Abstract
The development and application of high-throughput molecular profiling have transformed the study of human diseases. The problem of handling large, complex datasets has been facilitated by advances in complex computational analysis. In this review the recent literature regarding the application of transcriptional genomic information to renal transplantation, with specific reference to acute rejection, acute kidney injury in allografts, chronic allograft injury and tolerance is discussed, as is the current published data regarding other “omics” strategies- proteomics, metabolomics and the micro-RNA transcriptome. These data have shed new light on our understanding of the pathogenesis of specific disease conditions following renal transplantation but their utility as a biomarker of disease has been hampered by study design and sample size. This review aims to highlight the opportunities and obstacles that exist with genomics and other related technologies in order to better understand and predict renal allograft outcome.
Introduction
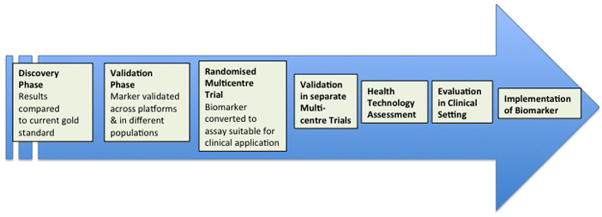
Interrogation of the genome to identify the consequences of static and dynamic genomic variation on allograft outcomes is being investigated actively in kidney transplantation (see reviews).1–4 This has been made possible by the combination of ease of access of sample procurement (allograft, blood or body fluid), the availability of high-throughput technologies, and complex computational analyses. Microarray technology has been used widely to study the pathogenesis of disease processes, although its translation to clinical practice has been limited.5 Nonetheless, the simultaneous detection of quantitative gene expression and the genome-wide expression profiles (GEPs) make it possible to obtain cross-sectional information regarding the physiological and cellular regulatory environment and distinguish this from pathological events such as acute rejection and graft fibrosis. Owing to the limitations of histological diagnosis in renal allograft biopsies,6 application of these analyses have been shown to improve upon,7,8 further classify9 and precede the histological appearance10,11 of disease. Transcriptional information obtained serially could help piece together sequential events that culminate in disease phenomena such as acute rejection, chronic allograft injury, tolerance and graft loss. Events identified in this fashion can be targeted earlier with the ultimate goal of altering the course of disease. At the very least, gene expression changes, once quantified and validated, could be used as biomarkers of simultaneous or impending disease states. Heritable genomic changes in either donor or recipient (e.g polymorphisms) are relatively easier to map from tissue or blood, and can be reproduced reliably in clinical settings. When such changes correlate with allograft outcomes, they have the potential to translate into clinical assays although most assays fall well short of a validated biomarker suitable for clinical use (Figure 1).12 Aside from transcripts of protein coding genes, microRNAs (miRNAs) have been identified as a regulatory layer demonstrating transcriptional and translational control on cellular protein expression and phenotype,13 and recently have been investigated in transplantation.14–16 Similarly, epigenetic changes (DNA- or histone-methylation patterns) have the potential to predict alterations in gene expression and correlate with specific pathogenic processes.17 In this paper, the current literature regarding the application of transcriptional genomic information to renal transplantation is reviewed with specific reference to what these studies bring to our understanding of acute renal allograft rejection and chronic allograft injury.
Figure 1.
Steps required to translate research findings predictive of a clinical outcome into a validated biomarker suitable for clinical use. Adapted from Willis and Lord. Nature Reviews Immunology. May 2015;15(5):323–32912.
Transcriptome assessments
Older genomic techniques confined the study of disease phenomena to one or a few candidate genes at a given time. These genes were selected based on biological plausibility 18,19 or animal experimentation and the studies were hypothesis-driven.3 With increased availability of high throughput technologies such as microarray and RNA-sequencing, it is possible to simultaneously identify the differential expression patterns of thousands of genes that associate with a particular outcome or disease state. Further, computational analysis of this genome-wide expression information, and its mapping to experimentally identified gene-expression information within large databases (http://www.broadinstitute.org/ or http://www.ncbi.nlm.nih.gov/geo/) can help reveal the cumulative signaling pathways operational in a pathologic state. This capability has given rise to a paradigm shift from traditional hypothesis-driven experiments toward large-scale hypothesis generation experiments with potential for testing through clinical trials.4
Important characteristics of commonly employed strategies to examine genomic information in the research settings in transplantation are summarized in Table-1. Microarrays work on the principle of DNA complementarity (see review).4 Reverse transcribed, amplified cDNA is labeled with fluorescent probes and hybridized into separate arrays. The fluorescence is quantified by image analysis software and the probe-IDs (corresponding to a sequence within a gene) are identified by their location on the predesigned platform. Data normalization and elimination of transcripts below a critical abundance threshold is followed by comparative analysis of fold changes of transcripts between control and disease conditions. Microarray results are expressed as relative fold changes in reference to a standard set of samples. Procurement and storage, RNA extraction methods and reverse transcription protocols, heterogeneity of gene chip assay used, fidelity of pre-array amplification techniques, and inaccuracy in identifying differences in low abundance transcripts are all sources of variation in the interpretation of microarray results. RNA-seq based on high throughput sequencing technologies offers key advantages over microarrays – single base resolution, absolute transcript quantification, need for lower amounts of total or fractionated RNA and a large dynamic range for detection of expression levels (8000-fold changes).20
The NanoString® nCounter system is a new technology for molecular profiling that uses a novel colour-coded barcode technology, with each barcode representing a single target molecule.21 It enables digital counting of individual molecules without the need for amplification. Up to 800 transcripts can be profiled in a single reaction, and has been applied to samples of blood, raw cells and urine. Its role in transcriptome profiling in other biomedical fields such as oncology has been prominent in recent years, but only recently been used in the setting of renal transplantation.22,23
Genomics of acute rejection
The pathogenesis of acute rejection (AR), cellular (ACR), antibody-mediated (ABMR) and mixed rejection, in renal allografts is well understood (see elegant reviews).24–26 Important features to emerge are the redundancy within the immune effector mechanisms mediating ACR, the need to discern co-existent ABMR due to its prognostic and therapeutic implications,27 and the paradigm of ongoing sub-clinical rejection and its impact.28–32 These compound the complexity of developing unified genomic biomarkers for AR and histological diagnosis remains the gold standard.4 Variability of the transcriptional milieu of ACR in the allograft and/or peripheral blood is also influenced by immunosuppressive agents, AR severity (borderline vs grade I or II), the timing of the rejection episode, the type/quality of organ, associated pathology and the representativeness of the sample used for transcriptome analysis. Most transcriptional studies have depended on clinical or for-cause biopsies for developing GEPs as biomarkers and may neglect transcriptional phenomenon in subclinical rejections. Transcriptional studies also frequently rely on “clean” phenotypes to develop and validate GEPs, ie specific pathology vs healthy controls – scenarios that are seldom encountered in pure form in routine clinical practice, making the information obtained difficult to both validate and/or to apply to patient care.33 Not withstanding these limitations, substantial progress has been made in understanding the peripheral blood and allograft transcriptomes of AR in renal transplantation.5,7,9,34–38 The reader is referred to comprehensive reviews that have recently tabulated the studies employing genomics in renal allograft rejection. 4,39
Prior to the widespread application of genomics, substantial progress had been made by using quantitative PCR (qPCR) to interrogate tissue from rejecting renal allografts for immune activation genes. Interestingly IL-2 gene upregulation was not associated with rejection but investigators have demonstrated significant upregulation of other cytokines especially IP-10, CXCR3 and CD3ε.18,19 Other markers said to be predictive of ACR include FOXP3, IFN-γ, T-cell receptor variable regions as well as cytotoxic T-lymphocyte effector molecules, such as perforin, granzyme B, Fas, and Fas ligand, and CD40 ligand.18,19,40 Recognizing the increased allograft infiltration and subsequent urinary appearance of lymphocytes during ACR episodes, pre-selected immune transcripts (T-cell/immune related) in urinary cell pellets measured by qPCR have been reported to non-invasively differentiate ACR from non-ACR.41–50 One study used 18S-ribosomal-RNA normalized 3-gene signature (CD3ε, IP-10 & 18S-rRNA) to identify ACR from non-ACR with high accuracy.47 The gene set was less effective at differentiating ACR from ABMR and borderline ACR (AUC – 0.78). More recently, the same investigators described the ability to distinguish AR from other causes of renal allograft dysfunction using a urinary gene signature, and subsequently applied a further transcript profile for ACR to distinguish ACR from ABMR within the AR group.49 A multicenter study with qPCRs performed independently at 6 centers for 7 genes on the same urine samples showed significant correlations (R >0.938) in mRNA levels of all 7 genes suggesting the feasibility of urinary mRNA levels as biomarkers.50 However, urine handling and storage for accurate transcript assessment has limited such studies to a small number of specialized research groups.
To date, many of the genomic studies have attempted to identify GEPs that can separate ACR from non-ACR. This has been possible within the context of the patients studied but the results have not been applicable to broader populations, either because the GEPs lacked the required specificity and sensitivity for general use, or alternatively they have not been studied beyond the initial reference populations. Furthermore, sample sizes were small and their general applicability, much less their diagnostic utility, were not confirmed. For instance, Akalin et al initially used high-density oligonucleotide array (Affymetrix GeneChip® Hu6800) to open-endedly analyze the allograft transcriptome of patients with ACR. They identified a panel of 4 transcripts that were present in all patients but only 7 patients were evaluated.5 However, the authors restricted their analysis only to those genes with a four fold or greater increased expression over baseline biopsies, and may have missed many transcripts that were changed consistently in ACR, albeit less than this threshold. Sarwal and colleagues used DNA microarrays in a systematic study of gene-expression patterns in biopsy samples from normal and dysfunctional renal allografts.9 By unsupervised hierarchical clustering, they identified four clusters of gene expression patterns, which generally corresponded to the histologically identified categories. An important finding of this study was that the gene expression pattern of the 26 ACR biopsies were classifiable into a further three clusters which were not differentiated by light microscopy, which as the authors suggest may reflect unidentified distinct mechanisms in the molecular pathogenesis of rejection and may be predictive of outcome. Again this early study lacked power and has not been corroborated by other investigators. Using gene array profiles obtained from renal transplant biopsies, Flechner et al also confirmed that it was possible to separate stable function with no rejection from graft dysfunction due to ACR and graft dysfunction not related to ACR.34 While AR and non-AR GEPs in PBMCS clustered separately from those of stable transplant recipients, there was very little overlap with GEPs identified within the allograft, which were associated with immune/inflammation mechanisms.
More recent studies have improved upon the design and technical problems that had prevented the development of this technology as a biomarker for AR. These problems include the lack of large cohorts to make firm conclusions, poor study design with a lack of prospective and systematic patient follow-up, heterogeneity across platforms and most significantly the lack of corroboration of findings in separate validation cohorts. Li et al used a novel approach of high throughput transcriptional profiling of total RNA from peripheral blood validated across 3 microarray platforms, followed by further selection by biological relevance and RTPCR validation, to identify a specific 5 gene-set that correlated well with AR/no AR status on simultaneously procured allograft biopsies.38 The 5-gene, qPCR-based, logistic regression model was built using samples from a single center, and validated in an independent multicenter cohort that included AR episodes from transplantation to 3 years of follow-up, of varying severity. Of note, the 5-gene set was ultimately selected based on a biological candidate gene approach ie genes with enrichment in cell–specific immune responses in AR, and not in an unbiased manner. Hence, the most differentially expressed genes by magnitude may not have been selected in the final panel. This gene set also did not distinguish ACR from ABMR, and a subsequent biopsy would be required in order to make treatment decisions. Based upon this and other publications,51,52 the same group of researchers has subsequently developed a 17-gene qPCR based assay (kidney solid organ response test (kSORT) assay) from peripheral blood of both adult and pediatric recipients that diagnosed AR with an AUC of 0.94 in the development cohort (n=143), and was validated with an AUC of 0.95 in a separate validation cohort (n=124).53 Prospective studies using this panel are ongoing. Reeve et al studied 403 allograft biopsies (divided into development and validation cohorts) and compared transcriptional profiles of histologically confirmed ACR (with/without ABMR lesions) to biopsies with other diagnosis in a supervised manner.7 They generated a T-cell mediated rejection score (TCMR-score) using the top 20 differentially regulated transcripts in each iteration. A median TCMR-score >0.1 identified pathologist-recognized ACR lesions with an accuracy of 89%. The strength of the TCMR-score was that it provided additive information to histology and allowed for accurate reclassification of ACR/non-ACR lesions, especially where there existed variation between pathologists such as when defining borderline changes of rejection (B-ACR). The major limitations of the TCMR-score were the absence of an external validation cohort, which limited generalizability of these findings, and the lack of a cohort with subclinical ACR (ie protocol biopsies).
Clinical and histological studies have identified that graft outcome for B-ACR is not as benign as originally thought with worse histological and functional 1 to 2 year outcomes than biopsies with non ACR.29–31,54 Larger transcriptional datasets of blood and allograft samples on exclusive or predominant B-ACR lesions, could help clarify whether this histological readout represents a dichotomy, a continuum or a distinct entity.
The development of a diagnostic GEP for AR has been further complicated by the difficulty to differentiate ACR, ABMR and mixed AR. Acute ABMR is diagnosed with histologic evidence of acute tissue injury (microvascular or endothelial injury and/or ATN), with evidence of current/recent antibody interaction with vascular endothelium (linear C4d staining in the peritubular capillaries), and serologic evidence of donor-specific antibodies (DSAs) (HLA or other antigens). In the recent Banff classification, ABMR has been subdivided to include biopsies that contain only one or two of these features.55 To date, the most detailed genomic evaluation of ABMR has been the INTERCOM cohort study, which contained 300 biopsies. Upon review, 41% of these biopsies had features of ABMR, that were not initially diagnosed by histology.56 Using an ABMR-transcriptome score developed by the same group these biopsies were diagnosed with an accuracy of 85%.36 Derived from 403 indication biopsies, allografts with high ABMR-scores had a significant over-expression of endothelium-associated transcripts and correlated strongly with the presence of DSA in the serum. The ABMR score discriminated ABMR cases from both ACR and other causes of microvascular injury. In response to these data, the Banff working group has added “increased expression of gene transcripts in the biopsy tissue indicative of endothelial injury, if thoroughly validated” as an ABMR diagnostic criteria under evidence of antibody-endothelium interaction.55 In both studies high ABMR scores had prognostic import and correlated independently with allograft loss.
Genomics of acute kidney injury in transplant
Information regarding the molecular phenotype of AKI is difficult to procure as most episodes are self- limited and performance of native kidney biopsies for AKI episodes are not clinically indicated. Kidney transplants experience AKI and are ideally positioned to study the GEP of AKI.57 cDNA microarrays were used to study GEP of 14 living-donor kidneys, 15 deceased donor kidneys with immediate function and 14 deceased donor kidneys with AKI (defined by dialysis in first week). GEPs of LD-kidneys were observed to cluster separately from deceased donor kidneys (132 genes). Further, 48 transcripts related to cell cycle regulation, cell growth/metabolism, and signal transduction were significantly differentially regulated in AKI-allografts compared to those with immediate function.58 In a separate study, the same authors compared GEPs of AKI allografts with carefully selected pristine 6-week protocol biopsies, and identified 394 transcripts that were differentially expressed in AKI.37 Using the geometric mean of the fold increase in the top 30 differentially regulated genes (compared with control nephrectomies), they devised an IRRAT (injury-repair-response-associated-transcript) score that correlated inversely with renal function at the time, and directly with eGFR improvement thereafter. IRRAT score was also significantly associated with DGF, deceased donor status, and interstitial inflammation but not histology. IRRAT scores have since been observed to be elevated in both ACR and ABMR suggesting the non-specific nature of this injury response signature.7,36 Of note, AKI related GEPs identified by different groups in transplantation have shown significant overlaps with both mouse orthologs identified in ischemia-reperfusion models,57,59 and with each other suggesting molecular homogeneity in this injury response.37,58,60
Genomics of chronic allograft injury (CAI)
Despite the significant improvement in acute rejection rates over the last two decades, this has not translated into improved long- term graft survival. Chronic allograft injury remains a major cause of long-term graft loss, and represents an irreversible pattern of injury resulting from a number of immunological (acute and chronic rejection, HLA mismatch, donor specific antibodies), and non-immunological factors (donor age, ischemia/reperfusion injury, infection, CNI toxicity). 61,62 There are differing opinions as to whether allograft loss is multifactorial, as some recent observational studies suggest that most cases of allograft loss can be attributed to a single disease entity 63,64 whilst other published findings suggests otherwise.65 Interstitial fibrosis and tubular atrophy (IF/TA) are the histopathologic hallmarks of CAI. It has subsequently been recognised that concurrent interstitial inflammation and IFTA changes on biopsy identifies recipients at greatest risk of graft loss.66,67 Despite greater understanding of the factors contributing to CAI/IFTA, the precise pathophysiological mechanisms are not fully understood. Consequently, the development of targeted therapeutic strategies has been limited. As with subclinical rejection, the presence of IFTA - a histologic diagnosis, predates the measured decline in renal function. The need for earlier detection to allow for timely therapeutic intervention has provided the impetus for numerous genome, transcriptome, and proteomic studies in the search for potential non-invasive, predictive biomarkers in CAI.
The application of microarray technology to evaluate the differential gene expression in CAI first emerged a decade ago.11 Since then, a number of studies utilizing microarrays have been published to characterize the genome and transcriptome profiles of allografts with IFTA although most have involved small cohorts of patients and non-prospective in design, amongst other limitations. The concept that early transcript changes may predate the subsequent histological phenotype of CAI has been an appealing hypothesis. Scherer et al68 concluded that transcriptome changes precede the observed histologic changes of IFTA, after profiling the gene expression of 3-month post transplant biopsies of 20 patients, and compared those that developed IFTA at 6 months versus those who did not. Using oligonucleotide microarray, differential transcript expression between the progressors and non- progressors was identified, with genes pertinent to T and B cell activation, the immune response profibrotic processes or in epithelial-to-mesenchymal transition featuring prominently in the progressor group. In contrast, a larger study of a retrospectively selected group of 107 allograft recipients who had their biopsy microarray data summarized into pathogenesis- based transcripts sets, found that the molecular phenotype in early (6-week post transplant) transcript expression mainly reflected the injury-repair responses to implantation stresses, without strong correlation to 6 month IFTA or renal function at 24 months.69 Naesens and colleagues also profiled the gene expression of 6 month biopsies from ‘progressors’ and ‘non-progressors’ in a pediatric cohort.70 These groups were defined based on 24- month CADI scores and 6 month protocol biopsies were interrogated. As a point of difference from other studies, only recipients with minimal or no chronic damage at implantation and no interval acute rejection episodes were included. This well- designed albeit highly selective study with small numbers, made a concerted effort to remove likely confounders of the molecular signature and study end point. In addition, the profiling of stable 6 month grafts seemed reasonable time point of study given that these should be devoid of the early inflammatory transcripts. 92 probe sets were significantly overexpressed in the ‘progressor’ group, not only in immunity related but non-immune biological functions, and shed light on the mechanistic pathways. Risk stratification for CAI based on an earlier molecular phenotype seems plausible. The disparities in these studies highlight inter-centre difference in time points of biopsy acquisition, even for ‘protocol’ samples, and study follow up, reinforcing the need for multicenter studies to produce more robust conclusions.
Other investigators have utilized gene arrays to interrogate protocol biopsies within the first year of transplantation.71,72 They showed that a combination of immunological and tissue remodeling gene patterns preceded the onset of fibrosis but specific gene pathways associated with expected biological process such as EMT and the TFG-β pathway were missing. The identification of macrophage-related genes in the 12 month protocol biopsies demonstrating the most fibrosis was of interest and led to a third study where the role of macrophages in renal transplant fibrosis was examined.73 In this study of 46 patients, two unexpected features were identified. Firstly the overwhelming majority of infiltrating macrophages were identified as M2 phenotype by immunohistochemistry. Paradoxically the gene expression patterns from arrays performed on their 12 month protocol biopsies demonstrated an alloimmune gene expression pattern with upregulation of interferon-gamma response genes in those grafts with progressive fibrosis despite there being no history of rejection and no evidence of it on histology. Whilst the study did show that there was a strong association of macrophages with fibrosis it did not clarify their role in the process. However it does highlight the importance of an ongoing subclinical alloimmune response as a driver of fibrosis in the first 12 months after transplant, even in those grafts where there was no histological evidence of it. In addition to immune related events, renal parenchymal factors have been identified as having an impact on renal graft fibrosis. In the prospective multicentre Genomics of Chronic Allograft Rejection (GoCAR) study expression of the SHROOM3 gene, which is found in tubular cells and podocytes in the kidney and encodes an actin- binding protein, in allografts at 3 months post transplant or the presence of the SHROOM3 risk allele in the donor, was associated with increased allograft fibrosis at 1 year.74
There have been few meta-analyses of genomic studies in chronic allograft injury,75–78 There are difficulties in undertaking such analyses as there are many confounders including the timing and reasons for the biopsy (protocol or for cause biopsies), the timing of clinical and histological follow up and the type of genomic platform used. One of the earlier meta-analysis76 utilized a novel non-parametric approach for combining data from multiple independent microarray studies, and pooled two published datasets comprising 40 transplant biopsies samples (27 with CAI and the remainder with normal histology).79,80 Both studies used Affymetrix microarray GeneChips, which eliminated cross-platform confounders. Amongst the identified differentially expressed genes for CAI, there was an over-representation of six KEGG pathways including oxidative phosphorylation, ATP synthesis, citrate cycle, reductive carboxylate cycle, methionine metabolism.76 More recently a meta-analysis of separate data sets were undertaken to evaluate the role of growth factors and integrin pathways in CAI.78 Three data sets were pooled and all were hydridized on Affymetrix arrays. Although the results were of interest with regard to the pathogenesis of CAI they were not robust in differentiating between mild, moderate and severe CAN/IFTA. IGF1 was the only growth factor pathway that was differentially expressed with progressive IFTA severity. Hepatocyte growth factor (HGF) and fibroblast growth factor (FGF) signaling pathways were significantly upregulated in early CAN/IFTA (Banff 0 vs 1) and were expressed in moderate/severe CAN/IFTA (Banff 0 vs Banff 2, 3). However, they were not able to differentiate between mild CAN/IFTA (Banff 1) and moderate and severe grades (Banff 2 and 3 respectively).
Studies in CAI that have put forth a limited gene set with potential as a non-invasive diagnostic biomarker panel have been limited. Of note, one such study aimed to build a multi-gene prediction model around vimentin, chosen based on other published work.81 In this cross-sectional investigation, indication or protocol biopsies of 114 patients were classified as having IFTA or normal histology, and urinary cell RNA was extracted from urine collected at the time of biopsy. Samples were divided prior to analysis at a 2:1 ratio into a discovery (76 patients, 32 with allograft fibrosis and 44 with normal histology) and validation set (38 patients, 16 with fibrosis and 22 normal histology). The levels of 22 selected mRNAs were measured using their pre-amplification enhanced kinetic quantitative PCR assay. Following adjustments only 2 mRNAs were found to be predictive of fibrosis, resulting in a 4- gene model comprised of vimentin, NKCC2, E-cadherin and 18S rRNA, which yielded a composite score that was highly associated with IF/TA. The gene set performed well in the validation cohort with an AUC of 0.89 (p<0.0001); specificity was 77.3% and sensitivity 87.5% for diagnosis of IFTA at the composite score cut-point (same as discovery set) of 4.5. However the timing of biopsies was heterogeneous, and results are potentially confounded by a statistically significant difference between the time to biopsy in the fibrosis and non-fibrosis groups. These results require confirmation in a more uniform study protocol. Further, whether this profile is predictive rather than diagnostic of fibrosis remains to be determined.
Recently studies examining the genome, transcriptome and/or proteome profile on the same biological samples in an integrated manner using high-throughput technologies have emerged. Kurian and colleagues82 used DNA microarrays along with tandem mass spectrometry to evaluate the gene and protein expression in the PBMCs of patients with biopsy proven IF/TA of varying severity. The aim was to identify diagnostic biomarkers of IF/TA in the peripheral blood. This proof-of-concept study identified transcript and protein signatures, which appeared to accurately stratify patients into the different IF/TA grades. However, as the authors identified, the patients were pooled from two independent sets, and were clinically heterogeneous with variation in the timing of sample collection and methods of transcript purification. In a separate study83 the proteome and genome expression was performed in parallel on allograft tissue from the same patients in the aforementioned study. Multiple sets of genes were mapped to different functional pathways, with levels appearing to correlate with histologic severity of IF/TA. These studies show that genomics has the ability to shed light on the pathogenesis of CAI whilst holding potential as an assay that is diagnostic and/or predictive of future fibrosis. However to date the utility of the findings is limited by small sample size, lack of homogeneity of the study population and were mostly non- prospective in design. Therefore carefully coordinated collaborative efforts between centers will be required to allow for robust validation settings.
Genomics of tolerance
The first study published on gene expression profiling in operationally tolerant renal graft recipients using microarray came from Brouard and colleagues.84 A subset of patients in their study was stratified into three clinical phenotypes: tolerant patients without immunosuppression for at least 2 years, chronic rejection and age-matched controls. Using a customized cDNA microarray platform (Lymphochip), a ‘tolerant footprint’ of 49 genes was identified from PBMCs. Thirty-three of these differentiated operationally tolerant from chronic rejection with 99% specificity and 86% sensitivity. There was under-expression of co-stimulatory genes and Th1/Th2 related cytokines in the tolerant group. In addition, 27% of these genes were regulated by TGF-β. In a later study from the same group, 20 of these were selected and formed a gene set which the authors proposed could potentially identify those patients at ‘minimal risk’ of rejection, where reduction of immunosuppression could be considered.85 In a cohort of 144 patients who were at least 5 years post-transplant with stable graft function, 3.5% displayed a tolerance profile in the peripheral blood based on this 20-gene set. However, these findings were not obtained in parallel with kidney biopsies to verify the absence of immune reactivity and to date no study has been published that tests their hypothesis.
The bias towards B cells in the tolerant state was also identified by multicenter studies by Newell86 and Sagoo.87 The patients in the tolerance cohort from each study were from the US Immune Tolerance Network (25 patients) and the European Indices of Tolerance consortium (11 patients). The two studies used samples from each other’s cohort for validation of their findings. Newell and colleagues used microarray for whole blood gene profiling, and found 30 genes over-expressed by at least two fold in 19 operationally tolerant patients (more than 12 months off immunosuppression) versus patients with stable graft function with ongoing triple immunosuppression. Twenty two of these genes were B-cell specific. However, this GEP could not differentiate between tolerant patients and healthy (non-transplanted) controls. Further analysis identified a three-gene signature that was able to distinguish operationally tolerant patients from those with stable function on immunosuppression with 100% sensitivity and 83% specificity in a test set of 12 patients divided equally into the two groups. The expansion of B-cell transcripts was not only identified in the blood, but also in the urine sediment, which exhibited upregulation of CD20 mRNA in the tolerant group. Using a different microarray platform Sagoo and colleagues was able to distinguish operationally tolerant recipients from chronic rejection, stable patients on immunosuppression and normal controls.87 Again a predominance of B-cell related genes and associated molecular pathways were identified in their tolerance footprint. However, it is possible that the strong representation of B cells in the aforementioned studies may reflect the absence of immunosuppression, rather than the state of tolerance per se. Newell was unable to identify any of the three genes in their predictive set in Sagoo’s platform, questioning the robustness of the findings. 86 More recently, Baron and colleagues88 compared the gene lists derived from the five tolerance-related blood transcriptomic studies, and were unable to identify a gene signature to differentiate tolerant subjects from stable controls on immunosuppression.84,86,87,89,90 To enhance the study power, the microarray data from the 96 operationally tolerant patients were integrated into one dataset. The authors made efforts to overcome systematic biases derived from merging microarray data obtained from different platforms by renormalization and standardization of the data prior to integration, and only the 1846 consensus genes were retained for analysis. Their meta-analysis identified a robust differential gene signature when compared to subjects who were stable on immunosuppression. The most discriminating gene clusters were linked to B-cells, monocytes and CD4 T cells at various stages of differentiation. Following cross validation, a selection of the 20 top-ranked genes with the majority overexpressed in tolerance and pertaining to B cells was revalidated in an independent cohort of 18 new tolerance samples of which 6 were new cases, with 91.7% accuracy (94.4% sensitivity, 90% specificity, 85% PPV and 96.4% NPV). This meta-analysis reaffirms the prominent role of B cells in the tolerant state as identified in earlier studies, but whether their strong presence is causative or consequent remains unanswered.
Other ‘omics: Proteomics, Transcriptomics and Metabolomics
A number of other biomic disciplines have emerged in the last decade. These disciplines, including proteomics, transcriptomics and metabolomics refer to the high throughput application to profile the proteome, transcriptome, and metabolome respectively.
Proteomics is the large-scale study of proteins. Previously, profiling of the proteome was limited by its complexity, but has gained significant momentum with technological advancements, particularly in relation to mass spectrometry (MS). Identification and quantitative strategies using MS have been aided by improvements in protein separation methods, both gel based (eg 2-dimensional differential in-gel electrophoresis (2D-DIGE)) and non-gel based (eg high performance liquid chromatography (HPLC)) whilst a range of isotope-based labelling methods allow for relative and absolute protein quantification. Urine has been frequently utilized in both non-targetted and targetted proteomic studies of the renal allograft, as has peripheral blood. 48,91–98
Earlier studies utilising mass spectrometry-based technology in urine proteomic profiling identified protein signals that were present in subjects with acute rejection, but did not further characterise the protein associated with these signals.99,100 In a proof of principle study,100 SELDI-TOF-MS was used to discover urine protein peaks associated with acute cellular rejection. A follow up study revealed that these were protein derivatives of non-tryptic cleaved forms of β-2 microglobulin.101 However, a multicenter study did not identify a statistically significant correlation of intact or cleaved β-2 microglobulin in urine in the presence of AR.48 This prospective targeted observational study conducted through the Clinical Trials in Organ Transplantation-01 (CTOT-1) protocol, aimed to evaluate and validate a panel of candidate urinary mRNA and protein biomarkers chosen from other published work for diagnosing and predicting acute rejection. These included CCR1, CCR5, CXCR3, CCL5 (RANTES), CXCL9, CXCL10, IL-8, perforin and granzyme B which were studied in this cohort of 280 adult and pediatric first transplant recipients. The most robust findings were for CXCL9, both its urinary mRNA and protein yielded PPVs of 61–67% for AR, but more impressively were the NPVs of 83% and 92% respectively. In addition, low urinary CXCL9 protein at 6 months identified those at low risk of subsequent AR. These findings suggest that urinary CXCL9 protein could aid clinical decision-making by ruling out AR in those with renal function impairment, and identify patients at low risk of subsequent AR where tapering of immunosuppression could therefore be considered. Other studies have applied a variety of proteomic techniques to identify protein expression in an unbiased high-throughput manner in allograft rejection, before utilizing targeted ELISA for validation of biomarker panels. 93,97,102 However as with the majority of promising proteomic studies to date, multicenter prospective trials are required to validate these candidate markers before meaningful conclusions can be made.
Transcriptomics is the study of the RNA transcripts encoded by the genome. Whilst much of the work in the last decade has been in mRNA evaluation, studies in miRNA expression profiling in the setting of renal transplantation, has increased exponentially in recent years. MiRNA are small, non-coding RNAs, which regulate gene expression of target mRNAs. Similar to genome and mRNA profiling, large scale sequencing of miRNAs has been dominated by microarray applications. One of the advantages of this technology is that there are far fewer miRNA than genes and hence the computational analysis is simpler and there remains the possibility that a more robust marker could be identified as a predictor of either AR or IF/TA. Scian and Mas103,104 have elegantly summarised miRNA profiling studies of the renal allograft in their reviews. To date no conclusive patterns of miRNA expression have been associated with AR14,105,106 or IF/TA.16,107,108
Metabolomics relates to the global profiling of small molecule (<1500 Da) metabolites. Since the genome, transcriptome and proteome can undergo further changes after translation, the metabolome reflects the downstream products of these processes. There have been advances in separation techniques, such as capillary electrophoresis and ultra-high pressure liquid chromatography (UPLC) systems, and improvements in analytic approaches, being mass spectrometry- based methods and high-resolution nuclear magnetic resonance (NMR) spectroscopy. This has enabled the simultaneous study of dozens of small molecule metabolites in tissue and biological fluids. Whilst the use of non-targeted metabolomic approaches in the study of renal diseases and drug metabolism have been well documented,109–111 studies demonstrating its potential role in monitoring kidney transplants are limited. It has been used to identify borderline tubulitis and acute TCMR in pediatric renal transplant recipients.112 Smaller studies have also been published evaluating the differential urinary and serum metabolomic profile in adult allograft recipients with AR using mass spectrometry based methods,113,114 and NMR spectroscopy was used to profile the urine metabolome in patients with IFTA versus IFTA with inflammation in the DeKAF study. 115
Future directions
The human genome project was the catalyst for what is now recognised as the ‘Omics revolution. Molecular profiling strategies have evolved at an unprecedented rate as a response to the demand for high throughput technology. This has been accompanied by significant advancements in mass spectrometry-based techniques in the field of proteomics, which has further contributed to our understanding of the biological complexities of the allograft recipient. These profiling strategies have the power, not only to shed light on the mechanistic pathways of disease, but also to identify potential diagnostic and predictive biological markers, as well as lead on to the development of therapeutic targets. However, it is also important to recognise the potential limitations of such tools. Beyond this, the major challenge of validating such biomarkers remains. There is a pressing need for multicentre collaboration to allow for large-scale, prospective studies, with better study design and comparative histological evaluation at defined time points. Only then can such discoveries transition from the bench to potentially become diagnostic and prognostic clinical tools, and ultimately achieve the common goal of improving graft longevity and patient survival.
Table 1.
Summary of advantages and disadvantages of current transcriptome assessment technologies
| Advantages | Disadvantages | |
|---|---|---|
| QUANTITATIVE RT-PCR |
|
|
| MICROARRAY |
|
|
| NEXT GENERATION SEQUENCING |
|
|
| NANOSTRING® nCOUNTER ANALYSIS SYSTEM |
|
|
Acknowledgments
Original work by the authors was funded by the NIH (grant 1U01AI070107) and National Health and Medical Research Council of Australia. KK is a recipient of the Jacquot Research Entry Scholarship from the Royal Australasian College of Physicians and the Australian and New Zealand Society of Nephrology. POC is a recipient of an NHMRC Practitioner Fellowship.
ABBREVIATIONS
- GEP
genome-wide expression profiles
- ABMR
antibody mediated rejection
- ACR
acute cellular rejection
- AR
acute rejection
- CAI
chronic allograft injury
- CAN
chronic allograft nephropathy
- DSA
donor specific antibodies
- IF/TA
interstitial fibrosis/tubular atrophy
- miRNA
microRNA
- MS
mass spectrometry
- PBMC
peripheral blood mononuclear cells
- qPCR
quantitative polymerase chain reaction
- TCMR
T-cell mediated rejection
Footnotes
All authors declare no conflicts of interest.
Assistant Professor Madhav C. Menon1, co-author of the paper.
Dr Karen L. Keung2, co-author of the paper.
Professor Barbara Murphy1, editing of the paper.
Professor Philip O’Connell2, contributed to the writing, and editing of the paper.
Contributor Information
Madhav C. Menon, Email: madhav.menon@mssm.edu.
Karen L. Keung, Email: kkeu6183@uni.sydney.edu.au.
Barbara Murphy, Email: barbara.murphy@mssm.edu.
Philip J. O’Connell, Email: philip.oconnell@sydney.edu.au.
References
- 1.Akalin E, Murphy B. Gene polymorphisms and transplantation. Curr Opin Immunol. 2001 Oct;13(5):572–576. doi: 10.1016/s0952-7915(00)00261-2. [DOI] [PubMed] [Google Scholar]
- 2.Gautreaux MD, Freedman BI. Genotypic variation and outcomes in kidney transplantation: donor and recipient effects. Kidney Int. 2013 Sep;84(3):431–433. doi: 10.1038/ki.2013.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akalin E, O'Connell PJ. Genomics of chronic allograft injury. Kidney Int Suppl. 2010 Dec;(119):S33–37. doi: 10.1038/ki.2010.420. [DOI] [PubMed] [Google Scholar]
- 4.Ying L, Sarwal M. In praise of arrays. Pediatr Nephrol. 2009 Sep;24(9):1643–1659. doi: 10.1007/s00467-008-0808-z. quiz 1655, 1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akalin E, Hendrix RC, Polavarapu RG, et al. Gene expression analysis in human renal allograft biopsy samples using high-density oligoarray technology. Transplantation. 2001 Sep 15;72(5):948–953. doi: 10.1097/00007890-200109150-00034. [DOI] [PubMed] [Google Scholar]
- 6.Furness PN, Taub N, Assmann KJ, et al. International variation in histologic grading is large, and persistent feedback does not improve reproducibility. Am J Surg Pathol. 2003 Jun;27(6):805–810. doi: 10.1097/00000478-200306000-00012. [DOI] [PubMed] [Google Scholar]
- 7.Reeve J, Sellares J, Mengel M, et al. Molecular diagnosis of T cell-mediated rejection in human kidney transplant biopsies. Am J Transplant. 2013 Mar;13(3):645–655. doi: 10.1111/ajt.12079. [DOI] [PubMed] [Google Scholar]
- 8.Reeve J, Einecke G, Mengel M, et al. Diagnosing rejection in renal transplants: a comparison of molecular- and histopathology-based approaches. Am J Transplant. 2009 Aug;9(8):1802–1810. doi: 10.1111/j.1600-6143.2009.02694.x. [DOI] [PubMed] [Google Scholar]
- 9.Sarwal M, Chua MS, Kambham N, et al. Molecular heterogeneity in acute renal allograft rejection identified by DNA microarray profiling. N Engl J Med. 2003 Jul 10;349(2):125–138. doi: 10.1056/NEJMoa035588. [DOI] [PubMed] [Google Scholar]
- 10.Park W, Griffin M, Grande JP, Cosio F, Stegall MD. Molecular evidence of injury and inflammation in normal and fibrotic renal allografts one year posttransplant. Transplantation. 2007 Jun 15;83(11):1466–1476. doi: 10.1097/01.tp.0000265501.33362.d3. [DOI] [PubMed] [Google Scholar]
- 11.Scherer A, Krause A, Walker JR, Korn A, Niese D, Raulf F. Early prognosis of the development of renal chronic allograft rejection by gene expression profiling of human protocol biopsies. Transplantation. 2003 Apr 27;75(8):1323–1330. doi: 10.1097/01.TP.0000068481.98801.10. [DOI] [PubMed] [Google Scholar]
- 12.Willis JC, Lord GM. Immune biomarkers: the promises and pitfalls of personalized medicine. Nat Rev Immunol. 2015 May;15(5):323–329. doi: 10.1038/nri3820. [DOI] [PubMed] [Google Scholar]
- 13.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008 Sep 4;455(7209):64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilflingseder J, Regele H, Perco P, et al. miRNA profiling discriminates types of rejection and injury in human renal allografts. Transplantation. 2013 Mar 27;95(6):835–841. doi: 10.1097/TP.0b013e318280b385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anglicheau D, Sharma VK, Ding R, et al. MicroRNA expression profiles predictive of human renal allograft status. Proc Natl Acad Sci U S A. 2009 Mar 31;106(13):5330–5335. doi: 10.1073/pnas.0813121106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ben-Dov IZ, Muthukumar T, Morozov P, Mueller FB, Tuschl T, Suthanthiran M. MicroRNA sequence profiles of human kidney allografts with or without tubulointerstitial fibrosis. Transplantation. 2012 Dec 15;94(11):1086–1094. doi: 10.1097/TP.0b013e3182751efd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Susztak K. Understanding the epigenetic syntax for the genetic alphabet in the kidney. J Am Soc Nephrol. 2014 Jan;25(1):10–17. doi: 10.1681/ASN.2013050461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Strehlau J, Pavlakis M, Lipman M, et al. Quantitative detection of immune activation transcripts as a diagnostic tool in kidney transplantation. Proc Natl Acad Sci U S A. 1997 Jan 21;94(2):695–700. doi: 10.1073/pnas.94.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharma VK, Bologa RM, Li B, et al. Molecular executors of cell death--differential intrarenal expression of Fas ligand, Fas, granzyme B, and perforin during acute and/or chronic rejection of human renal allografts. Transplantation. 1996 Dec 27;62(12):1860–1866. doi: 10.1097/00007890-199612270-00031. [DOI] [PubMed] [Google Scholar]
- 20.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009 Jan;10(1):57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geiss GK, Bumgarner RE, Birditt B, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotech. 2008;26(3):317–325. doi: 10.1038/nbt1385. 03//print. [DOI] [PubMed] [Google Scholar]
- 22.Shannon CPBR, Ng RT, Wilson-McManus JE, Keown P, et al. Two-stage, IN Silico Deconvolution of the Lymphocyte Compartment of the Peripheral Whole blood Transcriptome in the Context of Acute Kidney Allograft Rejection. PLoS One. 2014;9(4):e95244. doi: 10.1371/journal.pone.0095224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oghumu S1BA, Nori U, Maclean KH, Balada-Lasat JM, Brodsky S, Pelletier R, Henry M, Satoskar AR, Nadasdy T, Satoskar AA. Acute pyelonephritis in renal allografts: a new role for microRNAs? Transplantation. 2014;97(5):559–68. doi: 10.1097/01.TP.0000441322.95539.b3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suthanthiran M, Strom TB. Renal transplantation. N Engl J Med. 1994 Aug 11;331(6):365–376. doi: 10.1056/NEJM199408113310606. [DOI] [PubMed] [Google Scholar]
- 25.Nankivell BJ, Alexander SI. Rejection of the kidney allograft. N Engl J Med. 2010 Oct 7;363(15):1451–1462. doi: 10.1056/NEJMra0902927. [DOI] [PubMed] [Google Scholar]
- 26.Halloran PF. T cell-mediated rejection of kidney transplants: a personal viewpoint. Am J Transplant. 2010 May;10(5):1126–1134. doi: 10.1111/j.1600-6143.2010.03053.x. [DOI] [PubMed] [Google Scholar]
- 27.Racusen LC, Haas M. Antibody-mediated rejection in renal allografts: lessons from pathology. Clin J Am Soc Nephrol. 2006 May;1(3):415–420. doi: 10.2215/CJN.01881105. [DOI] [PubMed] [Google Scholar]
- 28.Rush DN, Henry SF, Jeffery JR, Schroeder TJ, Gough J. Histological findings in early routine biopsies of stable renal allograft recipients. Transplantation. 1994 Jan;57(2):208–211. doi: 10.1097/00007890-199401001-00009. [DOI] [PubMed] [Google Scholar]
- 29.Seron D, Moreso F, Bover J, et al. Early protocol renal allograft biopsies and graft outcome. Kidney Int. 1997 Jan;51(1):310–316. doi: 10.1038/ki.1997.38. [DOI] [PubMed] [Google Scholar]
- 30.El Ters M, Grande JP, Keddis MT, et al. Kidney allograft survival after acute rejection, the value of follow-up biopsies. Am J Transplant. 2013 Sep;13(9):2334–2341. doi: 10.1111/ajt.12370. [DOI] [PubMed] [Google Scholar]
- 31.Kee TY, Chapman JR, O'Connell PJ, et al. Treatment of subclinical rejection diagnosed by protocol biopsy of kidney transplants. Transplantation. 2006 Jul 15;82(1):36–42. doi: 10.1097/01.tp.0000225783.86950.c2. [DOI] [PubMed] [Google Scholar]
- 32.Nankivell BJ, Borrows RJ, Fung CL, O'Connell PJ, Allen RD, Chapman JR. Natural history, risk factors, and impact of subclinical rejection in kidney transplantation. Transplantation. 2004 Jul 27;78(2):242–249. doi: 10.1097/01.tp.0000128167.60172.cc. [DOI] [PubMed] [Google Scholar]
- 33.Reeve J, Halloran PF, Kaplan B. Common errors in the implementation and interpretation of microarray studies. Transplantation. 2015 Mar;99(3):470–475. doi: 10.1097/TP.0000000000000691. [DOI] [PubMed] [Google Scholar]
- 34.Flechner SM, Kurian SM, Head SR, et al. Kidney transplant rejection and tissue injury by gene profiling of biopsies and peripheral blood lymphocytes. Am J Transplant. 2004 Sep;4(9):1475–1489. doi: 10.1111/j.1600-6143.2004.00526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Freitas DG, Sellares J, Mengel M, et al. The nature of biopsies with "borderline rejection" and prospects for eliminating this category. Am J Transplant. 2012 Jan;12(1):191–201. doi: 10.1111/j.1600-6143.2011.03784.x. [DOI] [PubMed] [Google Scholar]
- 36.Sellares J, Reeve J, Loupy A, et al. Molecular diagnosis of antibody-mediated rejection in human kidney transplants. Am J Transplant. 2013 Apr;13(4):971–983. doi: 10.1111/ajt.12150. [DOI] [PubMed] [Google Scholar]
- 37.Famulski KS, de Freitas DG, Kreepala C, et al. Molecular phenotypes of acute kidney injury in kidney transplants. J Am Soc Nephrol. 2012 May;23(5):948–958. doi: 10.1681/ASN.2011090887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li L, Khatri P, Sigdel TK, et al. A peripheral blood diagnostic test for acute rejection in renal transplantation. Am J Transplant. 2012 Oct;12(10):2710–2718. doi: 10.1111/j.1600-6143.2012.04253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ong S, Mannon RB. Genomic and proteomic fingerprints of acute rejection in peripheral blood and urine. Transplant Rev (Orlando) 2015 Apr;29(2):60–67. doi: 10.1016/j.trre.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng XX, Schachter AD, Vasconcellos L, et al. Increased CD40 ligand gene expression during human renal and murine islet allograft rejection. Transplantation. 1998 Jun 15;65(11):1512–1515. doi: 10.1097/00007890-199806150-00022. [DOI] [PubMed] [Google Scholar]
- 41.Li B, Hartono C, Ding R, et al. Noninvasive diagnosis of renal-allograft rejection by measurement of messenger RNA for perforin and granzyme B in urine. N Engl J Med. 2001 Mar 29;344(13):947–954. doi: 10.1056/NEJM200103293441301. [DOI] [PubMed] [Google Scholar]
- 42.Ding R, Li B, Muthukumar T, et al. CD103 mRNA levels in urinary cells predict acute rejection of renal allografts. Transplantation. 2003 Apr 27;75(8):1307–1312. doi: 10.1097/01.TP.0000064210.92444.B5. [DOI] [PubMed] [Google Scholar]
- 43.Tatapudi RR, Muthukumar T, Dadhania D, et al. Noninvasive detection of renal allograft inflammation by measurements of mRNA for IP-10 and CXCR3 in urine. Kidney Int. 2004 Jun;65(6):2390–2397. doi: 10.1111/j.1523-1755.2004.00663.x. [DOI] [PubMed] [Google Scholar]
- 44.Kotsch K, Mashreghi MF, Bold G, et al. Enhanced granulysin mRNA expression in urinary sediment in early and delayed acute renal allograft rejection. Transplantation. 2004 Jun 27;77(12):1866–1875. doi: 10.1097/01.tp.0000131157.19937.3f. [DOI] [PubMed] [Google Scholar]
- 45.Muthukumar T, Dadhania D, Ding R, et al. Messenger RNA for FOXP3 in the urine of renal-allograft recipients. N Engl J Med. 2005 Dec 1;353(22):2342–2351. doi: 10.1056/NEJMoa051907. [DOI] [PubMed] [Google Scholar]
- 46.Afaneh C, Muthukumar T, Lubetzky M, et al. Urinary cell levels of mRNA for OX40, OX40L, PD-1, PD-L1, or PD-L2 and acute rejection of human renal allografts. Transplantation. 2010 Dec 27;90(12):1381–1387. doi: 10.1097/TP.0b013e3181ffbadd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suthanthiran M, Schwartz JE, Ding R, et al. Urinary-cell mRNA profile and acute cellular rejection in kidney allografts. N Engl J Med. 2013 Jul 4;369(1):20–31. doi: 10.1056/NEJMoa1215555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hricik DE, Nickerson P, Formica RN, et al. Multicenter Validation of Urinary CXCL9 as a Risk-Stratifying Biomarker for Kidney Transplant Injury. Am J Transplant. 2013;13(10):2634–2644. doi: 10.1111/ajt.12426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matignon M, Ding R, Dadhania DM, et al. Urinary cell mRNA profiles and differential diagnosis of acute kidney graft dysfunction. J Am Soc Nephrol. 2014 Jul;25(7):1586–1597. doi: 10.1681/ASN.2013080900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Keslar KS, Lin M, Zmijewska AA, et al. Multicenter evaluation of a standardized protocol for noninvasive gene expression profiling. Am J Transplant. 2013 Jul;13(7):1891–1897. doi: 10.1111/ajt.12284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Khatri P, Roedder S, Kimura N, et al. A common rejection module (CRM) for acute rejection across multiple organs identifies novel therapeutics for organ transplantation. J Exp Med. 2013 Oct 21;210(11):2205–2221. doi: 10.1084/jem.20122709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li L, Khush K, Hsieh SC, et al. Identification of common blood gene signatures for the diagnosis of renal and cardiac acute allograft rejection. PLoS One. 2013;8(12):e82153. doi: 10.1371/journal.pone.0082153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roedder S, Sigdel T, Salomonis N, et al. The kSORT assay to detect renal transplant patients at high risk for acute rejection: results of the multicenter AART study. PLoS Med. 2014 Nov;11(11):e1001759. doi: 10.1371/journal.pmed.1001759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dahan K, Audard V, Roudot-Thoraval F, et al. Renal allograft biopsies with borderline changes: predictive factors of clinical outcome. Am J Transplant. 2006 Jul;6(7):1725–1730. doi: 10.1111/j.1600-6143.2006.01348.x. [DOI] [PubMed] [Google Scholar]
- 55.Haas M, Sis B, Racusen LC, et al. Banff 2013 meeting report: inclusion of c4d-negative antibody-mediated rejection and antibody-associated arterial lesions. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2014 Feb;14(2):272–283. doi: 10.1111/ajt.12590. [DOI] [PubMed] [Google Scholar]
- 56.Halloran PF, Pereira AB, Chang J, et al. Microarray diagnosis of antibody-mediated rejection in kidney transplant biopsies: an international prospective study (INTERCOM) Am J Transplant. 2013 Nov;13(11):2865–2874. doi: 10.1111/ajt.12465. [DOI] [PubMed] [Google Scholar]
- 57.Perco P, Pleban C, Kainz A, Lukas A, Mayer B, Oberbauer R. Gene expression and biomarkers in renal transplant ischemia reperfusion injury. Transpl Int. 2007 Jan;20(1):2–11. doi: 10.1111/j.1432-2277.2006.00376.x. [DOI] [PubMed] [Google Scholar]
- 58.Hauser P, Schwarz C, Mitterbauer C, et al. Genome-wide gene-expression patterns of donor kidney biopsies distinguish primary allograft function. Lab Invest. 2004 Mar;84(3):353–361. doi: 10.1038/labinvest.3700037. [DOI] [PubMed] [Google Scholar]
- 59.Famulski KS, Broderick G, Einecke G, et al. Transcriptome analysis reveals heterogeneity in the injury response of kidney transplants. Am J Transplant. 2007 Nov;7(11):2483–2495. doi: 10.1111/j.1600-6143.2007.01980.x. [DOI] [PubMed] [Google Scholar]
- 60.Obeidat MA, Luyckx VA, Grebe SO, et al. Post-transplant nuclear renal scans correlate with renal injury biomarkers and early allograft outcomes. Nephrol Dial Transplant. 2011 Sep;26(9):3038–3045. doi: 10.1093/ndt/gfq814. [DOI] [PubMed] [Google Scholar]
- 61.Nankivell BJ, Borrows RJ, Fung CL, O'Connell PJ, Allen RD, Chapman JR. The natural history of chronic allograft nephropathy. N Engl J Med. 2003 Dec 11;349(24):2326–2333. doi: 10.1056/NEJMoa020009. [DOI] [PubMed] [Google Scholar]
- 62.Nankivell BJ, Chapman JR. Chronic allograft nephropathy: current concepts and future directions. Transplantation. 2006 Mar 15;81(5):643–654. doi: 10.1097/01.tp.0000190423.82154.01. [DOI] [PubMed] [Google Scholar]
- 63.El-Zoghby ZM, Stegall MD, Lager DJ, et al. Identifying specific causes of kidney allograft loss. Am J Transplant. 2009 Mar;9(3):527–535. doi: 10.1111/j.1600-6143.2008.02519.x. [DOI] [PubMed] [Google Scholar]
- 64.Einecke G, Sis B, Reeve J, et al. Antibody-mediated microcirculation injury is the major cause of late kidney transplant failure. Am J Transplant. 2009 Nov;9(11):2520–2531. doi: 10.1111/j.1600-6143.2009.02799.x. [DOI] [PubMed] [Google Scholar]
- 65.Naesens M, Kuypers DR, De Vusser K, et al. The histology of kidney transplant failure: a long-term follow-up study. Transplantation. 2014 Aug 27;98(4):427–435. doi: 10.1097/TP.0000000000000183. [DOI] [PubMed] [Google Scholar]
- 66.Park WD, Griffin MD, Cornell LD, Cosio FG, Stegall MD. Fibrosis with inflammation at one year predicts transplant functional decline. J Am Soc Nephrol. 2010 Nov;21(11):1987–1997. doi: 10.1681/ASN.2010010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Torres IB, Moreso F, Sarro E, Meseguer A. The Interplay between Inflammation and Fibrosis in Kidney Transplantation. 2014;2014:750602. doi: 10.1155/2014/750602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Scherer A, Gwinner W, Mengel M, et al. Transcriptome changes in renal allograft protocol biopsies at 3 months precede the onset of interstitial fibrosis/tubular atrophy (IF/TA) at 6 months. Nephrol Dial Transplant. 2009 Aug;24(8):2567–2575. doi: 10.1093/ndt/gfp183. [DOI] [PubMed] [Google Scholar]
- 69.Mengel M, Chang J, Kayser D, et al. The molecular phenotype of 6-week protocol biopsies from human renal allografts: reflections of prior injury but not future course. Am J Transplant. 2011 Apr;11(4):708–718. doi: 10.1111/j.1600-6143.2010.03339.x. [DOI] [PubMed] [Google Scholar]
- 70.Naesens M, Khatri P, Li L, et al. Progressive histological damage in renal allografts is associated with expression of innate and adaptive immunity genes. Kidney Int. 2011 Dec;80(12):1364–1376. doi: 10.1038/ki.2011.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vitalone MJ, O'Connell PJ, Jimenez-Vera E, et al. Epithelial-to-mesenchymal transition in early transplant tubulointerstitial damage. J Am Soc Nephrol. 2008 Aug;19(8):1571–1583. doi: 10.1681/ASN.2007050580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vitalone MJ, O'Connell PJ, Wavamunno M, Fung CL, Chapman JR, Nankivell BJ. Transcriptome changes of chronic tubulointerstitial damage in early kidney transplantation. Transplantation. 2010 Mar 15;89(5):537–547. doi: 10.1097/TP.0b013e3181ca7389. [DOI] [PubMed] [Google Scholar]
- 73.Toki D, Zhang W, Hor KL, et al. The role of macrophages in the development of human renal allograft fibrosis in the first year after transplantation. Am J Transplant. 2014 Sep;14(9):2126–2136. doi: 10.1111/ajt.12803. [DOI] [PubMed] [Google Scholar]
- 74.Menon MC, Chuang PY, Li Z, et al. Intronic locus determines SHROOM3 expression and potentiates renal allograft fibrosis. J Clin Invest. 2014 Dec 1; doi: 10.1172/JCI76902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Park WD, Stegall MD. A meta-analysis of kidney microarray datasets: investigation of cytokine gene detection and correlation with rt-PCR and detection thresholds. BMC Genomics. 2007;8:88. doi: 10.1186/1471-2164-8-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kong X, Mas V, Archer KJ. A non-parametric meta-analysis approach for combining independent microarray datasets: application using two microarray datasets pertaining to chronic allograft nephropathy. BMC Genomics. 2008;9:98. doi: 10.1186/1471-2164-9-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Saint-Mezard P, Berthier CC, Zhang H, et al. Analysis of independent microarray datasets of renal biopsies identifies a robust transcript signature of acute allograft rejection. Transpl Int. 2009 Mar;22(3):293–302. doi: 10.1111/j.1432-2277.2008.00790.x. [DOI] [PubMed] [Google Scholar]
- 78.Dosanjh A, Robison E, Mondala T, Head SR, Salomon DR, Kurian SM. Genomic meta-analysis of growth factor and integrin pathways in chronic kidney transplant injury. BMC Genomics. 2013;14:275. doi: 10.1186/1471-2164-14-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mas V, Maluf D, Archer K, et al. Establishing the molecular pathways involved in chronic allograft nephropathy for testing new noninvasive diagnostic markers. Transplantation. 2007 Feb 27;83(4):448–457. doi: 10.1097/01.tp.0000251373.17997.9a. [DOI] [PubMed] [Google Scholar]
- 80.Hotchkiss H, Chu TT, Hancock WW, et al. Differential expression of profibrotic and growth factors in chronic allograft nephropathy. Transplantation. 2006 Feb 15;81(3):342–349. doi: 10.1097/01.tp.0000195773.24217.95. [DOI] [PubMed] [Google Scholar]
- 81.Anglicheau D, Muthukumar T, Hummel A, et al. Discovery and validation of a molecular signature for the noninvasive diagnosis of human renal allograft fibrosis. Transplantation. 2012 Jun 15;93(11):1136–1146. doi: 10.1097/TP.0b013e31824ef181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kurian SM, Heilman R, Mondala TS, et al. Biomarkers for early and late stage chronic allograft nephropathy by proteogenomic profiling of peripheral blood. PLoS One. 2009;4(7):e6212. doi: 10.1371/journal.pone.0006212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nakorchevsky A, Hewel JA, Kurian SM, et al. Molecular mechanisms of chronic kidney transplant rejection via large-scale proteogenomic analysis of tissue biopsies. J Am Soc Nephrol. 2010 Feb;21(2):362–373. doi: 10.1681/ASN.2009060628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Brouard S, Mansfield E, Braud C, et al. Identification of a peripheral blood transcriptional biomarker panel associated with operational renal allograft tolerance. Proc Natl Acad Sci U S A. 2007 Sep 25;104(39):15448–15453. doi: 10.1073/pnas.0705834104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Brouard S, Le Bars A, Dufay A, et al. Identification of a gene expression profile associated with operational tolerance among a selected group of stable kidney transplant patients. Transpl Int. 2011 Jun;24(6):536–547. doi: 10.1111/j.1432-2277.2011.01251.x. [DOI] [PubMed] [Google Scholar]
- 86.Newell KA, Asare A, Kirk AD, et al. Identification of a B cell signature associated with renal transplant tolerance in humans. J Clin Invest. 2010 Jun;120(6):1836–1847. doi: 10.1172/JCI39933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sagoo P, Perucha E, Sawitzki B, et al. Development of a cross-platform biomarker signature to detect renal transplant tolerance in humans. J Clin Invest. 2010 Jun;120(6):1848–1861. doi: 10.1172/JCI39922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Baron D, Ramstein G, Chesneau M, et al. A common gene signature across multiple studies relate biomarkers and functional regulation in tolerance to renal allograft. Kidney Int. 2015 May;87(5):984–995. doi: 10.1038/ki.2014.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Braud C, Baeten D, Giral M, et al. Immunosuppressive drug-free operational immune tolerance in human kidney transplant recipients: Part I. Blood gene expression statistical analysis. J Cell Biochem. 2008 Apr 15;103(6):1681–1692. doi: 10.1002/jcb.21574. [DOI] [PubMed] [Google Scholar]
- 90.Lozano JJ, Pallier A, Martinez-Llordella M, et al. Comparison of transcriptional and blood cell-phenotypic markers between operationally tolerant liver and kidney recipients. Am J Transplant. 2011 Sep;11(9):1916–1926. doi: 10.1111/j.1600-6143.2011.03638.x. [DOI] [PubMed] [Google Scholar]
- 91.Quintana LF, Sole-Gonzalez A, Kalko SG, et al. Urine proteomics to detect biomarkers for chronic allograft dysfunction. J Am Soc Nephrol. 2009 Feb;20(2):428–435. doi: 10.1681/ASN.2007101137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Srivastava M, Eidelman O, Torosyan Y, Jozwik C, Mannon RB, Pollard HB. Elevated expression levels of ANXA11, integrins beta3 and alpha3, and TNF-alpha contribute to a candidate proteomic signature in urine for kidney allograft rejection. Proteomics Clin Appl. 2011 Jun;5(5–6):311–321. doi: 10.1002/prca.201000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sigdel TK, Kaushal A, Gritsenko M, et al. Shotgun proteomics identifies proteins specific for acute renal transplant rejection. Proteomics Clinical Applications. 2010;4(1):32–47. doi: 10.1002/prca.200900124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Banon-Maneus E, Diekmann F, Carrascal M, et al. Two-dimensional difference gel electrophoresis urinary proteomic profile in the search of nonimmune chronic allograft dysfunction biomarkers. Transplantation. 2010 Mar 15;89(5):548–558. doi: 10.1097/TP.0b013e3181c690e3. [DOI] [PubMed] [Google Scholar]
- 95.Wu D, Zhu D, Xu M, et al. Analysis of transcriptional factors and regulation networks in patients with acute renal allograft rejection. J Proteome Res. 2011;10(1):175–181. doi: 10.1021/pr100473w. [DOI] [PubMed] [Google Scholar]
- 96.Loftheim H, Midtvedt K, Hartmann A, et al. Urinary proteomic shotgun approach for identification of potential acute rejection biomarkers in renal transplant recipients. Transplant Res. 2012;1(1):9. doi: 10.1186/2047-1440-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sigdel TK, Salomonis N, Nicora CD, et al. The identification of novel potential injury mechanisms and candidate biomarkers in renal allograft rejection by quantitative proteomics. Mol Cell Proteomics. 2014 Feb;13(2):621–631. doi: 10.1074/mcp.M113.030577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Freue GV, Sasaki M, Meredith A, et al. Proteomic signatures in plasma during early acute renal allograft rejection. Mol Cell Proteomics. 2010 Sep;9(9):1954–1967. doi: 10.1074/mcp.M110.000554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.O'Riordan E, Orlova TN, Mei JJ, et al. Bioinformatic analysis of the urine proteome of acute allograft rejection. J Am Soc Nephrol. 2004;15(12):3240–3248. doi: 10.1097/01.ASN.0000145241.83482.68. [DOI] [PubMed] [Google Scholar]
- 100.Schaub S, Rush D, Wilkins J, et al. Proteomic-based detection of urine proteins associated with acute renal allograft rejection. J Am Soc Nephrol. 2004;15(1):219–227. doi: 10.1097/01.asn.0000101031.52826.be. [DOI] [PubMed] [Google Scholar]
- 101.Schaub S, Wilkins JA, Antonovici M, et al. Proteomic-based identification of cleaved urinary beta2-microglobulin as a potential marker for acute tubular injury in renal allografts. Am J Transplant. 2005 Apr;5(4 Pt 1):729–738. doi: 10.1111/j.1600-6143.2005.00766.x. [DOI] [PubMed] [Google Scholar]
- 102.Ling XB, Sigdel TK, Lau K, et al. Integrative urinary peptidomics in renal transplantation identifies biomarkers for acute rejection. J Am Soc Nephrol. 2010 Apr;21(4):646–653. doi: 10.1681/ASN.2009080876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Scian MJ, Maluf DG, Mas VR. MiRNAs in kidney transplantation: potential role as new biomarkers. Expert Rev Mol Diagn. 2013 Jan;13(1):93–104. doi: 10.1586/erm.12.131. [DOI] [PubMed] [Google Scholar]
- 104.Mas VR, Dumur CI, Scian MJ, Gehrau RC, Maluf DG. MicroRNAs as Biomarkers in Solid Organ Transplantation. Am J Transplant. 2013;13(1):11–19. doi: 10.1111/j.1600-6143.2012.04313.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sui W, Dai Y, Huang Y, Lan H, Yan Q, Huang H. Microarray analysis of MicroRNA expression in acute rejection after renal transplantation. Transpl Immunol. 2008 Apr;19(1):81–85. doi: 10.1016/j.trim.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 106.Lorenzen JM, Volkmann I, Fiedler J, et al. Urinary miR-210 as a mediator of acute T-cell mediated rejection in renal allograft recipients. Am J Transplant. 2011 Oct;11(10):2221–2227. doi: 10.1111/j.1600-6143.2011.03679.x. [DOI] [PubMed] [Google Scholar]
- 107.Scian MJ, Maluf DG, David KG, et al. MicroRNA Profiles in Allograft Tissues and Paired Urines Associate With Chronic Allograft Dysfunction With IF/TA. Am J Transplant. 2011;11(10):2110–2122. doi: 10.1111/j.1600-6143.2011.03666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Glowacki F, Savary G, Gnemmi V, et al. Increased circulating miR-21 levels are associated with kidney fibrosis. PLoS One. 2013;8(2):e58014. doi: 10.1371/journal.pone.0058014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Atzler D, Schwedhelm E, Zeller T. Integrated genomics and metabolomics in nephrology. Nephrol Dial Transplant. 2014 Aug;29(8):1467–1474. doi: 10.1093/ndt/gft492. [DOI] [PubMed] [Google Scholar]
- 110.Weiss RH, Kim K. Metabolomics in the study of kidney diseases. Nat Rev Nephrol. 2012 Jan;8(1):22–33. doi: 10.1038/nrneph.2011.152. [DOI] [PubMed] [Google Scholar]
- 111.Bohra R, Klepacki J, Klawitter J, Klawitter J, Thurman JM, Christians U. Proteomics and metabolomics in renal transplantation-quo vadis? Transpl Int. 2013;26(3):225–241. doi: 10.1111/tri.12003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Blydt-Hansen TD, Sharma A, Gibson IW, Mandal R, Wishart DS. Urinary metabolomics for noninvasive detection of borderline and acute T cell-mediated rejection in children after kidney transplantation. Am J Transplant. 2014 Oct;14(10):2339–2349. doi: 10.1111/ajt.12837. [DOI] [PubMed] [Google Scholar]
- 113.Wang JN, Zhou Y, Zhu TY, Wang X, Guo YL. Prediction of acute cellular renal allograft rejection by urinary metabolomics using MALDI-FTMS. J Proteome Res. 2008 Aug;7(8):3597–3601. doi: 10.1021/pr800092f. [DOI] [PubMed] [Google Scholar]
- 114.Zhao X, Chen J, Ye L, Xu G. Serum metabolomics study of the acute graft rejection in human renal transplantation based on liquid chromatography-mass spectrometry. J Proteome Res. 2014 May 2;13(5):2659–2667. doi: 10.1021/pr5001048. [DOI] [PubMed] [Google Scholar]
- 115.Gourishankar S, Leduc R, Connett J, et al. Pathological and clinical characterization of the 'troubled transplant': data from the DeKAF study. Am J Transplant. 2010 Feb;10(2):324–330. doi: 10.1111/j.1600-6143.2009.02954.x. [DOI] [PMC free article] [PubMed] [Google Scholar]