Abstract
Objective
Nearly 20%–29% of patients with colorectal cancer (CRC) succumb to liver or lung metastasis and there is a dire need for novel targets to improve the survival of patients with metastasis. The long isoform of the Carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1-L or CC1-L) is a key regulator of immune surveillance in primary CRC, but its role in metastasis remains largely unexplored. We have examined how CC1-L expression impacts on colon cancer liver metastasis.
Design
Murine MC38 transfected with CC1-L were evaluated in vitro for proliferation, migration and invasion, and for in vivo experimental liver metastasis. Using shRNA silencing or pharmacological inhibition, we delineated the role in liver metastasis of Chemokine (C-C motif) Ligand 2 (CCL2) and Signal Transducer and Activator of Transcription 3 (STAT3) downstream of CC1-L. We further assessed the clinical relevance of these findings in a cohort of patients with CRC.
Results
MC38-CC1-L-expressing cells exhibited significantly reduced in vivo liver metastasis and displayed decreased CCL2 chemokine secretion and reduced STAT3 activity. Down-modulation of CCL2 expression and pharmacological inhibition of STAT3 activity in MC38 cells led to reduced cell invasion capacity and decreased liver metastasis. The clinical relevance of our findings is illustrated by the fact that high CC1 expression in patients with CRC combined with some inflammation-regulated and STAT3-regulated genes correlate with improved 10-year survival.
Conclusions
CC1-L regulates inflammation and STAT3 signalling and contributes to the maintenance of a less-invasive CRC metastatic phenotype of poorly differentiated carcinomas.
Colorectal cancer (CRC) is a major disease affecting approximately 5% of the population in North America.1 Approximately 60% of patients survive more than 5 years, but the remaining 20%–29% (USA and Europe, respectively) develop fatal liver or lung metastasis.2 Novel molecular-based interventions or new surgical techniques for liver resections have been somewhat successful in prolonging life, but new molecular targets need to be identified for better therapeutic management. CarcinoEmbryonic Antigen Cell Adhesion Molecule 1 (CEACAM1, herein abbreviated CC1) is an intercellular adhesion molecule of the immunoglobulin superfamily and a carcinoembryonic antigen (CEA) family member.3 Many alternate splicing isoforms derived from the human and murine Ceacam1 gene are tethered to the cell membrane and include either short (S) or long (L) cytoplasmic domains.3 CC1-L contains two Tyr residues, positioned in Immunoreceptor Tyr Inhibition Motifs (ITIMs),4 both phosphorylated upon activation of the insulin receptor, the epidermal growth factor receptor, the granulocyte colony-stimulating factor receptor and Src-like kinases.3 Upon its Tyr phosphorylation and binding to the Src-Homology region 2 domain Tyr phosphatases SHP-14 or SHP-2,5 CC1-L downregulates regulatory signalling pathways,6 leading to intercellular adhesion regulation,7 insulin and lipid metabolism,8,9 angiogenesis,10 innate and adaptive immune responses11–13 and microbial and viral pathogen interactions.6
In tumours, CC1-L acts as tumour growth inhibitor in many early solid human neoplasms, including colon tumours.3 This effect is mediated by the 2 Tyr-phosphorylated residues binding to SHP-1 as shown in colon4,14,15 and prostate16 xenografts or allografts. In vivo, Cc1−/− mutant mice demonstrated augmented colon or intestinal tumour burden relative to controls upon azoxymethane treatment17 and in an Apc1638N/+ genetic background.18 However, in aggressive non-small-cell lung, thyroid, gastric, pancreatic cancers, malignant melanoma and metastatic colon cancer,3 CC1 overexpression correlates with increased invasiveness, metastatic spread and an unfavourable patient prognosis. In addition to its effects in epithelial cells, CC1-L signalling plays a significant role in invasive processes via endothelial cells,10,19 CD11b+Gr1+ immature myeloid cells,20,21 matrix metalloproteinase 9-positive leucocytes and stromal cells.22
Murine CRC MC38 cells null for CEACAM1 are highly invasive and metastatic,21 and thus the present study focuses on poorly differentiated CEACAM1-null colorectal adenocarcinomas. We have now addressed how the CC1-L isoform or a CC1-L mutant, disabled for Tyr phosphorylation (CC1-FF), expressed in CRC MC38 cells impacts on the development of syngeneic metastatic lesions in vivo. MC38-CC1-L cells demonstrated reduced in vitro proliferation, migration and invasion and reduced metastasis in vivo relative to CC1-negative MC38-Control (CT) cells in immunocompetent wild-type (WT) mice; CC1-L's reduced metastasis phenotype depended on CC1-L's ITIMs, as demonstrated using the MC38-CC1-FF cells. Investigation of the CC1-L-associated regulatory network in MC38 liver metastatic inhibition revealed that CC1-L expression impairs STAT3 activity, thus decreasing the synthesis and secretion of its CCL2 target23 that controls metastatic immune infiltration.24 Bioinformatics analysis of The Cancer Genome Atlas (TCGA) tumour samples of CRC patients for CC1 high-expressing tumours demonstrated statistical correlation with some inflammation-regulated and STAT3-regulated genes, with such patients showing improved survival.
MATERIALS AND METHODS
Cell lines and cell culture
Metastatic mouse MC38 colon cancer cells25 were separated through fluorescent activated cell sorting for CC1-negative expression and retrovirally infected with virions expressing the CC1-L WT and CC1-FF mutant constructs.15 CCL2-knockdown MC38-CT cells (CCL2-KD) were generated by infecting lentiviral shRNA constructs targeting CCL2 (Open Biosystems; Huntsville, Alabama, USA) under Hygromycin B selection (500 μg/mL). MC38-CTcells were treated with either the S3I-201 STAT3 inhibitor (100 μM in dimethyl sulfoxide (DMSO); Sigma-Aldrich, St-Louis, Missouri, USA), or the SH-0454 and SH-08100 STAT3 SH2 domain-binding inhibitors26 (1–10 μM in DMSO).
In vivo experiments and metastasis induction
All experimental animal procedures were conducted in compliance with McGill University and the Canadian Council on Animal Care guidelines. Eight-week-old to ten-week-old C57Bl/6 WT mice (Harlan; Frederick, Maryland, USA) and Ccr2−/− mice ( Jackson Labs; Bar Harbor, Maine, USA) were used for liver metastases development after intrasplenic injection of 2×105 viable MC38-derived cells in 50 μL of phosphate-buffered saline (PBS), followed by splenectomy 3 min after injections to prevent sepsis from splenic tumour growth. PBS-injected mice served as controls. Mice were sacrificed 14–21 days post-injection and liver tissue was excised and processed as described in online supplementary materials and methods. C57Bl/6 mice were intraperitoneally (IP)-injected with STAT3 inhibitors 3 days/week for 14 days using DMSO as a control, starting 2 days prior to cell injections.
Histology and immunofluorescence
We determined the number and average size of metastatic lesions, computed with ImageScope software, and reported as area fraction (ratio of surface nodule area/total surface area of liver).21 For detection on fluorescence or confocal microscopes,21 frozen sections of the same liver tissues were stained with primary antibodies (see online supplementary materials and methods), followed by incubation with FITC-conjugated or Texas Red-conjugated antibodies and with 4’,6-diamidino-2-phenylindole-containing mounting medium (DAKO; Burlington, Ontario, Canada).21
Proliferation, migration and invasion assays
MC38-CT or MC38-CC1-L or MC38-CC1-FF cells were plated in a 16-well E-Plate or CIM-Plate (ACEA Biosciences; San Diego, California, USA) and proliferation, migration and invasion were measured using a real-time xCELLigence instrument (Roche; Basel, Switzerland). Fetal bovine serum (10%) was used as a chemoattractant in migration and invasion assays, with serum-free medium (SFM) as a negative control. Invasion assays were performed using Matrigel-coated CIM-Plates.
Cytokine/chemokine immunoassays
Cell lysate proteins were prepared, the concentrations determined and proteins were subjected to Quansys 9, 16 cytokine/chemokine Multiplex ELISA arrays (Quansys Biosciences; Logan, Utah, USA) (n=3 independent experiments).21
Immunoblot analysis
Cell pellets were lysed21 and equal concentrations of proteins were resolved on SDS-PAGE gels, transferred to polyvinylidene fluoride membranes and immunoblotted with antibodies described in online supplementary materials and methods.
Analysis of patient samples using TCGA
The mRNA sequencing data were downloaded from the BROAD firehose website, using the September 2014 release, and analysed through the RNA-Seq by Expectation-Maximization (RSEM) processing pipeline. The TCGA database does not indicate whether the mRNA was obtained from the invasion front or the luminal side of tumours, which limits comparisons with the work of Ieda et al.27 Analysis of 409 colon and rectal carcinomas sequenced on both Illumina HiSeq and GA platforms was performed to assess the influence of gene signatures on survival. Details on normalisation methods are provided in online supplementary materials and methods. Linear regressions examined associations between clinicopathological variables and quantile-normalised gene expression. Cox proportional hazards analysis was performed to estimate the effect of each gene or signature on survival time and also to estimate whether there was evidence for interaction between high CEACAM1 expression and expression of other signature genes on survival.
Statistical analysis
Data were expressed as mean±SE. Statistical analysis was performed using GraphPad Prism 5 statistical software for Microsoft Windows and R (http://www.cran-r.project.org). The Student two-tailed t and analysis of variance with Bonferroni correction tests were used to determine the significance, and p values <0.05 were considered significant. Survival analysis used Cox proportional hazards models and Kaplan–Meier curves (see online supplementary materials and methods) with significance set at 0.1.
RESULTS
CC1-L expression in MC38 CRC cells decreases invasion in vitro, and liver metastasis in vivo
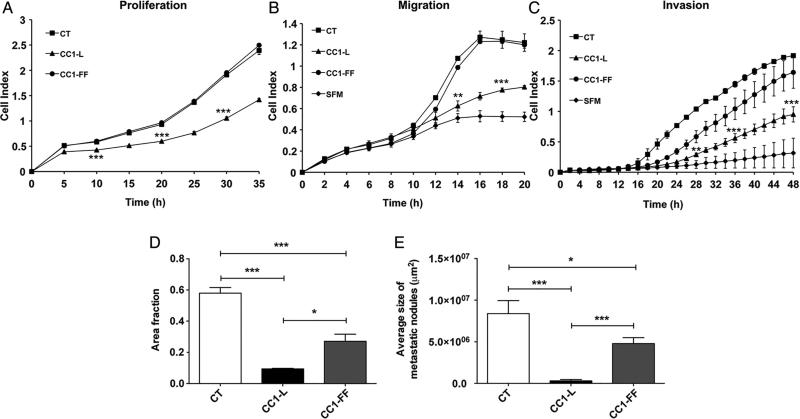
Depending on the genetic background of CRC cells, CC1 splice isoform expression results in different growth and migratory phenotypes.14,27 We have examined the behaviour of highly metastatic C57Bl/6 MC38 CC1-negative (CT) CRC cells21 and those expressing the mouse CC1-L isoform or a Tyr-phosphorylation-disabled CC1-L mutant (CC1-FF). MC38 cell-sorted CC1-L populations (see online supplementary figure S1A,B) consistently demonstrated reduced proliferation, migration and invasion (figure 1A–C; quantifications in online supplementary figure 1C–E) relative to CT or to CC1-FF cells. CRC cell chemotaxis was not induced in SFM (figure 1B, C). We performed MC38-mediated liver metastasis assays (intrasplenic injections) in WT B6 mice. MC38-CC1-L cells reduced metastatic burden relative to MC38 CT cells (83% or 96% decrease in number or size, respectively) (figure 1D, E). CC1-L Tyr residues within the ITIM motifs significantly contributed to this decrease as MC38-CC1-FF infectants partially reversed this phenotype with 50% reduction in metastatic burden compared with MC38-CT cells (figure 1D, E). Proliferation (Ki-67), vascular density (CD31) and immune infiltration (F4/80+ macrophages, CD11b+ monocytes and CD3+ and CD8+ T lymphocytes) detected via immunostaining of metastatic nodules indicated that these parameters were significantly diminished in the MC38-CC1-L-derived metastases and partially rescued in MC38-CC1-FF-induced nodules (see online supplementary figure S1F–K). Therefore, CC1-L expression modifies intrinsic MC38 signalling events through its two Tyr residues, leading to inhibition of liver metastatic development.
Figure 1.
Carcinoembryonic antigen-related cell adhesion molecule 1 (CC1) expression regulates metastatic capabilities of mouse MC38 colorectal cancer (CRC) cells. (A) Proliferation of MC38-CT, MC38-CC1-L or MC38-CC1-FF cells measured over 35 h using the xCELLigence system. (B) Migration of the same cells was measured over 20 h; serum-free medium (SFM) served as negative chemotaxis control. (C) Cell invasion in Matrigel-coated plates was measured over a 48 h time period with SFM as negative chemotaxis control. (D and E) Metastatic ability of the MC38-derived cells in vivo was examined 14 days after intrasplenic injection into C57Bl/6 mice. The number (D) and size (E) of metastatic nodules in the livers were quantified on H&E-stained step sections of livers. (ns p>0.05, *p<0.05, **p<0.01, ***p<0.001, two-tailed t test or analysis of variance with Bonferroni correction test). Data are presented as means±SEM with n=at least two independent sets of experiments. For in vivo studies, a minimum of 10 mice per group were used.
Relationship of the CCL2-CCR2 signalling axis and CC1-L-mediated decreased metastasis
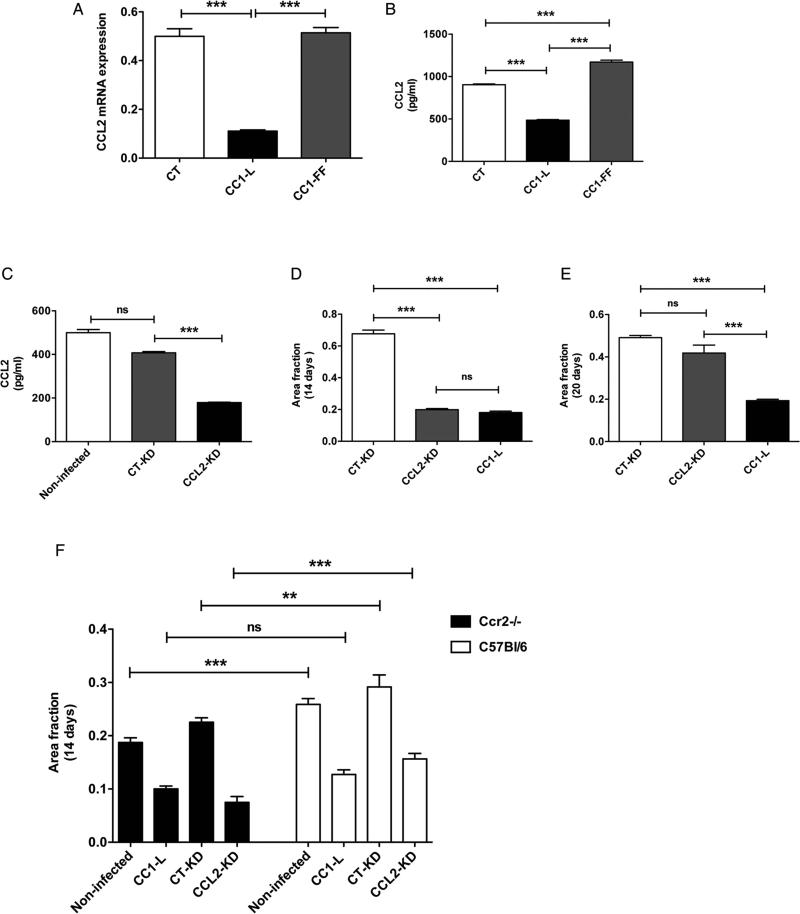
To examine CC1-L-driven contribution of MC38 tumour cells to immune infiltration, CT, CC1-L- and CC1-FF-transfected cell medium was assessed using ELISA for a panel of 16 cytokines/chemokines (see online supplementary figure S2A). Only CCL2 varied significantly between cell types. MC38-CT cells significantly expressed the CCL2 chemokine mRNA at 72 h post-plating relative to MC38-CC1-L cells (figure 2A). CCL2 was abundantly detected in MC38-CT cell medium, whereas CC1-L-expressing cells exhibited significantly decreased expression; CC1-FF cells had a 2.4-fold enhancement of CCL2 versus that of CC1-L cells (figure 2B), implying that CC1-L acts a negative regulator of CCL2-mediated immune recruitment in the metastatic process.
Figure 2.
CCL2-CCR2 signalling axis contributes to anti-metastatic function of CC1-L. (A) MC38-CT, MC38-CC1-L and MC38-CC1-FF cells were grown for 72 h, and CCL2 chemokine mRNA levels were measured using Q-PCR. (B) CCL2 protein levels in cell lysates were measured using ELISA 72 h post cell seeding. (C) MC38-CT cells were stably infected with virions expressing either CCL2 shRNA (CCL2-KD) or control shRNA (CT-KD). CCL2 protein levels in cell lysates were measured using ELISA in non-infected, CT-KD and CCL2-KD cells. (D and E) metastatic ability of MC38-CCL2-KD cells was evaluated in vivo in comparison with MC38-CT-KD and MC38-CC1-L cells, 14 (D) and 20 (E) days after intrasplenic injections into C57Bl/6 mice. (F) Liver metastasis formation was measured at day 14 in Ccr2−/− vs C57Bl/6 backgrounds. MC38-CCL2-KD, MC38-CT-KD, MC38 non-infected and MC38-CC1-L cells were used for experimental metastasis (ns p>0.05, *p<0.05, **p<0.01, *** p<0.001, two-tailed t test or analysis of variance with Bonferroni corrections). Data are presented as means±SEM and with n=at least two independent sets of experiments. For in vivo studies, a minimum of 10 mice per group were used.
CCL2 plays a key role in infiltration of CD11b+Gr1mid myeloid cells into the CRC hepatic metastatic microenvironment,24 and CCL2 expression in human stage 4 CRC correlates with higher metastatic burden, lower patient survival and recruitment of CCR2+Ly6C+ monocytes to lung metastases.28 To explore whether CCL2 was related to CC1-L signalling, we silenced Ccl2 mRNA in MC38-CT cells with a Ccl2-specific shRNA (CCL2-KD) relative to a control (CT-KD) shRNA, which resulted in approximately 60% CCL2 expression relative to those in MC38-CT or -CT-KD cells as defined by ELISA assays (figure 2C). The CCL2-KD cells produced 3.4-fold less metastases in vivo than the CT-KD cells, the former exhibiting similar levels to those of MC38-CC1-L cells (figure 2D). Immune cell profiling of metastatic livers indicated inhibition of F4/80+ macrophage infiltration, while CD11b+Gr1+ myeloid cell were significantly present in CCL2-KD metastases, suggesting decreased immunosuppression in response to CCL2 silencing (see online supplementary figure S2B,C). This may depend on lack of recruitment of myeloid cells as experimental metastasis performed on a Rag1−/− mouse background with CCL2-KD cells still produced a lower metastatic score relative to controls (see online supplementary figure S2D). Alternatively, both natural killer cells29 and dendritic cells,30 functional in normal Rag1−/− mice, may contribute to reduced metastasis formation in the context of CCL2-KD and CC1-L cells. However, as reported by Wolf et al,28 in later metastasis evaluation, mice injected with CCL2-KD cells did not show significant differences to those of CT-KD MC38 cells (figure 2E). Yet, mice injected with MC38-CC1-L cells still exhibited lower metastatic score than those injected with CT-KD cells (figure 2E), indicating that the CCL2 downregulated activity might be overcome by other targets during late hepatic metastasis such as CCL5, representing another STAT3 target (figure 3C, E), that contributes to in vivo CRC liver metastasis.31
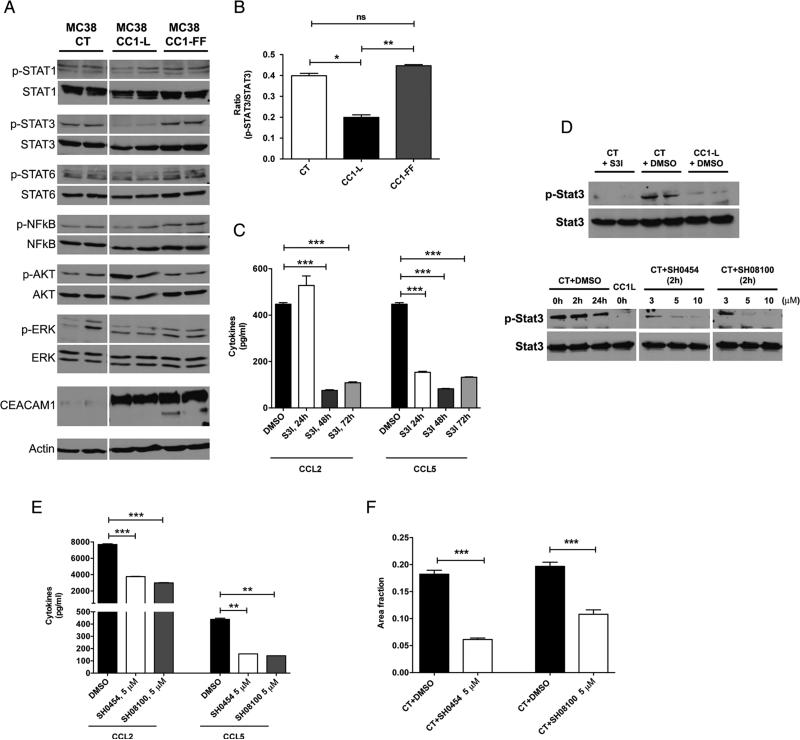
Figure 3.
STAT3 activity regulates liver metastasis downstream of CC1-L signalling and can be targeted with synthetic compounds. (A) Immunoblot analysis of several important signalling proteins in MC38-CT, MC38-CC1-L or MC38-CC1-FF cells. Actin was used as a loading control. (B) Quantification of STAT3 activity is shown as the ratio of p-Stat3 to Stat3. (C and D) MC38-CT cells (showing high level of STAT3 activity) were treated for 24, 48 or 72 h with either commercially available STAT3 inhibitor (S3I-201; 100 μM) or dimethyl sulfoxide (DMSO). (C) CCL2 and CCL5 chemokine levels in cell lysates were measured using ELISA after incubation with the inhibitor and (D, top panel) STAT3 activity was evaluated using immunoblotting. (E) Two new synthetic compounds specifically targeting STAT3 (SH-0454 and SH-08100) were examined for their dose-response inhibitory activity. Different concentrations (3, 5 and 10 μM) of each compound were applied to MC38-CT cells for 2 h; (D, bottom panel) STAT3 activity was measured by immunoblotting. STAT3 activity in CC1-L cells is shown for comparison purposes. (E) CCL2 and CCL5 levels were also measured using ELISA at median inhibitory concentration (5 μM) of each compound with DMSO used as vehicle control. (F) MC38-CT cells treated with either DMSO or 5 μM of each compound for 2 h were intrasplenically injected into C57Bl/6 mice and liver metastasis was evaluated 14 days post-injections. (ns p>0.05, *p<0.05, **p<0.01, ***p<0.001, two-tailed t test or analysis of variance with Bonferroni correction). Data are presented as means±SEM with n=at least two independent sets of experiments. For in vivo studies, a minimum of 10 mice per group were used.
The CCL2 receptor, CCR2, is expressed on both CD11b+Gr1+ myeloid cells and endothelium.24,28 To gauge whether the host CCL2-CCR2 signalling pathway might regulate CC1-L-mediated decreased metastasis, we performed metastasis experiments in Ccr2−/− and WT mice. MC38-CT and MC38-CC1-L-transfected cells expressed minimal amounts of Ccr2 mRNA and protein at their cell surface (see online supplementary figure S2E,F). As reported,28 MC38-CT cells (either non-infected or CT-KD) formed less metastases in the Ccr2−/− background than in WT mice (figure 2F). No apparent differences were noted between MC38-CC1-L cells in mice of either genetic background, whereas CCL2 silencing caused lower metastatic development in Ccr2-null animals (figure 2F). Hence, our results confirm that MC38-expressed CC1-L decreases CCL2 signalling, which contributes, in part, to lowered metastatic development.24,28
STAT3 contributes to CC1-L-mediated decreased metastasis
Pan and Shively demonstrated that CC1 deletion in neutrophils altered STAT3 activity and hampered granulopoiesis.13 Since activated STAT3 is associated with clinical outcomes of patients with adverse CRC,32 we examined STAT3-mediated signalling changes in MC38-transfected cells. STAT1, STAT6, NFκB, Akt or Erk activities were not different between these cell lines, as measured with specific antiphospho-antibodies (figure 3A). However, pSTAT3 activity followed the same pattern as CCL2 expression, that is, significantly decreased in MC38-CCL1-L cells and returned to baseline in MC38-CC1-FF cells relative to MC38-CT cells (figure 3A, B). These results suggested that a signalling loop might possibly exist between CC1-L as a phosphorylation target of, or coreceptor to, Tyr Kinases or cytokine receptors.3 Thus, CC1-L could negatively influence STAT3 transcriptional activity,13 which has been shown to positively regulate CCL2 expression by binding to the Ccl2 promoter.23 Treatment with novel STAT3 inhibitors targeting the STAT3 homodimerisation interface leads to breast cancer and glioma26 cell growth inhibition, apoptosis induction and inhibition of STAT3-regulated genes, including Ccl2.33 We thus treated MC38-CT cells with the S3I-102 STAT3 inhibitor, used mainly for in vitro studies, which considerably reduced both MC38-CT CCL2 and CCL5 chemokine expression at 48 or 72 h post-treatment (figure 3C). At these time points, pSTAT3 activity was completely abrogated in MC38-CT cells, as compared with vehicle-treated MC38-CC1-L cells (figure 3D, top panel). Therefore, reduced CCL2/CCL5 expression in MC38-CC1-L cells is likely caused by the decreased pSTAT3 activity observed.
To confirm that inhibition of STAT3 activity led to reduced CRC liver metastasis in vivo, MC38-CT cells were then treated with two potent and selective, direct-binding STAT3 derivatives of the BP-1-102 compound (SH-045426 and SH-08100, chemically modified for better potency) shown to inhibit human breast and lung cancer xenograft growth in vivo.34 Treatment of MC38-CT cells in vitro with either the SH-0454 or SH-08100 inhibitors (5 μM) led to dramatic reduction of STAT3 activity and CCL2/CCL5 chemokine secretion (figure 3D, bottom panel, and E) as well as decreases in cell migration in vitro (see online supplementary figure S3D), although lower concentrations did not modify MC38-CT cell proliferation or migration over several days (see online supplementary figure S3A, B), due to moderate or no reduction of pSTAT3 levels (figure 3D, bottom panel). These STAT3 inhibitors also exhibited a long-lasting effect in vitro: MC38-CT cells treated for 2 h with the inhibitors (5 μM), which were then completely removed, demonstrated no changes in MC38-CT proliferation but a significant reduction in migration over 30–45 h (see online supplementary figure S3C,D). MC38-CT cells treated for 2 h with either STAT3 inhibitors, then intrasplenically injected into C57Bl/6 mice, showed considerably reduced metastatic load (66% inhibition for SH-0454 and 45% inhibition for SH-08100) compared with mice bearing vehicle-treated MC38-CT cells (figure 3F). Additional thrice weekly injections (IP) of the SH-0454 inhibitor did not result in further decreases in in vivo metastasis (data not shown), most likely due to low blood concentrations of the inhibitor (313±8 nM, 30 min post-injection of 10 mg/kg).26 Hence, in vivo liver metastasis induced by MC38-CT cells is significantly reduced via decreased STAT3 activity, leading to attenuated CCL2/CCL5 expression.
Survival of patients with higher CC1-L expression
Our data suggested that CC1-L expression in some types of CRC tumours might confer favourable patient outcome through modulation of CCL2 and STAT3. CCL2 expression increases with CRC disease stages and high CCL2 levels significantly correlate with lower disease-free survival.35 Furthermore, CCL2/CCR2 expression promotes the recruitment of CD11b+Gr1+ myeloid cells to favour CRC metastatic development.24 In addition, STAT3 mRNA and both unphosphorylated and phosphorylated STAT3 protein expression in paired primary and invasive CRCs correlated with lymph node status.36 Finally, CRC STAT3 activation (pSTAT3) revealed by immunohisto-chemistry was positive in 54% (T3) and 35% (T4) of the large 724 CRC case study evaluated in two prospective cohorts.32
To corroborate whether CC1-L cooperated with CCL2/CCR2 and STAT3 in defining overall patient survival, we analysed 409 colon and rectal tumour samples of all stages from the TCGA RNA-Seq data for expression of key genes over a 10-year survival period. As STAT3 activation depends on Tyr750 phosphorylation of the protein and dimerisation resulting in nuclear translocation to activate transcription of target genes,37 we relied on STAT3 gene signatures to measure STAT3 activity in patient samples, (see online supplementary table S1).
Analysis of the associations between clinicopathological patient parameters and CEACAM1, CCL2, CCR2 and STAT3 gene signature expression was performed with univariate (see online supplementary table S2, available online only) and multivariate (table 1) linear regression models. The STAT3 expression signature was associated with more advanced age in both univariate and multivariate analyses. Although the presence of venous invasion was associated with CEACAM1, CCL2 and CCR2 when tumours of all stages were analysed in a univariate analysis, only CCR2 expression was associated with venous invasion when a multivariate model was applied, suggesting it as the dominant associated factor for this phenotype. In a subgroup analysis of patients over the age of 60 years, CEACAM1 expression showed association with primary lymphatic invasion (p=0.012 for stages T3–T4; p=0.039 for T1–T4).
Table 1.
p Values for multivariate analyses of associations between the listed clinicopathological variables and quantile-normalised expression of each of CEACAM1, CCL2, CCR2 and the STAT3 signature genes*
| CEACAM1 |
CCL2 |
CCR2 |
STAT3 signature |
|||||
|---|---|---|---|---|---|---|---|---|
| T3–T4 | T1–T4 | T3–T4 | T1–T4 | T3–T4 | T1–T4 | T3–T4 | T1–T4 | |
| Age | 0.17 | 0.039 | 0.788 | 0.411 | 0.686 | 0.285 | 0.035 | 0.017 |
| Gender (male vs. female) | 0.58 | 0.37 | 0.35 | 0.38 | 0.16 | 0.18 | 0.80 | 0.70 |
| Primary tumour site (colon vs rectum) | 0.23 | 0.23 | 0.67 | 0.65 | 0.88 | 0.87 | 0.34 | 0.35 |
| Primary lymphatic presentation | 0.092 | 0.11 | 0.68 | 0.51 | 0.53 | 0.34 | 0.42 | 0.52 |
| Lymphatic invasion | 0.86 | 0.75 | 0.45 | 0.70 | 0.87 | 0.61 | 0.54 | 0.83 |
| Venous invasion | 0.36 | 0.12 | 0.69 | 0.90 | 0.19 | 0.039 | 0.80 | 0.99 |
p Values are indicated for association in a multivariate linear model predicting gene expression as a function of the variables listed in the rows of the table. Separate models are fit for each column of the table; statistically suggestive values (<0.1) are indicated in italics.
We then queried whether the CC1, CCL2 and CCR2 gene expressions were predictive of overall survival; Kaplan–Meier survival analyses for high/low CC1, CCL2 and CCR2 expression did not reveal significant overall 10-year survival differences for patients with T3-T4 CRCs, although those with higher CCR2 expression exhibited a trend towards longer survival (P=0.08). Similarly, when all CRC stages were considered, patients with lower CCL2 expression tended towards longer 10-year survival (p=0.10) (see online supplementary figure S4A–C).
To gauge whether genes relating to STAT3 signatures (see online supplementary table S1) were linked to survival, Cox hazard proportional survival analyses were evaluated with and without an interaction with high/low CC1 expression, including all covariates presented in table 1. Hazard ratios (HR) from the Cox models for each gene (table 2 and see online supplementary table S3) indicate the probability of cancer-associated death. Values imply that higher (>1.0) or lower (<1.0) rates of death, respectively, are associated with higher gene expression of the indicated gene. For interaction models, the interpretation is similar, although the HRs can be viewed as the additional impact of high expression of the indicated gene when CEACAM1 is also high. For the Z-normalised gene expression values, a one-unit change is equivalent to a change of one SD, but for the quantile normalisation the interpretation will vary from gene to gene. Independent STAT3 signature genes included CCND1, FAS, IL6, IRF1, NPC1 and STAT1, suggesting that both inflammation-related and STAT3-related genes statistically influence survival, highlighted by the exponential of the survival coefficient (see online supplementary table S3). Other than IL6, all of these genes exhibited an HR (<1.0) indicative that lower probability of death was associated with higher expression.
Table 2.
Selected p values and HRs for tests of interaction between genes in any of the signatures and CEACAM1-high expression, from survival analysis models*
| Categories† | Gene | T3–T4 stage Quantile | HR coefficient | T1–T4 stage Quantile | HR coefficient |
|---|---|---|---|---|---|
| 2 | IRF1 | 0.042 | 0.44 | 0.122 | 0.57 |
| 3 | MCL1 | 0.057 | 0.36 | 0.005 | 0.23 |
| 3 | NPC1 | 0.013 | 0.17 | 0.027 | 0.24 |
| 2 | SOCS3 | 0.048 | 0.55 | 0.086 | 0.63 |
p Values for interaction are indicated and p<0.1 are highlighted.
Refers to gene categories in Supporting table 1. Gene expression was quantile-normalised, and all covariates in table 1 were included in the models. The HRs corresponding to each p value represent the estimated magnitude of the interactions between each gene and CEACAM1-high. Values <1 indicate lower probability of death and longer survival. Genes were selected for inclusion in this table if there was some (p<0.1) evidence for association with survival.
When these same gene signatures were analysed with the added interaction of CC1 high/low expression, four genes (IRF1, MCL1, NPC1 and SOCS3) showed statistical significance, emphasising that higher CC1 expression regulates the effect on time to death of genes regulating inflammation (IRF1) and STAT3 activity (MCL1, NPC1 and SOCS3) (table 2). However, these associations were not strong. We also performed permutation analyses and the number of small p values did not exceed what might have been expected by chance.
Nevertheless, patient cohorts with advanced CRC tumours exhibiting high CC1 expression significantly correlate with some inflammation-regulated and STAT3-regulated genes and the results are suggestive of longer survival.
DISCUSSION
CC1 is one of the members of the large CEA family of which three proteins (CEA, CEACAM6 and CC1) are known for their contribution to CRC development, progression and metastasis.3 Although CC1 is downregulated in benign CRC tumours,3 Ieda et al27 have shown that CC1 re-expression and, in particular, CC1-L isoform dominance over CC1-S at the invasion front correlates with CRC haematogenous metastasis. These data suggest that different CC1 isoforms expressed in 75 CRC patients in stages 3 and 4 over a 5-year survival period affect CRC metastasis. However, the CC1-L-governed signalling and mechanisms during in vivo metastasis have remained so far unexplored. We have now examined, using the murine MC38 CEACAM1-null cell line producing poorly differentiated adenocarcinoma, how CC1-L regulates a signalling network, operative in the context of a syngeneic mouse background.
CC1-L expression in MC38 cells resulted in a considerable decrease of CRC liver metastatic burden. In search of mechanisms responsible for the observed phenotype, we first identified that the CCL2 chemokine expression was decreased in MC38-CC1-L cells. CCL2 silencing led to significantly reduced metastatic development at early (14 d) but not late (20 d) time points. Similarly, ablation of the CCL2-CCR2 axis in Ccr2−/− mice only partially protected mice against liver metastasis, as described.24,28 It remains possible that CCL2 may also bind to the atypical chemokine receptors ACKR1 and R2 mediating CCL2 signalling in leucocytes,38 but this alternative pathway is yet to be investigated in liver metastasis. Importantly, CC1-L-mediated stromal effects are also implicated in the CRC metastatic phenotype with dampened CCL2 secretion leading to decreased CD11b+ and F4/80+ myeloid cell infiltration to the metastatic site.21 Inflammatory chemokines such as CCL2 and CCL5 play a significant role in driving CRC metastatic processes.39 In fact, discontinuation of an anti-CCL2 treatment in breast cancer models leads to accelerated metastasis through angiogenesis stimulation.40 Although high or low CC1 expression is not predictive of 10-year overall survival in patients with stages 3 and 4 CRC, as predicted by Kaplan–Meier survival curves, patients with low CCL2 or high CCR2 expression have a marginally better survival.
STAT3 is also associated with inflammatory CRC processes37 and CC1-L, possibly through the latter's role as a coinhibitory receptor to Tyr kinase,3,8 or to cytokine receptors regulating canonical STAT3 signaling13 acts on STAT3 inflammation-related signalling pathways. It remains possible that CC1-L might also signal through other unexplored non-canonical signalling pathways.37 The IL-6 receptor, as a major CRC STAT3 signalling cytokine receptor37 is not expressed in MC38 cells (data not shown), rendering its triggering by IL-6 non-operative. But it remains possible that IL-11 secretion by the MC38-CC1-L cells might activate the GP130/STAT3 signalling and reduce metastasis;41 this remains to be further evaluated.
Pharmacological inhibition of STAT3 activity in MC38-CT cells resulted in the abrogation of CCL2 and CCL5 chemokine secretion reduced in vivo liver metastatic development. These findings recapitulated the diminishment in STAT3 activity and CCL2/CCL5 chemokine levels observed in MC38-CC1-L cells (figure 3). Importantly, a 2 h exposure of MC38 cells to new STAT3 inhibitors demonstrated stable dampening of MC38 STAT3 activity in vivo. The low bio-availability of these compounds42 is currently being improved through chemical modifications. As STAT3 promotes resistance to widely used Tyr kinase inhibitors in lung cancer and CRC, and since its silencing reinstated drug sensitivity,43 these STAT3 inhibitors may constitute novel therapeutic compounds that are able to reverse drug resistance.
Previous findings have indicated that activated STAT3 represents a biomarker for poor survival outcome.32 In this analysis of TCGA CRC samples, we demonstrate that high CC1 expression combined with four of the STAT3 signature genes expressed during inflammation were predictive of longer survival times (table 2). Importantly, one of these genes is SOCS3, a known STAT3 inhibitor.37 The power of survival analysis rests on the number of deaths, and therefore power was limited in the TCGA data. Investigation of these relationships in a larger dataset would be worthwhile.
However, our data do not necessarily imply a direct regulation of these activities by CC1-L; this modulation could be executed through tiers of other events/molecules that interconnect CC1-L to other metastasis-promoting pathways. Furthermore, human CRC cell lines and tumour tissues often abundantly express two other CEACAM proteins, CEA and CEACAM6;3 these genes share 70%–79% of the proximal promoter sequences with that of CEACAM1. The hierarchy of CEACAM functions and predominance under static conditions has been partially addressed44 and this will need to be further pursued in plastic and modulated culture conditions in CRC cell lines with defined genetic backgrounds; this constitutes our challenge of the next few years.
Supplementary Material
Significance of this study.
What is already known on this subject?
▶ Liver metastasis of colorectal cancer (CRC) is a major cause of death in patients with cancer.
▶ Carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1) is downregulated in early colorectal tumours and CEACAM1-L is present at the invasion front of CRC cells. However, the mechanism through which CEACAM1 impacts metastatic dissemination of CRC cells to the liver has not been investigated to date.
▶ CCL2–CCR2 interactions and STAT3 activity influence breast and colon cancer metastases.
What are the new findings?
▶ CEACAM1-L expressed in some metastatic murine CRC cells significantly reduced experimental liver metastatic burden.
▶ CEACAM1-L expression in MC38 cells decreases STAT3 activity and CCL2 secretion, thus regulating inflammatory signalling networks and decreasing metastatic burden.
▶ Patients with stages 1–4 CRC expressing low levels of CCL2 or high CCR2 exhibit a better 10-year survival.
How might it impact on clinical practice in the foreseeable future?
▶ CEACAM1 plays an important role in colorectal carcinoma liver metastasis and may constitute a novel target for therapeutic intervention.
▶ Generation of new Stat3 inhibitors with improved potency and pharmacokinetic properties provide a promising prospect in prevention of cancer metastasis.
Acknowledgements
The authors wish to thank Ms Luisa DeMarte and the members of the Goodman Cancer Centre Histology core for expert technical assistance. They thank Dr. Kathryn V. Holmes for the kind gift of the Cc1 mAb. They also thank Dr. Pnina Brodt, Julie St-Pierre, Josie Ursini-Siegel and Peter Siegel for fruitful discussions on the manuscript.
Funding This study was supported by grants from the Canadian Institutes of Health Research MOP-86582 (NB, MS, and PG), the National Research Council of Canada, the Weekend to End Women's Cancers (CG), the National Institutes of Health (NIH) grants DK51362, DK44319, DK53056 and DK088199 (RSB), the Harvard Digestive Diseases Center (NIH P30DK034854)(RSB), the High Pointe Foundation (RSB), the “Fonds de Recherche du Québec en Santé” fellowship (AA) and the Canadian Institutes of Health Research studentship (JDC).
Footnotes
Contributors AA was responsible for acquisition of data, analysis and interpretation of data and drafting of the manuscript. JD-C, VB, SH, SY, CT provided technical and material support. KM, CMTG and UDA analysed the TCGA data. MS, RSB and PTG provided cells, reagents and inhibitors, and discussed study design. AA and NB were responsible for the study concept and design, and wrote the manuscript.
Additional material is published online only. To view please visit the journal online (http://dx.doi.org/10.1136/gutjnl-2014-308781).
Dedication: This manuscript is dedicated to the memory of Rosalind Goodman.
Competing interests None.
Provenance and peer review Not commissioned; externally peer reviewed.
Data sharing statement Statistical values for 10-year survival analysis are available from the corresponding author upon request.
REFERENCES
- 1.Parkin DM, Bray F, Ferlay J, et al. Global cancer statistics, 2002. Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Allemani C, Rachet B, Weir HK, et al. Colorectal cancer survival in the USA and Europe: a CONCORD high-resolution study. BMJ Open. 2013;3:e003055. doi: 10.1136/bmjopen-2013-003055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beauchemin N, Arabzadeh A. Carcinoembryonic Antigen-related Cell Adhesion Molecules (CEACAMs) in cancer progression and metastasis. Cancer Metastasis Rev. 2013;32:643–71. doi: 10.1007/s10555-013-9444-6. [DOI] [PubMed] [Google Scholar]
- 4.Beauchemin N, Kunath T, Robitaille J, et al. Association of biliary glycoprotein with protein tyrosine phosphatase SHP-1 in malignant colon epithelial cells. Oncogene. 1997;14:783–90. doi: 10.1038/sj.onc.1200888. [DOI] [PubMed] [Google Scholar]
- 5.Huber M, Izzi L, Grondin P, et al. The carboxyl-terminal region of biliary glycoprotein controls its tyrosine phosphorylation and association with protein-tyrosine phosphatases SHP-1 and SHP-2 in epithelial cells. J Biol Chem. 1999;274:335–44. doi: 10.1074/jbc.274.1.335. [DOI] [PubMed] [Google Scholar]
- 6.Gray-Owen SD, Blumberg RS. CEACAM1: contact-dependent control of immunity. Nat Rev Immunol. 2006;6:433–46. doi: 10.1038/nri1864. [DOI] [PubMed] [Google Scholar]
- 7.Müller MM, Klaile E, Vorontsova O, et al. Homophilic adhesion and CEACAM1-S regulate dimerization of CEACAM1-L and recruitment of SHP-2 and c-Src. J Cell Biol. 2009;187:569–81. doi: 10.1083/jcb.200904150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poy MN, Yang Y, Rezaei K, et al. CEACAM1 regulates insulin clearance in liver. Nat Genet. 2002;30:270–6. doi: 10.1038/ng840. [DOI] [PubMed] [Google Scholar]
- 9.Xu E, Leung N, Dubois MJ, et al. Targeted disruption of Ceacam1 promotes diet-induced hepatic steatosis and insulin resistance. Endocrinol. 2009;150:3503–12. doi: 10.1210/en.2008-1439. [DOI] [PubMed] [Google Scholar]
- 10.Ergün S, Kilic N, Ziegeler G, et al. CEA-Related cell adhesion molecule 1 (CEACAM1): a potent angiogenic factor and a major effector of vascular endothelial growth factor (VEGF). Mol. Cell. 2000;5:311–20. doi: 10.1016/s1097-2765(00)80426-8. [DOI] [PubMed] [Google Scholar]
- 11.Nagaishi T, Pao L, Lin SH, et al. SHP1 phosphatase-dependent T cell inhibition by CEACAM1 adhesion molecule isoforms. Immunity. 2006;25:769–81. doi: 10.1016/j.immuni.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 12.Horst A, Bickert T, Brewig N, et al. CEACAM1+ myeloid cells control angiogenesis in inflammation. Blood. 2009;113:6726–36. doi: 10.1182/blood-2008-10-184556. [DOI] [PubMed] [Google Scholar]
- 13.Pan H, Shively JE. Carcinoembryonic antigen-related cell adhesion molecule-1 regulates granulopoiesis by inhibition of granulocyte colony-stimulating factor receptor. Immunity. 2010;33:620–31. doi: 10.1016/j.immuni.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kunath T, Ordonez-Garcia C, Turbide C, et al. Inhibition of colonic tumor cell growth by biliary glycoprotein. Oncogene. 1995;11:2375–82. [PubMed] [Google Scholar]
- 15.Izzi L, Turbide C, Houde C, et al. cis-Determinants in the cytoplasmic domain of CEACAM1 responsible for its tumor inhibitory function. Oncogene. 1999;18:5563–72. doi: 10.1038/sj.onc.1202935. [DOI] [PubMed] [Google Scholar]
- 16.Hsieh J-T, Luo W, Song W, et al. Tumor suppressive role of an androgen-regulated epithelial cell adhesion molecule (C-CAM) in prostate carcinoma cell revealed by sense and antisense approaches. Cancer Res. 1995;55:190–7. [PubMed] [Google Scholar]
- 17.Leung N, Turbide C, Marcus V, et al. Carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1) contributes to progression of colon tumors. Oncogene. 2006;25:5527–36. doi: 10.1038/sj.onc.1209541. [DOI] [PubMed] [Google Scholar]
- 18.Leung N, Turbide C, Balachandra B, et al. Intestinal tumor progression is promoted by decreased apoptosis and dysregulated Wnt signaling in Ceacam1(−/−) mice. Oncogene. 2008;27:4943–53. doi: 10.1038/onc.2008.136. [DOI] [PubMed] [Google Scholar]
- 19.Nouvion AL, Oubaha M, LeBlanc S, et al. CEACAM1: a key regulator of vascular permeability. J Cell Sci. 2010;123:4221–30. doi: 10.1242/jcs.073635. [DOI] [PubMed] [Google Scholar]
- 20.Lu R, Kujawski M, Pan H, et al. Tumor Angiogenesis Mediated by Myeloid Cells Is Negatively Regulated by CEACAM1. Cancer Res. 2012;72:2239–50. doi: 10.1158/0008-5472.CAN-11-3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arabzadeh A, Chan C, Nouvion AL, et al. Host-related carcinoembryonic antigen cell adhesion molecule 1 promotes metastasis of colorectal cancer. Oncogene. 2013;32:849–60. doi: 10.1038/onc.2012.112. [DOI] [PubMed] [Google Scholar]
- 22.Gerstel D, Wegwitz F, Jannasch K, et al. CEACAM1 creates a pro-angiogenic tumor microenvironment that supports tumor vessel maturation. Oncogene. 2011;30:4275–88. doi: 10.1038/onc.2011.146. [DOI] [PubMed] [Google Scholar]
- 23.Potula HSK, Wang D, Quyen DV, et al. Src-dependent STAT3-mediated expression of monocyte chemoattractant protein-1 is required for 15(S)-hydroxyeicosatetraenoic acid-induced vascular smooth muscle cell migration. J Biol. Chem. 2009;284:31142–55. doi: 10.1074/jbc.M109.012526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao L, Lim SY, Gordon-Weeks AN, et al. Recruitment of a myeloid cell subset (CD11b/Gr1 mid) via CCL2/CCR2 promotes the development of colorectal cancer liver metastasis. Hepatology. 2013;57:829–39. doi: 10.1002/hep.26094. [DOI] [PubMed] [Google Scholar]
- 25.Wu Y, Brodt P, Sun H, et al. Insulin-like growth factor-I regulates the liver microenvironment in obese mice and promotes liver metastasis. Cancer Res. 2010;70:57–67. doi: 10.1158/0008-5472.CAN-09-2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haftchenary S, Luchman HA, Jouk AO, et al. Potent Targeting of the STAT3 Oncogene in Brain Cancer Stem Cells: A Promising Therapeutic Route for Treating Glioblastoma. ACS Med Chem Lett. 2013;4:1102–7. doi: 10.1021/ml4003138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ieda J, Yokoyama S, Tamura K, et al. Re-expression of CEACAM1 long cytoplasmic domain isoform is associated with invasion and migration of colorectal cancer. Int J Cancer. 2011;129:1351–61. doi: 10.1002/ijc.26072. [DOI] [PubMed] [Google Scholar]
- 28.Wolf MJ, Hoos A, Bauer J, et al. Endothelial CCR2 signaling induced by colon carcinoma cells enables extravasation via the JAK2-Stat5 and p38MAPK pathway. Cancer Cell. 2012;22:91–105. doi: 10.1016/j.ccr.2012.05.023. [DOI] [PubMed] [Google Scholar]
- 29.Hoechst B, Voigtlaender T, Ormandy L, et al. Myeloid derived suppressor cells inhibit natural killer cells in patients with hepatocellular carcinoma via the NKp30 receptor. Hepatology. 2009;50:799–807. doi: 10.1002/hep.23054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kudo-Saito C, Shirako H, Ohike M, et al. CCL2 is critical for immunosuppression to promote cancer metastasis. Clin Exp Metastasis. 2013;30:393–405. doi: 10.1007/s10585-012-9545-6. [DOI] [PubMed] [Google Scholar]
- 31.Cambien B, Richard-Fiardo P, Karimdjee BF, et al. CCL5 neutralization restricts cancer growth and potentiates the targeting of PDGFRβ in colorectal carcinoma. PLoS ONE. 2011;6:e28842. doi: 10.1371/journal.pone.0028842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morikawa T, Baba Y, Yamauchi M, et al. STAT3 expression, molecular features, inflammation patterns, and prognosis in a database of 724 colorectal cancers. Clin Cancer Res. 2011;17:1452–62. doi: 10.1158/1078-0432.CCR-10-2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsuyada A, Chow A, Wu J, et al. CCL2 mediates cross-talk between cancer cells and stromal fibroblasts that regulates breast cancer stem cells. Cancer Res. 2012;72:2768–79. doi: 10.1158/0008-5472.CAN-11-3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang X, Yue P, Page BD, et al. Orally bioavailable small-molecule inhibitor of transcription factor Stat3 regresses human breast and lung cancer xenografts. Proc Natl Acad Sci USA. 2012;109:9623–8. doi: 10.1073/pnas.1121606109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoshidome H, Kohno H, Shida T, et al. Significance of monocyte chemoattractant protein-1 in angiogenesis and survival in colorectal liver metastases. Int J Oncol. 2009;34:923–30. doi: 10.3892/ijo_00000218. [DOI] [PubMed] [Google Scholar]
- 36.Lassmann S, Schuster I, Walch A, et al. STAT3 mRNA and protein expression in colorectal cancer: effects on STAT3-inducible targets linked to cell survival and proliferation. J Clin Pathol. 2007;60:173–9. doi: 10.1136/jcp.2005.035113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu H, Lee H, Herrmann A, et al. Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat Rev Cancer. 2014;14:736–46. doi: 10.1038/nrc3818. [DOI] [PubMed] [Google Scholar]
- 38.Ford LB, Cerovic V, Milling SW, et al. Characterization of conventional and atypical receptors for the chemokine CCL2 on mouse leukocytes. J Immunol. 2014;193:400–11. doi: 10.4049/jimmunol.1303236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Borsig L, Wolf MJ, Roblek M, et al. Inflammatory chemokines and metastasis-tracing the accessory. Oncogene. 2014;33:3217–24. doi: 10.1038/onc.2013.272. [DOI] [PubMed] [Google Scholar]
- 40.Bonapace L, Coissieux MM, Wyckoff J, et al. Cessation of CCL2 inhibition accelerates breast cancer metastasis by promoting angiogenesis. Nature. 2014;515:130–3. doi: 10.1038/nature13862. [DOI] [PubMed] [Google Scholar]
- 41.Calon A, Espinet E, Palomo-Ponce S, et al. Dependency of colorectal cancer on a TGF-β-driven program in stromal cells for metastasis initiation. Cancer Cell. 2012;22:571–84. doi: 10.1016/j.ccr.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Page BDG, Fletcher S, Li Z, et al. Identification of a Nonphosphorylated, Cell Permeable, Small Molecule Ligand for the Stat3 SH2 Domain. Bioorg Med Chem Lett. 2011;21:5605–9. doi: 10.1016/j.bmcl.2011.06.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee HJ, Zhuang G, Cao Y, et al. Drug resistance via feedback activtion of Stat3 in oncogene-addicted cancer cells. Cancer Cell. 2014;26:207–21. doi: 10.1016/j.ccr.2014.05.019. [DOI] [PubMed] [Google Scholar]
- 44.Singer BB, Scheffrahn I, Kammerer R, et al. Deregulation of the CEACAM expression pattern causes undifferentiated cell growth in human lung adenocarcinoma cells. PLoS ONE. 2010;5:e8747. doi: 10.1371/journal.pone.0008747. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.