Figure 6A and Table S8). Moreover, in a panel of 15 heterologous Envs from multiple clades, CH235 UCA bound to 3/15 Envs and the introduction of only 3 mutations (W47L, G54W and S56R), which were selected based on the increase in surface area of interaction (G54W and S56R) or the reduction in clash score (W47L), increased this recognition (to 5/15 Envs), of which the dominant effect appeared to be reduction in clash (Figure 6B and Figure S5B).
Autoreactivity in the CH235 B cell lineage
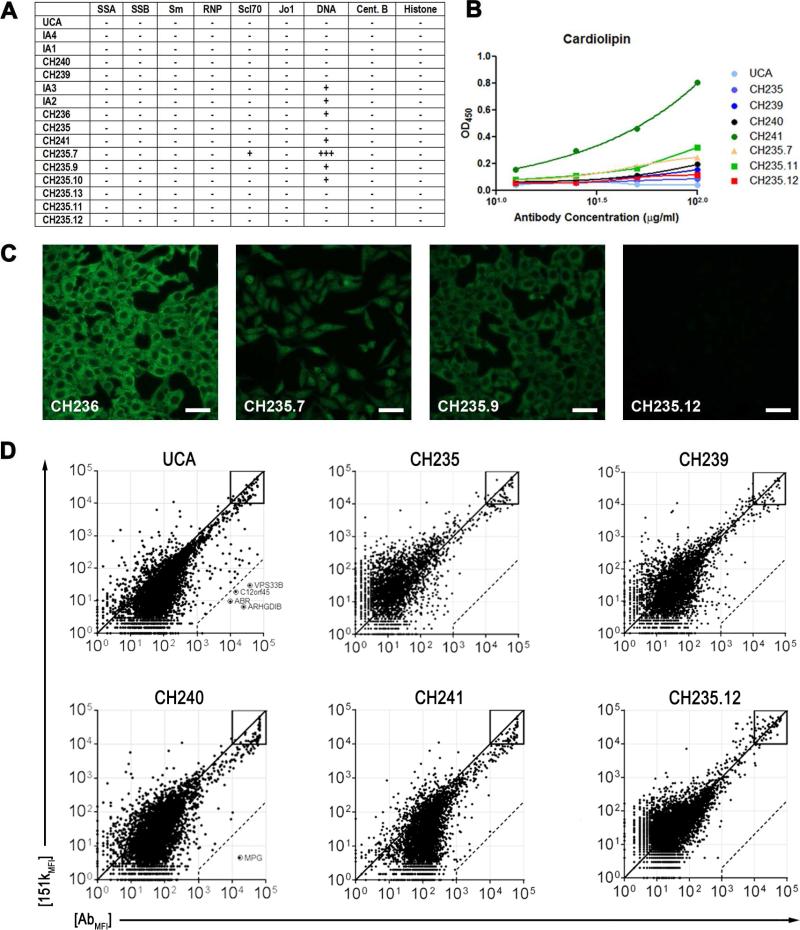
Development of auto- and polyreactivity during antibody maturation toward neutralization breadth is a critical aspect that may limit the ability of generating bnAbs during natural infection and upon vaccination (Bonsignori et al., 2014; Haynes et al., 2005; Haynes et al., 2012; Haynes and Verkoczy, 2014; Liu et al., 2015; Verkoczy et al., 2013; Verkoczy et al., 2010; Verkoczy et al., 2011). We had previously reported that in HIV-1-infected individual CH505, the CD4bs CH103 bnAb lineage was polyreactive and, similar to VRC01-class bnAbs, bound to human ubiquitin ligase E3A (UBE3A) with avidity correlated with neutralization (Liao et al., 2013; Liu et al., 2015). In addition, most of the mutations introduced in VRC07 - a somatic variant of VRC01 - that enhanced neutralizing activity also resulted in increased autoreactivity (Rudicell et al., 2014). Since CH235.12 is a potent and extremely broad CD4-mimic CD4bs bnAb, we compared the auto- and polyreactivity profile of CH235.12 with other members of the CH235 lineage. Most CH235 lineage antibodies displayed reactivity against DNA and sporadic reactivity with Scl70 (CH235.7) (Figure 7A). CH241 bound to cardiolipin (Figure 7B). In Hep-2 IF staining CH236, CH235.7 and CH235.9 were all cytoplasmic positive (Figure 7C). Conversely, CH235.12, despite being highly mutated and broadly neutralizing, did not display autoreactivity in any of these assays (Figure 7A-C) Of particular note, CH235 lineage antibodies, including CH235.12, did not react with UBE3A (Figure 7D).
Figure 7. CH235 Antibody Lineage Auto- and Polyreactivity.
(A) CH235 lineage antibody binding to ANA measured in ELISA. LogAUC was calculated from duplicate samples. Results representative of duplicate experiments. (B) Binding to cardiolipin was determined using Quanta Lite ACA IgG III ELISA Assay. (C) Hep2 cell IF staining. Size bars = 50 μm. (D) Measurement of polyreactivity against 9,400 human antigens using ProtoArray 5 microchip: CH235 lineage mAbs binding (x-axis) was compared to non-polyreactive control mAb 151K (y-axis). Polyreactivity is defined as 1 log stronger binding than 151k mAb to more than 90% of the test proteins. High affinity binding was measured as a >2 log increase in binding (dotted line) (Liu et al., 2015).
These data identify CH235.12 as an antibody that has developed neutralization breadth without being itself auto- and polyreactive, while less mutated precursor antibodies (CH235 is in the same clade of CH235.12) did develop autoreactivity. We conclude that in vivo decoupling of neutralization breadth of CD4 mimic CD4bs bnAbs from auto- and polyreactivity can occur, even for bnAb lineages that have developed autoreactivity during the course of their maturation and, therefore, inducing such bnAbs from such lineages through vaccination, though difficult, is an achievable goal.
DISCUSSION
Here we have traced the ontogeny of the CH235 VH1-46 8ANC131 class of CD4bs bnAbs from acute infection to chronic infection and defined both the structural and functional pathways of bnAb lineage induction. That the CH235 bnAb lineage that selected virus escape mutants that drove the CH103 CD4bs CDR H3-dependent bnAb lineage is itself an 8ANC131-class bnAb lineage and co-evolved with the CH103 bnAb is a remarkable demonstration of a bnAb-to virus-to bnAb interaction in the same HIV-1 infected individual. In addition, the similarity of VH1-46 8ANC131-like and VH1-2 VRC01 family CD4 supersite bnAbs demonstrates dramatic convergence of antibody structures to recognize the CD4 supersite. The CH235 lineage required over 20% SHM in heavy chain variable domain to achieve 90% breadth. Fortunately, a substantial portion of the VH-gene SHM was guided by the intrinsic mutability of the VH1-46 germline gene. Moreover, the CH235 lineage Ab that became broadly neutralizing acquired the ability to neutralize loop D mutants selected by early Ab lineage members (Gao et al., 2014) with a mechanism involving a compensatory mutation (T30N) in CDR H1, which allowed the formation of H-bonds with the HIV-1 gp120 C4 region, thus correcting the original loss of binding.
The driving forces of the CH235 lineage were the natural transmitted/founder and M5 Envs. In addition, despite near-complete autologous virus escape from CH235 lineage antibodies by wk 100, viruses arose later during the course of infection, which were sensitive to the more mature CH235 bnAb members and likely contributed to antigen drive. It is interesting to note that many of these late viruses were less sensitive to CH103 CDR H3 binder bnAbs prompting the hypothesis that the CH103 lineage may have the capacity for cooperation with the CH235 lineage after 5-6 years of co-development. Finally, the CH235.12 antibody that evolved late in CH235 development is an extraordinary broad and potent non-autoreactive antibody and is a candidate for preventive and therapeutic uses.
In summary, the acquisition of neutralization breadth in the CH235 VRC01-like VH1-46 CD4 mimic bnAb occurred with the sequence of transmitted/founder and early mutant-initiated antigen drive, selection of Env loop D mutants that cooperated with the CH103 bnAb lineage to drive it to bnAb breadth, followed by acquisition of the ability of the CH235 lineage itself to neutralize autologous loop D mutants coincident with potent neutralization of a broad array of heterologous HIV-1 isolates. Mapping these events points to a strategy for the simultaneous induction of both CDR H3 and VRC01-class CD4bs bnAbs, whereby sequential immunizations with transmitted founder Env followed by loop D mutant Envs comprise a rational immunization strategy.
EXPERIMENTAL PROCEDURES
Donor and sample information
Donor and sample information was previously reported (Liao et al., 2013) and is summarized in Supplemental Experimental Procedures. Memory B cell cultures were performed on PBMCs collected at 264 and 323 wks post-transmission. All work related to human subjects was in compliance with Institutional Review Board protocols approved by the Duke University Health System Institutional Review Board.
Preparation of libraries for 454 DNA pyrosequencing
454 DNA pyrosequencing was performed on genomic DNA template isolated with Qiagen kits from PBMCs collected at 6, 7, 8, 9, 14, 20, 22, 30, 41, 53, 66, 92, 100, 144 and 152 wks post-transmission as described in (Boyd et al., 2009) and in Supplemental Experimental Procedures. Only unique V-heavy rearrangements were included in the analysis to generate the phylogeny; in the case of duplicated sequences, the earliest occurrence was included in the analysis.
Phylogenetic analysis
For clonal phylogenetics, the UCA was inferred using Cloanalyst (Kepler, 2013), which simultaneously estimates the UCA and the phylogenetic tree relating the observed sequences to each other and to the UCA. Internally, Cloanalyst uses dnaml from the PHYLIP suite of phylogenetic software (Felsenstein, 2005). The CH235 antibody lineage clonogram was displayed using the ete2 Python package.
Isolation of CH235 Lineage Antibodies from Donor CH505
Fluorescence-activated cell sorting of antigen-specific IgG+ B cells from PBMC and the amplification and cloning of immunoglobulin genes were performed as described in (Bonsignori et al., 2011). CH505.TF gp120 Env-positive memory B cells were cultured as described in Supplemental Experimental Procedures.
Neutralization assays
Neutralization of donor CH235 mAbs were measured using single-round-of-infection HIV-1 Env pseudoviruses and TZM-bl target cells as described in Supplemental Experimental Procedures.
Neutralization signature
Antibody neutralization signatures were computed and compared as described in Supplemental Experimental Procedures.
Monoclonal Antibody and Antigen-Binding Fragment (Fab) Production
Ig genes of mAbs were amplified from RNA and expression plasmids for heavy and kappa chains were constructed. Expression and purification of recombinant IgG mAbs and preparation of Fab fragments are described in Supplemental Experimental Procedures.
Crystallization, X-Ray Data Collection, Structure Determination, and Refinement of Donor CH235 Antibodies in Complex with HIV-1 gp120
Purification, crystallization of antibody-gp120 complexes, data collection, structure solution, refinement and analysis are described in Supplemental Experimental Procedures. Diffraction data were integrated and scaled with the HKL2000 suite (Otwinowski and Minor, 1997).
Electron microscopy data collection and processing
BG505 SOSIP.664 and B41 SOSIP.664 gp140 trimers and donor CH235-derived Fab complex negative-stain electron microscopy images, analysis and visualization are described in the Supplemental Experimental Procedures.
Focused maturation and conformity analysis
Focused maturation and mAb conformity analysis are described in the Supplemental Experimental Procedures.
Surface Plasmon Resonance Affinity and Kinetics Measurements
MAb binding to autologous CH505 gp140 was measured using a BIAcore 3000 or BIAcore T200 instrument (GE Healthcare) as described in (Alam et al., 2007; Alam et al., 2009; Liao et al., 2013) and in Supplemental Experimental Procedures.
Direct-Binding ELISA
Direct-binding ELISAs were performed as described in Supplemental Experimental Procedures.
MAb CH235.9 Amino Acid Reversion
Site-directed mutagenesis of the CH235.9 mAb genes was performed using the Quikchange lightning multi-site-directed mutagenesis kit (Agilent) following manufacturer's protocol. Primers are listed in Supplemental Experimental Procedures.
Structural Modeling
Loop D mutations were structurally modeled using PyMOL with sidechains placed in the most frequently observed rotamer that did not result in steric clashing with neighboring residues. Hydrogen bonds were calculated using HBPLUS software (McDonald and Thornton, 1994).
Recombinant HIV-1 Proteins
HIV-1 genes of autologous CH505 Env were determined from samples collected from 4 to 323 wks post-infection by single genome amplification (Keele et al., 2008) and produced as described in (Liao et al., 2013).
Protein Array
MAbs were screened for binding on protein microarrays (ProtoArray) (PAH0525101; Invitrogen) pre-coated with 9,400 human proteins in duplicate and screened following manufacturer's instructions and as described in (Liu et al., 2015; Yang et al., 2013).
HEp-2 cell staining
Indirect immunofluorescence binding of mAbs or plasma to HEp-2 cells (Zeuss Scientific) was performed as previously described (Bonsignori et al., 2014; Haynes et al., 2005).
Supplementary Material
Highlights.
Developmental pathway of a CD4-mimicking antibody lineage from time of infection
Maturation involves focused recognition of CD4-binding site of vulnerability
Intrinsic VH1-46 gene mutability guides somatic mutation of CD4-binding site bnAbs
Study provided a strategy for inducing both CD4-mimicking and CDR H3-using bnAbs
ACKNOWLEDGMENTS
The authors thank K. Lloyd, A. Eaton, T. Von Holle, T. Gurley, L. Armand, D. Kozink, A. Cooper, F. Perrin, J. Pritchett, A. Foulger, G. Hernandez, S. Arora, R. Kauffman, A. Trama of the Duke Human Vaccine Institute, J. Whitesides, D. Marshall and the DHVI Flow Cytometry Core for technical assistance, and K. Soderberg for project coordination; members of the Structural Biology Section and Structural Bioinformatics Core, Vaccine Research Center, for discussions and comments on the manuscript; J. Baalwa, D. Ellenberger, K. Hong, J. Kim, F. McCutchan, L. Morris, J. Overbaugh, E. Sanders-Buell, R. Swanstrom, M. Thomson, S. Tovanabutra, C. Williamson, and L. Zhang for contributing HIV-1 Envelope plasmids. Supported by grant from the Division of AIDS, NIAID, NIH Center for HIV/AIDS Vaccine Immunology UM1 AI100645 (Duke CHAVI-ID), the Scripps CHAVI-ID (UM1 AI100663), the International AIDS Vaccine Initiative, and the Bill and Melinda Gates Foundation (CAVD) to ABW. Support for this work was provided by the Intramural Research Program of the Vaccine Research Center, NIAID, NIH, and by NIH grant P01-AI104722. Use of sector 22 (Southeast Region Collaborative Access team) at the Advanced Photon Source was supported by the US Department of Energy, Basic Energy Sciences, Office of Science (contract number W-31-109-Eng-38).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
MB, PDK, BFH conceived, designed and coordinated the study; MB, TZ, ZS, LC, MGJ, PDK, BFH wrote and revised manuscript and figures; JRM provided intellectual expertise and guidance; GO, GYC, CAS, KW, SMA, SDB, AZF, TBK, LS, ABW, JRM edited figures/text; Antibody discovery/screening: MB, MAG, KKH, SI, MCM, RJP, BFH; Crystal structures: TZ, LC, MGJ, PDK; Assisted with structure or protein production: PA, AC, AD, YDK, BZ, AZ; Gene mutability, epitope focusing and conformity: ZS, GYC, CAS, PDK; Clonal maturation analysis: MB, KW, TB, LS, TBK, BFH, PDK; Negative stain EM: GO, ABW; MAb and Env protein production: MB, TB, XL, KL, SI, HXL; Neutralization assays: MB, MCM, MKL, ISG, RTB, DCM, JRM; Single-cell sorts: MAM; Sequencing: FG, CAS, HS, BJH, RK, CS, NISC, JCM, DCD, SDB, AZF; Computational analysis: KW, LS, TBK; Antibody affinity measurement: SMA; Auto and polyreactivity: MB, GK, BFH; Autologous virus isolation, sequencing, analysis, selection and production: FG, AK, GMS, BHH, PTH, BTK. All authors read and approved the manuscript.
ACCESSION NUMBERS
Coordinates and structure factors for CH235, CH235.9 and CH235.12 in complex with HIV-1 gp120 have been deposited with the Protein Data Bank (PDB ID 5F9W, 5F9O and 5F96). Next-generation sequencing data have been deposited with the NCBI Sequence Reads Archive (SRP067168). Antibody heavy and light chains have been deposited with GenBank (KU570032-KU570053).
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, 5 figures and 8 tables.
REFERENCES
- Alam SM, McAdams M, Boren D, Rak M, Scearce RM, Gao F, Camacho ZT, Gewirth D, Kelsoe G, Chen P, et al. The role of antibody polyspecificity and lipid reactivity in binding of broadly neutralizing anti-HIV-1 envelope human monoclonal antibodies 2F5 and 4E10 to glycoprotein 41 membrane proximal envelope epitopes. J. Immunol. 2007;178:4424–4435. doi: 10.4049/jimmunol.178.7.4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam SM, Morelli M, Dennison SM, Liao HX, Zhang R, Xia SM, Rits-Volloch S, Sun L, Harrison SC, Haynes BF, et al. Role of HIV membrane in neutralization by two broadly neutralizing antibodies. Proc. Natl. Acad. Sci. U. S. A. 2009;106:20234–20239. doi: 10.1073/pnas.0908713106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonsignori M, Alam SM, Liao HX, Verkoczy L, Tomaras GD, Haynes BF, Moody MA. HIV-1 antibodies from infection and vaccination: insights for guiding vaccine design. Trends Microbiol. 2012;20:532–539. doi: 10.1016/j.tim.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonsignori M, Hwang KK, Chen X, Tsao CY, Morris L, Gray E, Marshall DJ, Crump JA, Kapiga SH, Sam NE, et al. Analysis of a clonal lineage of HIV-1 envelope V2/V3 conformational epitope-specific broadly neutralizing antibodies and their inferred unmutated common ancestors. J. Virol. 2011;85:9998–10009. doi: 10.1128/JVI.05045-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonsignori M, Wiehe K, Grimm SK, Lynch R, Yang G, Kozink DM, Perrin F, Cooper AJ, Hwang KK, Chen X, et al. An autoreactive antibody from an SLE/HIV-1 individual broadly neutralizes HIV-1. J. Clin. Invest. 2014;124:1835–1843. doi: 10.1172/JCI73441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd SD, Marshall EL, Merker JD, Maniar JM, Zhang LN, Sahaf B, Jones CD, Simen BB, Hanczaruk B, Nguyen KD, et al. Measurement and Clinical Monitoring of Human Lymphocyte Clonality by Massively Parallel V-D-J Pyrosequencing. Sci. Transl. Med. 2009;1 doi: 10.1126/scitranslmed.3000540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosenovic P, von Boehmer L, Escolano A, Jardine J, Freund NT, Gitlin AD, McGuire AT, Kulp DW, Oliveira T, Scharf L, et al. Immunization for HIV-1 Broadly Neutralizing Antibodies in Human Ig Knockin Mice. Cell. 2015;161:1505–1515. doi: 10.1016/j.cell.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP (Phylogeny Inference Package) 3.6a3 edn. Department of Genome Sciences, University of Washington; Seattle, WA: 2005. distributed by the author. [Google Scholar]
- Foote J, Milstein C. Kinetic maturation of an immune response. Nature. 1991;352:530–532. doi: 10.1038/352530a0. [DOI] [PubMed] [Google Scholar]
- Gao F, Bonsignori M, Liao HX, Kumar A, Xia SM, Lu X, Cai F, Hwang KK, Song H, Zhou T, et al. Cooperation of B cell lineages in induction of HIV-1-broadly neutralizing antibodies. Cell. 2014;158:481–491. doi: 10.1016/j.cell.2014.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiev IS, Doria-Rose NA, Zhou TQ, Do Kwon Y, Staupe RP, Moquin S, Chuang GY, Louder MK, Schmidt SD, Altae-Tran HR, et al. Delineating Antibody Recognition in Polyclonal Sera from Patterns of HIV-1 Isolate Neutralization. Science. 2013;340:751–756. doi: 10.1126/science.1233989. [DOI] [PubMed] [Google Scholar]
- Haynes BF. New approaches to HIV vaccine development. Curr. Opin. Immunol. 2015;35:39–47. doi: 10.1016/j.coi.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes BF, Bradley T. Broadly Neutralizing Antibodies and the Development of Vaccines. JAMA. 2015;313:2419–2420. doi: 10.1001/jama.2015.2427. [DOI] [PubMed] [Google Scholar]
- Haynes BF, Fleming J, St Clair EW, Katinger H, Stiegler G, Kunert R, Robinson J, Scearce RM, Plonk K, Staats HF, et al. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science. 2005;308:1906–1908. doi: 10.1126/science.1111781. [DOI] [PubMed] [Google Scholar]
- Haynes BF, Kelsoe G, Harrison SC, Kepler TB. B-cell-lineage immunogen design in vaccine development with HIV-1 as a case study. Nat. Biotechnol. 2012;30:423–433. doi: 10.1038/nbt.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes BF, Verkoczy L. AIDS/HIV. Host controls of HIV neutralizing antibodies. Science. 2014;344:588–589. doi: 10.1126/science.1254990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hraber P, Korber B, Wagh K, Giorgi EE, Bhattacharya T, Gnanakaran S, Lapedes AS, Learn GH, Kreider EF, Li Y, et al. Longitudinal Antigenic Sequences and Sites from Intra-Host Evolution (LASSIE) Identifies Immune-Selected HIV Variants. Viruses. 2015;7:5443–5475. doi: 10.3390/v7102881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hraber P, Seaman MS, Bailer RT, Mascola JR, Montefiori DC, Korber BT. Prevalence of broadly neutralizing antibody responses during chronic HIV-1 infection. AIDS. 2014;28:163–169. doi: 10.1097/QAD.0000000000000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardine J, Julien JP, Menis S, Ota T, Kalyuzhniy O, McGuire A, Sok D, Huang PS, MacPherson S, Jones M, et al. Rational HIV immunogen design to target specific germline B cell receptors. Science. 2013;340:711–716. doi: 10.1126/science.1234150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardine JG, Ota T, Sok D, Pauthner M, Kulp DW, Kalyuzhniy O, Skog PD, Thinnes TC, Bhullar D, Briney B, et al. HIV-1 VACCINES. Priming a broadly neutralizing antibody response to HIV-1 using a germline-targeting immunogen. Science. 2015;349:156–161. doi: 10.1126/science.aac5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, Sun C, Grayson T, Wang S, Li H, et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. U. S. A. 2008;105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepler TB. Reconstructing a B-cell clonal lineage. I. Statistical inference of unobserved ancestors. F1000Res. 2013;2:103. doi: 10.12688/f1000research.2-103.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepler TB, Munshaw S, Wiehe K, Zhang R, Yu JS, Woods CW, Denny TN, Tomaras GD, Alam SM, Moody MA, et al. Reconstructing a B-Cell Clonal Lineage. II. Mutation, Selection, and Affinity Maturation. Front. Immunol. 2014;5:170. doi: 10.3389/fimmu.2014.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F, Diskin R, Scheid JF, Gaebler C, Mouquet H, Georgiev IS, Pancera M, Zhou T, Incesu RB, Fu BZ, et al. Somatic mutations of the immunoglobulin framework are generally required for broad and potent HIV-1 neutralization. Cell. 2013;153:126–138. doi: 10.1016/j.cell.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao HX, Lynch R, Zhou T, Gao F, Alam SM, Boyd SD, Fire AZ, Roskin KM, Schramm CA, Zhang Z, et al. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature. 2013;496:469–476. doi: 10.1038/nature12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Yang G, Wiehe K, Nicely NI, Vandergrift NA, Rountree W, Bonsignori M, Alam SM, Gao J, Haynes BF, et al. Polyreactivity and autoreactivity among HIV-1 antibodies. J. Virol. 2015;89:784–798. doi: 10.1128/JVI.02378-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascola JR, Haynes BF. HIV-1 neutralizing antibodies: understanding nature's pathways. Immunol. Rev. 2013;254:225–244. doi: 10.1111/imr.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald IK, Thornton JM. Satisfying hydrogen bonding potential in proteins. J. Mol. Biol. 1994;238:777–793. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
- McGuire AT, Hoot S, Dreyer AM, Lippy A, Stuart A, Cohen KW, Jardine J, Menis S, Scheid JF, West AP, et al. Engineering HIV envelope protein to activate germline B cell receptors of broadly neutralizing anti-CD4 binding site antibodies. J. Exp. Med. 2013;210:655–663. doi: 10.1084/jem.20122824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pugach P, Ozorowski G, Cupo A, Ringe R, Yasmeen A, de Val N, Derking R, Kim HJ, Korzun J, Golabek M, et al. A native-like SOSIP.664 trimer based on an HIV-1 subtype B env gene. J. Virol. 2015;89:3380–3395. doi: 10.1128/JVI.03473-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudicell RS, Do Kwon Y, Ko SY, Pegu A, Louder MK, Georgiev IS, Wu XL, Zhu J, Boyington JC, Chen XJ, et al. Enhanced Potency of a Broadly Neutralizing HIV-1 Antibody In Vitro Improves Protection against Lentiviral Infection In Vivo. J. Virol. 2014;88:12669–12682. doi: 10.1128/JVI.02213-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders RW, Derking R, Cupo A, Julien JP, Yasmeen A, de Val N, Kim HJ, Blattner C, de la Pena AT, Korzun J, et al. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 2013;9:e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheid JF, Mouquet H, Feldhahn N, Seaman MS, Velinzon K, Pietzsch J, Ott RG, Anthony RM, Zebroski H, Hurley A, et al. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature. 2009;458:636–640. doi: 10.1038/nature07930. [DOI] [PubMed] [Google Scholar]
- Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Oliveira TY, Pietzsch J, Fenyo D, Abadir A, Velinzon K, et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui ZW, Chen QJ, Wu R, Zhang HB, Zheng M, Wang HZ, Chen Z. Cross-protection against influenza virus infection by intranasal administration of M2-based vaccine with chitosan as an adjuvant. Arch. Virol. 2010;155:535–544. doi: 10.1007/s00705-010-0621-4. [DOI] [PubMed] [Google Scholar]
- Verkoczy L, Chen Y, Zhang J, Bouton-Verville H, Newman A, Lockwood B, Scearce RM, Montefiori DC, Dennison SM, Xia SM, et al. Induction of HIV-1 broad neutralizing antibodies in 2F5 knock-in mice: selection against membrane proximal external region-associated autoreactivity limits T-dependent responses. J. Immunol. 2013;191:2538–2550. doi: 10.4049/jimmunol.1300971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkoczy L, Diaz M, Holl TM, Ouyang YB, Bouton-Verville H, Alam SM, Liao HX, Kelsoe G, Haynes BF. Autoreactivity in an HIV-1 broadly reactive neutralizing antibody variable region heavy chain induces immunologic tolerance. Proc. Natl. Acad. Sci. U. S. A. 2010;107:181–186. doi: 10.1073/pnas.0912914107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkoczy L, Kelsoe G, Moody MA, Haynes BF. Role of immune mechanisms in induction of HIV-1 broadly neutralizing antibodies. Curr. Opin. Immunol. 2011;23:383–390. doi: 10.1016/j.coi.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Zhou T, Zhu J, Zhang B, Georgiev I, Wang C, Chen X, Longo NS, Louder M, McKee K, et al. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333:1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Holl TM, Liu Y, Li Y, Lu X, Nicely NI, Kepler TB, Alam SM, Liao HX, Cain DW, et al. Identification of autoantigens recognized by the 2F5 and 4E10 broadly neutralizing HIV-1 antibodies. J. Exp. Med. 2013;210:241–256. doi: 10.1084/jem.20121977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, Kwon YD, Scheid JF, Shi W, Xu L, et al. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010;329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Lynch RM, Chen L, Acharya P, Wu X, Doria-Rose NA, Joyce MG, Lingwood D, Soto C, Bailer RT, et al. Structural Repertoire of HIV-1-Neutralizing Antibodies Targeting the CD4 Supersite in 14 Donors. Cell. 2015;161:1280–1292. doi: 10.1016/j.cell.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.