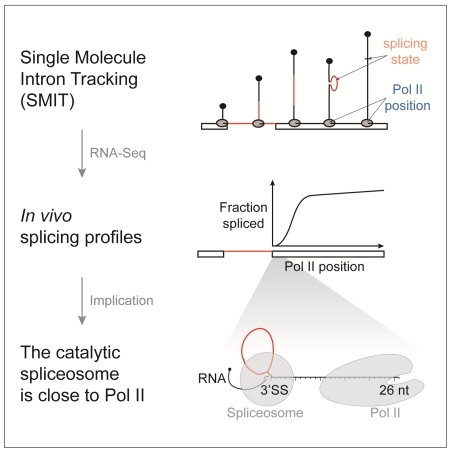
Summary
Protein-coding genes in eukaryotes are transcribed by RNA polymerase II (Pol II) and introns are removed from pre-mRNA by the spliceosome. Understanding the time lag between Pol II progression and splicing could provide mechanistic insights into the regulation of gene expression. Here we present two single molecule nascent RNA sequencing methods that directly determine the progress of splicing catalysis as a function of Pol II position. Endogenous genes were analyzed on a global scale in budding yeast. We show that splicing is 50% complete when Pol II is only 45nt downstream of introns, with the first spliced products observed as introns emerge from Pol II. Perturbations that slow the rate of spliceosome assembly or speed up the rate of transcription caused splicing delays, showing that regulation of both processes determines in vivo splicing profiles. We propose that matched rates streamline the gene expression pathway, while allowing regulation through kinetic competition.
Introduction
Transcription and pre-mRNA splicing are carried out by two macromolecular machines, RNA polymerase II (Pol II) and the spliceosome. Splicing begins while nascent RNA is still attached to the DNA axis by Pol II, i.e. “co-transcriptionally” (Brugiolo et al., 2013; Osheim et al., 1985), creating opportunities for cross-regulation between splicing and transcription. For example, transcriptional output is stimulated by the presence of introns within a gene (Ares et al., 1999; Bieberstein et al., 2012; Brinster et al., 1988). Moreover, the speed of elongating Pol II influences alternative splicing, such that a slower Pol II favors the recognition of weaker splice sites (Braunschweig et al., 2013; Carrillo Oesterreich et al., 2011; Luco et al., 2011). In addition to this “kinetic” principle, the proximity of nascent RNPs to the DNA axis creates the potential for interactions between the splicing machinery, chromatin, and Pol II (Luco et al., 2011; Wetterberg et al., 2001). Because of the enormous potential for regulation, how Pol II and the spliceosome interact in space and time is currently the subject of intense research. How close are Pol II and the spliceosome when splicing occurs?
Mounting evidence that cross-regulation between transcription and splicing impacts gene expression highlights the importance of determining the rates of these two processes in vivo. While substantial progress has been made in determining Pol II elongation rates genome-wide (Jonkers et al., 2014; Veloso et al., 2014), splicing rates are less well understood. Intron half-lives reflect both splicing and decay, and can be used to deduce splicing rates. Metabolic labeling has recently been used to compute intron half-lives on a global scale, yielding ranges of seconds to hours in fission yeast and human cells (Eser et al., 2015; Rabani et al., 2014), establishing a range of expected splicing rates that resemble those derived from RT-qPCR experiments in human cells (Pandya-Jones and Black, 2009; Singh and Padgett, 2009). Live cell fluorescence measurements have yielded intron half-lives of 20-30, 105, and 164 seconds for model human genes (Martin et al., 2013; Schmidt et al., 2011). These estimates are in a range consistent with a 15-30 second global splicing rate derived from human snRNP dynamics (Huranová et al., 2010), and a 30-60 second estimate from two different model genes in budding yeast (Alexander et al., 2010; Lacadie et al., 2006; Tardiff and Rosbash, 2006). Considering the reported shorter splicing rates (30 seconds) and average elongation rates (2 kb/minute), one can calculate that splicing may occur when Pol II is at least 1 kb downstream of introns. Consequently, the spliceosome may be tethered to Pol II by a ~1,000 nt long strand of RNA. Is this estimate accurate? How variable is the relationship between splicing and transcription from gene to gene?
Here we employ two single molecule RNA sequencing (RNA-Seq) strategies to quantify the transition of unspliced pre-mRNA to spliced product with respect to the position of Pol II at base-pair resolution. Nascent RNA contains both the information on splicing state and progression of Pol II and therefore enables a direct analysis of both processes, providing temporal and spatial information. We chose to profile co-transcriptional splicing kinetics in budding yeast, where the prevalence of single-intron genes avoids the complication of multiple introns and alternative splicing. First, we developed Single Molecule Intron Tracking (SMIT) to analyze thousands of individual nascent RNA molecules for each of 87 endogenous genes, yielding co-transcriptional splicing kinetics at base-pair resolution. Second, we employed long read sequencing to analyze full-length nascent RNAs genome-wide. We employ these methods to determine the earliest occurrence of splicing and the time window for splicing completion under wild-type conditions. Subsequently, we query the effects of splice site sequence variation and changes in transcription elongation rate to address regulation.
Results
Single Molecule Intron Tracking (SMIT) measures splicing progress as a function of Pol II position
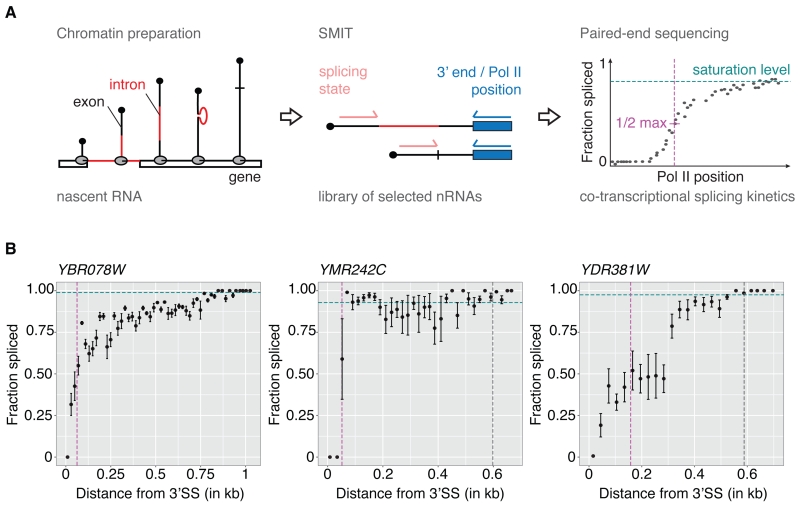
To measure both the position of Pol II and the splicing state of single nascent RNA molecules (i.e. the progression of splicing as a function of Pol II position), we developed Single Molecule Intron Tracking (SMIT; Figure 1A). We begin with the purification of nascent RNA from a chromatin preparation, which permits harsh washing due to the stability of the ternary complex between DNA, Pol II, and nascent RNA (Carrillo Oesterreich et al., 2010). 3’ end linker ligation marks the position of Pol II for each nascent RNA molecule (Churchman and Weissman, 2011; Weber et al., 2014). 3’ end linker ligation is followed by PCR using gene-specific primers located in first exons (Figure S1A-B) to selectively enrich nascent RNAs of interest, both spliced and unspliced. We use massively parallel paired-end sequencing to determine the splicing state (spliced or unspliced) and Pol II position (3’ ends) for millions of single nascent RNA molecules. SMIT was performed on 87 selected endogenous yeast genes (Table S1), representing ~30% of all intron-containing genes. The large number of nascent RNA observations (2 × 107) yielded an average of 2 × 105 reads per gene and 300 reads per Pol II position (Figure S1C-D). On average, each gene is represented by 329 distinct Pol II positions (Figure S1E). For most genes Pol II coverage extends to regions downstream of polyA sites, allowing us to analyze splicing along the entire terminal exon (Figure S1F). Both PCR and sequencing reactions involved in SMIT may generate a bias towards short reads. To quantify and correct for this bias, the insert length distribution for intronless genes was determined. This distribution can be well modeled by an exponential distribution (Figure S1G). This model allows us to correct for this bias in all the following analyses (see Supplementary Information for a detailed explanation). Representative profiles show that SMIT quantifies the transition of the pool of unspliced to spliced nascent RNA as a function of Pol II position (Figures 1B and S2).
Figure 1. Single molecule intron tracking (SMIT) measures co-transcriptional splicing kinetics.
(A) SMIT workflow: Nascent RNA is purified from chromatin (intron, red; exons, black). Nascent RNA molecules of interest are selected by PCR with a forward primer in exon 1. Pol II position (3’ end) and splicing state are determined by paired-end sequencing. Single molecule observations are corrected for PCR length bias, grouped by Pol II position and the fraction spliced is calculated. Saturation values (cyan), defining a statistically determined maximum value, allow calculation of ½ max Pol II positions (50% of saturation, magenta). For details see SI.
(B) SMIT profiles for three representative endogenous genes. Fraction spliced (mean ± SD) is plotted versus distance from 3’ splice sites (SSs). Co-transcriptional splicing saturation value (cyan dotted line), half-saturation Pol II position (magenta dotted line) and polyA site (grey dotted line, if in range) are indicated.
See Figures S1-S3 for characterization of SMIT and additional SMIT profiles.
SMIT profiles reveal gene-to-gene similarities and differences in the progress of the splicing reaction relative to Pol II position. We derive saturation levels (the fraction spliced at the end of the gene) and half-maxima (½ max, the Pol II position at which 50% of the saturation is reached) for each gene. See Figure 1A for a schematic representation and Experimental Procedures for a detailed description of the analysis. For most genes, the fraction spliced reaches saturation well before Pol II has reached the polyA cleavage site (Figures 1B and S2). Saturation levels ranged from 6 to 100% with an average saturation value of 82% (Figure S3A), which agrees well with genome-wide studies in other species (Brugiolo et al., 2013; Herzel and Neugebauer, 2015). Moreover, gene-specific saturation levels correlated well with a previous independent approach in yeast (Carrillo Oesterreich et al., 2010) (Figure S3B), validating the approach. Interestingly, for YDR381W (YRA1, see Figure 1B) spliced products were detected later during transcription (½ max = 158 nt), possibly due to a non-consensus branchpoint sequence previously associated with the inefficient splicing of YRA1 pre-mRNA (Preker and Guthrie, 2006). Despite this example, several genes with non-consensus splice site and branchpoint sequences displayed fast and complete splicing kinetics. Importantly, most genes exhibit a substantial fraction spliced transcripts at Pol II positions just downstream of 3’ splice sites (3’SSs, Figure 1B and Figure S2), indicating that splicing may occur unexpectedly early during transcription.
Long read sequencing confirms splicing close to the 3’SS detected by SMIT
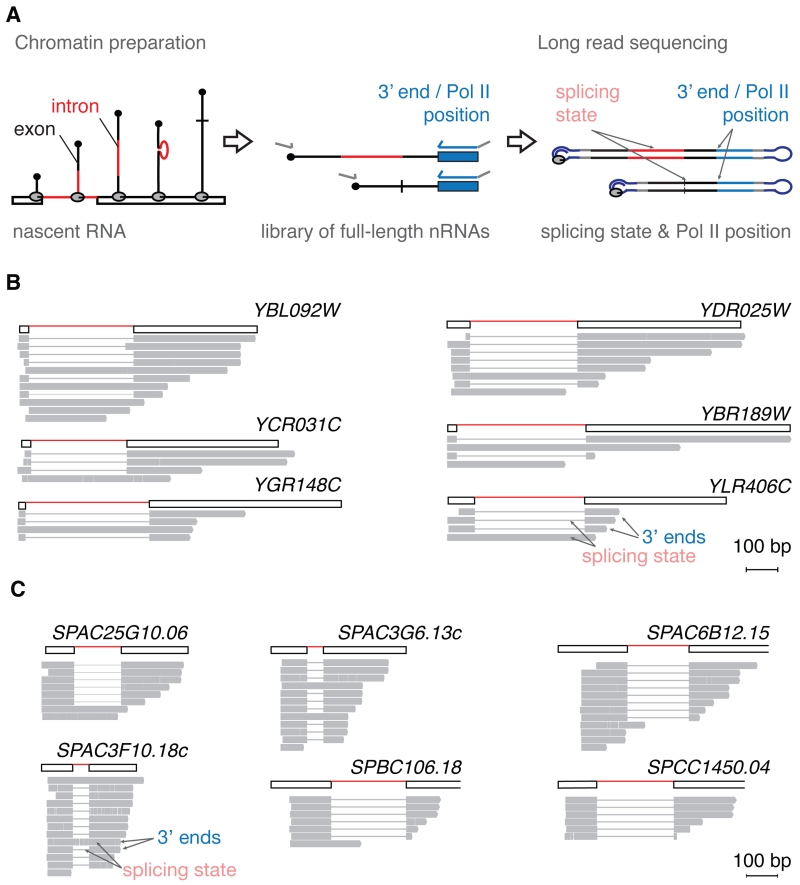
To validate the detection of spliced nascent RNA molecules with 3’ ends just downstream of 3’SSs, we developed a second experimental approach using long read sequencing of full-length nascent RNA to determine both the position of Pol II and the splicing state of single nascent RNA molecules (Figure 2A). As before, this method starts with nascent RNA purification, and 3’ ends are marked by linker ligation. In contrast to SMIT, however, minimal cDNA amplification is involved in library preparation; the forward PCR step serves to complete the cDNA for sequencing by DNA polymerase. Importantly, insert length bias is not observed in our data set; this point is discussed in detail below. Although this genome-wide method yields relatively few reads per gene, it clearly reveals spliced nascent transcripts with 3’ ends just downstream of 3’ splice sites (Figure 2B). Therefore, the long read sequencing recapitulates the observation made by SMIT. We extended the long read sequencing analysis to the distantly related fission yeast S. pombe and similarly detect splicing events close to 3’SSs (Figure 2C). These data confirm that splicing can occur unexpectedly early during transcription in both budding and fission yeasts.
Figure 2. Long read sequencing detects splicing of nascent RNA when Pol II is less than 100 nucleotides downstream of 3’SSs.
(A) Long read sequencing of nascent RNA workflow: Nascent RNA molecules are 3’ end ligated and reverse transcribed. A PCR targeting all cDNAs generates double-stranded cDNAs for long read sequencing.
(B) Long read sequencing observations of S. cerevisiae nascent RNA molecules for six representative genes. Presence (thick line) or absence (thin line) of intron sequence and 3’ end position indicate splicing state and Pol II position, respectively.
(C) Long read sequencing observations of S. pombe nascent RNA molecules is represented as described in (B).
Quantification of in vivo splicing kinetics
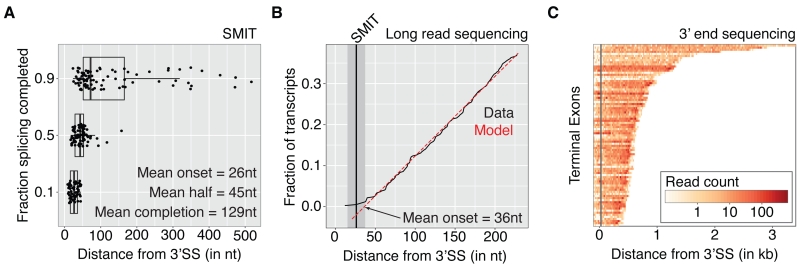
When exactly during transcription are the first spliced products detected? How quickly does the splicing reaction progress? To parameterize co-transcriptional splicing kinetics measured by SMIT, we determined the position of Pol II where 10% (onset), 50% (½ max) and 90% (completion) of the splicing saturation is reached (Figure 3A). For all 87 genes examined, onset is tightly clustered around 26 ± 1 nt (mean +/− SEM) downstream of 3’SSs, indicating that splicing can occur soon after intron synthesis. To validate these findings, we turned to the data obtained by long read sequencing. Gene-specific analysis is not possible, because of the low number of reads per gene (Figure S4A). However, we can take advantage of the genome-wide distribution of reads to infer kinetic parameters of co-transcriptional splicing as population averages. Pol II positions relative to the 3’SS were deduced for all spliced reads for intron-containing genes and visualized as a cumulative frequency distribution (Figures 3B and S4B). The x-intercept at 36 ± 11 nt (mean ± 95% confidence interval) maps the Pol II position at which detection of the first spliced products is predicted by the data (see Experimental Procedures). This mapping of splicing onset is not significantly different to that mapped by SMIT. Thus, two independent methods map the onset of spliced products to Pol II positions just downstream of the 3’SS.
Figure 3. Spliced nascent RNAs are first observed when Pol II is just downstream of 3’SSs.
(A) Pol II position (distance from 3’SS) at which 10%, 50% or 90% of splicing saturation is reached (n=87 endogenous genes). Gene specific values and summary statistics are shown.
(B) Cumulative distribution of Pol II positions of 158 spliced long read observations from 83 endogenous genes (black). A model assuming uniform sampling is indicated (red). Splicing onset inferred by the model (x-intercept: 36 ± 11 nt, mean ± 95% confidence interval) and measured by SMIT (grey line ± 95% confidence interval) is indicated. See also Figure S4.
(C) Pol II density over terminal exons of all 87 genes analyzed by SMIT. Densities were derived at nucleotide resolution by 3’ end sequencing of nascent RNA, grouped in 20 nt bins and encoded by color (Log10 scale, see color map). Rows represent individual terminal exons (extended by 100nt up- and downstream) sorted by length and aligned to the 3’SS. Distance from the 3’SS is indicated and grey line shows average Pol II position of splicing onset (10% of saturation value).
Spliced product at Pol II positions close to the 3’SS could either indicate that splicing is faster than anticipated or that Pol II pauses locally, allowing sufficient time for the splicing reaction to occur. As previously reported, local Pol II elongation rates correlate inversely with Pol II density (Carrillo Oesterreich et al., 2010). In order to determine Pol II density along genes, we sequenced 3’ ends of nascent RNAs isolated from chromatin, matching our strains and experimental conditions. We omitted the PCR necessary for the deduction of the splicing progression in SMIT, thereby avoiding the insert length bias described above. Pol II densities over terminal exons (extended by 100 nt up- and downstream) are shown for all 87 genes analyzed by SMIT (Figure 3C). In general, Pol II density does not increase at 3’SSs or the positions of splicing onset and/or saturation. This indicates that transcriptional pausing within gene regions where splicing occurs is not a major contributing factor to the immediacy of splicing when Pol II transcribes past introns. Note that terminal exon pausing occurs significantly further downstream, ~250 bp downstream of 3’SSs (Carrillo Oesterreich et al., 2010). Taken together, this analysis of Pol II density combined with SMIT analysis indicates that splicing occurs early in the transcription process with respect to time as well as distance.
Splicing kinetics is sensitive to splice site sequence variation
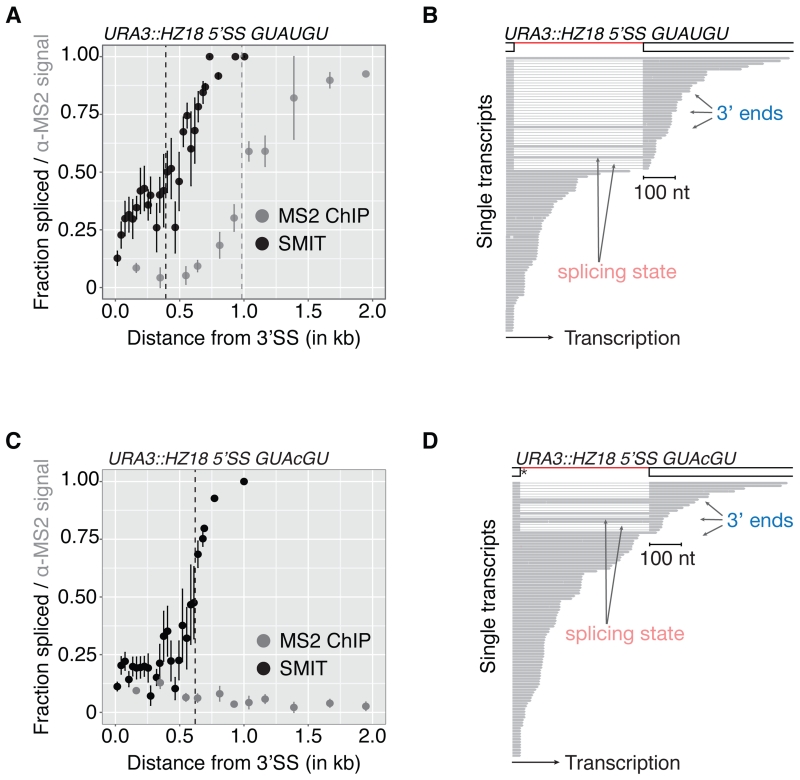
To address the differences between SMIT and previous findings inferred by ChIP, we applied SMIT analysis to the HZ18 reporter gene. HZ18 harbors two halves of an MS2 RNA stem-loop in its exons, such that the stem-loop forms after splicing (Lacadie et al., 2006). MS2-coat protein binds the stem-loop and served as a target in ChIP experiments that aimed to detect the Pol II position along the gene the moment splicing occurred. Splicing of this reporter is well characterized using a variety of assays, including the estimate from ChIP that splicing occurs when Pol II reaches 1 kb or more downstream of 3’SSs (Lacadie et al., 2006; Tardiff and Rosbash, 2006). We integrated HZ18 into the genome and performed MS2 ChIP and SMIT in parallel (Figure 4A). Although our MS2 ChIP experiments confirm those obtained previously, the MS2 ChIP and SMIT profiles differed significantly. While both exhibited a similar shape, the MS2 ChIP profile was delayed compared to the SMIT profile (½ max at ~1 kb vs. 400 nt downstream of the 3’SS). The SMIT profile of HZ18 was comparable to those observed for endogenous genes (e.g. YDR381W, Figure 1B). Long read sequencing was employed as an independent method for analysis of individual nascent HZ18 RNA molecules. Consistent with SMIT, splicing catalysis was observed in long read data when Pol II was immediately downstream of the 3’SS (Figure 4B). The long read sequencing data thereby confirm the SMIT profiles and support the conclusion that splicing of the HZ18 reporter occurs earlier during transcription than previously suggested.
Figure 4. 5’SS sequence determines co-transcriptional splicing kinetics.
(A) SMIT (black) and MS2 ChIP (grey) profiles (mean ± SD) for integrated HZ18 reporter with canonical 5’SS. Signals are plotted versus distance from 3’SSs.
(B) Long read sequencing observations from nascent RNA of integrated HZ18 reporter with consensus 5’SS. Gene architecture and representative single nascent RNA observations are shown. 5’SS sequence is indicated above the graph.
(C-D) SMIT (black), MS2 ChIP (grey) profiles (mean ± SD) as well as long read sequencing observations deduced from HZ18 reporter gene with mutated 5’SS splice-site are shown as described above.
See also Figure S5.
We took advantage of this well characterized reporter gene to test whether co-transcriptional splicing kinetics depend on splice site sequences that were previously shown to alter splicing kinetics in single molecule experiments in vitro (Hoskins et al., 2011; Shcherbakova et al., 2013). SMIT and MS2 ChIP were performed on HZ18 with a single nucleotide mutation in the 5’SS, which is present in 10% of all yeast intron-containing genes. Assayed by SMIT, this 5’SS mutation shifted the progression of co-transcriptional splicing 200 nt downstream (Figure 4C), consistent with the expectation that weakening 5’SS base-pairing with U1 snRNA delays spliceosome assembly. Consistent with SMIT, spliced transcripts were produced from the mutant HZ18 gene as determined by RT-PCR, and co-transcriptional spliceosome assembly was delayed but not abolished as determined by spliceosome ChIP (Figure S5). Remarkably, MS2 ChIP detected no co-transcriptional splicing (Figure 4C), yet detection of spliced nascent RNA shortly after synthesis of the 3’SS was confirmed by long read sequencing (Figure 4D). We conclude that non-consensus splicing signals that delay spliceosome assembly also delay splicing catalysis with respect to transcription in vivo.
Rapid transcription reveals that splicing can be rate-limiting
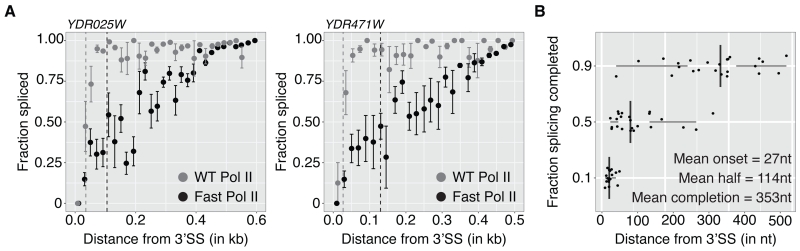
In addition to modulation through variation in RNA sequence as shown above, global changes in Pol II elongation rate may also contribute to co-transcriptional splicing kinetics. If Pol II elongation rate were increased sufficiently, one might expect to observe spliced products only when Pol II has reached positions significantly further downstream. To test this expectation directly, we employed a strain harboring single amino acid mutation (E1103G) that makes Pol II elongation rate 2.3 times faster than wild-type (Braberg et al., 2013; Kaplan et al., 2012) and determined co-transcriptional splicing kinetics by SMIT. Remarkably, SMIT profiles for “fast” Pol II are shifted to downstream gene regions compared to WT (Figure 5A and Figure S6). SMIT data for all genes examined show that splicing onset remained very close to WT values (27 nt downstream of the 3’SS). In contrast, ½ max and completion values were 2.5 and 2.7 times greater than in WT (compare Figures 5B to 3A), showing that co-transcriptional splicing kinetics are sensitive to changes in elongation rate. Thus, splicing can become rate-limiting when transcription is fast.
Figure 5. Pol II elongation rate is a determinant of co-transcriptional splicing kinetics.
(A) SMIT profiles (mean ± SD) for two representative endogenous genes with WT Pol II (grey) and mutated Pol II with increased elongations rates (fast Pol II, black). Fraction spliced is plotted versus distance from 3’SSs. Half-saturation Pol II positions are indicated.
(B) Pol II positions (distance from 3’SS) at which 10%, 50% or 90% of splicing saturation is reached (n=24 endogenous genes). Data points represent gene-specific values and boxplots summary statistics.
See also Figure S6.
Discussion
Although it is widely appreciated that transcription and splicing can occur simultaneously and influence one another, knowledge of the rate of splicing in vivo has been a missing link to our broader understanding of the underlying mechanisms. Here we have quantified the progress of splicing – from unspliced to spliced mRNA – as a function of Pol II position along the length of 87 endogenous yeast genes, using RNA-Seq strategies that yield single molecule information at base pair resolution. The data from Single Molecule Intron Tracking (SMIT) resemble kinetic curves that track biochemical reactions from precursor to product over time. The progression of splicing captured by SMIT thereby reveals the position of Pol II when the first spliced products are observed (onset) and a range of gene-specific dynamics as splicing saturates (completion). The range of Pol II positions when splicing onset and completion is achieved is 26 and 129 nt downstream of the 3’SS, respectively (Figure 6A). We show that significant transcriptional pausing within this window does not occur, allowing us to infer timing. Using an average transcription elongation rate in budding yeast (33 nt/sec (Mason and Struhl, 2005)) and average half-maximal values derived from SMIT, 50% of splicing is complete within ~1.4 seconds after 3’SS synthesis, at least an order of magnitude faster than previous estimates. The mechanistic implications of these findings cause us to reconsider previous assumptions on the physical and temporal relationships between transcription and splicing catalysis, the relative speed with which the spliceosome assembles, and how opportunities for regulation arise in vivo. These aspects are discussed below.
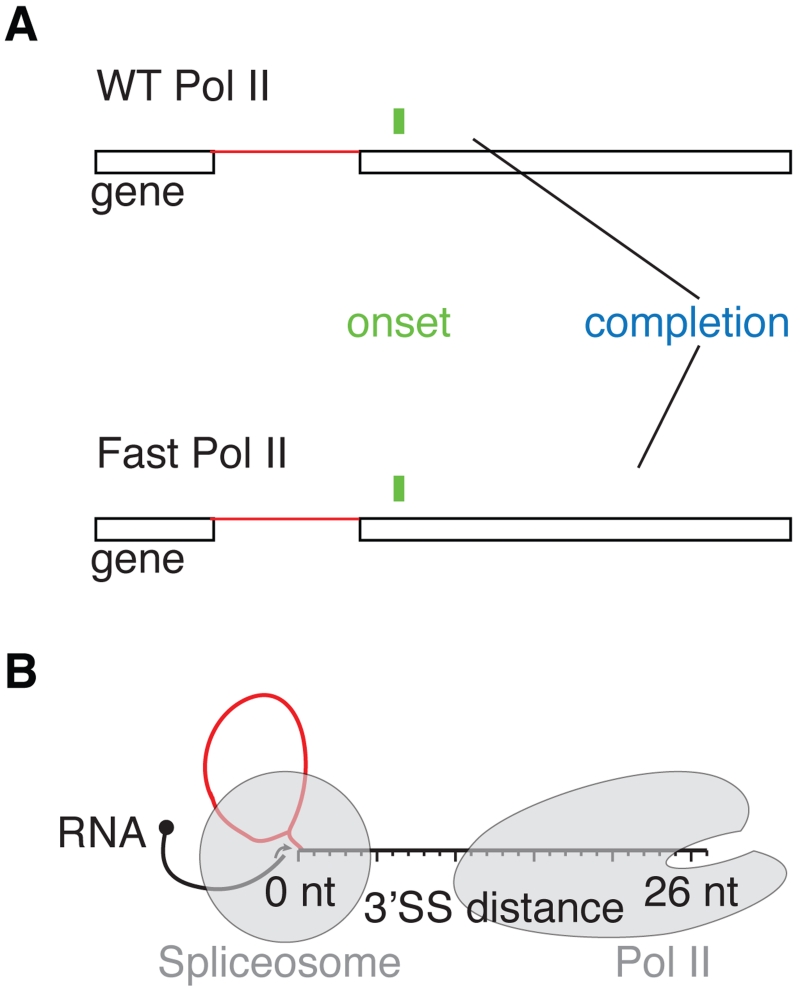
Figure 6. The onset of splicing coincides with intron exit from Pol II.
(A) Splicing catalysis starts when WT and Fast Pol II reach ~26 and 27 nt downstream of 3’SSs (green), respectively. Splicing saturation is reached within ~130 and 353 nt (blue), respectively. The spliceosome and mutant Pol II are separated by a longer stretch of nascent RNA at splicing completion.
(B) Schematic diagram showing that 15 nt of nascent RNA is protected by Pol II, and 9 nt is bound by the spliceosome (grey shades). At splicing onset 26–27 nt downstream of 3’SSs, the spliceosome and Pol II are separated by only 2–3 nt nascent RNA. Introns (red) and exons (black) are depicted.
The observation that splicing catalysis can occur within the time frame of transcription, with delays of only a few nucleotides, indicates that splicing and transcription rates are matched in budding and fission yeast. This interpretation is supported by the observation that splicing delays can be introduced by experimentally decreasing splicing rate or increasing transcription elongation rate. Fast transcription reveals that splicing can be rate-limiting. Interestingly, even when splicing completion is shifted further downstream by fast transcription, splicing onset remains early (Figure 6A), indicating the capacity of the spliceosome to assemble and function quickly. Moreover, these changes suggest that splicing and elongation rates are independent of one another. Consistent with these observations, a previous study showed that reducing Pol II elongation rate in budding yeast leads to greater inclusion of internal exons in genes with two introns (Howe et al., 2003). Our conclusions are also consistent with reports that Pol II elongation rates are inversely correlated with levels of spliced mRNA in yeast (Braberg et al., 2013; Moehle et al., 2014) and that elongation rates in mammalian cells are optimized for splicing (Fong et al., 2014). This observed matching of rates is reminiscent of the enzymatic coordination within metabolic pathways (Bar-Even et al., 2011).
Previous measurements of splicing duration in vivo range from 30 sec to 15 min and correspond to the time required for Pol II to transcribe 0.5-30 kb of DNA (see Introduction). These long estimates from a variety of reporters and model systems suggested that splicing is slower than simple enzymes. Our experiments utilizing the HZ18 reporter indicate that MS2 ChIP does not accurately report co-transcriptional splicing kinetics. A possible explanation for these differences is that MS2 RNA stem loop formation and/or release from spliceosomes delays stem-loop binding by MS2 coat protein. In contrast, the two independent single molecule methods presented here – SMIT and long read nascent RNA sequencing – report on co-transcriptional splicing kinetics directly with high resolution and sensitivity on endogenous genes. Both agree on fast co-transcriptional splicing in vivo with intron removal as soon as 3’SSs exit Pol II, indicating that Pol II and the spliceosome are physically closer during splicing catalysis than previously anticipated.
The immediacy of splicing with respect to transcription suggests potential spatial constraints on the splicing reaction. Our data also allow us to consider the proximity of the spliceosome to Pol II in terms of RNA length, which thereby range from 26-129 nt between the catalytic centers of both Pol II and the spliceosome. Splice sites cannot be utilized by the spliceosome until they exit the transcribing Pol II. Is the completion of splicing determined by the accessibility of the 3’SS to the spliceosome? Structural studies have shown that 15 nt of nascent RNA occupies the Pol II exit channel (Martinez-Rucobo et al., 2015). The spliceosome has currently only been visualized after splicing (Yan et al., 2015). From biochemical studies we can infer that at least 9 nt of downstream exonic RNA sequence is embedded in the spliceosome (Schwer, 2008). Thus, splicing is possible when Pol II has transcribed at least 24 nt downstream of the 3’SS. The detection of the first spliced products by SMIT (26 ± 1 nt) and long read sequencing (36 ± 11 nt) agrees well with this expected minimal RNA length. These values may not be absolute, given “structural intermingling” of colliding Pol II (Saeki and Svejstrup, 2009). Taken together, our findings indicate that Pol II and the spliceosome must be within a few RNA nucleotides of one another when splicing is first detected (Figure 6B).
If the spliceosome can act on introns as soon as they emerge from Pol II, spliceosome assembly must be complete by the time the end of the intron is transcribed. The spliceosome assembles in a step-wise fashion from individual small nuclear ribonucleoprotein particles (snRNPs) and non-snRNP components (Wahl et al., 2009), which could represent numerous rate-limiting steps (Hoskins et al., 2011). Indeed, experiments that monitor the stepwise assembly of the spliceosome suggested that spliceosomes assemble slowly. Specifically, U2 snRNP, U5 snRNP and nineteen complex components of the spliceosome exhibit peak signals ~600 bp downstream of introns (Görnemann et al., 2005; Gunderson and Johnson, 2009; Lacadie and Rosbash, 2005; Lacadie et al., 2006; Tardiff and Rosbash, 2006), and these peaks were loosely interpreted to indicate Pol II position at the time of splicing catalysis. However, these spliceosome components were also significantly detected at 3’SSs by ChIP, consistent with early function. Little is known about mRNA release from spliceosomes and/or co-transcriptional spliceosome disassembly. Combining our knowledge of these ChIP patterns with the present results from SMIT, we suggest that the peak ChIP accumulations further downstream of introns may reflect the duration of spliceosome association with nascent mRNA both during and after splicing.
In higher metazoans, intronic sequences required for splicing (5’SS, branchpoint, and 3’SS) are highly variable and often require trans-acting factors to assist splice site recognition. Indeed, interference with 3’SS recognition decreases splicing rates in human cells (Coulon et al., 2014). In budding yeast, 5’SS, branchpoint and 3’SS sequences mostly conform to consensus, while 25% of genes exhibit sequence variation that can significantly impact splicing kinetics in vitro (Shcherbakova et al., 2013). Thus, even small changes in sequence could change the timing of splicing, e.g. by altered spliceosome assembly or splicing rates. Consistent with this notion, we observed that a single, subtle mutation in 5’SS sequence delays splicing to later Pol II positions in the HZ18 reporter. Interestingly, numerous endogenous genes in our dataset (22/87) harbor non-consensus 5’SSs and are nevertheless spliced efficiently, suggesting there are compensatory mechanisms that enforce splicing rates in vivo. Some introns in yeast are highly structured (Gahura et al., 2011; Meyer et al., 2011); however, the potential effects of RNA folding on splicing kinetics in vivo have not been investigated. Detailed measurements of splicing for a large number of endogenous genes provide a starting point for using gene-specific diversity to pinpoint biologically relevant differences in the kinetics of gene expression.
Our observations that splicing kinetics can be regulated by gene-to-gene sequence variation as well as transcription elongation rates is especially relevant to metazoans, where fast kinetics might restrict alternative splicing. A wealth of data support the notion that splicing is in kinetic competition with transcription (Carrillo Oesterreich et al., 2010; Davis-Turak et al., 2015; Naftelberg et al., 2015). On the one hand, nucleosome positioning and histone post-translational modifications contribute physiologically relevant modulation of both transcription speed and splice site choice (Dujardin et al., 2014; Gunderson and Johnson, 2009; Moehle et al., 2014; Munoz et al., 2009; Schor et al., 2009). On the other hand, inhibition of splicing may open the window of opportunity for splice site choice, by allowing more nascent RNA to be synthesized before alternative splice sites are chosen. Abundant splicing repressors, such as PTB or hnRNP A1 (Fu and Ares, 2014), could play this unexpected role. Likely the combination of RNA sequence variation, changes in elongation rate as well as the expression of trans-acting splicing factors co-operate to tune splicing and transcription of intron-containing genes with different gene architectures under different gene expression programs. Thus the modulation of the kinetic competition between splicing and transcription is a major mechanistic feature of splicing regulation.
Experimental Procedures
3’ end ligation
600 ng DNase-treated, polyA- RNA (chromatin-associated RNA or total RNA, Supplemental Experimental Procedures) and 50 pmol 5N-barcoded 3′ end linker (/5rApp/NNNNNCTGTAGGCACCATCAAT/3ddC/, Integrated DNA Technologies) were combined to a final volume of 6 μl. After denaturation (65°C 5 min, >1 min on ice), the remaining components for 3’ end ligation were added (final 50 mM Tris-HCl, 10 mM MgCl2, 1 mM DTT, pH 7.5, 25% PEG 8000, 40 U RNaseOUT, 200 U T4 RNA ligase II (truncated K227Q, NEB)) and samples were incubated for 10 hours at 16°C. RNA column purification (RNA Clean & Concentrator-5 kit, Zymoresearch) was performed to remove unligated adaptor and enzyme.
Long read sequencing library preparation
3’ end ligated DNase treated, polyA+ RNA depleted nascent RNA was prepared as described in the Supplemental Experimental Procedures and by (Carrillo Oesterreich et al., 2010). rRNA was removed using the Ribo-Zero Gold rRNA Removal Kit (Yeast, Epicentre/Illumina). Nascent RNA was reversed transcribed (SMARTer PCR cDNA Synthesis Kit, Clontech). The included 3’ SMART CDS Primer II A was substituted with a custom primer (see Table S3). 1 μg of RNA was used per reaction. Double-stranded DNA was generated by a low cycle PCR (Advantage 2 PCR Kit, Clontech). For long read sequencing of nascent RNA from the HZ18 reporter a SMIT PCR with the gene-specific forward primer was done to increase sequencing read counts for this reporter (>10,000 unique transcripts sequenced). 1 μg of double-stranded cDNA was submitted for Pacific Biosciences library preparation and sequencing to the Yale Center for Genomic Analysis (YCGA) with standard protocols (SMRTbell Template Prep Kit 1.0).
SMIT Library preparation
3’ end ligated RNA was reverse transcribed with SuperScript III Reverse Transcriptase (Life technologies). Two PCRs, one to capture splicing state and 3’ ends for genes of interest (SMIT PCR) and a second to attach Illumina sequencing adaptors followed. In the SMIT PCR the forward primer was designed to anneal to a gene-specific sequence, whereas the reverse primer targets all 3’ ends of cDNAs. All primer sequences are given in Table S3. PCRs for individual genes were pooled, purified (MinElute PCR purification kit, Qiagen) and used as template for the second PCR. All PCR cycle numbers were optimized to obtain a cDNA smear in the size range expected for S. cerevisiae transcripts (~up to 2 kb, visualized by Agarose gel electrophoresis). A defined 31 nt RNA oligo was used as minimal size control in the SMIT protocol (Churchman and Weissman, 2012). cDNA was gel extracted for sequencing (Qiagen Gel Extraction kit). To avoid saturation of columns, extraction was performed for three size groups per lane separately and sampled pooled before sequencing. Gel extraction removed PCR amplicons without inserts (<200 bp). A second round of purification was performed with AMPure beads. Hiseq2000/2500 sequencing was performed at the Yale Center for Genome Analysis.
Determination of co-transcriptional splicing profiles and metrics
Spliced and unspliced splicing junction reads were paired to 3’ end reads by read-id. Read pairs were grouped by their 3’ end position, resembling grouping of nascent RNA molecules by Pol II position. For each Pol II position group the fraction of spliced molecules is calculated and grouped by gene identity to form raw SMIT profiles. PCR and sequencing steps of the SMIT protocol favor short sequences (Figure S1F) and can be well described by an exponential distribution. Normalized SMIT profiles were generated by calculating the insert-bias-corrected fraction of nascent RNA molecules spliced for each Pol II position. To this end, the exponential distribution was used to determine a probability for each observed insert length. For each Pol II position the observed sequencing read-count was scaled with this probability taken into account the splicing specific insert length (see Supplemental Experimental Procedures for details). Positions with at least 10 observations were grouped into 20 nt bins and represented by mean and standard deviation values. The last three bins containing at least 3 individual Pol II positions were used to determine the saturation values, given by the mean fraction spliced. To characterize splicing kinetics we determined the Pol II position at which 10%, 50% and 90% of saturation values were reached. Linear interpolation was done from the first Pol II position, which exceeds the specified threshold, to the previous position to approximate the crossing point (i.e. Pol II position). If the very first data-point exceeds the specified target fraction, no value was assigned.
Determination of onset of splicing using long read sequencing
Pol II positions for all spliced and unspliced nascent RNA molecules were determined relative to 3’SSs. The total number of observations is low and results in a sparse coverage of single Pol II positions for each gene (Figure S4A). To determine the average onset of splicing, we consider Pol II positions of spliced reads over all genes and the corresponding cumulative frequency distribution (Figure S4B). A linear phase is valid for all positions < 300 nt, which includes the minimal terminal exon length (230 nt) (Kolmogorov-Smirnov test with respect to uniform distribution in the window 26-230 nt, p-value = 0.56). To infer the onset of co-transcriptional we fit a linear model to the observed cumulative distribution within the window of minimal terminal exon length (R2=0.99, x-intercept of the linear model corresponds to onset of splicing 36 ± 11 nt 95% confidence interval). Confidence interval on the intercept was derived by bootstrapping.
Supplementary Material
Article Highlights.
-
-
Single molecule RNA-Seq method to derive in vivo splicing kinetics
-
-
Quantification of splicing profiles for endogenous yeast genes
-
-
Splicing occurs when Pol II is just downstream of introns
-
-
Splicing and transcription elongation rates are matched.
Acknowledgments
We thank members of our laboratory, Karen Adelman, Samie Jaffrey, Dieter Söll, Joan Steitz and Charles Query for discussions and critical comments on the manuscript. We are grateful to Craig Kaplan for the gift of strains as well as Guilin Wang and the Yale Center for Genome Analysis for excellent technical assistance and advice. The presented work was supported by funding from the MPI-CBG and by NIH R01GM112766 from the NIGMS. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession numbers
Data are deposited at GEO, accession number GSE70908.
Supplemental Information includes Supplemental Experimental Procedures, six figures and four tables, and can be found with this article online.
Author contributions
HZ18 ChIP experiments were performed by FCO and KH. SMIT experiments were done by LH and KS. LH performed long read sequencing and 3’ end sequencing experiments. Data analysis and figure preparation were done by FCO and LH. JH provided advice on analysis. FCO, LH and KN conceived the study and wrote the manuscript.
References
- Alexander RD, Innocente SA, Barrass JD, Beggs JD. Splicing-dependent RNA polymerase pausing in yeast. Mol Cell. 2010;40:582–593. doi: 10.1016/j.molcel.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ares M, Jr., Grate L, Pauling MH. A handful of intron-containing genes produces the lion’s share of yeast mRNA. RNA. 1999;5:1138–1139. doi: 10.1017/s1355838299991379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Even A, Noor E, Savir Y, Liebermeister W, Davidi D, Tawfik DS, Milo R. The moderately efficient enzyme: evolutionary and physicochemical trends shaping enzyme parameters. Biochemistry. 2011;50:4402–4410. doi: 10.1021/bi2002289. [DOI] [PubMed] [Google Scholar]
- Bieberstein NI, Oesterreich FC, Straube K, Neugebauer KM. First exon length controls active chromatin signatures and transcription. Cell Reports. 2012;2:62–68. doi: 10.1016/j.celrep.2012.05.019. [DOI] [PubMed] [Google Scholar]
- Braberg H, Jin H, Moehle EA, Chan YA, Wang S, Shales M, Benschop JJ, Morris JH, Qiu C, Hu F, et al. From structure to systems: high-resolution, quantitative genetic analysis of RNA polymerase II. Cell. 2013;154:775–788. doi: 10.1016/j.cell.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunschweig U, Gueroussov S, Plocik AM, Graveley BR, Blencowe BJ. Dynamic integration of splicing within gene regulatory pathways. Cell. 2013;152:1252–1269. doi: 10.1016/j.cell.2013.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinster RL, Allen JM, Behringer RR, Gelinas RE, Palmiter RD. Introns increase transcriptional efficiency in transgenic mice. Proc Natl Acad Sci U S A. 1988;85:836–840. doi: 10.1073/pnas.85.3.836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugiolo M, Herzel L, Neugebauer KM. Counting on co-transcriptional splicing. F1000Prime Reports. 2013;5:9. doi: 10.12703/P5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo Oesterreich F, Bieberstein N, Neugebauer KM. Pause locally, splice globally. Trends Cell Biol. 2011;21:328–335. doi: 10.1016/j.tcb.2011.03.002. [DOI] [PubMed] [Google Scholar]
- Carrillo Oesterreich F, Preibisch S, Neugebauer KM. Global analysis of nascent rna reveals transcriptional pausing in terminal exons. Molecular Cell. 2010;40:571–581. doi: 10.1016/j.molcel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- Churchman LS, Weissman JS. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 2011;469:368–373. doi: 10.1038/nature09652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchman LS, Weissman JS. Native elongating transcript sequencing (NET-seq) Curr Protoc Mol Biol. 2012;14:11–17. doi: 10.1002/0471142727.mb0414s98. Chapter 4, Unit 4. [DOI] [PubMed] [Google Scholar]
- Coulon A, Ferguson ML, de Turris V, Palangat M, Chow CC, Larson DR. Kinetic competition during the transcription cycle results in stochastic RNA processing. eLife. 2014;3:e03939. doi: 10.7554/eLife.03939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis-Turak JC, Allison K, Shokhirev MN, Ponomarenko P, Tsimring LS, Glass CK, Johnson TL, Hoffmann A. Considering the kinetics of mRNA synthesis in the analysis of the genome and epigenome reveals determinants of co-transcriptional splicing. Nucleic Acids Res. 2015;43:699–707. doi: 10.1093/nar/gku1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujardin G, Lafaille C, de la Mata M, Marasco LE, Munoz MJ, Le Jossic-Corcos C, Corcos L, Kornblihtt AR. How slow RNA polymerase II elongation favors alternative exon skipping. Mol Cell. 2014;54:683–690. doi: 10.1016/j.molcel.2014.03.044. [DOI] [PubMed] [Google Scholar]
- Eser P, Wachutka L, Maier KC, Demel C, Boroni M, Iyer S, Cramer P, Gagneur J. Determinants of RNA metabolism in the Schizosaccharomyces pombe genome. bioRxiv. 2015 doi: 10.15252/msb.20156526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong N, Kim H, Zhou Y, Ji X, Qiu J, Saldi T, Diener K, Jones K, Fu XD, Bentley DL. Pre-mRNA splicing is facilitated by an optimal RNA polymerase II elongation rate. Genes Dev. 2014;28:2663–2676. doi: 10.1101/gad.252106.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu XD, Ares M., Jr. Context-dependent control of alternative splicing by RNA-binding proteins. Nat Rev Genet. 2014;15:689–701. doi: 10.1038/nrg3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahura O, Hammann C, Valentova A, Puta F, Folk P. Secondary structure is required for 3′ splice site recognition in yeast. Nucleic Acids Res. 2011;39:9759–9767. doi: 10.1093/nar/gkr662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görnemann J, Kotovic KM, Hujer K, Neugebauer KM. Cotranscriptional spliceosome assembly occurs in a stepwise fashion and requires the cap binding complex. Molecular Cell. 2005;19:53–63. doi: 10.1016/j.molcel.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Gunderson FQ, Johnson TL. Acetylation by the transcriptional coactivator Gcn5 plays a novel role in co-transcriptional spliceosome assembly. PLoS Genet. 2009;5:e1000682. doi: 10.1371/journal.pgen.1000682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzel L, Neugebauer KM. Quantification of co-transcriptional splicing from RNA-Seq data. Methods. 2015;85:36–43. doi: 10.1016/j.ymeth.2015.04.024. [DOI] [PubMed] [Google Scholar]
- Hoskins AA, Friedman LJ, Gallagher SS, Crawford DJ, Anderson EG, Wombacher R, Ramirez N, Cornish VW, Gelles J, Moore MJ. Ordered and dynamic assembly of single spliceosomes. Science (New York, NY) 2011;331:1289–1295. doi: 10.1126/science.1198830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe KJ, Kane CM, Ares M., Jr. Perturbation of transcription elongation influences the fidelity of internal exon inclusion in Saccharomyces cerevisiae. RNA. 2003;9:993–1006. doi: 10.1261/rna.5390803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huranová M, Ivani I, Benda A, Poser I, Brody Y, Hof M, Shav-Tal Y, Neugebauer KM, Staněk D. The differential interaction of snRNPs with pre-mRNA reveals splicing kinetics in living cells. Journal of Cell Biology. 2010;191:75–86. doi: 10.1083/jcb.201004030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkers I, Kwak H, Lis JT. Genome-wide dynamics of Pol II elongation and its interplay with promoter proximal pausing, chromatin, and exons. eLife. 2014;2014:e02407. doi: 10.7554/eLife.02407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan CD, Jin H, Zhang IL, Belyanin A. Dissection of Pol II trigger loop function and Pol II activity-dependent control of start site selection in vivo. PLoS Genet. 2012;8:e1002627. doi: 10.1371/journal.pgen.1002627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacadie SA, Rosbash M. Cotranscriptional spliceosome assembly dynamics and the role of U1 snRNA:5′ss base pairing in yeast. Molecular Cell. 2005;19:65–75. doi: 10.1016/j.molcel.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Lacadie SA, Tardiff DF, Kadener S, Rosbash M. In vivo commitment to yeast cotranscriptional splicing is sensitive to transcription elongation mutants. Genes and Development. 2006;20:2055–2066. doi: 10.1101/gad.1434706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T. Epigenetics in alternative pre-mRNA splicing. Cell. 2011;144:16–26. doi: 10.1016/j.cell.2010.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin RM, Rino J, Carvalho C, Kirchhausen T, Carmo-Fonseca M. Live-cell visualization of pre-mRNA splicing with single-molecule sensitivity. Cell Rep. 2013;4:1144–1155. doi: 10.1016/j.celrep.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Rucobo, Fuensanta W, Kohler R, van de Waterbeemd M, Heck, Albert JR, Hemann M, Herzog F, Stark H, Cramer P. Molecular Basis of Transcription-Coupled Pre-mRNA Capping. Molecular Cell. 2015;58:1079–1089. doi: 10.1016/j.molcel.2015.04.004. [DOI] [PubMed] [Google Scholar]
- Mason PB, Struhl K. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol Cell. 2005;17:831–840. doi: 10.1016/j.molcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- Meyer M, Plass M, Perez-Valle J, Eyras E, Vilardell J. Deciphering 3′ss selection in the yeast genome reveals an RNA thermosensor that mediates alternative splicing. Mol Cell. 2011;43:1033–1039. doi: 10.1016/j.molcel.2011.07.030. [DOI] [PubMed] [Google Scholar]
- Moehle EA, Braberg H, Krogan NJ, Guthrie C. Adventures in time and space: splicing efficiency and RNA polymerase II elongation rate. RNA Biol. 2014;11:313–319. doi: 10.4161/rna.28646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz MJ, Perez Santangelo MS, Paronetto MP, de la Mata M, Pelisch F, Boireau S, Glover-Cutter K, Ben-Dov C, Blaustein M, Lozano JJ, et al. DNA damage regulates alternative splicing through inhibition of RNA polymerase II elongation. Cell. 2009;137:708–720. doi: 10.1016/j.cell.2009.03.010. [DOI] [PubMed] [Google Scholar]
- Naftelberg S, Schor IE, Ast G, Kornblihtt AR. Regulation of alternative splicing through coupling with transcription and chromatin structure. Annu Rev Biochem. 2015;84:165–198. doi: 10.1146/annurev-biochem-060614-034242. [DOI] [PubMed] [Google Scholar]
- Osheim YN, Miller OL, Jr., Beyer AL. RNP particles at splice junction sequences on Drosophila chorion transcripts. Cell. 1985;43:143–151. doi: 10.1016/0092-8674(85)90019-4. [DOI] [PubMed] [Google Scholar]
- Pandya-Jones A, Black DL. Co-transcriptional splicing of constitutive and alternative exons. RNA. 2009;15:1896–1908. doi: 10.1261/rna.1714509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preker PJ, Guthrie C. Autoregulation of the mRNA export factor Yra1p requires inefficient splicing of its pre-mRNA. RNA. 2006;12:994–1006. doi: 10.1261/rna.6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabani M, Raychowdhury R, Jovanovic M, Rooney M, Stumpo DJ, Pauli A, Hacohen N, Schier AF, Blackshear PJ, Friedman N, et al. High-resolution sequencing and modeling identifies distinct dynamic RNA regulatory strategies. Cell. 2014;159:1698–1710. doi: 10.1016/j.cell.2014.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeki H, Svejstrup JQ. Stability, flexibility, and dynamic interactions of colliding RNA polymerase II elongation complexes. Mol Cell. 2009;35:191–205. doi: 10.1016/j.molcel.2009.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt U, Basyuk E, Robert MC, Yoshida M, Villemin JP, Auboeuf D, Aitken S, Bertrand E. Real-time imaging of cotranscriptional splicing reveals a kinetic model that reduces noise: implications for alternative splicing regulation. J Cell Biol. 2011;193:819–829. doi: 10.1083/jcb.201009012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schor IE, Rascovan N, Pelisch F, Allo M, Kornblihtt AR. Neuronal cell depolarization induces intragenic chromatin modifications affecting NCAM alternative splicing. Proc Natl Acad Sci U S A. 2009;106:4325–4330. doi: 10.1073/pnas.0810666106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B. A conformational rearrangement in the spliceosome sets the stage for Prp22-dependent mRNA release. Mol Cell. 2008;30:743–754. doi: 10.1016/j.molcel.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbakova I, Hoskins AA, Friedman LJ, Serebrov V, Corrêa IR, Xu MQ, Gelles J, Moore MJ. Alternative Spliceosome Assembly Pathways Revealed by Single-Molecule Fluorescence Microscopy. Cell Reports. 2013;5:151–165. doi: 10.1016/j.celrep.2013.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J, Padgett RA. Rates of in situ transcription and splicing in large human genes. Nature structural & molecular biology. 2009;16:1128–1133. doi: 10.1038/nsmb.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardiff DF, Rosbash M. Arrested yeast splicing complexes indicate stepwise snRNP recruitment during in vivo spliceosome assembly. RNA. 2006;12:968–979. doi: 10.1261/rna.50506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veloso A, Kirkconnell KS, Magnuson B, Biewen B, Paulsen MT, Wilson TE, Ljungman M. Rate of elongation by RNA polymerase II is associated with specific gene features and epigenetic modifications. Genome Research. 2014;24:896–905. doi: 10.1101/gr.171405.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Lührmann R. The Spliceosome: Design Principles of a Dynamic RNP Machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- Weber CM, Ramachandran S, Henikoff S. Nucleosomes are context-specific, H2A.Z-modulated barriers to RNA polymerase. Mol Cell. 2014;53:819–830. doi: 10.1016/j.molcel.2014.02.014. [DOI] [PubMed] [Google Scholar]
- Wetterberg I, Zhao J, Masich S, Wieslander L, Skoglund U. In situ transcription and splicing in the Balbiani ring 3 gene. EMBO J. 2001;20:2564–2574. doi: 10.1093/emboj/20.10.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C, Hang J, Wan R, Huang M, Wong CC, Shi Y. Structure of a yeast spliceosome at 3.6-angstrom resolution. Science. 2015;349:1182–1191. doi: 10.1126/science.aac7629. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.