Abstract
Although dynamic nuclear polarization (DNP) has dramatically enhanced solid-state NMR spectral sensitivities of many synthetic materials and some biological macromolecules, recent studies of membrane-protein DNP using exogenously doped paramagnetic radicals as polarizing agents have reported varied and sometimes surprisingly limited enhancement factors. This motivated us to carry out a systematic evaluation of sample preparation protocols for optimizing the sensitivity of DNP NMR spectra of membrane-bound peptides and proteins at cryogenic temperatures of ~110 K. We show that mixing the radical with the membrane by direct titration instead of centrifugation gives a significant boost to DNP enhancement. We quantify the relative sensitivity enhancement between AMUPol and TOTAPOL, two commonly used radicals, and between deuterated and protonated lipid membranes. AMUPol shows ~4 fold higher sensitivity enhancement than TOTAPOL, while deuterated lipid membrane does not give net higher sensitivity for the membrane peptides than protonated membrane. Overall, a ~100 fold enhancement between the microwave-on and microwave-off spectra can be achieved on lipid-rich membranes containing conformationally disordered peptides, and absolute sensitivity gains of 105–160 can be obtained between low-temperature DNP spectra and high-temperature non-DNP spectra. We also measured the paramagnetic relaxation enhancement of lipid signals by TOTAPOL and AMUPol, to determine the depths of these two radicals in the lipid bilayer. Our data indicate a bimodal distribution of both radicals, a surface-bound fraction and a membrane-bound fraction where the nitroxides lie at ~10 Å from the membrane surface. TOTAPOL appears to have a higher membrane-embedded fraction than AMUPol. These results should be useful for membrane-protein solid-state NMR studies under DNP conditions and provide insights into how biradicals interact with phospholipid membranes.
Introduction
High-field dynamic nuclear polarization (DNP) is a powerful technique to enhance the sensitivity of solid-state NMR (SSNMR) spectroscopy (Can et al., 2015; Hall et al., 1997; Maly et al., 2008; Ni et al., 2013). By microwave irradiation of electron paramagnetic resonance (EPR) transitions of stable radicals, the large electron-spin polarization is transferred to the surrounding nuclear spins, resulting in sensitivity enhancements that are theoretically equal to the ratio of the electron and nuclear spin gyromagnetic ratios (Carver and Slichter, 1956). Thus, for 13C NMR spectra measured with 1H-13C cross polarization (CP), the maximum enhancement factor is ~660, the ratio of the electron and proton gyromagnetic ratios. Experimentally, enhancement factors of 250–300 have been obtained on model compounds (Matsuki et al., 2009). These enhancement factors are empirically measured as the intensity ratios of spectra obtained with and without microwave (MW) irradiation. The two-orders-of-magnitude enhancements are achieved in practice using a number of crucial elements: a paramagnetic polarizing agent in the form of a stable radical, a high-power and high-frequency microwave source (Bajaj et al., 2007; Barnes et al., 2008; Becerra et al., 1993; Gerfen et al., 1995; Rosay et al., 2010), and low temperature to slow down electron and nuclear spin relaxation. A wide variety of mono- and bi-radicals have been designed and synthesized (Kubicki et al., 2016; Michaelis et al., 2014), with the two most commonly used ones being TOTAPOL and AMUPol, which contain two nitroxide radicals separated by ~13 Å via intervening functional groups with varying lengths, rigidity and polarity (Hu et al., 2008; Hu et al., 2004; Sauvee et al., 2013; Song et al., 2006). At low temperatures of 90–120 K commonly used for DNP SSNMR experiments, a cryoprotecting solution is often used to distribute the exogenous radical uniformly in the sample and to prevent ice formation at low temperature in hydrated biological samples. The most common DNP cryoprotectant solution consists of d6-glycerol/D2O/H2O (60/30/10 by volume), but other compounds such as DMSO and different concentrations of the individual components have also been used. For mostly dry compounds, wetting the sample with the radical without a cryoprotecting solution has been shown to be effective (Takahashi et al., 2012).
The two-orders-of-magnitude sensitivity gain opens up a wide range of previously inaccessible biological macromolecules (Akbey et al., 2013; Sergeyev et al., 2011) and chemical systems (Rossini et al., 2013) for investigation by SSNMR. Membrane proteins represent a major class of molecules that stand to benefit from this sensitivity enhancement (Cheng and Han, 2013), since dilution of membrane proteins in the lipid matrix limits the sensitivity of conventional SSNMR experiments. However, recent reports of DNP applications to membrane-bound peptides and proteins have found enhancement factors that are often, surprisingly, well below those of non-membrane systems. With the exception of bacteriorhodopsin and channel rhodopsin, most membrane proteins yielded enhancement factors of ~2 to ~30, measured on commercial DNP spectrometers with 1H Larmor frequencies of 400–800 MHz. For example, enhancement factors are 1.7–3.5 for a lung surfactant peptide bound to a 50% deuterated DPPC/POPG membrane with 40 mM TOTAPOL as the polarizing agent (Smith et al., 2015). The potassium channel KcsA bound to asolectin membranes showed enhancement factors of 3–8 on an 800 MHz DNP spectrometer, with 5 mM TOTAPOL or 25 mM AMUPol as the polarizing agent (Koers et al., 2014). A neurotoxin bound to the nicotinic acetylcholine receptor in native membranes gave a sensitivity enhancement of ~12 at optimized TOTAPOL concentrations (Linden et al., 2011). A signal peptide bound to the Sec translocon in E. coli lipids yielded an enhancement factor of ~32, where 20 mM TOTAPOL was used as the polarizing agent (Reggie et al., 2011). Whole cells, cell envelopes, and native E. coli membranes enriched in specific membrane proteins showed enhancement factors of 20–30 (Jacso et al., 2012; Renault et al., 2012). 15N NMR spectra of oriented membranes without cryoprotectant showed an enhancement factor of ~18 (Salnikov et al., 2010).
The highest DNP sensitivity gain among membrane proteins is so far found for bacteriorhodopspin (Bajaj et al., 2009) and channel rhodopsin (Becker-Baldus et al., 2015), with enhancement factors of 43–62. Both proteins exist in dense and highly ordered arrays in lipid membranes, thus their high enhancement factors may be related to the special nature of these protein-rich assemblies. On the other hand, a recent DNP study of sensory rhodopsin using a 2:1 protein/lipid mass ratio gave an enhancement factor of ~15 (Voinov et al., 2015), more comparable to results of other membrane peptides and proteins.
Apart from the distribution of an exogenous polarizing agent to the target molecules via a cryoprotecting solution, paramagnetic dopants have also been covalently attached to the protein or lipid to produce site-specific sensitivity enhancements and to avoid the use of cryoprotectants, which take up sample volume and may be incompatible with the compounds of interest. However, the sensitivity gains using site-specifically tagged polarizing agents are so far not higher than the exogenously doped samples. For example, enhancement factors are 12–15 for MTSSL-tagged KcsA (van der Cruijsen et al., 2015), 3.5–6 for nitroxide-tagged gramicidin (Wylie et al., 2015), up to ~10 for spin-labeled lipids (Smith et al., 2015), and up to ~15 for ToSMTSL-tagged sensory rhodopspin (Voinov et al., 2015).
The significant variation and the often limited DNP enhancement factors of membrane proteins are generally believed to be partly due to unoptimal sample preparation protocols. Experimental parameters that may affect the DNP enhancement include the type and concentration of the polarizing agent, the composition of the cryoprotecting solution, the extent of mixing of the polarizing agent with the target molecule, deuteration level of the cryoprotectant and membrane matrix, and the extent of conformational disorder of the protein at low temperature, which affects linewidths and hence sensitivity. Among these factors, the mixing of the radical-containing cryoprotectant solution with the membrane merits particular attention. In a few studies, the lipid, protein, cryoprotectant and radical were mixed directly and centrifuged to obtain the membrane pellet (Smith et al., 2015). However, glycerol and deuterated water have significantly higher densities than protonated water. Thus, most proteoliposomes cannot be centrifuged down in the typical glycerol-rich DNP solution (Smith et al., 2015; Voinov et al., 2015), which would reduce radical distribution to the membrane. Many studies partly circumvented this problem by preforming the proteoliposomes in regular aqueous solution, then washing or incubating the hydrated membrane pellets in the high-density cryoprotectant-radical mixture, followed by a second centrifugation step to collect the radical-bound membrane (Andreas et al., 2013; Bajaj et al., 2009; Becker-Baldus et al., 2015; Mak-Jurkauskas et al., 2008).
In this study, we examine the effects of five sample preparation conditions on the sensitivity and resolution of DNP NMR spectra, with the goal of optimizing both. The five parameters are the radical-membrane mixing protocol, the membrane deuteration level, the relative merit of AMUPol and TOTAPOL, the relative merit of glycerol and DMSO as the cryoprotectant, and comparison between phosphocholine (PC) and phosphoethanolamine (PE) lipids (Lee and Hong, 2014). We next investigate the location of TOTAPOL and AMUPol with respect to the membrane using paramagnetic relaxation enhancement (PRE) effects at ambient temperature. Our data show that, with careful optimization of sample preparation protocols, one can obtain enhancement factors of ~100 in 13C CP-MAS spectra of lipid-rich membrane peptides that do not have very high structural order. When DNP spectra measured at ~110 K are compared with spectra measured at 243 K without polarizing agents or cryoprotectants, total sensitivity gains of 105–160 were found. We show that lipid deuteration does not have a net beneficial effect on the absolute sensitivity of the DNP spectra. Finally, we show that TOTAPOL and AMUPol both partition to the membrane at ~10 Å from the surface, but TOTAPOL has a higher inserted fraction, consistent with the different chemical structures and three-dimensional structures of these two radicals.
Materials and Method
Lipid membranes and membrane peptides
Several lipid membranes were used in this study: 1,2-dimyristoylsn-glycero-3-phosphocholine (DMPC), d54-DMPC, 1,2-dilauroyl-sn-glycero-3-phosphoethanolamine (DLPE), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), and a virus-mimetic (VM+) membrane mixture (Cady et al., 2011). The VM+ membrane contains 1-palmitoyl-2-oleoyl-sn-glycero-3-phophocholine (POPC), POPE, egg sphingomyelin (SM) and cholesterol (Chol) at molar ratios of 25.6% : 25.6% : 25.6% : 23%.
For single-component membranes, the phospholipids were suspended in pH 7.5 HEPES buffer (10 mM HEPES, 1mM EDTA, 0.1 mM NaN3) and freeze-thawed seven times between liquid nitrogen temperature and ambient temperature to produce homogeneous vesicles. The vesicle solution was centrifuged at 40,000 rpm using a Beckman SW60Ti rotor at 277 K overnight to form membrane pellets. For the VM+ membrane mixture, phospholipids and cholesterol were dissolved in chloroform while SM was dissolved in a chloroform/methanol mixture. The two solutions were mixed, most organic solvents were removed under nitrogen gas, and the mixture was vacuum-dried overnight. The dried lipid mixture was suspended in the pH 7.5 HEPES buffer, then subjected to the same freeze-thaw cycles and ultracentrifugation to obtain membrane pellets.
A D44A mutant of the influenza A M2 transmembrane peptide (M2TM, residues 22–46) was synthesized using Fmoc chemistry by PrimmBiotech (Cambridge, MA). Uniformly 13C, 15N-labeled residues were incorporated at L26, V27, S31, G34, and A44. The peptide was dissolved in octyl-β-D-glucoside (OG) and mixed with DMPC vesicles in 10 mM pH 7.5 HEPES buffer. The resulting proteoliposomes were incubated at room temperature for ~3 h, then dialyzed against 10 mM pH 7.5 HEPES buffer for 3 days with 2 buffer changes per day to remove OG. The dialyzed proteoliposomes were centrifuged at 40,000 rpm overnight to obtain membrane pellets.
DNP sample preparation
Stock solutions of d8-glycerol/D2O/H2O (60/30/10 by volume) and d6-DMSO/D2O/H2O (60/30/10 by volume) containing 10 mM of TOTAPOL or AMUPol were prepared. Two methods, centrifugation and direct titration, were used to prepare membrane samples for DNP. The centrifugation method was used to prepare the DMPC-bound M2TM and DMPC-bound ROCKER samples (samples 4 and 6 in Table 1). Briefly, 100 μL of stock solution was added to the membrane pellets. The pellets were allowed to equilibrate at room temperature for an hour, then spun at 7,000 rpm for 5 minutes using a desktop centrifuge. The bulk solution was pipetted out, and the membrane sample was incubated in a desiccator until it reached 40wt% hydration. The titration method was used to prepare all other samples (samples 1–3, 5 and 7–9). Briefly, small aliquots of the stock solution were directly titrated into the proteoliposome pellets. The pellets were vortexed to ensure uniform distribution of the radicals. An appropriate amount of D2O was added to the pellet to reach the desired D2O/H2O ratio of 3 : 1. Excess water was then removed by 3 times of 10-minute lyophilization to reach a hydration level of ~40wt%.
Table 1.
Measured DNP enhancement factors εC,CP at 400 MHz.
| Samples | Varied parameters | Sample number | Cryoprotectant and biradical | εC,CP ≡ IMW on/IMW off |
|---|---|---|---|---|
| M2TM in DMPC | d54-DMPC | 1 | d8-glycerol, 10 mM AMUPol |
80 (C′), 100 (Cα), 69 (glycerol) 60 (C′), 56 (Cα), 56 (lipid CH2), 78 (glycerol) |
| 1H-DMPC | 2 | |||
| M2TM in 1H-DMPC | d8-glycerol | 2 | 10 mM AMUPol |
60 (C′), 56 (Cα), 56 (lipid CH2), 78 (glycerol) 39 (C′), 39 (Cα), 38 (lipid CH2), 63 (DMSO) |
| d6-DMSO | 3 | |||
| M2TM in 1H-DMPC | Titration | 3 | d6-DMSO, 10 mM AMUPol |
39 (C′), 39 (Cα), 63 (DMSO) 22 (C′), 27 (Cα), 50 (DMSO) |
| Centrifugation | 4 | |||
| ROCKER in 1H-DMPC | Titration | 5 | d6-DMSO, 10 mM AMUPol |
26 (C′), 40 (Cα), 45 (lipid CH2), 85 (DMSO) 19 (C′), 14 (Cα), 13 (lipid CH2), 22 (DMSO) |
| Centrifugation | 6 | |||
| VM+ membrane | 10 mM AMUPol | 7 | d8-glycerol | 42 (lipid CH2), 60 (glycerol) 10 (lipid CH2), 20 (glycerol) |
| 10 mM TOTAPOL | 8 | |||
| M2TM | 1H-DMPC | 2 | d8-glycerol, 10 mM AMUPol |
60 (C′), 56 (Cα), 56 (lipid CH2), 78 (glycerol) |
| 1H-DLPE | 9 |
58 (C′), 75 (Cα), 64 (lipid CH2), 78 (glycerol) |
A previously prepared DMPC-bound ROCKER sample (Joh et al., 2014) was converted for DNP experiments in two steps. First, the hydrated membrane was resuspended in 100 μL d6-DMSO/D2O/H2O containing 10 mM AMUPol, then spun at 7,000 rpm for 5 minutes to obtain a radical-bound sample (sample 6). After DNP experiments, this sample was resuspended in 4 mL of 10 mM pH 7.5 HEPES buffer and centrifuged at 40,000 rpm at 277 K overnight to remove most of the radical and cryoprotectant. After washing, ~5 μL of the same d6-DMSO/D2O/H2O solution containing 10 mM AMUPol was titrated into the pellet and the sample was measured again (sample 5). The two samples were compared to assess the titration and centrifugation methods for mixing the radical with the membrane (Table 1).
Several M2TM samples were prepared, differing in the cryoprotectants (d8-glycerol or d6-DMSO), radical-mixing protocols (titration or centrifugation), lipid deuteration, and lipid headgroup structure (samples 1–4 and 9). One M2TM sample was reconstituted into the d54-DMPC membrane, and d8-glycerol/D2O/H2O containing 10 mM AMUPol was titrated into the membrane (sample 1). The second sample was bound to protonated DMPC, and d8-glycerol/D2O/H2O containing 10 mM AMUPol was titrated into the membrane (sample 2). The third and fourth samples involved protonated DMPC, d6-DMSO/D2O/H2O as the cryoprotectant, AMUPol as the polarizing agent, and the titration method and centrifugation method for radical mixing were compared (samples 3 and 4). The fifth sample bound M2TM in DLPE membranes, and used d8-glycerol/D2O/H2O as the cryoprotectant and AMUPol as the polarizing agent (sample 9).
Solid-state NMR experiments with and without DNP
Low-temperature DNP experiments were performed on a 400 MHz and 600 MHz wide-bore SSNMR spectrometer equipped with a 263 GHz and 395 GHz gyrotron, respectively (Bruker, Billerica). The cathode currents of the gyrotron were 120–140 mA. All spectra were measured using a 3.2 mm 1H/13C/15N MAS probe with an MAS frequency of 8 kHz. Unless otherwise specified, the sample temperatures were ~105 K with the MW off and 113–120 K with the MW on. 1H T1 relaxation times were measured using the inversion recovery experiment. The recycle delay was 5 s for all 1D experiments and 3 s for 2D measurements. Thus, the enhancement factors reported here are steady-state values. Most 1D 13C spectra comparing the MW on and off conditions were measured using 128 scans.
Ambient-temperature 1D 13C and 1H MAS spectra for PRE studies of radical localization were measured on a 400 MHz NMR spectrometer. The spinning frequency was 5 kHz for DMPC and 7 kHz for VM+ samples. Typical radiofrequency field strengths were 83 kHz for 1H and 62.5 kHz for 13C. All 13C chemical shifts were externally referenced to the adamantane CH2 peak at 38.48 ppm on the TMS scale.
2D 13C-13C INADEQUATE spectra and 2D 15N-13C correlation spectra were measured without DNP on DMPC-bound ROCKER (Joh et al., 2014) and DMPC-bound M2TM samples. These samples do not contain cryoprotectants or radicals. The ROCKER spectra were measured on a 600 MHz spectrometer at 233 K under 11 kHz MAS while the M2TM spectra were measured on a 400 MHz spectrometer at 283 K and 203 K under 7 kHz MAS.
Results and discussion
Factors that increase the DNP sensitivity gain
We examined five factors in membrane sample preparation to maximize the DNP sensitivity enhancement. The first parameter is the method of radical mixing with the membrane. So far most DNP studies of membrane proteins used samples prepared by washing preformed membrane pellets in a highly deuterated cryoprotectant solution containing the polarizing agent. This deuterated cryoprotectant mixture serves the purposes of distributing the radicals uniformly to the membrane and minimizing ice formation at low temperature. The solution is then centrifuged to collect the membrane pellet. However, there are two limitations to this sample preparation method. First, the efficiency of radical mixing with the membrane under centrifugation is not high and is likely sample-dependent. Glycerol, the most commonly used cryoprotectant, has a density of 1.26 g/cm3, while deuterated water has a density of 1.1 g/cm3, both of which are larger than the density of protonated water. Thus, lipid membranes with a low protein concentration may not be possible to spin down in this high-density cryoprotectant solution. Indeed, the membrane-on-top and cryoprotectant-at-bottom phenomenon has been reported (Andreas et al., 2013; Voinov et al., 2015). Second, the centrifugation approach makes it difficult to quantify the radical concentration, the hydration level and the solvent composition of the final sample. Thus, we explored the alternative method of direct titration of the cryoprotectant solution to the membrane pellet. Additional D2O was added to the hydrated membrane first to yield the desired D2O/H2O ratio. After titration, the membrane mixture was vigorously vortexed and then incubated for an hour to allow homogeneous mixing. The membrane was then subjected to short periods of lyophilization to reach 40 wt% water and a radical concentration of 10 mM. A 31P static spectrum of one of these samples shows a uniaxial powder pattern expected for lamellar bilayers, indicating that the short lyophilization periods and low-temperature experiments do not disrupt the membrane integrity (data not shown).
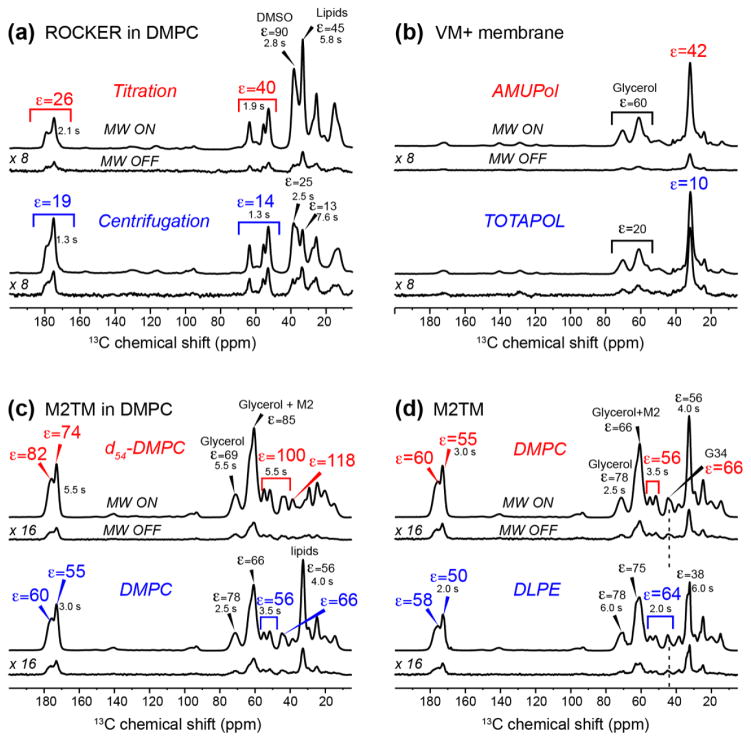
Fig. 1a shows the 13C CP-MAS spectra of DMPC-bound ROCKER sample prepared by the direct titration or centrifugation methods for preformed vesicles (samples 5 and 6). The DMPC membrane is protonated, and d6-DMSO/D2O/H2O containing 10 mM AMUPol was used as the cryoprotectant and polarizing agent, respectively. The enhancement factor εC,CP ≡ IMW on IMW off is 26–45 for the titration method and 14–19 for the centrifugation method (Table 1). Thus, the titration method gives 1.5–2.5 higher sensitivity gain than the centrifugation method. The sensitivities of the MW-off 13C spectra are not appreciably different between the centrifuged and titrated samples, thus ruling out low sensitivity of the MW-off spectra as the reason for the higher enhancement factor of the titration method. A similar increase of 1.5 fold by titration over centrifugation was also observed for membrane-bound M2TM (data not shown). The two ROCKER spectra showed much higher enhancement factors for the DMSO signals than the peptide signals, indicating that the radical is not fully dispersed to the lipid membrane and remains significantly confined to the cryoprotectant solution.
Figure 1.
1D 13C CP-MAS spectra of lipid membrane with or without peptides to determine the optimal sample preparation methods for DNP sensitivity enhancement. 1H T1 values are given for key signals. (a) Effects of the radical mixing protocol on sensitivity enhancement. ROCKER peptide bound to 1H-DMPC protected with d6-DMSO and containing 10 mM AMUPol was examined (samples 5 and 6). (b) Comparison of AMUPol and TOTAPOL for sensitivity enhancement. VM+ membrane protected with d8-glycerol and containing 10 mM AMUPol or TOTAPOL are compared (samples 7 and 8). (c) Effects of lipid deuteration on sensitivity enhancement. The spectra of D44A-M2TM bound to protonated DMPC membrane versus d54-DMPC membrane are compared. Both samples are protected with d8-glycerol and contain 10 mM AMUPol (samples 1 and 2). (d) Effects of lipid headgroup structure on spectral resolution. M2TM in DMPC or DLPE membranes protected with d8-glycerol and containing 10 mM AMUPol are compared (samples 2 and 9).
The second factor we examined is the relative enhancement by AMUPol versus TOTAPOL. AMUPol was designed to have longer electron relaxation times, larger electron-electron dipole couplings, and higher aqueous solubility than TOTAPOL, which facilitate polarization transfer (Sauvee et al., 2013). Initial demonstration on proline confirmed the design principle. We compared the enhancement factors due to AMUPol and TOTAPOL by observing the natural abundance 13C CP-MAS spectra of the VM+ membrane protected by d8-glycerol/D2O/H2O (Fig. 1b, samples 7 and 8). The lipid CH2 signal exhibited an εC,CP of 10 for the TOTAPOL-doped sample and 42 for the AMUPol-doped sample, corresponding to a 4-fold larger enhancement, in good agreement with the model compound results (Sauvee et al., 2013). Comparison of the MW-off spectra of the two samples again shows similar sensitivities, thus the higher εC,CP of AMUPol reflects true increase in the sensitivity of the MW-on spectra rather than low sensitivity of the MW-off spectra. This result differs from a recent study of the nuclear depolarization effects by these two radicals (Mentink-Vigier et al., 2015), as measured on the model compound urea. That study found that AMUPol caused a 2-fold large depolarization (i.e. lower sensitivity of the MW-off spectra) than TOTAPOL, so that the actual sensitivity of the MW-on spectra of the AMUPol-bound urea is 2-fold rather than 4-fold higher than that of TOTAPOL-bound urea. We attribute our finding that the MW-on spectra of AMUPol-bound membranes have ~4-fold higher sensitivity than TOTAPOL-bound membranes to the fact that for phospholipid membranes, other factors such as radical mixing with the membrane significantly come into play to produce the total spectral sensitivity, and TOTAPOL may perform less well in these other aspects.
The third factor we investigated for sensitivity enhancement is the 1H density of the heterogeneous system comprising the cryoprotectant, water, lipids, and the protein. For a given concentration of the polarizing agent, the sensitivity enhancement of 13C CP-MAS spectra depends on the 1H density of the system. An insufficient 1H concentration may compromise 1H-1H spin diffusion that relays the electron polarization to the nuclei, while an excessive 1H concentration in the solvent and lipids may reduce the amount of polarization transferred to the protein (Hu et al., 2008; Wylie et al., 2015). To investigate the effect of the environmental protonation level on the DNP enhancement, we measured the 13C CP-MAS spectra of M2TM bound to chain-perdeuterated DMPC (d54-DMPC) versus regular protonated DMPC (samples 1 and 2). The same protein and lipid masses of 2 mg and 10 mg were used in the two samples. Fig. 1c shows M2TM εC,CP values of up to ~118 for the deuterated membrane and up to ~66 for the protonated membrane. To our knowledge, the former is the highest enhancement factor reported so far for a membrane peptide at magnetic fields of 400 MHz or higher. However, when the MW-on spectra are compared, the deuterated and protonated samples show similar sensitivities, while the MW-off spectrum of the protonated DMPC sample shows 2-fold higher peptide 13C intensities compared to the deuterated sample. Since the peptide mass is similar in the two samples, this result indicates that the deuterated DMPC reduces the 1H-13C CP intensities of the bound peptide in the MW-off spectrum, thus giving rise to the higher εC,CP. In other words, while the deuterated membrane facilitated electron polarization transfer to the peptide, the lower lipid 1H density reduced the 1H-13C CP efficiency, thus the total sensitivity of the MW-on spectra of the peptide in the deuterated membrane is similar to that in the protonated membrane. The implication is that lipid 1H spins increases the 13C sensitivities of the embedded peptides by transferring their magnetization to the peptide protons during CP. The lipid CH2 signals confirm that lipid deuteration does not improve the DNP sensitivity enhancement. For example, a protonated VM+ membrane gave an enhancement factor of ~42 for the lipid CH2, while a partially deuterated virus-mimetic membrane containing d31-POPE and d31-POPC gave a moderately lower enhancement factor of ~30 (data not shown).
The fourth factor we investigated is the relative merit of PC and PE lipids for maintaining spectral resolution at low temperature. Recently we showed that down to ~200 K, PE lipids give narrower linewidths than PC and saturated-chain lipids give higher spectral resolution than unsaturated lipids (Lee and Hong, 2014). To investigate whether this resolution difference persists to ~100 K, we compared the 13C CP-MAS spectra of M2TM bound to DMPC and DLPE membranes (samples 2 and 9). Both membranes were protected with glycerol and used AMUPol as the polarizing agent. Fig. 1d shows that similar enhancement factors of ~60 were obtained for the peptide signals, and most 13C linewidths are also similar, except for the G34 Cα signal, which is narrower in the DLPE sample. G34 is known to be a conformational switch in M2TM and exhibits multiple chemical shifts depending on the membrane thickness, drug binding, and pH (Cady et al., 2011; Hu et al., 2011; Luo et al., 2009). The narrower linewidth of G34 in the DLPE membrane thus indicates the beneficial effect of this membrane for reducing protein conformational distribution.
Finally, we compared the merits of glycerol and DMSO for membrane cryoprotection and DNP sensitivity enhancement (Yu and Quinn, 1998). At or above 200 K, our recent study showed that DMSO resulted in much narrower lipid NMR lines than glycerol (Lee and Hong, 2014). We prepared two DMPC-bound M2TM samples (samples 2 and 3), one protected with d8-glycerol/D2O/H2O (60/30/10 by volume) and the other with d6-DMSO/D2O/H2O (40/50/10 by volume). Enhancement factors of ~55 were found for the glycerol-protected sample and ~40 for the DMSO-protected sample (Table 1). The poorer performance of DMSO can be attributed to the higher viscosity of the DMSO-protected membrane, even though the DMSO/water solution itself is more fluid than the glycerol/water mixture. Thus, while DMSO better maintains the conformational homogeneity of lipid membranes, it has the disadvantage of interfering with radical distribution. This is also supported by the fact that the measured 1H T1 values are less homogeneous in the DMSO-protected compared to the T1 values measured in glycerol-protected membranes (Fig. 1a, c).
While intensity ratios εC,CP of MW-on and MW-off spectra provide a simple way of reporting DNP sensitivity enhancement, it is now recognized that these values can be artificially elevated by reduced sensitivity of the MW-off spectra compared to undoped samples due to paramagnetic quenching and nuclear depolarization under MAS (Mentink-Vigier et al., 2015; Thurber and Tycko, 2014). Further, higher-temperature spectra generally have narrower linewidths, which benefit sensitivity. To evaluate the true sensitivity gains compared to conventional SSNMR experiments (Rossini et al., 2013), we compared DNP spectra measured at 110–120 K with non-DNP spectra measured at 243 K on unprotected and undoped membranes. The spectral intensities were normalized to the same protein mass and number of scans. Table 2 shows that at 400 MHz, the absolute sensitivity gains, Σlow T/high T, are 105–160 for glycerol-protected membranes, and 42–62 for DMSO-protected samples. When the field strength increased to 600 MHz, Σlow T/high T is less pronounced but still high, 62–68. The reduction of sensitivity gain from 400 to 600 MHz is consistent with the known dependence of the main DNP mechanism, the cross effect, on the field strength (Can et al., 2015).
Table 2.
Sensitivity enhancements Σlow T/high T of 13C CP-MAS spectra measured at 113–120 K with DNP compared to spectra measured at 243 K without DNP, cryoprotectants nor radicals.
| Samples | Σlow T/high T | 1H Larmor frequency |
|---|---|---|
| M2TM in d54-DMPC with d8-glycerol and 10 mM AMUPol, compared to M2TM in 1H-DMPC |
105 – 127 62 – 68 |
400 MHz 600 MHz |
| M2TM in 1H-DMPC with d8-glycerol and 10 mM AMUPol, compared to M2TM in 1H-DMPC |
112 – 160 | 400 MHz |
| M2TM in 1H-DMPC with d6-DMSO and 10 mM AMUPol, compared to M2TM in 1H-DMPC |
42 – 62 | 400 MHz |
Resolution of DNP spectra at low temperature
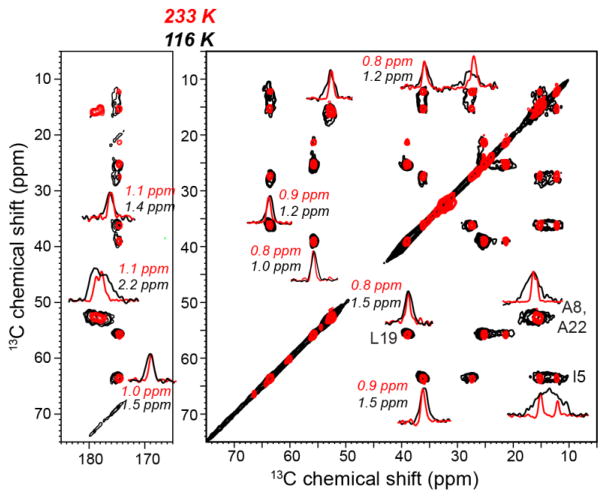
Fig. 2 compares the 2D 13C-13C PDSD correlation spectra of DMPC-bound ROCKER at 116 K with DNP and at 233 K without DNP. The 233 K spectrum was measured on a sample without radical and cryoprotectant. ROCKER forms a four-helix bundle in lipid bilayers to co-transport Zn2+ and H+ (Joh et al., 2014). 1D cross sections of representative cross peaks are shown. The linewidths of non-methyl Cα and Cβ peaks broadened from 0.8–1.1 ppm at 233 K to 1.0–1.5 ppm at 116 K, while the two Ala methyl 13C signals broadened more significantly, from 0.7–1.2 ppm at 233 K to 3.3 ppm at 116 K. This methyl broadening is due to the well-known phenomenon of methyl rotation occurring at rates that are comparable to the 1H decoupling field strength, thus interfering with 1H decoupling (Bajaj et al., 2009; Franks et al., 2005). Thus, the line broadening due to increased static conformational disorder is 0.2–0.7 ppm.
Figure 2.
2D 13C-13C PDSD spectra of DMPC-bound ROCKER at 116 K with DNP (black) and at 233 K without DNP (red). The spectra were measured on a 600 MHz spectrometer on sample 5. Selected 1D cross sections of cross peaks are shown. Typical linewidths at 116 K are 1.0–1.5 ppm, which are 0.2–0.7 ppm broader than the linewidths at 233 K. The 2D spectra are plotted using Topspin contour parameters of lev0=5, nlev=16, and toplev=100.
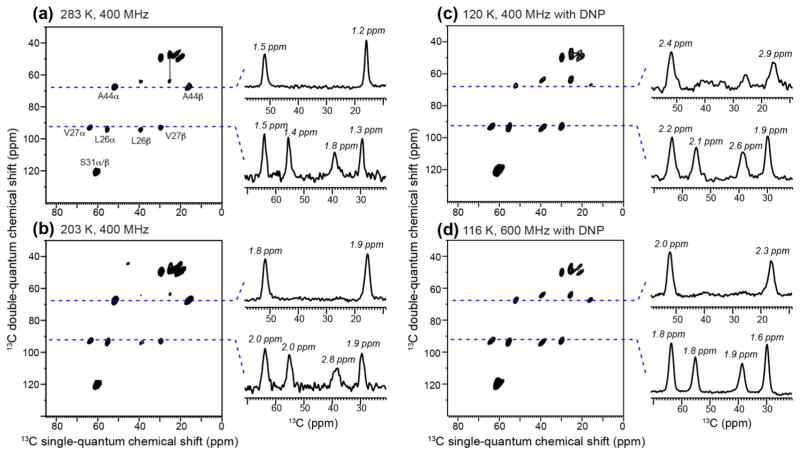
Fig. 3 compares the 2D 13C-13C dipolar INADEQUATE spectra of M2TM bound to DMPC bilayers measured at different temperatures and magnetic field strengths. At 283 K on a 400 MHz spectrometer, 13C linewidths of 1.2–1.8 ppm were observed. Decreasing the temperature to 203 K increased the linewidths to 1.8–2.8 ppm, and the largest line broadening is seen at the L26 Cβ signal. Decreasing the temperature further to 116 K while increasing the magnetic field strength to 600 MHz resulted in similar linewidths to those of the 203 K 400 MHz spectrum, except for the A44 Cβ methyl signal, which broadened due to motional interference. Interestingly, the L26 Cβ peak is sharper at 116 K and 600 MHz than at 203 K and 400 MHz. Thus, the low-temperature line broadening due to conformational heterogeneity is offset by the use of higher field strengths, indicating that the field-independent line broadening mechanisms are significant at 400 MHz. Finally, at 120 K on a 400 MHz DNP spectrometer, the non-methyl 13C linewidths range from 1.9 to 2.6 ppm, corresponding to a line broadening of 0.7–1.0 ppm compared to the 283 K situation.
Figure 3.
2D 13C-13C dipolar INADEQUATE spectra of D44A-M2TM in DMPC bilayers. (a) Spectrum measured at 283 K on a 400 MHz spectrometer. (b) Spectrum measured at 203 K on a 400 MHz spectrometer. (c) Spectrum measured at 120 K using DNP on a 400 MHz spectrometer. (d) Spectrum measured at 116 K using DNP on a 600 MHz spectrometer. The sample for (a–b) does not contain cryoprotectant or radicals and used protonated DMPC. The sample for (c–d) used glycerol-protected d54-DMPC and 10 mM AMUPol (sample 1). The lowest contour level for all 2D spectra is at 15% of the highest intensity of each spectrum, and 24 contour levels are shown. Representative 1D cross sections are extracted to compare 13C linewidths. The non-methyl 13C signals broadened by 0.1–0.4 ppm from the 283 K 400 MHz spectrum to the 116 K 600 MHz DNP spectrum.
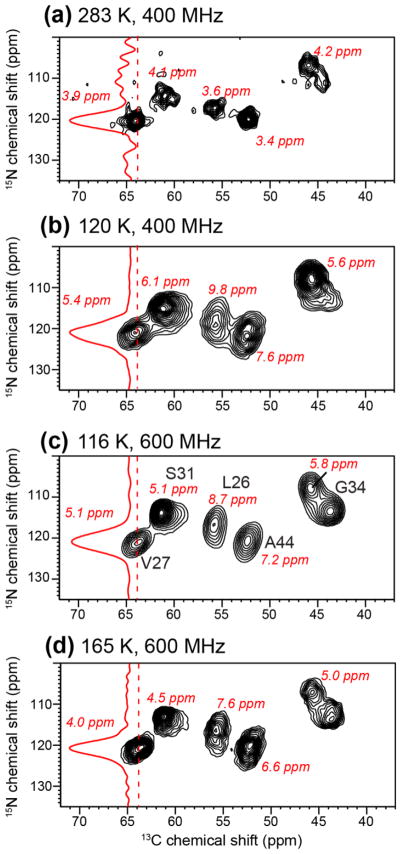
It is well known that 15N chemical shifts are more sensitive than 13C chemical shifts to conformational disorder. To examine the effects of cryogenic temperature on 15N linewidths, we compared the 2D 15N-13C correlation spectra of membrane-bound M2TM at different temperatures and field strengths with and without DNP. Fig. 4a shows 15N linewidths of 3.4–4.3 ppm at 283 K on a 400 MHz spectrometer in the absence of radicals. Decreasing the temperatures to 120 K broadened the linewidths to 5.4–9.8 ppm (Fig. 4b). The extent of line broadening is variable and residue-specific. Increasing the field strength to 600 MHz did not significantly improve the 15N resolution (Fig. 4c), giving linewidths of 5.1–8.7 ppm, indicating that the main contribution to 15N linewidths is roughly constant in ppm and thus field-dependent. Increasing the temperature to 165 K while maintaining the 600 MHz field reduced the 15N linewidths to 4.1–7.6 ppm, which are still larger than the linewidths measured at 283 K at lower field. The pore-facing V27 and G34 residues showed less line broadening between 165 K and 283 K, while the lipid-facing and interfacial residues, L26 and A44, showed the largest line broadening at low temperature. These results suggest that lipid disorder is the largest source of line broadening to membrane peptides, while disorder at the water-protein interface appears to be smaller. Since M2TM is a small four-helix bundle with a significant lipid interface, this result suggests that larger ion channels and membrane protein complexes with large protein-protein interfaces should better maintain the spectral resolution at ~110 K.
Figure 4.
2D 15N-13C correlation spectra of membrane-bound M2TM under different conditions to examine 15N spectral resolution. (a) Spectrum of DMPC-bound M2TM at 283 K without DNP on a 400 MHz spectrometer. (b) Spectrum of DLPE-bound M2TM at 120 K measured on a 400 MHz DNP spectrometer (sample 9). (c) Spectrum of d54-DMPC bound M2TM at 116 K measured on a 600 MHz DNP spectrometer (sample 1). (d) Spectrum of d54-DMPC bound M2TM at 165 K measured on a 600 MHz DNP spectrometer. The samples for (b–d) are cryoprotected with glycerol containing 10 mM AMUPol. All spectra were plotted with the lowest contour at 20% of the highest peak in each spectrum and 16 contour levels are plotted.
Radical distribution in lipid membranes
While AMUPol and TOTAPOL are extensively used for DNP SSNMR experiments, to our knowledge, the locations of these radicals in lipid membranes have not been reported. In principle, these paramagnetic organic radicals may be distributed non-uniformly in two ways: they may exhibit a concentration gradient along the bilayer normal due to their amphipathicity, and they may laterally cluster to certain molecules in a multi-component lipid membrane. Understanding the spatial distribution of these radicals is important for optimizing the DNP sensitivity enhancement.
We investigated the TOTAPOL and AMUPol distribution in lipid membranes by measuring the lipid 1H and 13C spectral intensities at ambient temperature to observe distance-dependent PRE (Buffy relaxation due to the biradical broadens the NMR et al., 2003; Chu et al., 2010). Faster nuclear-spin T2 signals and reduces peak intensities. For 13C CP-MAS experiments, the 13C intensities depend not only on 13C T2 relaxation times but also 1H T1ρ: enhanced T1ρ relaxation reduces the CP intensity. The intensity ratios, S/S0, between radical-bound samples (S) and radical-free samples (S0), give qualitative information about the distance of the radical from the lipid functional groups. Quantitatively, PRE, defined as the relaxation rate difference between the paramagnetic sample and the diamagnetic sample, is proportional to the inverse of the electron-nuclear distance to the 6th power, the electron-spin relaxation time, and the square of the nuclear spin gyromagnetic ratio γ (Bloembergen, 1957; Solomon, 1955). Thus, for 1H and 13C DP experiments, which reflect 1H and 13C T2 PRE, respectively, the 1H PRE effect should be ~16 fold larger than 13C for the same distances, thus causing much lower S/S0 values. In comparison, the S/S0 values from 13C CP spectra should be more comparable to the 1H S/S0 values, since both 1H T1ρ and 13C T2 PREs contribute to the 13C intensity reduction.
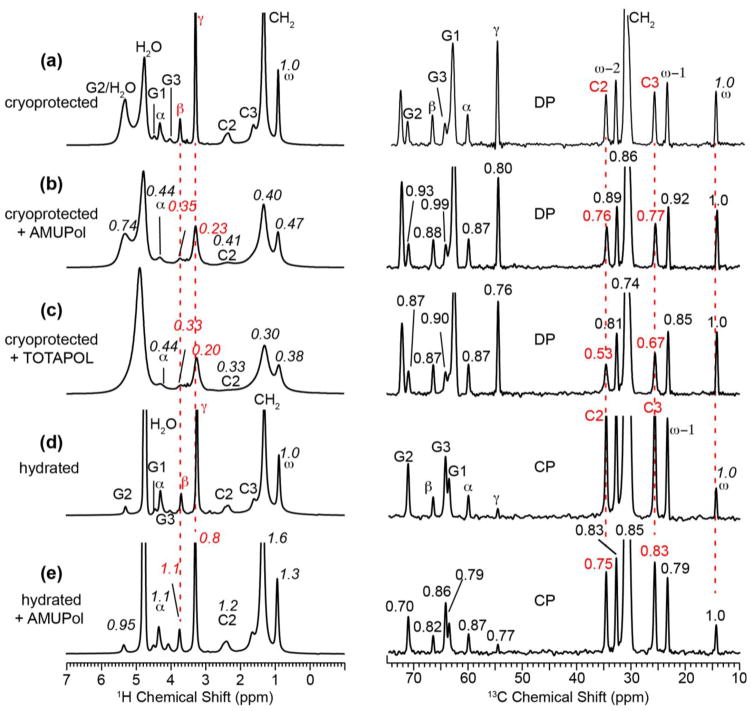
Fig. 5 shows the 1H and 13C MAS spectra of DMPC membranes with and without glycerol protection. The glycerol-protected spectra, measured with direct polarization (DP) for both 1H and 13C (Fig. 5a–c), give information about radical distribution in the presence of cryoprotectants, while the unprotected samples (Fig. 5d–e), measured with CP for 13C, give information about radical distribution without potential perturbation by cryoprotectants. The 1H S/S0 ratios are directly read off from the radical-bound and radical-free spectra, while the 13C S/S0 values are subjected to an additional normalization with respect to the maximum S/S0 value, which is found for the chain-end ω. The ω S/S0 value is slightly higher than 1 in some samples, indicating slightly different amounts of the various samples.
Figure 5.
Radical-induced PRE of DMPC membranes. Left column: 1H MAS spectra. Right column: 13C MAS spectra measured with DP (a–c) and CP (d–e). All spectra were measured at 298 K on a 400 MHz spectrometer. (a–c) Glycerol-protected DMPC membranes without radical (a), with 10 mM AMUPol (b), and with 10 mM TOTAPOL (c). (d–e) Hydrated DMPC membranes without cryoprotectants. (d) Without radical. (e) With 10 mM AMUPol. The S/S0 values in (b, c) are calculated with respect to the control spectra in (a), while the S/S0 values in (e) are calculated with respect to (d). The 13C S/S0 values are further normalized with respect to the chain-end ω peak at 14.2 ppm.
For the glycerol-protected membranes, AMUPol and TOTAPOL caused the largest intensity reduction to the lipid headgroup signals in the 1H spectra and the top of the acyl chains (C2 and C3) in the 13C spectra. For the 1H spectra, the intensity reduction is consistent with line broadening. For example, AMUPol broadened the headgroup Hβ and Hα signals by ~100 Hz and the acyl chain end ω signal by ~40 Hz, while the TOTAPOL sample broadened the headgroup Hβ and Hα signals by ~80 Hz and the ω signal by ~90 Hz (Fig. 5a,b). Overall, TOTAPOL caused stronger PRE to the acyl chains than AMUPol, as shown by the lower S/S0 values for TOTAPOL than AMUPol for the resolved acyl chain signals in the 13C DP spectra.
In the absence of glycerol, the residual intensities in the 1H spectra of the AMUPol-containing sample (Fig. 5e) are much higher than those of the glycerol-protected sample (Fig. 5b), indicating that AMUPol is less bound to the membrane in the absence of glycerol. Thus, glycerol facilitates radical mixing with the membrane. While this effect is expected at low temperature due to the antifreezing ability of glycerol, it is not immediately obvious at ambient temperature. We attribute this result to the ability of glycerol to partition to the membrane-water interface (Lee and Hong, 2014), thus carrying the radicals with it to the membrane.
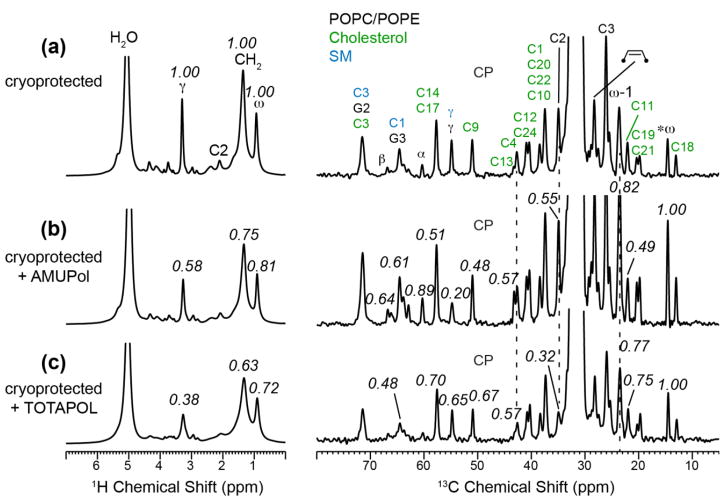
We also measured the PRE effect of glycerol-protected VM+ membrane to investigate if radical binding to the membrane depends on the lipid composition. Fig. 6 shows the 1H spectra and 13C CP-MAS spectra. The former reports 1H T2 PRE while the latter reports both 13C T2 PRE and 1H T1ρ PRE. Qualitatively, the observed S/S0 values are similar between the 1H and 13C spectra. For the AMUPol-bound sample, the lowest S/S0 value is observed for headgroup sites, while for the TOTAPOL-bound membrane, the lowest S/S0 values are found for the top of the acyl chains and the headgroup, similar to the result of TOTAPOL in DMPC membranes.
Figure 6.
Radical-induced PRE of glycerol-protected VM+ membranes. Left column: 1H spectra. Right column: 13C CP-MAS spectra. (a) Without radical. (b) With 10 mM AMUPol. (c) With 10 mM TOTAPOL. Peak assignments are given in (a) for phospholipids (black), SM (blue), and cholesterol (green). The 1H S/S0 values are with respect to the control spectrum (a), while the 13C S/S0 values are further normalized with respect to the ω peak.
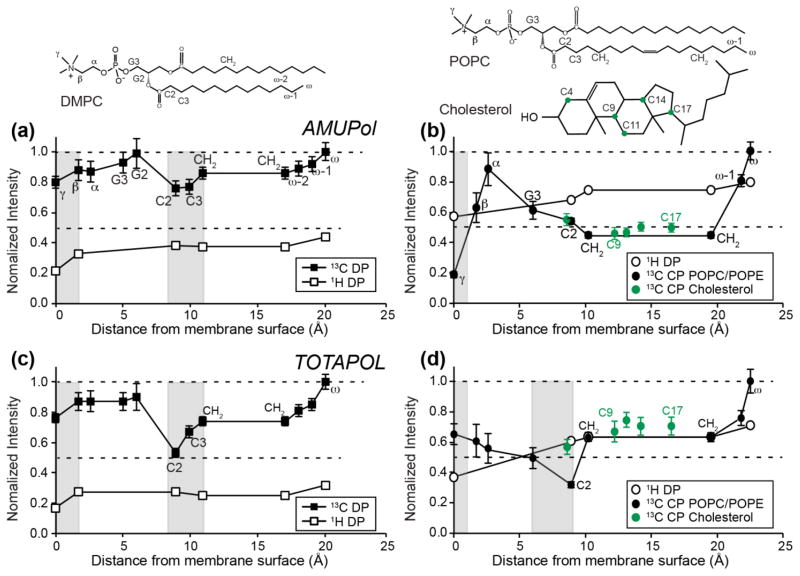
Fig. 7 summarizes the 1H and 13C S/S0 values of DMPC and VM+ membranes for AMUPol and TOTAPOL. The S/S0 values are plotted as a function of the distances of lipid functional groups from the membrane surface, using information obtained from joint analysis of the X-ray and neutron diffraction data of lipid membranes (White and Wimley, 1999). For the DMPC membrane, the 1H dephasing values are 0.2–0.4 while the 13C S/S0 values range from 0.5 to 1.0. This difference is qualitatively consistent with the lower γ of 13C spins than 1H and the dependence of the PRE on γ2. Two local minima in 13C S/S0 values are observed, one at the acyl chain C2 and C3, and the other at the headgroup γ. The 1H data showed one minimum, in the headgroup region. However, the 1H signals of C2 and C3 groups are not well resolved from the dominant CH2 peak in the radical-containing samples (Fig. 5b, c), thus these 1H intensities are not accurate.
Figure 7.
Residual intensities, S/S0, of radical-containing membranes. (a, c) DMPC membrane. (b, d) VM+ membrane. (a, b) 1H and 13C S/S0 values of membranes containing 10 mM AMUPol. (c, d) 1H and 13C S/S0 values of membranes containing 10 mM TOTAPOL. The x-axis shows the depths of lipid and cholesterol functional groups (Craven, 1976; Kucerka et al., 2005; Nagle and Tristram-Nagle, 2000), with the chemical structures of lipids and cholesterol drawn to scale along the x-axis. Shaded vertical bars indicate the positions of local S/S0 minima, which indicate the most likely radical positions. The error bars of 1H data points are less than 2%, thus are not plotted here.
Both AMUPol and TOTAPOL show two minima, but the TOTAPOL-bound sample has much lower C2 and C3 S/S0 values than AMUPol (Fig. 7a, c). Overall, TOTAPOL preferentially dephases the lipid chain signals more than the headgroup signals. Together, these data suggest that both radicals partition bimodally, with one fraction at ~10 Å from the membrane surface, where C2 and C3 lie, and the other fraction residing on the membrane surface. But TOTAPOL has a larger fraction inside the membrane than AMUPol, consistent with the higher solubility of AMUPol due to its tetraethylene glycol sidechain.
The glycerol-protected and AMUPol-doped VM+ membrane (Fig. 7b) shows a different 1H PRE profile from the corresponding DMPC sample. The lowest intensities are found for the headgroup Cγ and chain CH2 signals, but the Cγ intensities are lower than the CH2 intensities, indicating that more AMUPol is bound to the VM+ surface than to the VM+ interior. Between cholesterol and phospholipids, the 13C S/S0 values are similar, indicating an absence of lateral clustering of AMUPol. For the TOTAPOL-bound VM+ membrane, the 13C intensities are the lowest for the acyl chain C2, similar to the situation of the DMPC sample, indicating that TOTAPOL preferentially binds to the middle of the membrane, ~10 Å from the membrane surface. However, the headgroup Cγ S/S0 values are much higher in the 13C spectra than in the 1H spectra. Since 13C CP intensity of the mobile Cγ is sensitive to fluctuations in the spin-lock field strengths, the 1H S/S0 value is more reliable, thus we conclude that a fraction of TOTAPOL remains bound to the VM+ surface. Between the 1H PRE profiles of DMPC and VM+ membranes, the VM+ membrane exhibits higher residual intensities than the DMPC membrane, indicating that the radicals are on average less inserted into the complex membrane, suggesting that the higher viscosity of the cholesterol-containing membrane may obstruct radical insertion.
While the 1H spectra of the lipid membranes show clear line broadening that is consistent with the intensity reduction, the 13C linewidths are much less affected by the radical (Fig. 5, 6). This situation differs from the PRE effects of Mn2+ ions bound to the membrane surface (Buffy et al., 2003), where the 13C linewidths increased concomitantly to intensity reduction. We do not yet fully understand the limited 13C line broadening. One possible explanation is that at the low concentration of biradicals (10 mM) used, the average distances between each biradical molecule and the lipids are sufficiently long that only the 1H signals are uniformly affected by the PRE while the 13C spins may experience heterogeneous PRE, with the signals of lipids in the vicinity of the biradicals being suppressed while lipids far away from the biradicals being unaffected and thus manifesting signals with narrow linewidths similar to those of the diamagnetic sample. At ~40 wt% hydration, the estimated radical to lipid molar ratio is about 1 : 150, which is much lower than the Mn2+ to lipid molar ratios used in previous studies (Buffy et al., 2003). In addition to this dilution issue, the radical distribution in the membrane may be somewhat heterogeneous, which may impact the 13C PRE more than the 1H PRE.
To fully understand the membrane partitioning of TOTAPOL and AMUPol, one needs to take into account the conformations of these two molecules. Each compound contains two nitroxides with ROO distances of ~13 Å (Hu et al., 2008; Sauvee et al., 2013), as estimated from DFT calculations and EPR measurements. Depending on how these two molecules are oriented in the membrane, the two nitroxide spin labels may or may not lie at the same depths with respect to the membrane surface. Intuitively, we expect the polar tetraethylene glycol sidechain in AMUPol to “snorkel” to the membrane surface while the hydrophobic backbone lies inside the membrane, roughly parallel to the membrane plane. Molecular dynamics simulations may provide insights into the energetically favorable orientation and depth of these two molecules in the lipid membrane.
Conclusions
The data presented here show that DNP of membrane peptides and proteins can achieve MW on-off sensitivity enhancements εC,CP of ~100 fold and overall sensitivity enhancements Σlow T/high T of 105–160 fold under optimized sample preparation methods. The main protocols (Table 3) include titration of the radical-containing cryoprotectant solution to preformed membrane pellets, use of AMUPol in place of TOTAPOL, and use of glycerol in place of DMSO as the cryoprotectant. Deuterated lipids do not increase the absolute sensitivity of the MW-on spectra, as the benefit of targeting the electron polarization to the protonated protein is roughly offset by the lower 1H-13C CP efficiency of the protein due to the lower 1H density of the lipid matrix. Glycerol distributes the radicals to the membrane better than DMSO, even though at temperatures higher than ~200 K DMSO gives higher spectral resolution. PE lipids give better resolution than PC for disordered residues in proteins. Resolution remains a limitation in spectra measured from ~105 K to ~165 K, and improvements may require different freezing protocols.
Table 3.
Summary of the effects of sample preparation protocols on DNP sensitivity enhancement of membrane peptides. The optimal conditions are in italic.
| Parameters | Relative sensitivity enhancement |
|---|---|
| AMUPol vs TOTAPOL | AMUPol yields 4-fold higher εC,CP. |
| Titration vs centrifugation | Direct titration yields 1.5–2.5 fold higher εC,CP due to better mixing of the radical with the membrane. |
| Glycerol vs DMSO | Glycerol yields 1.5-fold higher ε C,CP. |
| Deuterated lipids vs protonated lipids | Perdeuterated lipids yield 1.5–2.0 times higher peptide εC,CP. However, perdeuterated lipids cause lower peptide 13C CP signals in the MW-off spectra, thus the sensitivities of the MW-on spectra are similar between protonated and deuterated membranes. |
| DMPC vs DLPE | DLPE gives sharper signals for disordered peptide residues. |
1H and 13C spectra of radical-bound membranes at ambient temperature indicate that AMUPol and TOTAPOL partition bimodally, with one fraction at ~10 Å from the membrane surface and the other fraction on the membrane surface. Based on the residual 1H and 13C intensities, a higher fraction of TOTAPOL binds inside the membrane than AMUPol. Radical binding to cholesterol-containing membranes is weaker than to simple PC membranes. More detailed information about the orientations and depths of these paramagnetic radicals in the lipid membrane will require further experiments, and may benefit from molecular dynamics simulations.
Acknowledgments
The authors thank Prof. Robert Griffin, Eric Keeler and Vladmir Michaelis for stimulating discussions. This work is supported by National Institutes of Health grants GM088204 and GM066976 to M.H.
References
- Akbey U, Franks WT, Linden A, Orwick-Rydmark M, Lange S, Oschkinat H. Dynamic nuclear polarization enhanced NMR in the solid-state. Top Curr Chem. 2013;338:181–228. doi: 10.1007/128_2013_436. [DOI] [PubMed] [Google Scholar]
- Andreas LB, Barnes AB, Corzilius B, Chou JJ, Miller EA, Caporini M, Rosay M, Griffin RG. Dynamic Nuclear Polarization Study of Inhibitor Binding to the M2(18–60) Proton Transporter from Influenza A. Biochemistry. 2013;52:2774–2782. doi: 10.1021/bi400150x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajaj VS, Hornstein MK, Kreischer KE, Sirigiri JR, Woskov PP, Mak-Jurkauskas ML, Herzfeld J, Temkin RJ, Griffin RG. 250 GHz CW gyrotron oscillator for dynamic nuclear polarization in biological solid state NMR. J Magn Reson. 2007;189:251–279. doi: 10.1016/j.jmr.2007.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajaj VS, Mak-Jurkauskas ML, Belenky M, Herzfeld J, Griffin RG. Functional and shunt states of bacteriorhodopsin resolved by 250 GHz dynamic nuclear polarization-enhanced solid-state NMR. Proc Natl Acad Sci USA. 2009;106:9244–9249. doi: 10.1073/pnas.0900908106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes AB, De Paepe G, van der Wel PCA, Hu KN, Joo CG, Bajaj VS, Mak-Jurkauskas ML, Sirigiri JR, Herzfeld J, Temkin RJ, Griffin RG. High-field dynamic nuclear polarization for solid and solution biological NMR. Appl Magn Reson. 2008;34:237–263. doi: 10.1007/s00723-008-0129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becerra LR, Gerfen GJ, Temkin RJ, Singel DJ, Griffin RG. Dynamic Nuclear-Polarization with a Cyclotron-Resonance Maser at 5 T. Phys Rev Lett. 1993;71:3561–3564. doi: 10.1103/PhysRevLett.71.3561. [DOI] [PubMed] [Google Scholar]
- Becker-Baldus J, Bamann C, Saxena K, Gustmann H, Brown LJ, Brown RCD, Reiter C, Bamberg E, Wachtveitl J, Schwalbe H, Glaubitz C. Enlightening the photoactive site of channelrhodopsin-2 by DNP-enhanced solid-state NMR spectroscopy. Proc Natl Acad Sci USA. 2015;112:9896–9901. doi: 10.1073/pnas.1507713112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloembergen N. Proton Relaxation Times in Paramagnetic Solutions - Comment. J of Chem Phys. 1957;27:595–596. [Google Scholar]
- Buffy JJ, Hong T, Yamaguchi S, Waring AJ, Lehrer RI, Hong M. Solid-state NMR investigation of the depth of insertion of protegrin-1 in lipid bilayers using paramagnetic Mn2+ Biophys J. 2003;85:2363–2373. doi: 10.1016/s0006-3495(03)74660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cady SD, Wang J, Wu Y, DeGrado WF, Hong M. Specific binding of adamantane drugs and direction of their polar amines in the pore of the influenza M2 transmembrane domain in lipid bilayers and dodecylphosphocholine micelles determined by NMR spectroscopy. J Am Chem Soc. 2011;133:4274–4284. doi: 10.1021/ja102581n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cady SD, Wang T, Hong M. Membrane-dependent effects of a cytoplasmic helix on the structure and drug binding of the influenza virus M2 protein. J Am Chem Soc. 2011;133:11572–11579. doi: 10.1021/ja202051n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Can TV, Ni QZ, Griffin RG. Mechanisms of dynamic nuclear polarization in insulating solids. J Magn Reson. 2015;253:23–35. doi: 10.1016/j.jmr.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver TR, Slichter CP. Experimental Verification of the Overhauser Nuclear Polarization Effect. Phys Rev. 1956;102:975–980. [Google Scholar]
- Cheng CY, Han SI. Dynamic Nuclear Polarization Methods in Solids and Solutions to Explore Membrane Proteins and Membrane Systems. Ann Rev Phys Chem. 2013;64:507–532. doi: 10.1146/annurev-physchem-040412-110028. [DOI] [PubMed] [Google Scholar]
- Chu S, Maltsev S, Emwas AH, Lorigan GA. Solid-state NMR paramagnetic relaxation enhancement immersion depth studies in phospholipid bilayers. J Magn Reson. 2010;207:89–94. doi: 10.1016/j.jmr.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven BM. Crystal–Structure of Cholesterol Monohydrate. Nature. 1976;260:727–729. doi: 10.1038/260727a0. [DOI] [PubMed] [Google Scholar]
- Franks WT, Zhou DH, Wylie BJ, Money BG, Graesser DT, Frericks HL, Sahota G, Rienstra CM. Magic-angle spinning solid-state NMR spectroscopy of the beta1 immunoglobulin binding domain of protein G (GB1): 15N and 13C chemical shift assignments and conformational analysis. J Am Chem Soc. 2005;127:12291–12305. doi: 10.1021/ja044497e. [DOI] [PubMed] [Google Scholar]
- Gerfen GJ, Becerra LR, Hall DA, Griffin RG, Temkin RJ, Singel DJ. High-Frequency (140 Ghz) Dynamic Nuclear-Polarization - Polarization Transfer to a Solute in Frozen Aqueous-Solution. J Chem Phys. 1995;102:9494–9497. [Google Scholar]
- Hall DA, Maus DC, Gerfen GJ, Inati SJ, Becerra LR, Dahlquist FW, Griffin RG. Polarization-enhanced NMR spectroscopy of biomolecules in frozen solution. Science. 1997;276:930–932. doi: 10.1126/science.276.5314.930. [DOI] [PubMed] [Google Scholar]
- Hu FH, Luo WB, Cady SD, Hong M. Conformational plasticity of the influenza A M2 transmembrane helix in lipid bilayers under varying pH, drug binding, and membrane thickness. Biochim Biophys Acta. 2011;1808:415–423. doi: 10.1016/j.bbamem.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu KN, Song C, Yu HH, Swager TM, Griffin RG. High-frequency dynamic nuclear polarization using biradicals: a multifrequency EPR lineshape analysis. J Chem Phys. 2008;128:052302. doi: 10.1063/1.2816783. [DOI] [PubMed] [Google Scholar]
- Hu KN, Yu HH, Swager TM, Griffin RG. Dynamic nuclear polarization with biradicals. J Am Chem Soc. 2004;126:10844–10845. doi: 10.1021/ja039749a. [DOI] [PubMed] [Google Scholar]
- Jacso T, Franks WT, Rose H, Fink U, Broecker J, Keller S, Oschkinat H, Reif B. Characterization of membrane proteins in isolated native cellular membranes by dynamic nuclear polarization solid-state NMR spectroscopy without purification and reconstitution. Angew Chem Int Ed Engl. 2012;51:432–435. doi: 10.1002/anie.201104987. [DOI] [PubMed] [Google Scholar]
- Joh NH, Wang T, Bhate MP, Acharya R, Wu Y, Grabe M, Hong M, Grigoryan G, DeGrado WF. De novo design of a transmembrane Zn2+-transporting four-helix bundle. Science. 2014;346:1520–1524. doi: 10.1126/science.1261172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koers EJ, van der Cruijsen EAW, Rosay M, Weingarth M, Prokofyev A, Sauvee C, Ouari O, van der Zwan J, Pongs O, Tordo P, Maas WE, Baldus M. NMR-based structural biology enhanced by dynamic nuclear polarization at high magnetic field. J Biomol NMR. 2014;60:157–168. doi: 10.1007/s10858-014-9865-8. [DOI] [PubMed] [Google Scholar]
- Kosen PA. Spin labeling of proteins. Methods Enzymol. 1989;177:86–121. doi: 10.1016/0076-6879(89)77007-5. [DOI] [PubMed] [Google Scholar]
- Kubicki DJ, Casano G, Schwarzwalder M, Abel S, Sauvee C, Ganesan K, Yulikov M, Rossini AJ, Jeschke G, Coperet C, Lesage A, Tordo P, Ouari O, Emsley L. Rational design of dinitroxide biradicals for efficient cross-effect dynamic nuclear polarization. Chem Sci. 2016 doi: 10.1039/c5sc02921j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucerka N, Liu YF, Chu NJ, Petrache HI, Tristram-Nagle ST, Nagle JF. Structure of fully hydrated fluid phase DMPC and DLPC lipid bilayers using X-ray scattering from oriented multilamellar arrays and from unilamellar vesicles. Biophys J. 2005;88:2626–2637. doi: 10.1529/biophysj.104.056606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M, Hong M. Cryoprotection of lipid membranes for high-resolution solid-state NMR studies of membrane peptides and proteins at low temperature. J Biomol NMR. 2014;59:263–277. doi: 10.1007/s10858-014-9845-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linden AH, Lange S, Franks WT, Akbey U, Specker E, van Rossum BJ, Oschkinat H. Neurotoxin II bound to acetylcholine receptors in native membranes studied by dynamic nuclear polarization NMR. J Am Chem Soc. 2011;133:19266–19269. doi: 10.1021/ja206999c. [DOI] [PubMed] [Google Scholar]
- Luo W, Cady SD, Hong M. Immobilization of the Influenza A M2 Transmembrane Peptide in Virus-Envelope Mimetic Lipid Membranes: A Solid-State NMR Investigation. Biochemistry. 2009;48:6361–6368. doi: 10.1021/bi900716s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak-Jurkauskas ML, Bajaj VS, Hornstein MK, Belenky M, Griffin RG, Herzfeld J. Energy transformations early in the bacteriorhodopsin photocycle revealed by DNP-enhanced solid-state NMR. Proc Natl Acad Sci USA. 2008;105:883–888. doi: 10.1073/pnas.0706156105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maly T, Debelouchina GT, Bajaj VS, Hu KN, Joo CG, Mak-Jurkauskas ML, Sirigiri JR, van der Wel PCA, Herzfeld J, Temkin RJ, Griffin RG. Dynamic nuclear polarization at high magnetic fields. J Chem Phys. 2008:128. doi: 10.1063/1.2833582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuki Y, Maly T, Ouari O, Karoui H, Le Moigne F, Rizzato E, Lyubenova S, Herzfeld J, Prisner T, Tordo P, Griffin RG. Dynamic Nuclear Polarization with a Rigid Biradical. Angew Chem Int Ed Engl. 2009;48:4996–5000. doi: 10.1002/anie.200805940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mentink-Vigier F, Paul S, Lee D, Feintuch A, Hediger S, Vega S, De Paëpe G. Nuclear depolarization and absolute sensitivity in magic-angle spinning cross effect dynamic nuclear polarization. Phys Chem Chem Phys. 2015;17:21824–21836. doi: 10.1039/c5cp03457d. [DOI] [PubMed] [Google Scholar]
- Michaelis VK, Ong TC, Kiesewetter MK, Frantz DK, Walish JJ, Ravera E, Luchinat C, Swager TM, Griffin RG. Topical Developments in High-Field Dynamic Nuclear Polarization. Isr J Chem. 2014;54:207–221. doi: 10.1002/ijch.201300126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadaud PS, Helmus JJ, Hofer N, Jaroniec CP. Long-range structural restraints in spin-labeled proteins probed by solid-state nuclear magnetic resonance spectroscopy. J Am Chem Soc. 2007;129:7502–7503. doi: 10.1021/ja072349t. [DOI] [PubMed] [Google Scholar]
- Nagle JF, Tristram-Nagle S. Lipid bilayer structure. Curr Opin Struct Biol. 2000;10:474–480. doi: 10.1016/s0959-440x(00)00117-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni QZ, Daviso E, Can TV, Markhasin E, Jawla SK, Swager TM, Temkin RJ, Herzfeld J, Griffin RG. High Frequency Dynamic Nuclear Polarization. Acc Chem Res. 2013 doi: 10.1021/ar300348n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggie L, Lopez JJ, Collinson I, Glaubitz C, Lorch M. Dynamic nuclear polarization-enhanced solid-state NMR of a 13C-labeled signal peptide bound to lipid-reconstituted Sec translocon. J Am Chem Soc. 2011;133:19084–19086. doi: 10.1021/ja209378h. [DOI] [PubMed] [Google Scholar]
- Renault M, Pawsey S, Bos MP, Koers EJ, Nand D, Tommassen-van Boxtel R, Rosay M, Tommassen J, Maas WE, Baldus M. Solid-state NMR spectroscopy on cellular preparations enhanced by dynamic nuclear polarization. Angew Chem Int Ed Engl. 2012;51:2998–3001. doi: 10.1002/anie.201105984. [DOI] [PubMed] [Google Scholar]
- Rosay M, Tometich L, Pawsey S, Bader R, Schauwecker R, Blank M, Borchard PM, Cauffman SR, Felch KL, Weber RT, Temkin RJ, Griffin RG, Maas WE. Solid-state dynamic nuclear polarization at 263 GHz: spectrometer design and experimental results. Phys Chem Chem Phys. 2010;12:5850–5860. doi: 10.1039/c003685b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossini AJ, Zagdoun A, Lelli M, Lesage A, Coperet C, Emsley L. Dynamic Nuclear Polarization Surface Enhanced NMR Spectroscopy. Acc Chem Res. 2013 doi: 10.1021/ar300322x. [DOI] [PubMed] [Google Scholar]
- Salnikov E, Rosay M, Pawsey S, Ouari O, Tordo P, Bechinger B. Solid-state NMR spectroscopy of oriented membrane polypeptides at 100 K with signal enhancement by dynamic nuclear polarization. J Am Chem Soc. 2010;132:5940–5941. doi: 10.1021/ja1007646. [DOI] [PubMed] [Google Scholar]
- Sauvee C, Rosay M, Casano G, Aussenac F, Weber RT, Ouari O, Tordo P. Highly Efficient, Water-Soluble Polarizing Agents for Dynamic Nuclear Polarization at High Frequency. Angew Chem Int Ed Engl. 2013;52:10858–10861. doi: 10.1002/anie.201304657. [DOI] [PubMed] [Google Scholar]
- Sergeyev IV, Day LA, Goldbourt A, McDermott AE. Chemical shifts for the unusual DNA structure in Pf1 bacteriophage from dynamic-nuclear-polarization-enhanced solid-state NMR spectroscopy. J Am Chem Soc. 2011;133:20208–20217. doi: 10.1021/ja2043062. [DOI] [PubMed] [Google Scholar]
- Smith AN, Caporini MA, Fanucci GE, Long JR. A Method for Dynamic Nuclear Polarization Enhancement of Membrane Proteins. Angew Chem Int Ed Engl. 2015;54:1542–1546. doi: 10.1002/anie.201410249. [DOI] [PubMed] [Google Scholar]
- Solomon I. Relaxation Processes in a System of 2 Spins. Phy Rev. 1955;99:559–565. [Google Scholar]
- Song CS, Hu KN, Joo CG, Swager TM, Griffin RG. TOTAPOL: A biradical polarizing agent for dynamic nuclear polarization experiments in aqueous media. J Am Chem Soc. 2006;128:11385–11390. doi: 10.1021/ja061284b. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Lee D, Dubois L, Bardet M, Hediger S, De Paepe G. Rapid Natural-Abundance 2D 13C-13C Correlation Spectroscopy Using Dynamic Nuclear Polarization Enhanced Solid-State NMR and Matrix-Free Sample Preparation. Angew Chem Int Ed Engl. 2012;51:11766–11769. doi: 10.1002/anie.201206102. [DOI] [PubMed] [Google Scholar]
- Thurber KR, Tycko R. Perturbation of nuclear spin polarizations in solid state NMR of nitroxide-doped samples by magic-angle spinning without microwaves. J Chem Phys. 2014;140:184201. doi: 10.1063/1.4874341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Cruijsen EAW, Koers EJ, Sauvee C, Hulse RE, Weingarth M, Ouari O, Perozo E, Tordo P, Baldus M. Biomolecular DNP-Supported NMR Spectroscopy using Site-Directed Spin Labeling. Chem Eur J. 2015;21:12971–12977. doi: 10.1002/chem.201501376. [DOI] [PubMed] [Google Scholar]
- Voinov MA, Good DB, Ward ME, Milikisiyants S, Marek A, Caporini MA, Rosay M, Munro RA, Ljumovic M, Brown LS, Ladizhansky V, Smirnov AI. Cysteine-Specific Labeling of Proteins with a Nitroxide Biradical for Dynamic Nuclear Polarization NMR. J Phys Chem B. 2015;119:10180–10190. doi: 10.1021/acs.jpcb.5b05230. [DOI] [PubMed] [Google Scholar]
- White SH, Wimley WC. Membrane protein folding and stability: physical principles. Annu Rev Biophys Biomol Struct. 1999;28:319–365. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]
- Wylie BJ, Dzikovski BG, Pawsey S, Caporini M, Rosay M, Freed JH, McDermott AE. Dynamic nuclear polarization of membrane proteins: covalently bound spin-labels at protein-protein interfaces. J Biomol NMR. 2015;61:361–367. doi: 10.1007/s10858-015-9919-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu ZW, Quinn PJ. The modulation of membrane structure and stability by dimethyl sulphoxide (Review) Mol Membr Biol. 1998;15:59–68. doi: 10.3109/09687689809027519. [DOI] [PubMed] [Google Scholar]