Abstract
BACKGROUND
Myocyte enhancer factor 2 (MEF2) transcription factors play critical roles in diverse cellular processes during CNS development. Studies attempting to address the role of MEF2 in brain have largely relied on overexpression of a constitutive MEF2 construct that impairs memory formation or knockdown of MEF2 function that increases spine numbers and enhances memory formation. Genetic deletion of individual MEF2 isoforms in brain during embryogenesis demonstrated Mef2c loss negatively regulates spine numbers resulting in learning and memory deficits, possibly due to its essential role in development.
METHODS
To further investigate Mef2c function in brain, we genetically deleted Mef2c during postnatal development in mice. We characterized these conditional Mef2c knockout mice in an array of behavioral paradigms and examined the impact of postnatal loss of Mef2c on long-term potentiation.
RESULTS
We observed increased spine numbers in hippocampus of the conditional Mef2c knockout mice. However, the postnatal loss of Mef2c did not impact learning and memory, long-term potentiation, or social and repetitive behaviors.
CONCLUSIONS
Our findings demonstrate a critical role for MEF2c in the regulation of spine numbers with a dissociation on learning and memory, synaptic plasticity and measures of autism-related behaviors in postnatal brain.
Keywords: MEF2C, behavior, synaptic plasticity, fear conditioning, autism, postnatal brain
INTRODUCTION
Myocyte enhancer factor 2 (MEF2) transcription factors are crucial for cell differentiation and development in peripheral tissues as well as brain (1). MEF2 transcription factors are well-known regulators of gene expression integrating extracellular stimuli that lead to specific post-translational modifications and the recruitment of transcription cofactors. In the mammalian genome the MEF2 family is comprised of 4 gene isoforms, Mef2a–Mef2d, that share high homology with the family of MAD (MCM1-Agamous-Deficiens-Serum response factor) box transcription factors. The Mef2a, c, and d genes are each expressed in a distinctive manner in the mouse brain (2). Mef2c is expressed the earliest in embryonic brain development and maintains high levels of expression in adult brain including the cortex, striatum and hippocampus, suggesting potential roles in embryonic as well as adult brain function (2, 3). Indeed, previous work has demonstrated a key role for MEF2C during the development of several lineages including craniofacial and central nervous system (1, 4). Human genetic studies have also implicated MEF2C as a critical factor in neurodevelopmental disorders, including autism spectrum disorders. However, there is currently a gap in the understanding of the role of endogenous MEF2C in adult brain.
Initial work examining the role of MEF2 in neurons in vitro used an RNA interference construct to knockdown Mef2a and Mef2d in primary hippocampal neuronal cultures and found MEF2 transcription factors negatively regulate excitatory synapse formation in an activity dependent manner (5). The link between MEF2 and synapse formation triggered interest in whether MEF2 also impacts learning and memory processes. Expression of a constitutively active form of MEF2 in adult hippocampus was found to disrupt memory formation while shRNAi directed knockdown of MEF2 or expression of a dominant negative MEF2 construct in adult hippocampus enabled memory formation (6) suggesting that MEF2 activity in the adult brain constrains memory formation (7). However, these studies have relied on constructs that broadly impact MEF2 without examining the contribution of individual isoforms. Constitutive Mef2a null mice and constitutive Mef2c null mice have early lethality (8, 9), thus to examine the function of the individual MEF2 isoforms in behavior has required the generation of conditional knockout mice. Mef2c conditional knockout mice that targeted the deletion selectively to the brain during embryogenesis revealed that Mef2c is the critical isoform in negatively regulating synapse number while conditional deletion of Mef2a and Mef2d has no impact on spine numbers (10, 11). The embryonic deletion of Mef2c in mouse brain resulted in a robust increase in spine density accompanied by increased potentiation of basal and evoked synaptic transmission and impaired hippocampal-dependent learning and memory (11). However, given the critical role of MEF2C in development, it is important to consider that Mef2c deletion during embryogenesis could result in developmental effects that ultimately impact structural and functional phenotypes.
To circumvent this potential limitation and directly examine a role for Mef2c in postnatal brain, we generated conditional Mef2c knockout mice using the calcium/calmodulin-dependent protein kinase II (CaMKII)-Cre93 line that targets the gene deletion in broad forebrain regions including hippocampus at postnatal day 10–14 (12–14). We found that postnatal deletion of Mef2c in the brain results in a significant increase in spine numbers in the hippocampus. The conditional Mef2c knockout mice also display alterations in locomotor activity and motor coordination deficits. However, Mef2c deletion in postnatal brain did not impact learning and memory, measurements of synaptic plasticity, or several behavioral measures suggested to recapitulate aspects of autism spectrum disorders in mice. These data suggest that the effects of MEF2C as a regulator of spine numbers is independent of embryonic and postnatal stages of development presumably due to a continual role of MEF2C in the negative regulation of synaptogenesis. However, the role of MEF2C as a critical mediator of learning and memory, synaptic plasticity, and behavioral measures of ASDs is dependent on MEF2C expression during embryonic development and is not tied unequivocally to changes in spine numbers per se. These data provides fundamental new insight into the importance of developmentally specific genetic manipulation of endogenous Mef2c expression as an essential consideration for elucidating the role of MEF2 in CNS function.
MATERIALS AND METHODS
Generation of Conditional Mef2C KO mice in postnatal brain
Conditional Mef2C knockout (KO) mice were generated by mating floxed Mef2c (fMef2c) mice, which have been previously described (15, 16), to mice transgenic for Cre recombinase under the control of the CaMKII promoter (CaMKII-Cre93) (13).
Quantitative PCR (qPCR)
The qPCR was carried out as previously described (17). The fold change in Mef2c expression relative to Gapdh was calculated as mean ± SEM.
Quantification of Dendritic Spine Density
Golgi staining was performed using the FD rapid Golgi Stain Kit (NeuroTechnologies).
Behavioral Overview
Mice were allowed to habituate for at least 1 hour in the behavioral room prior to testing. All experiments including the scoring of the behavior and data analysis were conducted in a manner blind to genotype. Data were expressed as mean ± SEM, and analyzed by Student t-test unless otherwise noted with significance set at p < 0.05.
Locomotor Activity and Rotarod performance were examined as described previously (18).
Hindlimb Clasping Test An animal was suspended from the tail and an observer blind to the genotype scored the hindlimb clasping, based on the following scale; 0 = normal movement, 1 = intermittent and/or intermediate clasping, 2 = both hindlimbs are fully drawn to the abdomen (19).
Fear Conditioning was examined as previously described (11).
Pain Sensitivity Test Foot shock threshold was measured 1 week after the context-dependent fear conditioning test.
Novelty Object Recognition Test was performed as described in supplementary methods.
Social Interaction Tests were carried out as previously described (17, 20).
Olfaction Test was composed of three consecutive sessions and performed in red light. The time spent sniffing the cotton swabs during each presentation was measured.
Nest Building Test An individual mouse was placed in a fresh cage containing one nestlet and left undisturbed, height and quality of the nests was measured 1 and 4 hrs later.
Grooming Test Each animal was placed in a fresh cage under red light between 10am and 1pm to minimize potential circadian-related behavioral changes and videotaped for 30 min.
Field EPSP Recording were performed as previously described (10).
Please see supplementary methods for a detailed description of the methods.
RESULTS
Postnatal-neuron-specific deletion of Mef2C affects spine density
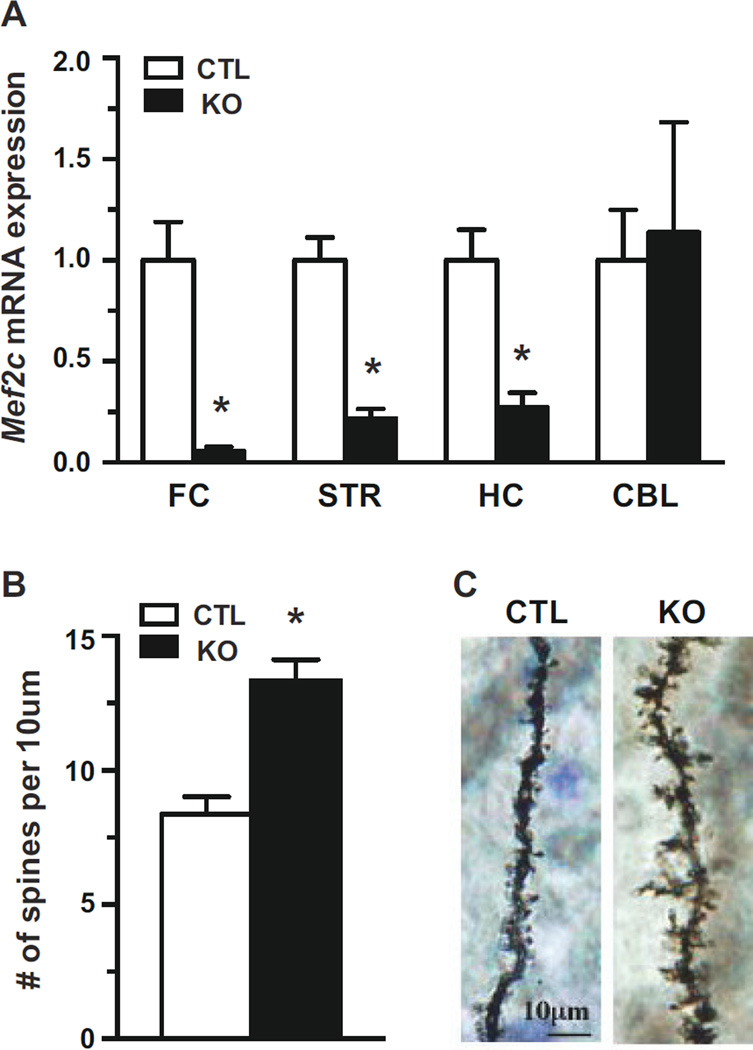
Conditional Mef2c postnatal knockouts (KO) were viable and indistinguishable in appearance from littermate control (CTL) mice. To confirm recombination of the Mef2c allele, mRNA levels were examined in several regions of adult brain by quantitative PCR (qPCR). In conditional KOs, Mef2c mRNA expression was effectively reduced 70–90% in frontal cortex, striatum, and hippocampus compared to CTLs (Figure 1A). However, Mef2c expression was not altered in the cerebellum of Mef2c KOs, in agreement with previous work demonstrating that CaMKII-Cre93 mediates recombination in broad forebrain regions but not the cerebellum (13).
Figure 1.
Forebrain specific knockout of Mef2c leads to increased spine density. (A) Mef2c mRNA expression in frontal cortex (FC), striatum (STR), hippocampus (HC), and cerebellum (CBL) of conditional Mef2c knockout (KO) mice generated using the CaMKII-Cre93 driver line and littermate control (CTL) mice. Levels of Mef2c mRNA were normalized to Gapdh relative to CTL mice. (B) Spine density of granular neurons in the dentate gyrus as assessed by Golgi staining (n=5 mice for both CTL and KO). (C) Representative images of granular neurons from the dentate gyrus (*p < 0.05).
MEF2C has been shown to negatively regulate spine numbers in the hippocampus following embryonic deletion of the gene in brain (11). Therefore, we examined whether postnatal deletion of Mef2c alters spine density in the hippocampus of Mef2c KOs. Golgi staining of granular cells in dentate gyrus revealed a significant increase in the number of spines in the Mef2c KOs (Figure 1B, C), similar to the significant increase observed following deletion of Mef2c in embryonic brain (11).
Mef2c KO mice are hyperactive and display abnormal motor coordination
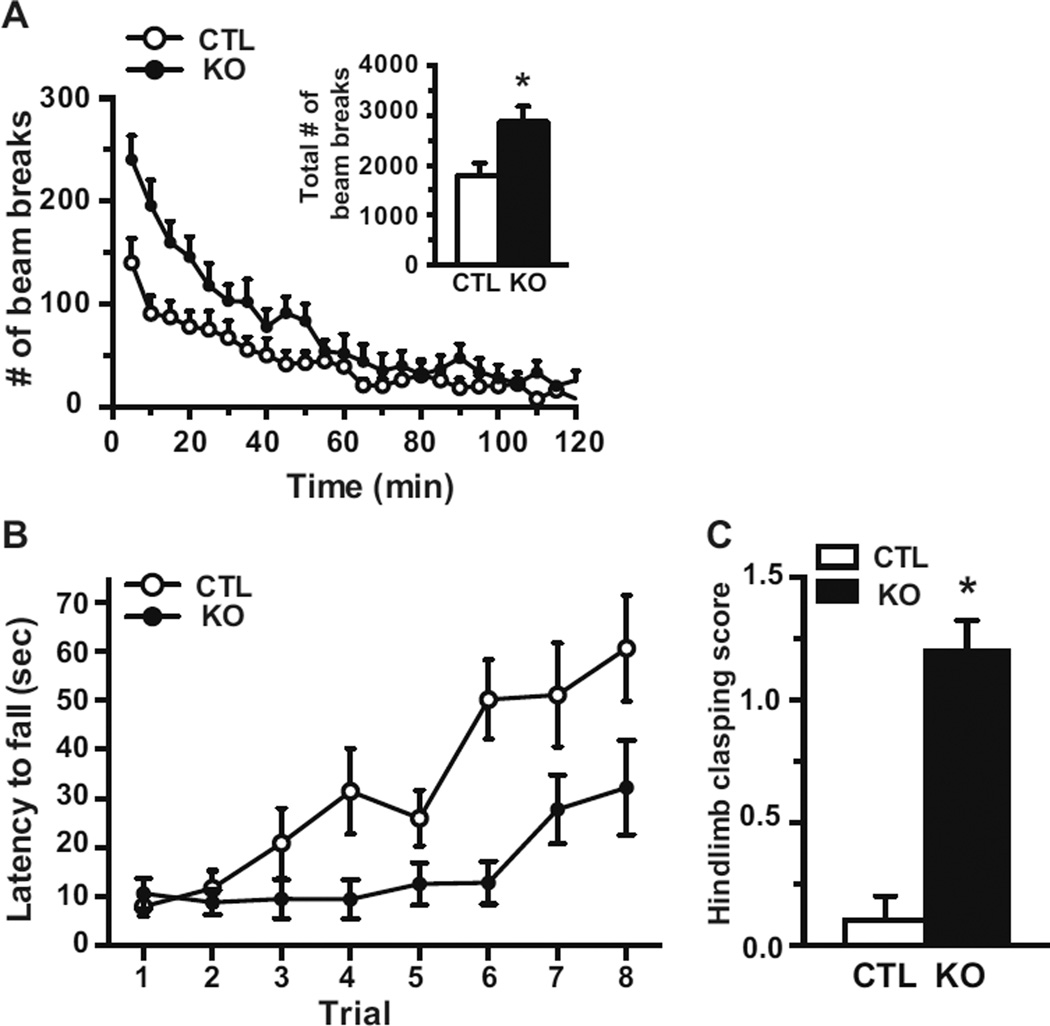
To investigate the impact of brain specific postnatal deletion of Mef2c on behavior, 4–6 month old mice were assessed in several paradigms. Locomotor activity was examined by monitoring mice individually in a fresh home cage for 2 hours and recording the number of horizontal beam breaks. The total activity during the testing period was significantly increased in Mef2c KOs in comparison to CTLs (Figure 2A, inset). Locomotor data analyzed in 5-minute epochs showed Mef2c KOs were significantly hyperactivity upon placement in the boxes and for most of the initial hour (Figure 2A).
Figure 2.
Mef2c KO mice are hyperactive and have impaired motor coordination. (A) Locomotor activity was measured over 2 hrs and the number of beam breaks is presented in 5 min blocks of time. Inset, the total number of beam breaks for the 2 hr time period (CTL, n=9; KO, n=7). (B) Motor coordination was assessed in the rotarod test. Mice performed 4 trials per day for 2 days and the latency to fall from the accelerating rotarod was measured (CTL, n=9; KO, n=8). (C) The severity of hindlimb clasping was scored in mice at 5 months of age (CTL, n=5; KO, n=5) (*p < 0.05).
Mice were next placed on the accelerating rotarod for 4 trials/day for two days until they fell off or until a maximum of 5 minutes/trial. Mef2c KOs had a significantly shorter latency to fall off than CTLs, suggesting deficits in motor coordination (Figure 2B). Of note, the Mef2c KOs had a similar relationship between the latency to fall and the trial number as assessed by the slope of the curve compared to CTLs suggesting a similar learning profile. At ~16 weeks of age the Mef2c KOs displayed an overt hindlimb clasping phenotype that we quantitated using a 3 point scale (19) and presented moderate hindlimb clasping when suspended from the tail (Figure 2C).
Characterization of Mef2c KO mice and autism-related behaviors
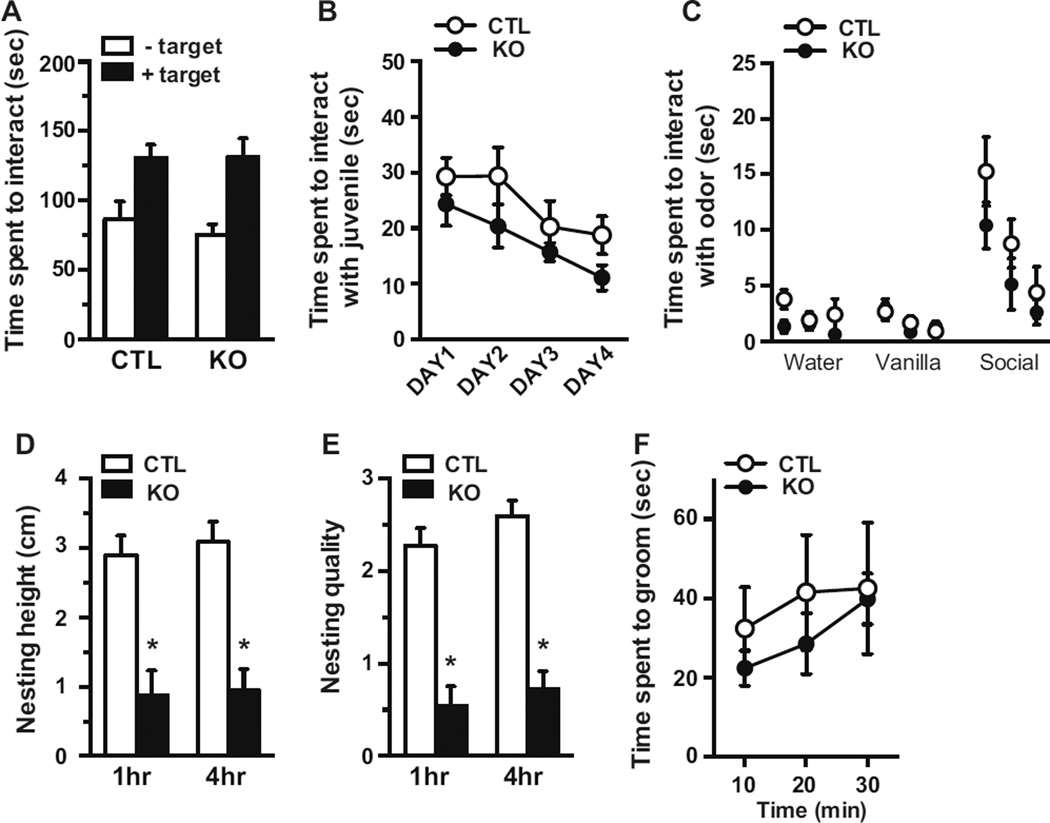
Recent human genetic studies have identified rare variants in the MEF2C gene from subjects with autism spectrum disorders (ASDs) (21, 22). We therefore examined conditional Mef2c KOs in behavioral paradigms suggested to recapitulate aspects of ASDs. Social interaction was assessed by quantifying the amount of time an experimental adult mouse interacted with another adult mouse. In this paradigm, an experimental animal is allowed to explore in an open arena that has an empty wire mesh cage located against one of the walls. After 5 minutes, an adult C57BL/6 is placed behind the wire mesh and the amount of time the experimental animal interacts with the C57BL/6 is measured during a 5-minute period. As expected, CTLs spend significantly more time near the wire mesh when a C57BL/6 mouse is present due to their preference for social interaction. Similarly, conditional Mef2c KOs also spend more time near the wire mesh when a C57BL/6 mouse is present rather than in its absence. Examination of the total interaction time with the C57BL/6 mouse was indistinguishable between Mef2c KOs and CTLs (Figure 3A). To further examine social interactions, we assessed the amount of time an experimental animal directly interacts with a particular juvenile mouse in a 5-minute time period over 4 consecutive days (Day 1 through Day 4). Conditional Mef2c KOs spend a similar amount of time interacting with a juvenile mouse as CTLs for the 4 consecutive days (Figure 3B). Moreover, regardless of genotype, the mice spend less time interacting with the juvenile on Day 4 compared to Day 1, suggesting the mice recognize the social target they have been interacting with in the 4-day test. To rule out potential deficits in olfaction, we measured the amount of time Mef2c KOs spend interacting with cotton balls scented with various scents including social odors, and found no difference between the genotypes (Figure 3C).
Figure 3.
Mef2c KO mice does not alter social interactions. (A) Adult test mice were assessed for their ability to interact with another adult mouse. In the adult social interaction test, the time a mouse spent in the interaction zone was measured in the absence and presence of a target animal placed behind the wire-mesh (CTL, n=11; KO, n=10). (B) Adult mice were also examined for their ability to interact with a juvenile mouse. In the juvenile social interaction test, an adult mouse was presented with the same juvenile mouse on day 1 through day 4 and the time the adult animal interacted with the juvenile animal was recorded. (C) An olfaction test was carried out to assess their sensitivity to odor. The mice were presented with 3 types of scents (water, vanilla, and social odor) three times for each scent. (CTL, n=8; KO, n=10). (D) The animals’ ability to make nests was examined by measuring the height (D) and the quality (E) of the nest using a 5 point scale (CTL, n=11; KO, n=10). (F) The time a mouse spent grooming was measured in 10 min time periods over a 30 min period. (CTL, n=6; KO, n=6)(*p < 0.05).
Patients with ASD often engage in repetitive and obsessive-compulsive-like behaviors. To examine such behavioral phenotypes in rodents, we assessed nest building and grooming behavior, which are thought to be biological characteristics of motor stereotypes (23). As expected, CTL mice were able to build a nest with a high round wall in 1 hour (Figure 3D, E). In contrast, conditional Mef2c KOs were not able to build a nest of the same height or quality in the time period suggesting a deficit in this behavioral measure. To examine grooming behavior, mice were placed in a novel home cage for a 30-minute time period under red light and behavior recorded and then scored. There were no significant differences in the amount of time spent grooming by the Mefc2 KOs compared to CTLs (Figure 3F).
Mef2c KO mice have normal context and cue dependent learning and memory
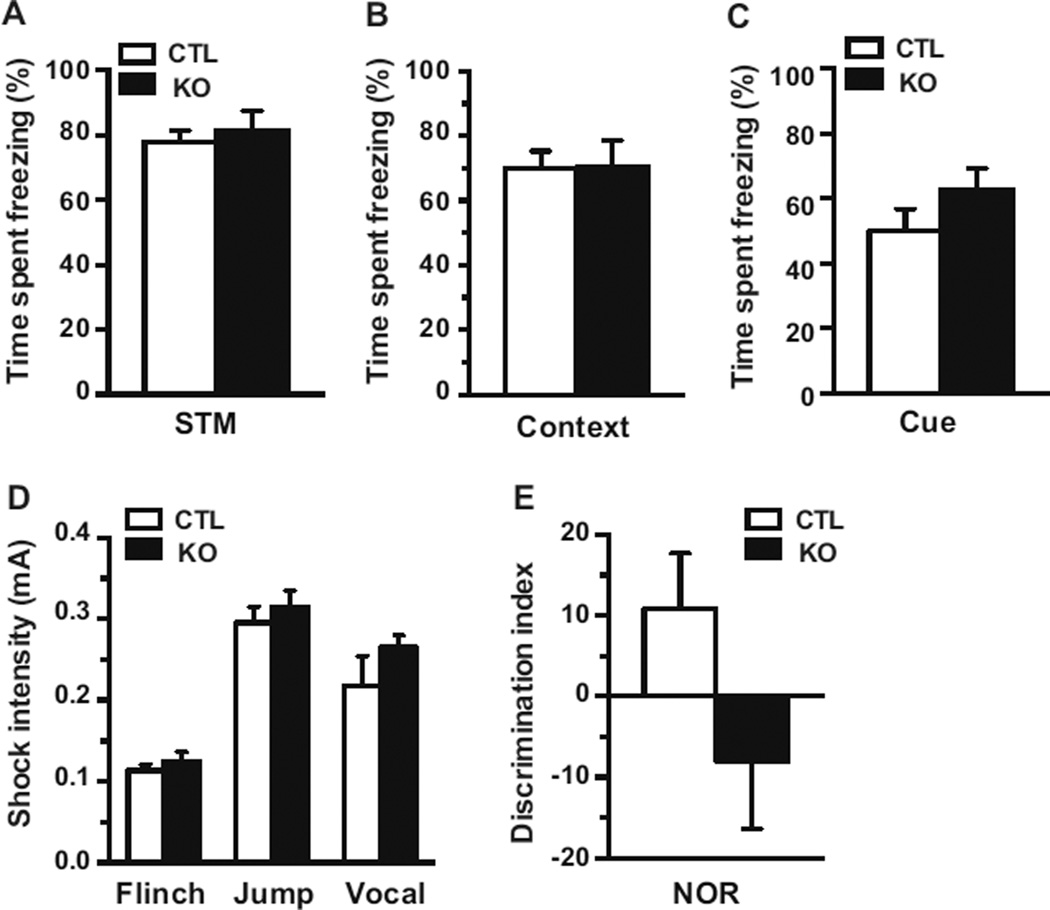
Given that the embryonic deletion of Mef2c in the brain results in impairments in context dependent fear conditioning, we examined the conditional Mef2c KOs in this paradigm. Baseline freezing of the Mef2c KOs was indistinguishable with CTLs during the training period (data not shown). Short-term memory assessed 90 minutes after training revealed similar levels of freezing with the Mef2c KOs compared to CTLs (Figure 4A). Long-term memory assessed 24 hours after training rather surprisingly demonstrated that Mef2c KOs displayed a similar level of freezing as CTLs, suggesting normal context-dependent memory (Figure 4B). A separate cohort of animals was assessed for cue-dependent fear conditioning. Cue-dependent memory 24 hours after training was also indistinguishable between Mef2C KO and CTL mice (Figure 4C). There was no difference between the genotypes in foot shock thresholds required for the mice to flinch, vocalize, and jump ruling out differences in nociception as a confounding variable (Figure 4D).
Figure 4.
Postnatal deletion of Mef2c does not impact learning and memory. (A) Short term memory of the conditional Mef2c KO mice was indistinguishable from littermate controls 90 minutes after training (CTL, n=11; KO, n=10). (B) Context-dependent fear conditioning was assessed 24 hrs after training (CTL, n=11; KO, n=10). (C) Cue-dependent fear conditioning was examined 24 hrs after the training (CTL, n=9; KO, n=7). (D) Pain sensitivity was assessed by measuring thresholds of foot shock intensities to flinch, jump, and vocalize. (CTL, n=11; KO, n=10). (E) Mice were assessed in their performance in the novelty recognition test 24 hrs after the training (CTL, N=9; KO, N=7).
To mice were also tested in the novel object recognition paradigm, a single trial memory test in which the animal has to discriminate between a familiar and unfamiliar object. Mice are allowed to familiarize with two objects in an open field, and then 24 hours later the mice are presented with one familiar and one novel object in the same arena. The CTLs spent more time with the novel object than the familiar object as indicated by a positive value for the discrimination index. The conditional Mef2c KOs had no significant difference in the discrimination index relative to CTLs.
Conditional Mef2c KO mice have normal hippocampal LTP
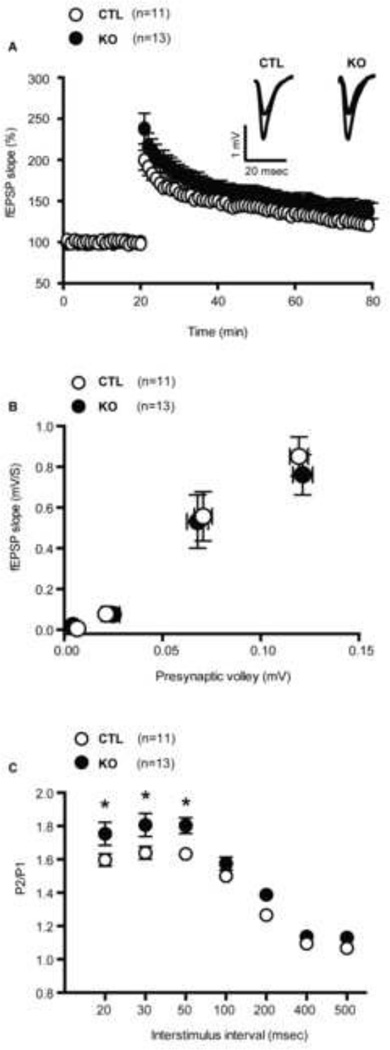
We performed field recordings on hippocampal slices to examine whether postnatal loss of Mef2c was associated with changes in long-term potentiation (LTP). LTP was induced in the CA1 region of the hippocampus by stimulating the Schaffer-collateral pathway with theta burst stimulation (TBS). There were no significant difference in TBS-induced LTP between Mef2c KO and CTLs for the duration of the recordings (Figure 5A), suggesting postnatal loss of Mef2c does not impact long-term synaptic plasticity. We examined whether the conditional Mef2c KOs have alterations in baseline synaptic transmission and short-term plasticity as assessed by input/output (I/O) relationships and paired pulse facilitation (PPF), respectively. The I/O curve of the presynaptic volley amplitude versus the fEPSP slope constructed from hippocampal slices of Mef2c KOs were similar to CTLs (Figure 5B) suggesting that basal synaptic transmission is unaltered following the postnatal loss of Mef2c. However, PPF was significantly augmented in the Mef2c KOs at interstimulation intervals of 20, 30 and 50 ms (Figure 5C) consistent with a decrease in presynaptic release probability.
Figure 5.
Conditional Mef2c knockouts have unimpaired long term potentiation. (A) LTP in the hippocampus induced by theta burst was normal in conditional Mef2c KO mice in comparison to control mice. (B) Input-output curves were measured in hippocampal slices from CTL and Mef2c KO mice. Field potential slopes were plotted as a function of presynaptic volley at 0.01, 0.025, 0.075, 0.125 mV stimulation intensities. (C) Paired pulse ratio of the first 2 responses elicited by various stimulation frequencies at Schaeffer collateral synapses. Mef2c KO mice exhibited significantly augmented PPF at Interstimulus intervals of 20 ms (t(22) = 1.9, p < 0.05), 30 ms (t(22) = 2.1, p < 0.05) and 50 ms (t(22) = 2.9, p < 0.05), indicating decreased probability of neurotransmitter release (CTL, n=11; KO, n=13 for all the experiments)(*p < 0.05).
DISCUSSION
Our results reveal a previously unknown role for MEF2C as a negative regulator of synaptogenesis in postnatal development in isolation from its putative role in learning and memory, LTP and social interaction behaviors. Conditional Mef2c KO generated using the CaMKII-Cre93 driver line to delete Mef2c postnatally in broad forebrain regions resulted in a significant increase in spine numbers in hippocampus. However, the increase in spine numbers was not correlated with alterations in learning and memory, measures of LTP, or behavioral alterations in social interaction in the conditional Mef2c KO mice. These data are in striking contrast to previous observations using mice with an embryonic deletion of Mef2c in the brain, which show an increase in the number of spines in the hippocampus and significant deficits in learning and memory as well as synaptic transmission (11). These data reveal that MEF2C plays a critical role in the negative regulation of synaptogenesis during postnatal development as well as embryogenesis as previously reported. However, the differing effect on learning and memory, LTP and behavioral measures of autism related behaviors suggests that MEF2C plays a critical role during embryonic development that goes beyond its regulation of synapse number. Taken together, these data also substantiate the critical importance of specific genetic manipulation of endogenous Mef2c expression to elucidate its precise role in the brain.
The postnatal deletion of Mef2c resulted in a significant increase in the number of spines in granular neurons of the dentate gyrus. The expression of MEF2C is highly enriched in the dentate gyrus with only low level of expression in the CA1 region of the hippocampus (2, 3), thus our focus on the contribution of MEF2C on spines in the dentate gyrus. The significant increase in the number of spines in the conditional Mef2c knockout mice in which the deletion was induced postnatally was similar to that observed following embryonic deletion of Mef2c demonstrating that MEF2C is a key regulator of synapse number in the hippocampus (11). In our analysis we focused on male Mef2c knockout mice as estrogen levels are known to enhance dendritic spines on hippocampal pyramidal cells, long-term potentiation and learning and memory, all processes that were being investigated in the present study (24–26). Collectively, the current data extend previous findings on the role of MEF2C as a critical negative regulator of spine number in the hippocampus in postnatal brain.
The conditional Mef2c KO mice were hyperactive and presented deficits in the rotarod test similar to the phenotypes observed in mice in which Mef2c was selectively deleted in embryonic brain (11). While there is a learning component to the rotarod test, the Mef2c KOs had a similar relationship between the latency to fall and the trial number as assessed by the slope of the curve compared to CTLs suggesting a similar learning profile. The postnatal deletion of Mef2c did not impact context dependent fear conditioning which is in contrast to that observed in the embryonic brain specific Mef2c KOs. The conditional Mef2c KOs performance was also indistinguishable from CTLs in the novel object recognition test. Taken together, our data suggests that postnatal deletion of Mef2c in the brain does not impact learning and memory in contrast to embryonic knockout of Mef2c. While the present study utilized a forebrain specific knockout to identify the role of MEF2C in postnatal brain, it is possible that the differences in learning and memory phenotypes observed in the embryonic knockout of Mef2c could be influenced by the regional loss of MEF2C. However, both the embryonic and postnatal lines had loss of Mef2c in the hippocampus, a region involved in learning and memory, and a subsequent increase in synaptogenesis suggesting a differential developmental role for MEF2C and its influence on learning and memory. In previous work we analyzed the patterns of Cre-mediated recombination in the CNS of the CaMKII-Cre (used in the present study) and hGFAP-Cre (used to generate the embryonic KO) transgenic lines by crossing to reporter mice and found both lines mediate recombination in a spatially restricted expression pattern in the forebrain that is restricted to neurons (27). These data support work showing that neuroblasts and new neurons are derived from GFAP-expressing progenitor cells (28). MEF2 gene expression is very low in glial (29) suggesting that the phenotypes reported in the previous hGFAP-Cre conditional KOs involve loss of Mef2c in neurons similar to the CaMKII-Cre conditional KOs.
The postnatal deletion of Mef2c did not impact LTP recorded from the CA1 region of hippocampus following theta burst stimulation of the Schaffer collateral pathway. There was also no change in the input-output curve constructed from hippocampal slices of Mef2c KOs suggesting that basal synaptic transmission is unaltered. However, PPF, an indicator of presynaptic neurotransmitter release probability, was increased in postnatal Mef2c KOs suggesting alterations in short-term plasticity. Interestingly, in contrast to our current observations, in the earlier study we observed a decrease in PPF in the dentate gyrus after loss of Mef2c during embryogenesis (11). PPF is a measure of short-term enhancement of synaptic activity in response to the second of two paired stimuli due to residual Ca2+ in the presynaptic terminal after the initial stimulus. A decrease in this form of presynaptic plasticity is associated with an increase in the probability of neurotransmitter release. Therefore, this putative presynaptic function for MEF2C dependent transcription may have been a key determinant of the learning and memory deficits observed earlier (11). While the embryonic Mef2c KOs had significant learning and memory deficits in previous work, LTP was not assessed leaving it an open question as to the impact of the embryonic deletion on measures of LTP and synaptic plasticity.
Microdeletion of human chromosome 5q14.3, the region encompassing the MEF2C gene, or haploinsufficiency of MEF2C, has been detected in individuals with severe mental retardation, epilepsy, stereotypic behavior, and muscle hypotonia (22, 30–37). MEF2 transcription factors have also been implicated in the altered gene expression patterns that are associated with ASDs (38). Therefore, we examined whether loss of Mef2c in postnatal neurons impacts social behavior and others measures attributed to model aspects of autistic like behavior in mice. The conditional Mef2c KOs interacted with another adult mouse, as well as with a juvenile mouse, similar to CTLs suggesting that postnatal loss of Mef2c did not impact social behavior. The Mef2c KOs did have significant impairments in nest building in regards to the height and quality of the nest compared to CTLs. However, we cannot fully exclude the possibility that this impairment could be due to abnormal motor dysfunction observed in the postnatal Mef2c KOs. The postnatal Mef2c KOs displayed normal grooming behavior, a measure of repetitive type behavior that is a feature of ASD. Collectively, these data suggest that the postnatal deletion of Mef2c in brain does not impact social interactions or most other measures of ASD related behaviors examined in this study. Conditional deletion of Mef2c in embryonic neural progenitor stem cells in mice results in deficits in learning as well as behavioral abnormalities suggestive of autistic-like behavior (39). Given that ASDs are neurodevelopmental disorders, these data suggest that at least in regards to MEF2C its loss during embryonic development may be important in initiating autistic-like behavior with only mild alterations observed following postnatal forebrain deletion. These data further suggest that while changes in synapse number, both increases and decreases have been associated with certain ASDs, changes in synapse number that occur later in life may be dissociated from the behavioral phenotypes of ASD.
Studies over the past few years have suggested that MEF2 activity constrains memory formation (7). Increasing MEF2 activity in the hippocampus through the use of a constitutive active form of MEF2 disrupted memory formation (6). Conversely, shRNAi directed knock down of MEF2 or expression of a dominant negative MEF2 construct in the hippocampus enabled memory formation (6). Studies expressing a constitutive active form of MEF2 in other brain regions including the amygdala (6) and anterior cingulate cortex (40) have also demonstrated impairments in memory formation. The expression of a constitutive active form of MEF2 in the nucleus accumbens has been shown to impact cocaine mediated sensitization (41). Together, these data have supported a critical role for MEF2 in memory processes, however these studies did not examine the role of endogenous MEF2 isoforms but instead relied on overexpression, dominant negative, or knockdown constructs that broadly impact MEF2 function. In the current study we used a genetic approach to selectively delete Mef2c in the forebrain, and in agreement with other studies examining the genetic loss of Mef2 isoforms in the brain, including Mef2c, do not find an enabling of memory formation (10, 11). These data highlight the importance genetic approaches in the study of endogenous MEF2 isoforms in the brain to prevent potential confounds and off-target effects that may result from other approaches.
The results of the present study demonstrate that MEF2C is a critical negative regulator of synapse numbers in the hippocampus even into postnatal development. These data support previous work for a critical role of MEF2C mediating transcription processes in controlling synapse number (11). However, these changes in synapse number did not impact learning and memory, LTP or most behavioral measures of autism-related phenotypes suggesting a dissociation between structural changes and these behavioral and functional processes later in development. The postnatal loss of Mef2c triggered increases in the number of spines in the hippocampus may have negligible effects on further structural plasticity or activity-mediated synaptogenesis although this will have to be further explored in future experiments. It should be noted that the Mef2c forebrain deletion occurred postnatally but at a time of rapid brain development, postnatal 10–14 and that mice were not tested until they were adults. Therefore the resulting phenotypes may be due to lack of Mef2c during adolescent development that could impact the behavioral analysis of adults. Collectively, these results provide novel information regarding the dysregulation of synapse number caused by the postnatal loss of MEF2C and the demonstration that these structural changes do not necessarily contribute to deficits in activity dependent processes such as learning and memory later in life, suggesting key developmental components that are involved in mediating these effects.
Supplementary Material
ACKNOWLEDGMENTS
We thank M. Mahgoub and E.T. Kavalali for critical comments, members of the Monteggia laboratory for help and suggestions, and E.N. Olson for floxed Mef2c mice. This work was supported by MH081060 to L.M.M.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
COMPETING FINANCIAL INTERESTS
The authors declare no biomedical financial interests or potential conflicts of interest.
REFERENCES
- 1.Potthoff MJ, Olson EN. MEF2: a central regulator of diverse developmental programs. Development. 2007;134:4131–4140. doi: 10.1242/dev.008367. [DOI] [PubMed] [Google Scholar]
- 2.Lyons GE, Micales BK, Schwarz J, Martin JF, Olson EN. Expression of mef2 genes in the mouse central nervous system suggests a role in neuronal maturation. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1995;15:5727–5738. doi: 10.1523/JNEUROSCI.15-08-05727.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leifer D, Golden J, Kowall NW. Myocyte-specific enhancer binding factor 2C expression in human brain development. Neuroscience. 1994;63:1067–1079. doi: 10.1016/0306-4522(94)90573-8. [DOI] [PubMed] [Google Scholar]
- 4.Black B, Cripps R. Myocyte enhancer factor-2 transcription factors in heart development and disease. Heart Development and Regeneration. 2010;2:673–699. [Google Scholar]
- 5.Flavell SW, Cowan CW, Kim TK, Greer PL, Lin Y, Paradis S, et al. Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science. 2006;311:1008–1012. doi: 10.1126/science.1122511. [DOI] [PubMed] [Google Scholar]
- 6.Cole CJ, Mercaldo V, Restivo L, Yiu AP, Sekeres MJ, Han JH, et al. MEF2 negatively regulates learning-induced structural plasticity and memory formation. Nature neuroscience. 2012;15:1255–1264. doi: 10.1038/nn.3189. [DOI] [PubMed] [Google Scholar]
- 7.Rashid AJ, Cole CJ, Josselyn SA. Emerging roles for MEF2 transcription factors in memory. Genes Brain Behav. 2014;13:118–125. doi: 10.1111/gbb.12058. [DOI] [PubMed] [Google Scholar]
- 8.Naya FJ, Black BL, Wu H, Bassel-Duby R, Richardson JA, Hill JA, et al. Mitochondrial deficiency and cardiac sudden death in mice lacking the MEF2A transcription factor. Nat Med. 2002;8:1303–1309. doi: 10.1038/nm789. [DOI] [PubMed] [Google Scholar]
- 9.Phan D, Rasmussen TL, Nakagawa O, McAnally J, Gottlieb PD, Tucker PW, et al. BOP, a regulator of right ventricular heart development, is a direct transcriptional target of MEF2C in the developing heart. Development. 2005;132:2669–2678. doi: 10.1242/dev.01849. [DOI] [PubMed] [Google Scholar]
- 10.Akhtar MW, Kim MS, Adachi M, Morris MJ, Qi X, Richardson JA, et al. In vivo analysis of MEF2 transcription factors in synapse regulation and neuronal survival. PLoS One. 2012;7:e34863. doi: 10.1371/journal.pone.0034863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barbosa AC, Kim MS, Ertunc M, Adachi M, Nelson ED, McAnally J, et al. MEF2C, a transcription factor that facilitates learning and memory by negative regulation of synapse numbers and function. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:9391–9396. doi: 10.1073/pnas.0802679105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akbarian S, Rios M, Liu RJ, Gold SJ, Fong HF, Zeiler S, et al. Brain-derived neurotrophic factor is essential for opiate-induced plasticity of noradrenergic neurons. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2002;22:4153–4162. doi: 10.1523/JNEUROSCI.22-10-04153.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen RZ, Akbarian S, Tudor M, Jaenisch R. Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nat Genet. 2001;27:327–331. doi: 10.1038/85906. [DOI] [PubMed] [Google Scholar]
- 14.Luikart BW, Nef S, Virmani T, Lush ME, Liu Y, Kavalali ET, et al. TrkB has a cell-autonomous role in the establishment of hippocampal Schaffer collateral synapses. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2005;25:3774–3786. doi: 10.1523/JNEUROSCI.0041-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arnold MA, Kim Y, Czubryt MP, Phan D, McAnally J, Qi X, et al. MEF2C transcription factor controls chondrocyte hypertrophy and bone development. Dev Cell. 2007;12:377–389. doi: 10.1016/j.devcel.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 16.Lin Q, Schwarz J, Bucana C, Olson EN. Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C. Science. 1997;276:1404–1407. doi: 10.1126/science.276.5317.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adachi M, Autry AE, Covington HE, 3rd, Monteggia LM. MeCP2-mediated transcription repression in the basolateral amygdala may underlie heightened anxiety in a mouse model of Rett syndrome. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2009;29:4218–4227. doi: 10.1523/JNEUROSCI.4225-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gemelli T, Berton O, Nelson ED, Perrotti LI, Jaenisch R, Monteggia LM. Postnatal loss of methyl-CpG binding protein 2 in the forebrain is sufficient to mediate behavioral aspects of Rett syndrome in mice. Biol Psychiatry. 2006;59:468–476. doi: 10.1016/j.biopsych.2005.07.025. [DOI] [PubMed] [Google Scholar]
- 19.Lange N, Hamann M, Shashidharan P, Richter A. Behavioural and pharmacological examinations in a transgenic mouse model of early-onset torsion dystonia. Pharmacol Biochem Behav. 2011;97:647–655. doi: 10.1016/j.pbb.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 20.Berton O, McClung CA, Dileone RJ, Krishnan V, Renthal W, Russo SJ, et al. Essential role of BDNF in the mesolimbic dopamine pathway in social defeat stress. Science. 2006;311:864–868. doi: 10.1126/science.1120972. [DOI] [PubMed] [Google Scholar]
- 21.Neale BM, Kou Y, Liu L, Ma'ayan A, Samocha KE, Sabo A, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Novara F, Beri S, Giorda R, Ortibus E, Nageshappa S, Darra F, et al. Refining the phenotype associated with MEF2C haploinsufficiency. Clin Genet. 2010;78:471–477. doi: 10.1111/j.1399-0004.2010.01413.x. [DOI] [PubMed] [Google Scholar]
- 23.Chao HT, Chen H, Samaco RC, Xue M, Chahrour M, Yoo J, et al. Dysfunction in GABA signalling mediates autism-like stereotypies and Rett syndrome phenotypes. Nature. 2010;468:263–269. doi: 10.1038/nature09582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bi R, Foy MR, Vouimba RM, Thompson RF, Baudry M. Cyclic changes in estradiol regulate synaptic plasticity through the MAP kinase pathway. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:13391–13395. doi: 10.1073/pnas.241507698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gould E, Woolley CS, Frankfurt M, McEwen BS. Gonadal steroids regulate dendritic spine density in hippocampal pyramidal cells in adulthood. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1990;10:1286–1291. doi: 10.1523/JNEUROSCI.10-04-01286.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith CC, McMahon LL. Estrogen-induced increase in the magnitude of long-term potentiation occurs only when the ratio of NMDA transmission to AMPA transmission is increased. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2005;25:7780–7791. doi: 10.1523/JNEUROSCI.0762-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monteggia LM, Barrot M, Powell CM, Berton O, Galanis V, Gemelli T, et al. Essential role of brain-derived neurotrophic factor in adult hippocampal function. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:10827–10832. doi: 10.1073/pnas.0402141101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garcia AD, Doan NB, Imura T, Bush TG, Sofroniew MV. GFAP-expressing progenitors are the principal source of constitutive neurogenesis in adult mouse forebrain. Nature neuroscience. 2004;7:1233–1241. doi: 10.1038/nn1340. [DOI] [PubMed] [Google Scholar]
- 29.Lin X, Shah S, Bulleit RF. The expression of MEF2 genes is implicated in CNS neuronal differentiation. Brain research Molecular brain research. 1996;42:307–316. doi: 10.1016/s0169-328x(96)00135-0. [DOI] [PubMed] [Google Scholar]
- 30.Berland S, Houge G. Late-onset gain of skills and peculiar jugular pit in an 11-year-old girl with 5q14.3 microdeletion including MEF2C. Clin Dysmorphol. 2010;19:222–224. doi: 10.1097/MCD.0b013e32833dc589. [DOI] [PubMed] [Google Scholar]
- 31.Carr CW, Zimmerman HH, Martin CL, Vikkula M, Byrd AC, Abdul-Rahman OA. 5q14.3 neurocutaneous syndrome: a novel continguous gene syndrome caused by simultaneous deletion of RASA1 and MEF2C. Am J Med Genet A. 2011;155A:1640–1645. doi: 10.1002/ajmg.a.34059. [DOI] [PubMed] [Google Scholar]
- 32.Engels H, Wohlleber E, Zink A, Hoyer J, Ludwig KU, Brockschmidt FF, et al. A novel microdeletion syndrome involving 5q14.3-q15: clinical and molecular cytogenetic characterization of three patients. Eur J Hum Genet. 2009;17:1592–1599. doi: 10.1038/ejhg.2009.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Le Meur N, Holder-Espinasse M, Jaillard S, Goldenberg A, Joriot S, Amati-Bonneau P, et al. MEF2C haploinsufficiency caused by either microdeletion of the 5q14.3 region or mutation is responsible for severe mental retardation with stereotypic movements, epilepsy and/or cerebral malformations. J Med Genet. 2010;47:22–29. doi: 10.1136/jmg.2009.069732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mikhail FM, Lose EJ, Robin NH, Descartes MD, Rutledge KD, Rutledge SL, et al. Clinically relevant single gene or intragenic deletions encompassing critical neurodevelopmental genes in patients with developmental delay, mental retardation, and/or autism spectrum disorders. Am J Med Genet A. 2011;155A:2386–2396. doi: 10.1002/ajmg.a.34177. [DOI] [PubMed] [Google Scholar]
- 35.Nowakowska BA, Obersztyn E, Szymanska K, Bekiesinska-Figatowska M, Xia Z, Ricks CB, et al. Severe mental retardation, seizures, and hypotonia due to deletions of MEF2C. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:1042–1051. doi: 10.1002/ajmg.b.31071. [DOI] [PubMed] [Google Scholar]
- 36.Paciorkowski AR, Traylor RN, Rosenfeld JA, Hoover JM, Harris CJ, Winter S, et al. MEF2C Haploinsufficiency features consistent hyperkinesis, variable epilepsy, and has a role in dorsal and ventral neuronal developmental pathways. Neurogenetics. 2013;14:99–111. doi: 10.1007/s10048-013-0356-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zweier M, Gregor A, Zweier C, Engels H, Sticht H, Wohlleber E, et al. Mutations in MEF2C from the 5q14.3q15 microdeletion syndrome region are a frequent cause of severe mental retardation and diminish MECP2 and CDKL5 expression. Hum Mutat. 2010;31:722–733. doi: 10.1002/humu.21253. [DOI] [PubMed] [Google Scholar]
- 38.Morrow EM, Yoo SY, Flavell SW, Kim TK, Lin Y, Hill RS, et al. Identifying autism loci and genes by tracing recent shared ancestry. Science. 2008;321:218–223. doi: 10.1126/science.1157657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li H, Radford JC, Ragusa MJ, Shea KL, McKercher SR, Zaremba JD, et al. Transcription factor MEF2C influences neural stem/progenitor cell differentiation and maturation in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:9397–9402. doi: 10.1073/pnas.0802876105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vetere G, Restivo L, Cole CJ, Ross PJ, Ammassari-Teule M, Josselyn SA, et al. Spine growth in the anterior cingulate cortex is necessary for the consolidation of contextual fear memory. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:8456–8460. doi: 10.1073/pnas.1016275108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pulipparacharuvil S, Renthal W, Hale CF, Taniguchi M, Xiao G, Kumar A, et al. Cocaine regulates MEF2 to control synaptic and behavioral plasticity. Neuron. 2008;59:621–633. doi: 10.1016/j.neuron.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.