Ten eleven translocation (TET) genes, and especially TET2, are frequently mutated in various cancers, but how the TET proteins contribute to the onset and maintenance of these malignancies is largely unknown. In this review, Rasmussen and Helin highlight recent advances in understanding the physiological function of the TET proteins and their role in regulating DNA methylation and transcription.
Keywords: DNA methylation, DNA demethylation, TET; hydroxymethyl cytosine, hematopoiesis, leukemia
Abstract
The pattern of DNA methylation at cytosine bases in the genome is tightly linked to gene expression, and DNA methylation abnormalities are often observed in diseases. The ten eleven translocation (TET) enzymes oxidize 5-methylcytosines (5mCs) and promote locus-specific reversal of DNA methylation. TET genes, and especially TET2, are frequently mutated in various cancers, but how the TET proteins contribute to prevent the onset and maintenance of these malignancies is largely unknown. Here, we highlight recent advances in understanding the physiological function of the TET proteins and their role in regulating DNA methylation and transcription. In addition, we discuss some of the key outstanding questions in the field.
Pertinent questions
How are TET proteins recruited to genomic elements?
Does deposition of 5-hydroxymethylcytosine (5hmC) inhibit the maintenance of DNA methylation in vivo?
Do 5hmC/5-formylcytosine (5fC)/5-carboxylcytosine (5caC) have roles as epigenetic marks?
What is the link between TET2 inactivation, transcriptional changes, and premalignant and malignant hematopoiesis?
Are fully transformed tumor cells dependent on the initial TET mutation?
Do the biological functions of the TET proteins depend on their catalytic function?
The vertebrate genome is bundled into a highly organized chromatin structure fundamental for precise gene regulation and maintenance of genome integrity. It is widely believed that dynamic chromatin states guide cells through development and impart an epigenetic “memory” to maintain cell type-specific transcriptional programs. Along with post-translational modifications of histone proteins, the direct methylation of cytosines in CG dinucleotides in DNA (called CpG sites) is one of several layers of regulatory information that determines chromatin states. Until recently, DNA methylation was believed to be an irreversible epigenetic event associated with gene repression, which could only be alleviated through DNA replication. Thus, it was remarkable when ten eleven translocation protein 1 (TET1) was discovered and shown to be able to modify methylcytosine and potentially erase DNA methylation (Tahiliani et al. 2009). TET1 belongs to a family of three proteins—namely, TET1, TET2, and TET3—that catalyze the successive oxidation of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC) (He et al. 2011; Ito et al. 2011). These 5mC oxidation products were promptly implicated as intermediates in the conversion of 5mC to unmodified cytosines, potentially providing the first steps in a pathway for active DNA demethylation and implying that DNA methylation patterns are not as static as previously assumed.
Disruption of epigenetic landscapes, including DNA methylation patterns, is a hallmark of cancer. Somatic mutations in genes (e.g., in DNMT3A) that encode for the machinery that establishes DNA methylation have been causally linked to malignant transformation. Interestingly, the activity of TET enzymes, which is involved in removing this epigenetic mark, has also emerged as an important tumor suppressor mechanism in cancer. TET2 is one of the most frequently mutated genes in hematopoietic malignancies, and its disruption is an early event in the onset of disease. It is now known that all three TET genes are mutated and show reduced expression, and the proteins have impaired activity in a wide range of different cancer types. Thus, precise regulation of DNA methylation patterns, which is partly mediated by TET enzymes, is important for normal development and provides a fundamental protection against cellular transformation.
TET proteins and their role in regulation of DNA methylation patterns
TET proteins are large (∼180- to 230-kDa) multidomain enzymes (Fig. 1). All TET proteins contain a conserved double-stranded β-helix (DSBH) domain, a cysteine-rich domain, and binding sites for the cofactors Fe(II) and 2-oxoglutarate (2-OG) that together form the core catalytic region in the C terminus. Structural studies suggest that this core catalytic region preferentially binds cytosines in a CpG context but does not interact with surrounding DNA bases and shows little or no specificity for flanking DNA sequences (Hu et al. 2013, 2015; Hashimoto et al. 2014b). In addition to their catalytic domain, TET1 and TET3 have an N-terminal CXXC zinc finger domain that can bind DNA (Fig. 1; Zhang et al. 2010; Xu et al. 2011c, 2012).
Figure 1.
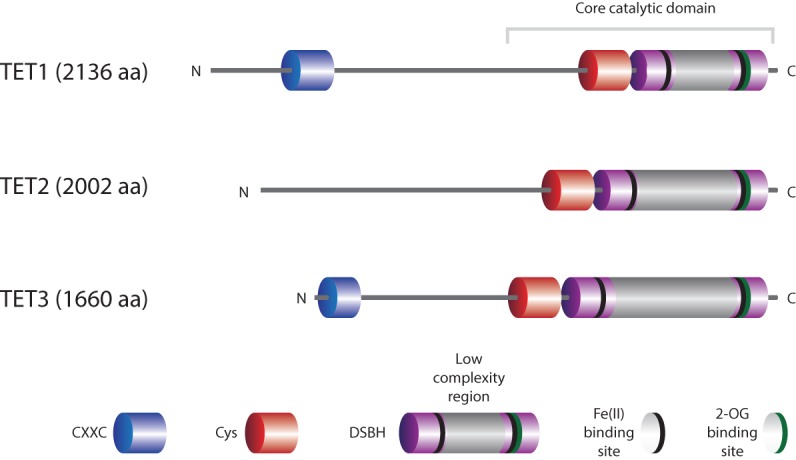
Domain structure of TET proteins. The C-terminal core catalytic domain shared by all TET enzymes consists of the DSBH domain, a cysteine-rich (Cys) domain, and binding sites for the Fe(II) and 2-OG cofactors. The DSBH domain contains a large low-complexity region of unknown function. TET1 and TET3 have an N-terminal CXXC domain that can bind directly to DNA and facilitate recruitment to genomic target sites.
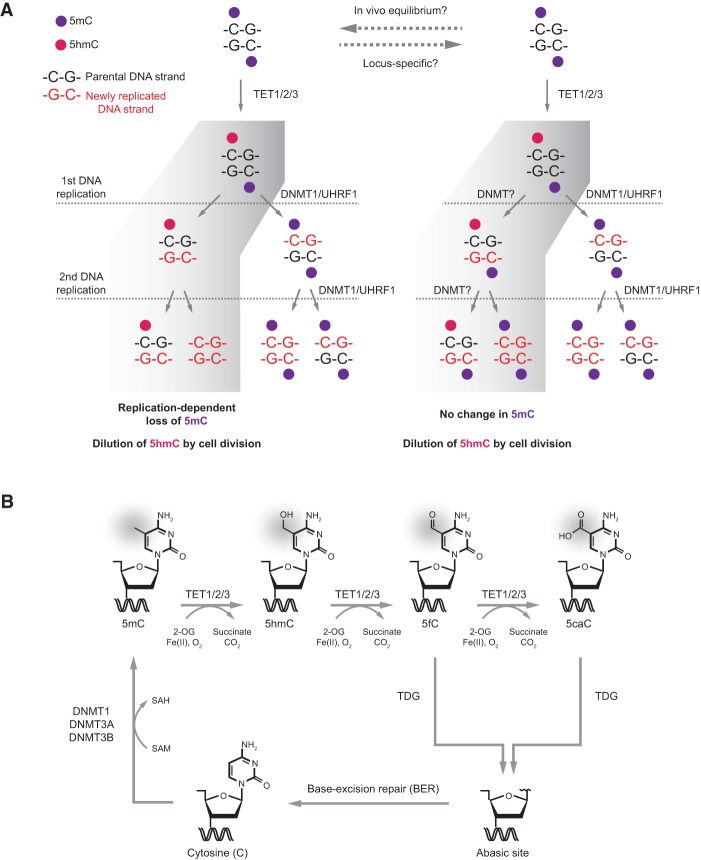
The deposition and removal of DNA methylation occur in a cyclical manner and require the enzymatic activity of multiple proteins. The majority of CpG sites are symmetrically methylated such that both strands carry the cytosine modification. During S phase, the SRA domain of UHRF1 recognizes and binds newly replicated hemimethylated DNA (Jones and Liang 2009). This interaction promotes recruitment of the “maintenance” DNA methyltransferase DNMT1 that re-establishes symmetry by copying the DNA methylation pattern of the template strand onto the newly synthesized strand. This mechanism is responsible for faithful inheritance of the bulk of DNA methylation patterns across cell division (Jones and Liang 2009). Once methylated, the modified cytosine can be the substrate of a stepwise TET-mediated oxidation process that forms 5hmC, 5fC, and 5caC in a Fe(II)- and 2-OG-dependent manner. In contrast to 5mC, the majority of these oxidation products is asymmetrically deposited on a given CpG site, suggesting that mitotic inheritance, if any, is achieved by a different mechanism (Jones and Liang 2009; Yu et al. 2012; Wu et al. 2014).
The conversion of 5mC and its oxidized derivatives back to the unmodified state has been proposed to occur by either “passive” or “active” demethylation. “Passive” DNA demethylation refers to the failure to maintain DNA methylation patterns across cell divisions and is believed to result in replication-dependent dilution of 5mC. In addition to down-regulation or cytoplasmic retention of the DNA methylation machinery, the deposition of 5hmC can have a role in inducing passive DNA demethylation. In vitro studies have shown that DNMT1 activity is dramatically reduced (up to 60-fold) on a DNA substrate containing 5hmC (Valinluck and Sowers 2007; Hashimoto et al. 2012; Ji et al. 2014). TET-mediated 5hmC deposition may therefore trigger passive replication-dependent DNA demethylation on the opposite DNA strand and be important to counter sporadic accumulation of aberrant DNA methylation. However, more recent data suggest that the role of 5hmC could be more complex. It has been shown that the DNMT1 interaction partners UHRF1 and UHRF2 can bind 5hmC DNA (Frauer et al. 2011; Hashimoto et al. 2012; Iurlaro et al. 2013; Spruijt et al. 2013; Zhou et al. 2014). It is therefore possible that one or both of these promote maintenance of DNA methylation by recruiting DNMT1 to hemihydroxymethylated sites in vivo. Moreover, the two “de novo” DNA methyltransferases DNMT3A2 and DNMT3B2 have also been implicated in the maintenance of DNA methylation (Chen et al. 2003; Jones and Liang 2009), and these enzymes readily methylate a DNA substrate containing 5hmC (Hashimoto et al. 2012; Ji et al. 2014). Widespread deposition of 5hmC has also been proposed to induce passive DNA demethylation in preimplantation embryos and primordial germ cells (for review, see Hill et al. 2014). However, it is now becoming apparent that the global demethylation observed in these cell types can proceed independently of 5hmC production. For instance, DNMT1 is excluded from the nucleus in early embryos (Seisenberger et al. 2013), and the rate of demethylation of both paternal and maternal genomes cannot be explained by a passive demethylation pathway (both TET-dependent and TET-independent) alone (Wang et al. 2014). Thus, the overall contribution of 5hmC to global demethylation processes is still not fully understood (Hill et al. 2014), and further investigations are necessary to discern the effect of 5hmC on the maintenance of DNA methylation in vivo in cells with intact DNA methylation machinery (Fig. 2A).
Figure 2.
Pathways of DNA demethylation mediated by TET enzymes. (A) Model of passive replication-dependent DNA demethylation. The diagrams illustrate the two opposite fates of a hemihydroxymethylated CpG dinucleotide through two rounds of DNA replication. The left panel shows the replication-dependent DNA demethylation, in which DNA methylation is lost on the DNA strand opposite to 5hmC. The right panel shows the reverse outcome, in which maintenance of DNA methylation is achieved by DNMT1 in complex with UHRF1/2 or DNMT3A/B. The relative extent of these two opposing outcomes of 5hmC deposition in vivo remains to be determined and is likely influenced by global as well as locus-specific factors. In both cases, newly replicated DNA dilutes 5hmC during cell division. (B) Model of active DNA demethylation by a TET/TDG (thymine–DNA–glycosylase)/BER (base excision repair)-dependent pathway. A cytosine base can be methylated by the DNA methylation machinery (DNMT1 or DNMT3A/B) to form 5mC, which in turn can be iteratively oxidized by TET enzymes to produce 5hmC, 5fC, and 5caC. TDG then recognizes 5fC and 5caC, and the oxidized cytosine base is excised. This yields an abasic site that is repaired by BER and results in restoration of the unmodified cytosine state. Additional pathways of active DNA demethylation have been suggested (for review, see Wu and Zhang 2014). (SAH) S-adenosyl-homocysteine; (SAM) S-adenosyl-methionine.
“Active” DNA demethylation refers to an enzymatic process in which 5mC bases, also in their oxidized forms, are replaced by unmodified cytosines in a replication-independent manner. Although several mechanisms have been proposed (for review, see Wu and Zhang 2014), the pathway involving thymine–DNA–glycosylase (TDG)-catalyzed base excision and DNA base excision repair (BER) is now believed to be the major driver of active DNA demethylation. TDG is a DNA mismatch repair enzyme that binds and excises mismatched pyrimidines in G:U and G:T base pairs (Lindahl and Wood 1999). Subsequent to the discovery of oxidized 5mC bases, it was demonstrated that TDG has no effect on 5hmC but shows robust recruitment and in vitro catalytic activity toward 5fC and 5caC (He et al. 2011; Maiti and Drohat 2011; Spruijt et al. 2013). TDG-mediated base excision yields an abasic site that is then replaced by an unmodified cytosine by BER, thus resulting in net DNA demethylation (Fig. 2B; He et al. 2011; Zhang et al. 2012). In support of this mechanism, it has been shown that depletion or genetic disruption of Tdg in embryonic stem cells (ESCs) leads to a fivefold to 10-fold increase of 5fC and 5caC levels (He et al. 2011; Shen et al. 2013; Song et al. 2013a). Conversely, overexpression of TDG in HEK293 cells results in depletion of 5fC and 5caC with little or no change of 5mC and 5hmC (Nabel et al. 2012). These data therefore suggest that 5fC and 5caC are constantly being purged from the genome by a TDG/BER-dependent pathway and that this constitutes an active, TET-mediated DNA demethylation process in vivo.
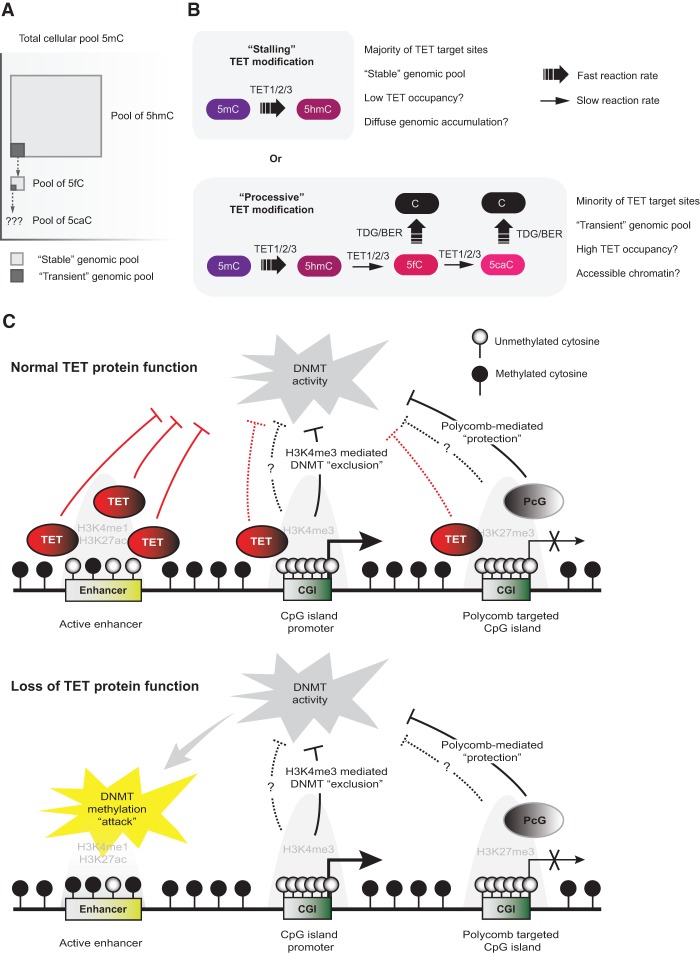
Although oxidation of 5mC to 5hmC is the first enzymatic step leading to restoration of unmodified cytosine by a TET/TDG/BER-dependent pathway, recent reports have revealed an apparent uncoupling of global 5hmC levels and active DNA demethylation. Experiments using isotope labeling combined with sensitive detection methods show that hydroxymethylation (Bachman et al. 2014) and low levels of formylation (Bachman et al. 2015) are present as relatively stable cytosine modifications in genomic DNA of both dividing and nondividing cells. This means that the majority of observable 5hmC and 5fC is not short-lived transient intermediates in an enzymatic process leading to DNA demethylation but is instead integrated and remains in genomic DNA for an extended period of time. Moreover, the steady-state levels of stable 5hmC and 5fC must be considerably larger than a corresponding pool of transient oxidized cytosines associated with active DNA demethylation processes (Fig. 3A). In agreement with this, kinetic analyses of TET catalytic activity suggest that the rate of cytosine oxidation is significantly reduced for the 5hmC (4.9-fold to 6.3-fold) and 5fC (7.8-fold to 12.6-fold) substrates compared with the initial oxidation reaction of 5mC (Ito et al. 2011; Hu et al. 2015). A comparison of crystal structures of human TET2 in complex with all three substrates indicates that the substrate preference is due to subtle differences in the catalytic cavity (Hu et al. 2013; 2015). The position of the cytosine is unchanged between the three substrates, but the hydroxy and carbonyl groups of 5hmC and 5fC form intermolecular and intramolecular hydrogen bonds and adopt a more restrained conformation than the methyl group of 5mC. These changes as well as a further distance to the Fe(II) cofactor lead to a decreased hydrogen abstraction efficiency and overall lower catalytic activity (Hu et al. 2015). Thus, the TET proteins have intrinsic properties to restrain their ability to form 5fC and 5caC that may lead to stalling of TET-mediated oxidation after 5hmC formation and diffuse accumulation of stable hydroxymethylated cytosines in the genome (Fig. 3B, top panel).
Figure 3.
The dynamics of TET-mediated cytosine oxidation and its impact on DNA methylation patterns on regulatory elements in vivo. (A) Diagram illustrating the relative abundance of cytosines in the genome with “stable” levels of modifications as well as the small “transient” fractions that undergo rapid turnover and removal by active DNA demethylation processes. The stable levels of genomic 5caC have not been experimentally determined, as they are often under the detection limit. (B) Model illustrating the dual role of TET enzymes. The rate constant of TET enzymes to produce 5hmC from 5mC is fast, whereas the rate constants for further oxidation to 5fC and 5caC are considerably slower. On a majority of TET target sites, 5mC is oxidized and persists as “stable” 5hmC, likely due to low TET residence time (“stalling TET modification”). On a minority of sites, preferably located in open chromatin associated with regulatory elements, 5mC is oxidized iteratively to 5fC and 5caC and results in net DNA demethylation by the TET/TDG/BER pathway (“processive TET modification”). (C) Model illustrating the impact of loss of TET protein function on different regulatory elements. The protection of active and inactive CpG island promoters by the TET proteins is only one of several mechanisms by which they are protected against ectopic DNA methylation. In contrast, distal regulatory elements such as active enhancers are vulnerable to aberrant DNMT activity and become DNA hypermethylated upon loss of TET activity. (PcG) Polycomb group.
Apart from their function in 5mC turnover, TET proteins have been shown to catalyze the oxidation of thymine bases in T:A base pairs to form 5-hydroxymethyluracil (5hmU). This DNA modification is present at exceedingly low amounts in ESCs and is efficiently cleaved and repaired by the DNA glycosylases TDG and SMUG1 followed by BER (Cortellino et al. 2011; Pfaffeneder et al. 2014). As such, 5hmU could have an independent function in the recruitment of chromatin readers but has also been proposed as a by-product of high TET activity to increase local DNA repair processes associated with TDG/BER-mediated DNA demethylation. However, the functional relevance of 5hmU as an epigenetic mark and its link to TET-mediated DNA demethylation remain to be determined.
Roles of 5mC, 5hmC, 5fC, and 5caC
Methylation of CpG sites is the most abundant DNA modification in the vertebrate genome, and its role in directing development and gene regulation has been extensively studied (for review, see Baylin and Jones 2011). DNA methylation is found throughout the genome (70%–80% of all CpGs are methylated) and is faithfully maintained during cell division by replication-coupled methylation machinery. Notable exceptions to this pervasive genomic methylation are gene regulatory regions such as CpG-rich promoter elements, also known as CpG islands, and active enhancers that are often associated with little or no DNA methylation. The DNA methylation landscapes across different human and mouse tissues are remarkably stable. In both species, only a fraction of genomic CpG sites (7%–22% of 28 million autosomal CpGs) changes between a methylated and an unmethylated state even in distantly related cell types (Hon et al. 2013; Ziller et al. 2013). Detailed analysis of these dynamically regulated sites has revealed that the appearance and disappearance of tissue-specific hypomethylated regions colocalize with promoters, enhancers, and transcription factor (TF)-binding sites and that these correlate with expression of neighboring genes. Thus, in contrast to a relatively static DNA methylation environment, the gain and loss of DNA methylation at regulatory regions are prominent features of normal development (Hon et al. 2013; Ziller et al. 2013).
Early studies using sensitive mass spectrometry analysis revealed that 5mC oxidation products were present in all examined tissues, albeit in low and highly variable amounts. Whereas 5mC is found in all tissues, corresponding to ∼4%–5% of all cytosines, the most abundant of the 5mC oxidation products, 5hmC, is present at a range of 0.3%–0.7% in the CNS down to as little as 0.03%–0.06% in the spleen and testes. By comparison, the products of further TET-mediated oxidation, 5fC and 5caC, are observed at ∼10-fold to 100-fold lower levels than 5hmC and often close to or under the detection limit (Kriaucionis and Heintz 2009; Globisch et al. 2010; Szwagierczak et al. 2010; He et al. 2011; Ito et al. 2011; Pfaffeneder et al. 2011; Bachman et al. 2015).
There has been substantial interest in mapping cytosine oxidation products in the genome with the aim of understanding their role in gene regulation and DNA methylation dynamics (Pastor et al. 2011; Stroud et al. 2011; Szulwach et al. 2011; Williams et al. 2011; Wu et al. 2011a; Xu et al. 2011c; Sérandour et al. 2012). Various enzymatic and chemical treatments of DNA combined with whole-genome sequencing now provide the possibility of producing genome-wide maps of 5hmC, 5fC, and 5caC (for review, see Yardimci and Zhang 2015). Although these techniques are prone to artifacts due to incomplete conversion or protection of distinct cytosine modifications, they offer important quantitative insights. A whole-genome and base-resolution map of 5hmC in ESCs identified ∼2 million 5hmC sites with an average modification rate of ∼20%. 5hmC enrichment was found in regions with low CpG content, including promoters associated with lowly expressed genes, gene bodies, and intragenic regions as well as distal regulatory regions such as DNase I hypersensitivity sites, p300-marked enhancers, and CTCF-bound insulator sites (Yu et al. 2012). By comparison, whole-genome sequencing combined with simultaneous detection of 5fC and 5caC identified ∼0.33 million modified sites with an average modification rate of 8%–10% (Neri et al. 2015). Thus, in accordance with isotope labeling and mass spectrometry analysis, the measurements imply that methylated cytosines successively oxidized to 5fC/5caC are significantly less abundant than 5hmC-modified cytosines. 5fC and 5caC are mostly located in or close to distal regulatory elements (DNase I hypersensitivity sites, enhancers, and CTCF-bound insulator sites) but can also be detected on transcription start sites of active genes (Wu et al. 2014; Neri et al. 2015). However, it should be noted that the overall distribution as well as enrichment of 5fC and 5caC in Tdg-depleted cells show that distal regulatory elements are the primary site of 5fC/5caC deposition (Wu et al. 2014). Interestingly, overlap of these base-resolution maps additionally shows that the vast majority of 5hmC and 5fC/5caC is found on distinct cytosine bases in the genome (out of ∼22 million CpGs covered in both maps, only 7.5% of 5hmC-modified and 18.8% 5fC/5caC-modified cytosine bases showed accumulation of the other modification). Although sensitivity of detection could be an issue, this lack of positional overlap implies that most 5hmC is not converted to higher oxidation levels. Conversely, the failure to detect 5hmC does not preclude the existence of active DNA demethylation at a given CpG site, as this can occur by fast and processive TET-mediated oxidation to 5fC and 5caC followed by TDG-dependent excision.
The overall abundance of oxidized cytosine species, the discordant genomic localization of 5hmC and 5fC/5caC at the nucleotide level, and the catalytic properties of TET enzymes are consistent with a dual role of TET proteins in the regulation of DNA methylation (Fig. 3B): On a majority of TET genomic target sites, TET oxidizes 5mC to 5hmC but stalls after this initial reaction (“stalling” TET modification). This results in a relatively high steady-state level of genomic 5hmC that on some sites could promote passive DNA demethylation and serve an important role to prevent faulty DNA methylation. It is also possible that 5hmC functions as a stable epigenetic mark; however, the low average modification rate in ESCs is incompatible with such a role on the majority of modified sites. Conversely, on a small fraction of TET genomic target sites mostly located in regulatory regions, high TET occupancy or stimulation of TET enzymatic activity results in processive oxidation of 5mC to 5fC or 5caC (“processive” TET modification). These highly oxidized bases are then recognized and excised by TDG/BER and result in net DNA demethylation of the cytosine base. The intrinsic catalytic constraint of TET enzymes as well as robust excision of TDG/BER contribute to the exceedingly low steady-state levels of 5fC and 5caC in the genome. Interestingly, sites with 5fC and 5caC are enriched in open chromatin and loci with TF occupancy (Wu et al. 2014). Thus, TF binding and chromatin remodeling may play a role in making sites permissive for processive TET-mediated 5fC and 5caC formation and promote active DNA demethylation. However, a detailed characterization of the conditions that favor processive TET activity at some, but not all, target sites is an important subject of future investigations. Together, these dual roles of TET enzymes are likely to promote DNA methylation fidelity, as has previously been suggested (Williams et al. 2012), and may execute a direct regulatory function at specific genomic target sites located in regulatory regions.
Cytosine oxidation products as independent epigenetic marks?
Considering the well-characterized role of a methylated cytosine as an inheritable epigenetic mark, it is tempting to speculate that the newly discovered cytosine derivatives also might have functions in the genome independently of their role in DNA methylation turnover. Most notably, the high abundance of 5hmC in post-mitotic brain tissues makes this a possible candidate as an epigenetic mark. However, attempts to systematically identify chromatin-associated “reader” proteins interacting with 5mC-, 5hmC-, 5fC-, or 5caC-containing DNA oligos yielded surprisingly few 5hmC-specific binders. Instead, the much less abundant cytosine species 5fC and 5caC appear to interact with numerous proteins, including TDG, p53, DNA repair factors, chromatin remodeling factors, and forkhead box TFs (Iurlaro et al. 2013; Spruijt et al. 2013). While some of these may be recruited as a result of DNA repair associated with active DNA demethylation processes, the biochemical and physiological relevance of the interactions is unclear.
Although the effects of cytosine modifications are likely to be highly context- and sequence-specific, the failure to identify proteins enriched at 5hmC and a biological role of these may be indicative of a general negative impact of this cytosine modification on chromatin binders. Indeed, proteins that are known to interact with methylated cytosines, such as MBD1, MBD2, and MBD4, cannot bind 5hmC-containing DNA or bind with lower affinity (Jin et al. 2010; Hashimoto et al. 2012). In addition, oxidation of cytosines in the DNA-binding motif of the TF Wilms tumor 1 (WT1; to 5hmC and 5fC but not 5caC) effectively disrupts protein–DNA interaction (Hashimoto et al. 2014a). Thus, oxidation of TF-binding sites in the genome can potentially lead to displacement of some proteins from chromatin and modulate downstream gene function. Whether such an effect or perhaps the opposite can be observed in the context of other DNA-binding factors remains to be investigated.
Despite a general lack of in vivo validation, the amounts of specific 5fC and 5caC binders observed in the in vitro pull-down experiments support a potential role of these cytosine species beyond functioning as transient intermediates in active DNA demethylation. In line with this, 5fC and 5caC have been shown to interfere with transcription by RNA polymerase II (Pol II) (Kellinger et al. 2012). Structural analysis of Pol II in complex with 5caC indicates that the interaction between Pol II residues and the 5mC derivative groups induces a positional shift that places the incoming GTP nucleotide in a less favorable position for addition to the growing RNA strand (Wang et al. 2015a). Thus, the presence of 5fC and 5caC, but presumably not 5mC or 5hmC, in the template DNA strand is equivalent to “speed bumps” and induces transient Pol II pausing and a reduced elongation rate both in vitro and in vivo (Kellinger et al. 2012; Wang et al. 2015a). It is therefore possible that controlled deposition of 5fC and 5caC can fine-tune the elongation rate of specific genes and indirectly affect gene expression; however, the physiological significance of this remains unknown. Interestingly, a recent report identified mutants of the TET1 protein in the amoeba Naegleria that primarily generate 5hmC with little or no further oxidation to 5fC and 5caC (Hashimoto et al. 2015). Detailed analysis of such mutants in mammalian cells may help to elucidate the specific role of 5hmC in chromatin as well as the biological role of 5fC and 5caC as epigenetic marks.
Impact of TET proteins on DNA methylation in vivo
Although genome-wide maps of 5mC oxidation products provide a detailed snapshot of the affected genomic sites, the significance of TET proteins in the maintenance of DNA methylation patterns can most accurately be assessed by functional studies. Genetic disruption of all three Tet proteins in mouse ESCs (Tet triple-knockout cells) is compatible with viability despite complete absence of 5hmC and a small but consistent increase in global DNA methylation level (Dawlaty et al. 2014; Lu et al. 2014). Mapping of DNA methylation changes by whole-genome bisulfite sequencing revealed that DNA methylation predominantly accumulates on enhancers and DNase I hypersensitivity sites in Tet triple-knockout as well as Tet2 knockout cells (Hon et al. 2014; Lu et al. 2014). In fact, ∼15%–25% of all mapped ESC enhancers undergo significant DNA hypermethylation, underlining the genome-wide scale of Tet protein function. In contrast, little or no change of DNA methylation levels was observed at promoter elements and their associated CpG islands. This is in agreement with previous results demonstrating that several mechanisms exist that protect CpG island promoters from DNA methylation, including the binding of Tet1 (Williams et al. 2011; Wu et al. 2011b) and the exclusion of de novo DNA methyltransferases by H3K4me3 methylation (Ooi et al. 2007; Otani et al. 2009; Guo et al. 2015; Noh et al. 2015). In addition, transcriptionally repressed promoters and CpG islands (associated with PRC2 and marked by H3K27me3) also appear to be protected from ectopic DNA methylation by the polycomb-associated protein Fbxl10 (Boulard et al. 2015).
Although ESCs represent a well-established model system, it remains important to confirm these findings in differentiated cell lineages and in cells in which the TET proteins are known to have important regulatory roles. Initial studies of DNA methylation patterns in TET2 mutated patients with hematological disorders reported conflicting results. In some studies, promoter DNA methylation was found to be unchanged (Yamazaki et al. 2012) or predominantly DNA-hypomethylated (Ko et al. 2010; Pérez et al. 2012), whereas others reported increased levels of DNA methylation (Figueroa et al. 2010; Rampal et al. 2014). These discrepancies are possibly due to different experimental protocols and patient groups as well as control cells studied. Moreover, DNA methylation changes were assessed at only a relatively small number of CpG sites predominantly located in promoters.
An analysis of genome-wide DNA methylation as well as 5hmC levels in fetal and adult frontal cortices showed that differential DNA methylation is predominantly found in distal regulatory elements (Lister et al. 2013). Interestingly, the elements that became active (DNA hypomethylated and associated with increased levels of H3K4me1 and H3K27ac) in the transition from the fetal to the adult brain were already enriched with 5hmC in the fetal brain. Thus, 5hmC may play a role in marking enhancers that become activated later in development. Moreover, a large fraction of these enhancers (19.7%) had slightly increased levels of DNA methylation in the frontal cortices of Tet2 knockout mice. Although the increase in DNA methylation was rather modest, this study suggests that Tet2 is important for establishing the DNA-hypomethylated state of tissue-specific enhancers during development of neuronal tissues (Lister et al. 2013).
In a more recent study, Yamazaki et al. (2015) analyzed differentially methylated sites in a cohort of chronic myelomonocytic leukemia (CMML) patients with or without TET2 mutations. Here the investigators detected significant DNA hypermethylation in enhancer-like regions. In contrast, they did not detect changes in DNA methylation at CpG islands (Yamazaki et al. 2015). Finally, in a genetic tractable mouse model in which loss of Tet2 contributes to acute myeloid leukemia (AML), we could show that Tet2 maintains low DNA methylation levels at tissue-specific enhancers to prevent leukemic transformation. In addition, we were able to uncover a distinct enhancer DNA hypermethylation phenotype in human AML patients carrying TET2 mutations. The DNA methylation levels at CpG islands were unchanged in both mouse and human AML cells (Rasmussen et al. 2015).
Together, data from ESCs as well as differentiated tissues and malignant cells support a model in which TET-mediated DNA demethylation plays an essential role in maintaining low DNA methylation levels at distal regulatory elements. In contrast, CpG islands and promoters are largely resistant to ectopic DNA methylation due to several different layers of control (Fig. 3C).
Recruitment of TET proteins to DNA and regulation of enzymatic activity
It has become increasingly clear that, besides dynamic transcriptional control, TET protein activity and recruitment are regulated at multiple levels that together determine the final effect on DNA methylation patterns. For instance, the activity of TET enzymes can be stimulated or inhibited in the presence of distinct metabolites, cofactors, and post-translational modifications. In addition, interaction with multiple binding partners both stably and transiently is likely to affect genomic localization as well as protein stability.
TET1 and TET3 are most likely recruited to genomic target sites through direct binding of their respective CXXC domains to DNA (Fig. 1). In vitro binding assays confirm that these domains can bind CpG-rich oligonucleotides with a slight preference for unmethylated versus methylated substrates (Zhang et al. 2010; Xu et al. 2011c, 2012). In addition, in vivo chromatin immunoprecipitation of Tet1 shows that it is highly enriched on CpG islands, most of which are associated with promoters with intermediate and high CpG content, and that this enrichment is lost upon introduction of a mutation in the CXXC domain (Williams et al. 2011; Wu et al. 2011b; Xu et al. 2011c). Structural studies of CXXC domains in other proteins (Cierpicki et al. 2010; Song et al. 2011; Xu et al. 2011a) have shown that these can bind to a single CpG site. The observed enrichment of Tet1 at CpG islands may therefore reflect the higher number of potential binding sites in these elements. In agreement with the fact that the CXXC domain interacts with only one CpG, Tet1 has been shown to be associated with distal regulatory elements as well as genome-wide diffuse 5hmC deposition (Stadler et al. 2011; Putiri et al. 2014), and Tet3-mediated cytosine oxidation in the developing embryo occurs at a wide range of genomic elements (Wang et al. 2014). Thus, the CXXC domain may be involved in the recruitment of Tet1 and Tet3 to CpG sites irrespective of the local CpG density, and this is consistent with a model in which the Tet proteins, like the DNMTs, are binding throughout the genome.
In contrast to TET1 and TET3, the structure of TET2 does not reveal any discernable domains reported to bind directly to DNA, and it is therefore potentially recruited to genomic DNA by a distinct CXXC domain-independent mechanism. To date, it has not been possible to produce a reliable genome-wide map of TET2 chromatin occupancy due to nonspecific retention of signal even in genetic knockout cells (Jain et al. 2015; our unpublished results). Although several genome-wide data sets have been published (Chen et al. 2013; Deplus et al. 2013), none of them have systematically tested signal specificity. Thus, further studies are required to accurately map TET2 occupancy to understand how and where the protein is recruited to chromatin.
Considering the potential regulatory role of TET2 at distal regulatory elements, it is possible that TET2 recruitment can be facilitated through direct binding of DNA targeting partners. In fact, the TET2 protein, either endogenous or overexpressed, has been shown to bind tissue-specific TFs such as early B-cell factor 1 (EBF1) (Guilhamon et al. 2013) and WT1 (Rampal et al. 2014; Wang et al. 2015b). However, it should be noted that it is unlikely that one or a few factors, such as those mentioned above, can account for the entire spectrum of TET2 genomic target sites. Instead, it is possible that dynamic expression of DNA-binding factors as well as interaction with TET2 can confer tissue-specific and temporal modulation of TET activity on a limited set of genomic loci.
Recruitment by DNA-binding factors can also result in TET2 protein degradation. The TET2-interacting protein IDAX/CXXC4 that is believed to contain the ancestral CXXC domain of TET2 binds unmethylated DNA and is located predominantly at CpG islands. Analysis of cell lines with combined ectopic expression of these proteins indicated that IDAX/CXXC4 interacts with and activates caspase-dependent degradation of TET2 by a mechanism dependent on DNA binding (Ko et al. 2013). Thus, in agreement with the minor effect of TET2 disruption on these genomic elements, it is possible that IDAX/CXXC4 has evolved to negatively regulate TET2 activity on CpG islands with little or no effect outside of them. Although this represents a plausible model, IDAX/CXXC4 and TET2 are not coexpressed in the cells in which the studies were performed, and the interaction remains to be validated on endogenous protein.
Apart from site-specific recruitment, the activity of TET enzymes can be stimulated or inhibited at a global level in the presence of distinct metabolites, cofactors, and post-translational modifications. Mutations in the genes coding for the metabolic enzymes isocitrate dehydrogenases 1 and 2 (IDH1/2), succinate dehydrogenase (SDH), and fumarate hydratase (FH) have been reported in a wide variety of liquid and solid tumors (Oermann et al. 2012). These mutations, either deleterious (SDH and FH) or gain of function (IDH1/2), result in aberrant accumulation of metabolites such as 2-hydroxyglutarate (2-HG), succinate, and fumarate that inhibit TET protein enzymatic activity along with a large number (>60) of additional enzymes (Xu et al. 2011b; Xiao et al. 2012; Losman and Kaelin 2013). Interestingly, IDH1/2 mutations were found to be mutually exclusive with TET2 mutations in AML patients and have been associated with DNA hypermethylation in brain tumors and leukemias (Figueroa et al. 2010; Turcan et al. 2012). Despite the pleiotropic effect of these metabolites, the oncogenic effect of IDH, SDH, and FH mutations can be ascribed at least partly to the global inhibition of TET-mediated cytosine oxidation.
In contrast, increasing the levels of vitamin C (ascorbic acid) has been shown to stimulate TET protein enzymatic activity in cultured cells as well as mouse tissues (Blaschke et al. 2013; Minor et al. 2013; Yin et al. 2013; Bachman et al. 2014). This can be detected as increased levels of the cytosine oxidation products 5hmC, 5fC, and 5caC as well as a small reduction of global DNA methylation in the absence of changes in TET expression levels (Yin et al. 2013; Wu et al. 2014). Although the precise mechanism is unknown, it is likely that vitamin C interacts directly with the catalytic domain of TET proteins and provides a local reducing environment that increases recycling efficiency of the Fe(II) cofactor (Yin et al. 2013).
Finally, it has been shown that all three TET enzymes can be monoubiquitinylated at a conserved lysine residue (K1299 in TET2) by the E3 ubiquitin ligase complex CLR4VprBP (Yu et al. 2013; Nakagawa et al. 2015). This modification does not affect in vitro catalytic activity but potentially induces a conformational change that promotes TET binding to DNA. It is therefore possible that monoubiquitinylation is important to stabilize interactions with DNA-binding partners or alter the DNA-binding affinity of the TET proteins themselves. Interestingly, several TET2 missense mutations observed in AML patients affect K1299 or the surrounding region (Nakagawa et al. 2015). This suggests that TET protein activity can be modulated on a global level by CLR4VprBP-mediated monoubiquitinylation and that this regulation has direct implications for tumorigenesis.
Physiological functions of TET proteins
The elucidation of the physiological role of the TET protein family has received and continues to receive considerable attention. Even though they share catalytic activity, genetic studies strongly suggest that TET proteins are functionally nonredundant, most likely due to varying expression levels and recruitment mechanisms. For instance, expression of Tet3 is high in mouse oocytes and early preimplantation embryos but decreases rapidly as the cells progress to the inner cell mass (ICM) stage in blastocysts. Instead, mouse Tet1 and Tet2 show high expression in the ICM and ESCs, and, while Tet1 is gradually down-regulated, Tet2 and Tet3 remain constant or are significantly up-regulated during differentiation to the three germ layers. Finally, Tet2 (and, to some degree, also Tet3) shows robust expression in a wide range of adult tissues, including hematopoietic and neuronal lineages. It should be noted that the relative importance of each member within a specific cell type remains a topic of continuing debate, as there have been few attempts to systematically evaluate absolute protein levels in different tissues and developmental stages. Moreover, in some cases, deletion of one or several of the TET enzymes can be compensated for by increased expression or activity of the other TET family members (Dawlaty et al. 2013).
The physiological relevance of the TET proteins in development has been investigated using genetic knockout mouse models. Constitutive deletion of Tet3 leads to neonatal lethality with 100% penetrance (Gu et al. 2011; Kang et al. 2015), whereas deletions of either Tet1 (Dawlaty et al. 2011) or Tet2 (Moran-Crusio et al. 2011; Quivoron et al. 2011) do not result in any detectable developmental phenotype. This suggests that Tet3 has a unique role during embryogenesis that cannot be compensated for by other Tet enzymes. In contrast, when generating compound mutant mice deficient for both Tet1 and Tet2, it was noted that, while some survive and develop normally, the majority die perinatally and show a wide spectrum of defects, including exencephaly, growth retardation, and compromised imprinting (Dawlaty et al. 2013). The complexity of this phenotype (both partially penetrant and highly variable) strongly suggests that the defects cannot be explained by disruption of a single process or pathway. Instead, combined Tet1 and Tet2 deficiency is likely to result in epigenetic abnormalities that increase developmental stochasticity and derail embryogenesis at multiple levels (Dawlaty et al. 2013). Finally, the successful generation and characterization of Tet1/2/3 triple-knockout ESCs have shown that combined TET activity is essential for supporting embryonic development (Dawlaty et al. 2014).
The fact that single-knockout and some double-knockout mice survive until adulthood has prompted investigators to search for late-onset phenotypes in conditional Tet knockout mice. Interestingly, the disruption of any of the Tet genes in adult hematopoietic tissues leads to increased hematopoietic stem cell (HSC) function as measured in competitive repopulation assays (Ko et al. 2011, 2015; Moran-Crusio et al. 2011; Quivoron et al. 2011; Cimmino et al. 2015), and both myeloid and lymphoid alterations are observed to varying degrees. Loss of Tet2 results in age-associated extramedullary hematopoiesis and expansion of monocytes and granulocytes in the spleen (Ko et al. 2011; Li et al. 2011; Moran-Crusio et al. 2011; Quivoron et al. 2011; Shide et al. 2012), whereas Tet1 deficiency leads to an overall lymphoid bias and dysregulated B-cell lineage differentiation (Cimmino et al. 2015). This is further accompanied by a gradual expansion of myeloid-biased HSCs in Tet2-deficient mice versus lymphoid-primed multipotent progenitors (LMPPs) in Tet1 mutant mice, thus reflecting an early skewing of myeloid or lymphoid differentiation (Quivoron et al. 2011; Cimmino et al. 2015). Preliminary experiments using a hematopoietic-specific deletion of Tet3 failed to detect expansion of the HSC compartment or disrupted hematopoietic differentiation (Ko et al. 2015). Nonetheless, these data show that HSCs are uniquely sensitive to changes in TET protein activity and suggest that other family members cannot fully compensate for loss of an individual TET protein. Furthermore, terminal hematopoietic lineage differentiation is differentially affected upon disruption of either Tet1 or Tet2, which could be a reflection of distinct lineage-specific expression patterns and protein function. In agreement with these murine studies, enhanced engraftment potential and myeloid skewing of differentiation have also been observed in human hematopoietic CD34+ stem and progenitor cells with disruption or lack of TET2 expression (Delhommeau et al. 2009; Pronier et al. 2011; Itzykson et al. 2013).
Apart from a role in embryonic development and hematopoietic differentiation, Tet proteins and DNA demethylation has been implicated in several processes in differentiation and development. For instance, loss of Tet activity is associated with altered neuronal activity and has been implicated in learning and memory processes as well as cocaine addiction (Hahn et al. 2013; Rudenko et al. 2013; Feng et al. 2015). In addition, TET activity plays an important role in induced pluripotent stem cell formation (Doege et al. 2012; Costa et al. 2013) as well as smooth muscle cell differentiation (Liu et al. 2013b). Recently, Tet2 was further implicated in the regulation of cytokine expression during innate as well as T-cell-mediated immune responses. Upon myeloid cell-specific deletion, it was found that Tet2-deficient cells fail to efficiently repress proinflammatory cytokines after LPS-induced stimulation in a histone deacetylase (HDAC)-dependent manner (Zhang et al. 2015). Similarly, Tet2 deficiency induced in CD2+ T cells is associated with aberrant cytokine production and leads to increased autoimmunity in a mouse model of human multiple sclerosis (Ichiyama et al. 2015). Thus, Tet activity is likely an important factor in numerous developmental stages, and disruption can lead to failure of cell type-specific functions.
TET proteins in hematological cancer
Among the different cancer types studied, the role of TET proteins in hematological malignancies has received the most experimental support. The family of TET proteins was originally named due to the characterization of TET1 as a fusion partner in mixed-lineage leukemia (MLL)-rearranged AML (Ono et al. 2002; Lorsbach et al. 2003). However, this translocation is rare (0.3% of MLL fusions) (Ittel et al. 2013), and its precise significance in leukemogenesis has yet to be investigated. In contrast, the TET2 gene was independently unearthed in a study of myelodysplastic syndrome (MDS) and AML patients with 4q24 alterations, where it was the only ORF found in a minimally deleted region (Delhommeau et al. 2009; Langemeijer et al. 2009). Follow-up studies in diverse patient groups quickly identified not just chromosomal aberrations but also frequent point mutations—including insertions, deletions, missense, nonsense, and frameshift mutations—that often result in truncations or decreased enzymatic activity of TET2 (Weissmann et al. 2012). These mutations are most often heterozygous even though loss of the wild-type allele (loss of heterozygosity [LOH]) and biallelic mutations are commonly observed (e.g., up to 26% of MDS cases) (Bejar et al. 2011). The global levels of 5hmC are significantly reduced in patients that carry either homozygous or heterozygous TET2 disruption when compared with patients with wild-type TET2 (Ko et al. 2010), suggesting that these are true haploinsufficient loss-of-function mutations.
Somatic alterations in TET2 are observed to varying degrees in a wide range of hematological disease, including both myeloid and lymphoid malignancies (for review, see Scourzic et al. 2015). TET2 mutations are the most common genetic abnormality observed in patients with MDS (6%–26%) and CMML (20%–58%) but is also frequently observed in primary and secondary AML (12%–32%), blastic plasmacytoid dendritic neoplasm (25%–54%), and myeloproliferative neoplasms (MPNs) such as polycythemia vera, primary myelofibrosis, and essential thrombocytosis (2%–20%). In addition, TET2 alterations are found in B-cell (2%–12%) and T-cell (20%–83%) lymphomas, including diffuse large B-cell lymphoma (DLBCL), follicular lymphoma (FL), angioimmunoblastic T-cell lymphoma (AITL), and peripheral T-cell lymphoma (PTCL). Thus, TET2 mutations are not limited to a specific disease subtype, as seen for other oncogenic drivers in hematopoiesis. Instead, the mutational landscape suggests that these alterations can be involved in very different disease processes.
There are now several lines of evidence that point to a role of TET2 alterations as one of the first genetic aberrations in the onset of hematological malignancies. First, TET2 mutations can be found in premalignant HSCs in MDS and AML patients and are preserved in the tumor bulk along with additional oncogenic mutations (Jan et al. 2012; Chan and Majeti 2013; Itzykson et al. 2013). Second, TET2 mutations are frequently observed in aged healthy individuals with clonal hematopoiesis and increase the propensity to develop hematopoietic malignancies (Busque et al. 2012; Genovese et al. 2014; Jaiswal et al. 2014; Xie et al. 2014). Finally, disruption of Tet2 in mouse models increases HSC proliferation and activity, but malignant transformation is rare and occurs only with low penetrance and long latency, suggesting that other oncogenic events are necessary for development of disease. Thus, these data support a model in which TET2 alterations occur in HSCs and impart a clonal advantage that lead to a myeloid differentiation bias and expansion of the stem cell pool. These premalignant stem cell clones are then prone to acquiring additional oncogenic mutations that, together with TET2 disruption, induce the disease phenotype observed (Fig. 4). Further studies aimed at deciphering the role of TET2 disruption and its impact on DNA methylation patterns in premalignant HSCs as well as the significance in the onset of hematological diseases are needed.
Figure 4.
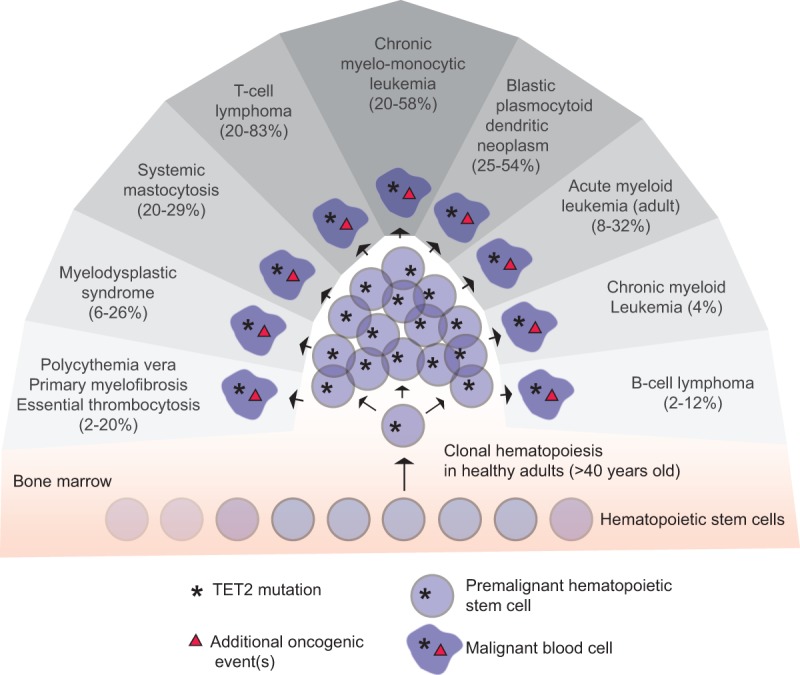
Illustration of the mutational landscape of TET2 in hematological diseases. A somatic mutation in TET2 results in premalignant hematopoiesis and clonal expansion. Additional oncogenic events cooperate with the initial TET2 mutation to drive the onset of a wide variety of hematopoietic malignancies. The frequencies of TET2 mutations in the different patient groups are indicated (for review, see Scourzic et al. 2015).
Although analysis of patients and Tet2 knockout mice supports a cooperative role of TET2 loss in leukemogenesis, it remains important to validate such a model in vivo. Several whole-genome and whole-exome sequencing studies have mapped and identified comutated genes in patients carrying TET2 alterations. These mutational patterns offer a chance to search for statistical co-occurrences or mutual exclusivity to identify combinations that may play a direct role in disease onset. Accordingly, with statistical significance, TET2 mutations are associated with SF3B1, ZRSR2, and ASXL1 mutations in MDS patients (Haferlach et al. 2014); STAG2, DNAH9, and serine–threonine kinase mutations in AML patients (Cancer Genome Atlas Research Network 2013); and RHOA, IDH2, and DNMT3A mutations in AITL patients (Palomero et al. 2014; Sakata-Yanagimoto et al. 2014). However, the functional relevance of these as well as the vast majority of additional co-occurring genetic alterations is largely unknown. In general, it is not clear how and to what extent they genetically interact with TET2 mutations or whether some are merely inconsequential “passenger” events fixed in a clonal or subclonal population.
To directly address this, several groups have generated mouse models with defined genetic alterations and assessed the phenotypic outcome in the hematopoietic system (Table 1). The combination of Tet2 disruption with either Asxl1null, Ezh2null, or Jak2V617F mutant alleles results in MDS and MPN-like disease phenotypes (Abdel-Wahab et al. 2013; Muto et al. 2013; Chen et al. 2015; Kameda et al. 2015), whereas expression of hyperactive KITD814V leads to systemic mastocytosis (Soucie et al. 2012; De Vita et al. 2014). In contrast, inactivation of the Notch pathway (Lobry et al. 2013) or expression of AML1-ETO or FLT3-ITD (two common oncogenic alterations) in Tet2-depleted hematopoietic cells results in complete leukemogenic transformation into AML-like diseases (Rasmussen et al. 2015; Shih et al. 2015; Hatlen et al. 2016). Depending on the secondary mutation, Tet2 inactivation is necessary for disease development or, alternatively, accelerates and worsens an already existing malignancy. Notably, several of these mouse models have a considerable latency between introductions of the genetic alterations to disease manifestation. It is therefore possible that acquisition of additional oncogenic events besides the ones introduced may play a role in transformation. Despite this, these mouse models of Tet2 loss of function offer compelling evidence that directly supports a role of TET2 mutations in a wide range of hematopoietic malignancies and begins to elucidate the minimal genetic events necessary for the onset of disease.
Table 1.
Overview of mouse models of hematological disease with TET protein loss of function
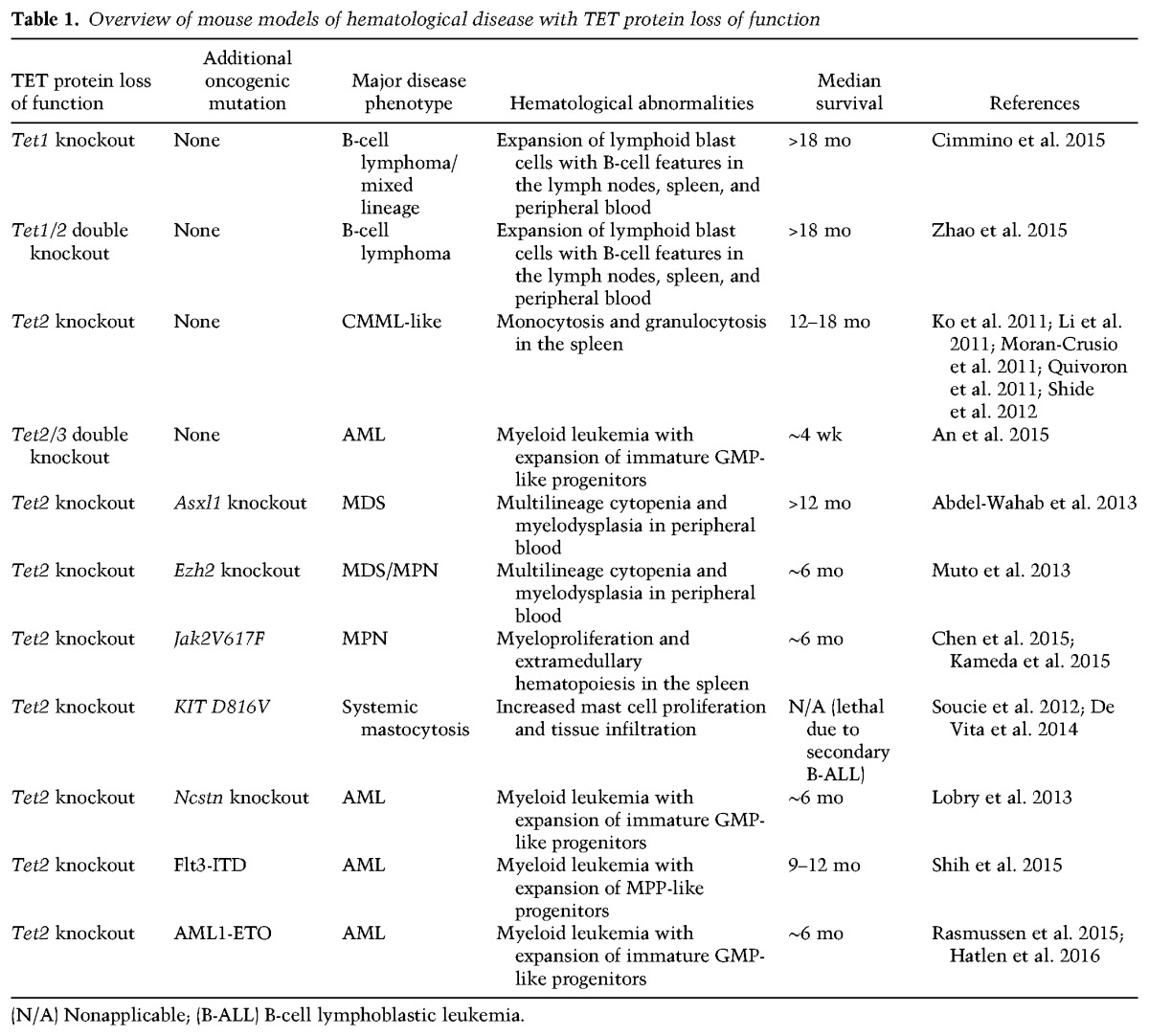
Compared with TET2 mutations, somatic alterations of TET1 and TET3 are exceedingly rare in patients with hematological diseases. Despite this, a role for Tet1 as well as Tet3 in the development of hematopoietic disease has recently been uncovered (Table 1). It was shown that mice with constitutive deletion of Tet1 (Cimmino et al. 2015) as well as combined Tet1 and Tet2 deficiency (Tet1/2 double knockout) (Zhao et al. 2015) are prone to developing late-onset B-cell lymphoma. In support of this, exome sequencing of Tet1-deficient tumors revealed recurrent mutations in genes altered in human DLBCL and FL, and the investigators could detect DNA hypermethylation of the TET1 promoter as well as decreased expression levels in patient samples. Thus, these observations support a role of TET1 as a tumor suppressor in lymphoid hematopoiesis and suggest that silencing of TET1 could be an overlooked mechanism in the onset of B-cell lymphomas (Cimmino et al. 2015; Zhao et al. 2015). In another study, the effects of combined Tet2 and Tet3 disruption (Tet2/3 double knockout) in the hematopoietic system were investigated (An et al. 2015). In contrast to the relatively mild phenotype of Tet2 knockout animals, these compound mutant animals developed a rapid and fully penetrant myeloid leukemia within 7 wk. Transformation of Tet2/3 double-knockout cells was associated with impaired DNA repair and chromosomal translocations, suggesting that Tet2 and Tet3 act redundantly to preserve genomic integrity and prevent leukemogenesis (An et al. 2015). Further investigations are needed to understand the link between loss of TET activity and DNA damage in hematopoietic cells and how the findings in Tet2/3 double-knockout animals are relevant to patients with hematological diseases.
TET proteins in solid tumors
Mutations of the TET genes or decreased expression of TET proteins are not limited to malignant hematopoiesis but have also been observed in a wide variety of human solid tumors. However, much less is known regarding the role of TET enzymes in disease onset and the maintenance of these. Missense and truncating mutations in the TET genes are observed in nearly all tumor types with relatively low frequency (0.1%–10% of cases). In some studies, up to 20% of patients (e.g., melanoma and colorectal cancer) were found to carry mutations in one or more of the TET genes (http://www.cBioPortal.org). However, it should be noted that the types of mutations that accumulate in solid tumors are often missense mutations with unknown significance that may have relative little effect on TET activity (Scourzic et al. 2015). In addition, the large number of additional somatic alterations observed in these tumor types makes it difficult to ascertain whether TET mutations confer a selective advantage or represent inconsequential “passenger” alterations.
Nonetheless, decreased expression of TET proteins and lower 5hmC levels are general hallmarks of many cancer types, including gastric, prostate, liver, lung, and breast cancer as well as glioblastoma and melanoma (Kudo et al. 2012; Lian et al. 2012; Turcan et al. 2012; Liu et al. 2013a; Yang et al. 2013). Due to the extent of this phenomenon, it is unlikely that the observed reduction in 5hmC is solely an effect of deleterious TET mutations. Interestingly, evaluation of the growth rate and 5hmC content of a wide range of tissues has revealed that the proliferation rate shows a significant negative correlation with global 5hmC levels (Bachman et al. 2014). Conversely, arresting the cancer cell lines MCF7 and HCT116 in G1/S phase artificially increased 5hmC levels (Bachman et al. 2014). Thus, it is likely that at least part of the decrease in 5hmC levels can be attributed to the high proliferation rate of cancer cells that causes a general failure to maintain tissue-specific 5hmC levels. In addition to this cell cycle-dependent effect, there are several proposed mechanisms that could lead to a decrease in TET protein activity. The TET2-interacting proteins IDAX/CXXC4 and RINF/CXXC5 are overexpressed in several solid tumors, including breast and colorectal cancer (Knappskog et al. 2011), and are reported to have a negative impact on the stability and function of TET2 (Ko et al. 2013). Moreover, several microRNAs, some of which are overexpressed in cancer, have been reported to directly target TET proteins (Cheng et al. 2013; Fu et al. 2013; Morita et al. 2013; Chuang et al. 2015). In particular, miR-22 was found to negatively regulate all three TET family members, and its expression correlates with poor clinical outcome of breast cancer patients (Song et al. 2013b). Finally, mutations in the metabolic genes IDH1/2, SDH, and FH are found in a wide variety of solid tumors (Oermann et al. 2012) and are likely to have a profound impact on DNA methylation patterns through simultaneous inhibition of all three TET enzymes. Accordingly, neomorphic IDH1 mutations that result in high levels of 2-HG and impairment of TET enzymes in gliomas are tightly associated with aberrant DNA hypermethylation (Noushmehr et al. 2010; Turcan et al. 2012).
In summary, these observations suggest that a combination of mutations, high proliferation rate, and alterations in regulators of TET proteins can result in epigenetic degradation of 5hmC and 5mC patterns. However, the precise impact of altered TET activity on the transformation, progression, and maintenance of these tumors is largely unknown and remains a topic of active research.
Perspectives
Since their discovery, many important advances in our basic and mechanistic understanding of TET enzymes have been reported. The enzymatic processes leading to the formation of 5hmC, 5fC, and 5caC have been thoroughly characterized, and high-resolution cocrystal structures of human TET2 and its substrates have provided structural insights into the enzymatic activities of the TET enzymes. In addition, there is evidence supporting the existence of an active, replication-independent DNA demethylation pathway that proceeds by a TET/TDG/BER-dependent axis. Finally, TET-mediated cytosine oxidation products have been mapped at the nucleotide level in ESCs and yielded a detailed insight into TET enzymatic activity in vivo.
However, several fundamental aspects of TET protein function remain unknown. First, despite the fact that TET mutations as well as catalytic impairment by IDH mutations have been linked to disruption of DNA methylation patterns, it has yet to be demonstrated that the catalytic activity of the TET proteins is required for biological function. TET protein occupancy alone and the corecruitment of auxiliary factors to genomic target sites could account for a considerable part, if not all, of TET physiological functions. Second, recent data suggest that 5mC oxidation by TET enzymes stalls after deposition of 5hmC at many genomic sites. The relative importance of genomic 5hmC deposition and its potential function in passive replication-dependent DNA demethylation versus active DNA demethylation by the TET/TDG/BER pathway is currently unknown. Third, the mechanisms that govern recruitment of TET enzymes to DNA are not fully understood. Although CXXC domain-dependent mechanisms may account for a large part of TET genomic localization, other factors may also play a role. Addressing these questions will be an important part of future investigations.
There is also a considerable gap between the mechanistic understanding of TET enzymes and the impact of TET2 alterations in normal development, premalignant hematopoiesis, and leukemogenesis. Recent studies of normal and malignant hematopoietic cells have uncovered a genome-wide role of TET2 in the maintenance of a hypomethylated state at enhancers. Increased DNA methylation is negatively correlated with chromatin accessibility and histone marks associated with enhancers (Hon et al. 2014; Taberlay et al. 2014; Rasmussen et al. 2015). Thus, TET2 mutations and the concomitant increase in DNA methylation can potentially attenuate enhancer activity and alter the gene regulatory network of the cell. However, the direct connections between increased DNA methylation at enhancers, transcriptional output, and predisposition to hematopoietic diseases have yet to be established. It is possible that altered TET activity on one or a few crucial genomic sites can trigger cellular changes that ultimately promote transformation. However, initial attempts to identify them as well as the genes they regulate have not been successful. At the other extreme, concomitant DNA methylation changes on a large number of enhancers may destabilize gene regulatory networks, increase cellular plasticity, and elevate the risk of cellular transformation. A subtle but global epigenomic disturbance of enhancer function may also explain why mutations or impairment of TET enzymes are associated with such a broad range of cancer types. Further investigations are needed to confirm or disprove these two disparate models and will most likely uncover both global and locus-specific effects.
Irrespective of the underlying molecular events, TET2 mutations lead to altered DNA methylation patterns and confer a competitive advantage to HSCs that, over time, often result in malignant transformation. Therapeutic intervention against this early aberration in AML, MDS, and CMML patients using demethylating agents (azacytidine and decitabine) have yielded inconclusive results (Braun et al. 2011; Itzykson et al. 2011; Bejar et al. 2014; Meldi et al. 2015). Although some studies indicate that TET2 mutations correlate with a better clinical response, treatment does not result in increased survival. In fact, considering the discrete DNA methylation changes observed in mouse studies, it is unlikely that a global therapeutic reduction of DNA methylation can efficiently counteract the effect of TET2 alterations. In addition, it is not known whether fully transformed cells are dependent on the initial TET2 mutation. In order to exploit TET2 mutations therapeutically, a better understanding of the molecular mechanisms leading to the premalignant and malignant transformation of TET2 mutated cells is needed.
Acknowledgments
We thank members of the Helin laboratory for discussions. K.D.R. is supported by a post-doctoral fellowship from the Danish Medical Research Council (1333-00120B). The work in the Helin laboratory is supported by the European Research Council (294666_DNAMET), the Danish Cancer Society, the Danish National Research Foundation (DNRF 82), the Danish Council for Strategic Research, the Novo Nordisk Foundation, and the Lundbeck Foundation and through a center grant from the Novo Nordisk Foundation (the Novo Nordisk Foundation Section for Stem Cell Biology in Human Disease).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.276568.115.
References
- Abdel-Wahab O, Gao J, Adli M, Dey A, Trimarchi T, Chung YR, Kuscu C, Hricik T, Ndiaye-Lobry D, LaFave LM, et al. 2013. Deletion of Asxl1 results in myelodysplasia and severe developmental defects in vivo. J Exp Med 210: 2641–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An J, González-Avalos E, Chawla A, Jeong M, López-Moyado IF, Li W, Goodell MA, Chavez L, Ko M, Rao A. 2015. Acute loss of TET function results in aggressive myeloid cancer in mice. Nat Commun 6: 10071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachman M, Uribe-Lewis S, Yang X, Williams M, Murrell A, Balasubramanian S. 2014. 5-hydroxymethylcytosine is a predominantly stable DNA modification. Nat Chem 6: 1049–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachman M, Uribe-Lewis S, Yang X, Burgess HE, Iurlaro M, Reik W, Murrell A, Balasubramanian S. 2015. 5-formylcytosine can be a stable DNA modification in mammals. Nat Chem Biol 11: 555–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylin SB, Jones PA. 2011. A decade of exploring the cancer epigenome—biological and translational implications. Nat Rev Cancer 11: 726–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejar R, Stevenson K, Abdel-Wahab O, Galili N, Nilsson B, Garcia-Manero G, Kantarjian H, Raza A, Levine RL, Neuberg D, et al. 2011. Clinical effect of point mutations in myelodysplastic syndromes. N Engl J Med 364: 2496–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejar R, Lord A, Stevenson K, Bar-Natan M, Pérez-Ladaga A, Zaneveld J, Wang H, Caughey B, Stojanov P, Getz G, et al. 2014. TET2 mutations predict response to hypomethylating agents in myelodysplastic syndrome patients. Blood 124: 2705–2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaschke K, Ebata KT, Karimi MM, Zepeda-Martínez JA, Goyal P, Mahapatra S, Tam A, Laird DJ, Hirst M, Rao A, et al. 2013. Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature 500: 222–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulard M, Edwards JR, Bestor TH. 2015. FBXL10 protects Polycomb-bound genes from hypermethylation. Nat Genet 47: 479–485. [DOI] [PubMed] [Google Scholar]
- Braun T, Itzykson R, Renneville A, de Renzis B, Dreyfus F, Laribi K, Bouabdallah K, Vey N, Toma A, Recher C, et al. 2011. Molecular predictors of response to decitabine in advanced chronic myelomonocytic leukemia: a phase 2 trial. Blood 118: 3824–3831. [DOI] [PubMed] [Google Scholar]
- Busque L, Patel JP, Figueroa ME, Vasanthakumar A, Provost S, Hamilou Z, Mollica L, Li J, Viale A, Heguy A, et al. 2012. Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nat Genet 44: 1179–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. 2013. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 368: 2059–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SM, Majeti R. 2013. Role of DNMT3A, TET2, and IDH1/2 mutations in pre-leukemic stem cells in acute myeloid leukemia. Int J Hematol 98: 648–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Ueda Y, Dodge JE, Wang Z, Li E. 2003. Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol Cell Biol 23: 5594–5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Chen Y, Bian C, Fujiki R, Yu X. 2013. TET2 promotes histone O-GlcNAcylation during gene transcription. Nature 493: 561–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E, Schneider RK, Breyfogle LJ, Rosen EA, Poveromo L, Elf S, Ko A, Brumme K, Levine R, Ebert BL, et al. 2015. Distinct effects of concomitant Jak2V617F expression and Tet2 loss in mice promote disease progression in myeloproliferative neoplasms. Blood 125: 327–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Guo S, Chen S, Mastriano SJ, Liu C, D'Alessio AC, Hysolli E, Guo Y, Yao H, Megyola CM, et al. 2013. An extensive network of TET2-targeting MicroRNAs regulates malignant hematopoiesis. Cell Rep 5: 471–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang K-H, Whitney-Miller CL, Chu C-Y, Zhou Z, Dokus MK, Schmit S, Barry CT. 2015. MicroRNA-494 is a master epigenetic regulator of multiple invasion-suppressor microRNAs by targeting ten eleven translocation 1 in invasive human hepatocellular carcinoma tumors. Hepatology 62: 466–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cierpicki T, Risner LE, Grembecka J, Lukasik SM, Popovic R, Omonkowska M, Shultis DD, Zeleznik-Le NJ, Bushweller JH. 2010. Structure of the MLL CXXC domain–DNA complex and its functional role in MLL-AF9 leukemia. Nat Struct Mol Biol 17: 62–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimmino L, Dawlaty MM, Ndiaye-Lobry D, Yap YS, Bakogianni S, Yu Y, Bhattacharyya S, Shaknovich R, Geng H, Lobry C, et al. 2015. TET1 is a tumor suppressor of hematopoietic malignancy. Nat Immunol 16: 653–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, Le Coz M, Devarajan K, Wessels A, Soprano D, et al. 2011. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell 146: 67–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa Y, Ding J, Theunissen TW, Faiola F, Hore TA, Shliaha PV, Fidalgo M, Saunders A, Lawrence M, Dietmann S, et al. 2013. NANOG-dependent function of TET1 and TET2 in establishment of pluripotency. Nature 495: 370–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawlaty MM, Ganz K, Powell BE, Hu Y-C, Markoulaki S, Cheng AW, Gao Q, Kim J, Choi S-W, Page DC, et al. 2011. Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Stem Cell 9: 166–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawlaty MM, Breiling A, Le T, Raddatz G, Barrasa MI, Cheng AW, Gao Q, Powell BE, Li Z, Xu M, et al. 2013. Combined deficiency of Tet1 and Tet2 causes epigenetic abnormalities but is compatible with postnatal development. Dev Cell 24: 310–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawlaty MM, Breiling A, Le T, Barrasa MI, Raddatz G, Gao Q, Powell BE, Cheng AW, Faull KF, Lyko F, et al. 2014. Loss of Tet enzymes compromises proper differentiation of embryonic stem cells. Dev Cell 29: 102–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhommeau F, Dupont S, Valle Della V, James C, Trannoy S, Massé A, Kosmider O, Le Couédic J-P, Robert F, Alberdi A, et al. 2009. Mutation in TET2 in myeloid cancers. N Engl J Med 360: 2289–2301. [DOI] [PubMed] [Google Scholar]
- Deplus R, Delatte B, Schwinn MK, Defrance M, Méndez J, Murphy N, Dawson MA, Volkmar M, Putmans P, Calonne E, et al. 2013. TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J 32: 645–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vita S, Schneider RK, Garcia M, Wood J, Gavillet M, Ebert BL, Gerbaulet A, Roers A, Levine RL, Mullally A, et al. 2014. Loss of function of TET2 cooperates with constitutively active KIT in murine and human models of mastocytosis. PLoS One 9: e96209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doege CA, Inoue K, Yamashita T, Rhee DB, Travis S, Fujita R, Guarnieri P, Bhagat G, Vanti WB, Shih A, et al. 2012. Early-stage epigenetic modification during somatic cell reprogramming by Parp1 and Tet2. Nature 488: 652–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Shao N, Szulwach KE, Vialou V, Huynh J, Zhong C, Le T, Ferguson D, Cahill ME, Li Y, et al. 2015. Role of Tet1 and 5-hydroxymethylcytosine in cocaine action. Nat Neurosci 18: 536–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, et al. 2010. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 18: 553–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frauer C, Hoffmann T, Bultmann S, Casa V, Cardoso MC, Antes I, Leonhardt H. 2011. Recognition of 5-hydroxymethylcytosine by the Uhrf1 SRA domain. PLoS One 6: e21306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X, Jin L, Wang X, Luo A, Hu J, Zheng X, Tsark WM, Riggs AD, Ku HT, Huang W. 2013. MicroRNA-26a targets ten eleven translocation enzymes and is regulated during pancreatic cell differentiation. Proc Natl Acad Sci 110: 17892–17897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovese G, Kähler AK, Handsaker RE, Lindberg J, Rose SA, Bakhoum SF, Chambert K, Mick E, Neale BM, Fromer M, et al. 2014. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med 371: 2477–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Globisch D, Münzel M, Müller M, Michalakis S, Wagner M, Koch S, Brückl T, Biel M, Carell T. 2010. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS One 5: e15367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu T-P, Guo F, Yang H, Wu H-P, Xu G-F, Liu W, Xie Z-G, Shi L, He X, Jin S-G, et al. 2011. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature 477: 606–610. [DOI] [PubMed] [Google Scholar]
- Guilhamon P, Eskandarpour M, Halai D, Wilson GA, Feber A, Teschendorff AE, Gomez V, Hergovich A, Tirabosco R, Fernanda Amary M, et al. 2013. Meta-analysis of IDH-mutant cancers identifies EBF1 as an interaction partner for TET2. Nat Commun 4: 2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Wang L, Li J, Ding Z, Xiao J, Yin X, He S, Shi P, Dong L, Li G, et al. 2015. Structural insight into autoinhibition and histone H3-induced activation of DNMT3A. Nature 517: 640–644. [DOI] [PubMed] [Google Scholar]
- Haferlach T, Nagata Y, Grossmann V, Okuno Y, Bacher U, Nagae G, Schnittger S, Sanada M, Kon A, Alpermann T, et al. 2014. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia 28: 241–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MA, Qiu R, Wu X, Li AX, Zhang H, Wang J, Jui J, Jin S-G, Jiang Y, Pfeifer GP, et al. 2013. Dynamics of 5-hydroxymethylcytosine and chromatin marks in mammalian neurogenesis. Cell Rep 3: 291–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Liu Y, Upadhyay AK, Chang Y, Howerton SB, Vertino PM, Zhang X, Cheng X. 2012. Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic Acids Res 40: 4841–4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Olanrewaju YO, Zheng Y, Wilson GG, Zhang X, Cheng X. 2014a. Wilms tumor protein recognizes 5-carboxylcytosine within a specific DNA sequence. Genes Dev 28: 2304–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Pais JE, Zhang X, Saleh L, Fu Z-Q, Dai N, Corrêa IR, Zheng Y, Cheng X. 2014b. Structure of a Naegleria Tet-like dioxygenase in complex with 5-methylcytosine DNA. Nature 506: 391–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Pais JE, Dai N, Corrêa IR, Zhang X, Zheng Y, Cheng X. 2015. Structure of Naegleria Tet-like dioxygenase (NgTet1) in complexes with a reaction intermediate 5-hydroxymethylcytosine DNA. Nucleic Acids Res 43: 10713–10721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatlen MA, Arora K, Vacic V, Grabowska EA, Liao W, Riley-Gillis B, Oschwald DM, Wang L, Joergens JE, Shih AH, et al. 2016. Integrative genetic analysis of mouse and human AML identifies cooperating disease alleles. J Exp Med 213: 25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y-F, Li B-Z, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, et al. 2011. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333: 1303–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill PWS, Amouroux R, Hajkova P. 2014. DNA demethylation, Tet proteins and 5-hydroxymethylcytosine in epigenetic reprogramming: an emerging complex story. Genomics 104: 324–333. [DOI] [PubMed] [Google Scholar]
- Hon GC, Rajagopal N, Shen Y, McCleary DF, Yue F, Dang MD, Ren B. 2013. Epigenetic memory at embryonic enhancers identified in DNA methylation maps from adult mouse tissues. Nat Genet 45: 1198–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon GC, Song C-X, Du T, Jin F, Selvaraj S, Lee AY, Yen C-A, Ye Z, Mao S-Q, Wang B-A, et al. 2014. 5mC oxidation by Tet2 modulates enhancer activity and timing of transcriptome reprogramming during differentiation. Mol Cell 56: 286–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L, Li Z, Cheng J, Rao Q, Gong W, Liu M, Shi YG, Zhu J, Wang P, Xu Y. 2013. Crystal structure of TET2–DNA complex: insight into TET-mediated 5mC oxidation. Cell 155: 1545–1555. [DOI] [PubMed] [Google Scholar]
- Hu L, Lu J, Cheng J, Rao Q, Li Z, Hou H, Lou Z, Zhang L, Li W, Gong W, et al. 2015. Structural insight into substrate preference for TET-mediated oxidation. Nature 527: 118–122. [DOI] [PubMed] [Google Scholar]
- Ichiyama K, Chen T, Wang X, Yan X, Kim B-S, Tanaka S, Ndiaye-Lobry D, Deng Y, Zou Y, Zheng P, et al. 2015. The methylcytosine dioxygenase Tet2 promotes DNA demethylation and activation of cytokine gene expression in T cells. Immunity 42: 613–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. 2011. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333: 1300–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ittel A, Jeandidier E, Helias C, Perrusson N, Humbrecht C, Lioure B, Mazurier I, Mayeur-Rousse C, Lavaux A, Thiebault S, et al. 2013. First description of the t(10;11)(q22;q23)/MLL–TET1 translocation in a T-cell lymphoblastic lymphoma, with subsequent lineage switch to acute myelomonocytic myeloid leukemia. Haematologica 98: e166–e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzykson R, Kosmider O, Cluzeau T, Mansat-De Mas V, Dreyfus F, Beyne-Rauzy O, Quesnel B, Vey N, Gelsi-Boyer V, Raynaud S, et al. 2011. Impact of TET2 mutations on response rate to azacitidine in myelodysplastic syndromes and low blast count acute myeloid leukemias. Leukemia 25: 1147–1152. [DOI] [PubMed] [Google Scholar]
- Itzykson R, Kosmider O, Renneville A, Morabito M, Preudhomme C, Berthon C, Adès L, Fenaux P, Platzbecker U, Gagey O, et al. 2013. Clonal architecture of chronic myelomonocytic leukemias. Blood 121: 2186–2198. [DOI] [PubMed] [Google Scholar]
- Iurlaro M, Ficz G, Oxley D, Raiber E-A, Bachman M, Booth MJ, Andrews S, Balasubramanian S, Reik W. 2013. A screen for hydroxymethylcytosine and formylcytosine binding proteins suggests functions in transcription and chromatin regulation. Genome Biol 14: R119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain D, Baldi S, Zabel A, Straub T, Becker PB. 2015. Active promoters give rise to false positive ‘phantom peaks’ in ChIP-seq experiments. Nucleic Acids Res 43: 6959–6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, Lindsley RC, Mermel CH, Burtt N, Chavez A, et al. 2014. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med 371: 2488–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan M, Snyder TM, Corces-Zimmerman MR, Vyas P, Weissman IL, Quake SR, Majeti R. 2012. Clonal evolution of preleukemic hematopoietic stem cells precedes human acute myeloid leukemia. Sci Transl Med 4: 149ra118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji D, Lin K, Song J, Wang Y. 2014. Effects of Tet-induced oxidation products of 5-methylcytosine on Dnmt1- and DNMT3a-mediated cytosine methylation. Mol Biosyst 10: 1749–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S-G, Kadam S, Pfeifer GP. 2010. Examination of the specificity of DNA methylation profiling techniques towards 5-methylcytosine and 5-hydroxymethylcytosine. Nucleic Acids Res 38: e125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA, Liang G. 2009. Rethinking how DNA methylation patterns are maintained. Nat Rev Genet 10: 805–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kameda T, Shide K, Yamaji T, Kamiunten A, Sekine M, Taniguchi Y, Hidaka T, Kubuki Y, Shimoda H, Marutsuka K, et al. 2015. Loss of TET2 has dual roles in murine myeloproliferative neoplasms: disease sustainer and disease accelerator. Blood 125: 304–315. [DOI] [PubMed] [Google Scholar]
- Kang J, Lienhard M, Pastor WA, Chawla A, Novotny M, Tsagaratou A, Lasken RS, Thompson EC, Surani MA, Koralov SB, et al. 2015. Simultaneous deletion of the methylcytosine oxidases Tet1 and Tet3 increases transcriptome variability in early embryogenesis. Proc Natl Acad Sci 112: E4236–E4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellinger MW, Song C-X, Chong J, Lu X-Y, He C, Wang D. 2012. 5-formylcytosine and 5-carboxylcytosine reduce the rate and substrate specificity of RNA polymerase II transcription. Nat Struct Mol Biol 19: 831–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knappskog S, Myklebust LM, Busch C, Aloysius T, Varhaug JE, Lønning PE, Lillehaug JR, Pendino F. 2011. RINF (CXXC5) is overexpressed in solid tumors and is an unfavorable prognostic factor in breast cancer. Ann Oncol 22: 2208–2215. [DOI] [PubMed] [Google Scholar]
- Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, An J, Lamperti ED, Koh KP, Ganetzky R, et al. 2010. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature 468: 839–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M, Bandukwala HS, An J, Lamperti ED, Thompson EC, Hastie R, Tsangaratou A, Rajewsky K, Koralov SB, Rao A. 2011. Ten-eleven-translocation 2 (TET2) negatively regulates homeostasis and differentiation of hematopoietic stem cells in mice. Proc Natl Acad Sci 108: 14566–14571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M, An J, Bandukwala HS, Chavez L, Aijö T, Pastor WA, Segal MF, Li H, Koh KP, Lähdesmäki H, et al. 2013. Modulation of TET2 expression and 5-methylcytosine oxidation by the CXXC domain protein IDAX. Nature 497: 122–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M, An J, Pastor WA, Koralov SB, Rajewsky K, Rao A. 2015. TET proteins and 5-methylcytosine oxidation in hematological cancers. Immunol Rev 263: 6–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. 2009. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324: 929–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo Y, Tateishi K, Yamamoto K, Yamamoto S, Asaoka Y, Ijichi H, Nagae G, Yoshida H, Aburatani H, Koike K. 2012. Loss of 5-hydroxymethylcytosine is accompanied with malignant cellular transformation. Cancer Sci 103: 670–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langemeijer SMC, Kuiper RP, Berends M, Knops R, Aslanyan MG, Massop M, Stevens-Linders E, van Hoogen P, van Kessel AG, Raymakers RAP, et al. 2009. Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat Genet 41: 838–842. [DOI] [PubMed] [Google Scholar]
- Li Z, Cai X, Cai CL, Wang J, Zhang W, Petersen BE, Yang FC, Xu M. 2011. Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood 118: 4509–4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian CG, Xu Y, Ceol C, Wu F, Larson A, Dresser K, Xu W, Tan L, Hu Y, Zhan Q, et al. 2012. Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell 150: 1135–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T, Wood RD. 1999. Quality control by DNA repair. Science 286: 1897–1905. [DOI] [PubMed] [Google Scholar]
- Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, Lucero J, Huang Y, Dwork AJ, Schultz MD, et al. 2013. Global epigenomic reconfiguration during mammalian brain development. Science 341: 1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Liu L, Chen X, Shen J, Shan J, Xu Y, Yang Z, Wu L, Xia F, Bie P, et al. 2013a. Decrease of 5-hydroxymethylcytosine is associated with progression of hepatocellular carcinoma through downregulation of TET1. PLoS One 8: e62828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Jin Y, Tang WH, Qin L, Zhang X, Tellides G, Hwa J, Yu J, Martin KA. 2013b. Ten-eleven translocation-2 (TET2) is a master regulator of smooth muscle cell plasticity. Circulation 128: 2047–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobry C, Ntziachristos P, Ndiaye-Lobry D, Oh P, Cimmino L, Zhu N, Araldi E, Hu W, Freund J, Abdel-Wahab O, et al. 2013. Notch pathway activation targets AML-initiating cell homeostasis and differentiation. J Exp Med 210: 301–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorsbach RB, Moore J, Mathew S, Raimondi SC, Mukatira ST, Downing JR. 2003. TET1, a member of a novel protein family, is fused to MLL in acute myeloid leukemia containing the t(10;11)(q22;q23). Leukemia 17: 637–641. [DOI] [PubMed] [Google Scholar]
- Losman J-A, Kaelin WG. 2013. What a difference a hydroxyl makes: mutant IDH, (R)-2-hydroxyglutarate, and cancer. Genes Dev 27: 836–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F, Liu Y, Jiang L, Yamaguchi S, Zhang Y. 2014. Role of Tet proteins in enhancer activity and telomere elongation. Genes Dev 28: 2103–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti A, Drohat AC. 2011. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites. J Biol Chem 286: 35334–35338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meldi K, Qin T, Buchi F, Droin N, Sotzen J, Micol J-B, Selimoglu-Buet D, Masala E, Allione B, Gioia D, et al. 2015. Specific molecular signatures predict decitabine response in chronic myelomonocytic leukemia. J Clin Invest 125: 1857–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minor EA, Court BL, Young JI, Wang G. 2013. Ascorbate induces ten-eleven translocation (Tet) methylcytosine dioxygenase-mediated generation of 5-hydroxymethylcytosine. J Biol Chem 288: 13669–13674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, Figueroa ME, Vasanthakumar A, Patel J, Zhao X, et al. 2011. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell 20: 11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita S, Horii T, Kimura M, Ochiya T, Tajima S, Hatada I. 2013. miR-29 represses the activities of DNA methyltransferases and DNA demethylases. Int J Mol Sci 14: 14647–14658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto T, Sashida G, Oshima M, Wendt GR, Mochizuki-Kashio M, Nagata Y, Sanada M, Miyagi S, Saraya A, Kamio A, et al. 2013. Concurrent loss of Ezh2 and Tet2 cooperates in the pathogenesis of myelodysplastic disorders. J Exp Med 210: 2627–2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabel CS, Jia H, Ye Y, Shen L, Goldschmidt HL, Stivers JT, Zhang Y, Kohli RM. 2012. AID/APOBEC deaminases disfavor modified cytosines implicated in DNA demethylation. Nat Chem Biol 8: 751–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Lv L, Nakagawa M, Yu Y, Yu C, D'Alessio AC, Nakayama K, Fan H-Y, Chen X, Xiong Y. 2015. CRL4(VprBP) E3 ligase promotes monoubiquitylation and chromatin binding of TET dioxygenases. Mol Cell 57: 247–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neri F, Incarnato D, Krepelova A, Rapelli S, Anselmi F, Parlato C, Medana C, Dal Bello F, Oliviero S. 2015. Single-base resolution analysis of 5-formyl and 5-carboxyl cytosine reveals promoter DNA methylation dynamics. Cell Rep 10: 674–683. [DOI] [PubMed] [Google Scholar]
- Noh K-M, Wang H, Kim HR, Wenderski W, Fang F, Li CH, Dewell S, Hughes SH, Melnick AM, Patel DJ, et al. 2015. Engineering of a histone-recognition domain in Dnmt3a alters the epigenetic landscape and phenotypic features of mouse ESCs. Mol Cell 59: 89–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, et al. 2010. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell 17: 510–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oermann EK, Wu J, Guan K-L, Xiong Y. 2012. Alterations of metabolic genes and metabolites in cancer. Semin Cell Dev Biol 23: 370–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono R, Taki T, Taketani T, Taniwaki M, Kobayashi H, Hayashi Y. 2002. LCX, leukemia-associated protein with a CXXC domain, is fused to MLL in acute myeloid leukemia with trilineage dysplasia having t(10;11)(q22;q23). Cancer Res 62: 4075–4080. [PubMed] [Google Scholar]
- Ooi SKT, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin S-P, Allis CD, et al. 2007. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 448: 714–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otani J, Nankumo T, Arita K, Inamoto S, Ariyoshi M, Shirakawa M. 2009. Structural basis for recognition of H3K4 methylation status by the DNA methyltransferase 3A ATRX–DNMT3–DNMT3L domain. EMBO Rep 10: 1235–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomero T, Couronné L, Khiabanian H, Kim M-Y, Ambesi-Impiombato A, Perez-Garcia A, Carpenter Z, Abate F, Allegretta M, Haydu JE, et al. 2014. Recurrent mutations in epigenetic regulators, RHOA and FYN kinase in peripheral T cell lymphomas. Nat Genet 46: 166–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor WA, Pape UJ, Huang Y, Henderson HR, Lister R, Ko M, McLoughlin EM, Brudno Y, Mahapatra S, Kapranov P, et al. 2011. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature 473: 394–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez C, Martínez-Calle N, Martín-Subero JI, Segura V, Delabesse E, Fernandez-Mercado M, Garate L, Alvarez S, Rifon J, Varea S, et al. 2012. TET2 mutations are associated with specific 5-methylcytosine and 5-hydroxymethylcytosine profiles in patients with chronic myelomonocytic leukemia. PLoS One 7: e31605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffeneder T, Hackner B, Truss M, Münzel M, Müller M, Deiml CA, Hagemeier C, Carell T. 2011. The discovery of 5-formylcytosine in embryonic stem cell DNA. Angew Chem Int Ed Engl 50: 7008–7012. [DOI] [PubMed] [Google Scholar]
- Pfaffeneder T, Spada F, Wagner M, Brandmayr C, Laube SK, Eisen D, Truss M, Steinbacher J, Hackner B, Kotljarova O, et al. 2014. Tet oxidizes thymine to 5-hydroxymethyluracil in mouse embryonic stem cell DNA. Nat Chem Biol 10: 574–581. [DOI] [PubMed] [Google Scholar]
- Pronier E, Almire C, Mokrani H, Vasanthakumar A, Simon A, da Costa Reis Monte Mor B, Massé A, Le Couédic J-P, Pendino F, Carbonne B, et al. 2011. Inhibition of TET2-mediated conversion of 5-methylcytosine to 5-hydroxymethylcytosine disturbs erythroid and granulomonocytic differentiation of human hematopoietic progenitors. Blood 118: 2551–2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putiri EL, Tiedemann RL, Thompson JJ, Liu C, Ho T, Choi J-H, Robertson KD. 2014. Distinct and overlapping control of 5-methylcytosine and 5-hydroxymethylcytosine by the TET proteins in human cancer cells. Genome Biol 15: R81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quivoron C, Couronné L, Valle Della V, Lopez CK, Plo I, Wagner-Ballon O, Do Cruzeiro M, Delhommeau F, Arnulf B, Stern M-H, et al. 2011. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell 20: 25–38. [DOI] [PubMed] [Google Scholar]
- Rampal R, Alkalin A, Madzo J, Vasanthakumar A, Pronier E, Patel J, Li Y, Ahn J, Abdel-Wahab O, Shih A, et al. 2014. DNA hydroxymethylation profiling reveals that WT1 mutations result in loss of TET2 function in acute myeloid leukemia. Cell Rep 9: 1841–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen KD, Jia G, Johansen JV, Pedersen MT, Rapin N, Bagger FO, Porse BT, Bernard OA, Christensen J, Helin K. 2015. Loss of TET2 in hematopoietic cells leads to DNA hypermethylation of active enhancers and induction of leukemogenesis. Genes Dev 29: 910–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudenko A, Dawlaty MM, Seo J, Cheng AW, Meng J, Le T, Faull KF, Jaenisch R, Tsai L-H. 2013. Tet1 is critical for neuronal activity-regulated gene expression and memory extinction. Neuron 79: 1109–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakata-Yanagimoto M, Enami T, Yoshida K, Shiraishi Y, Ishii R, Miyake Y, Muto H, Tsuyama N, Sato-Otsubo A, Okuno Y, et al. 2014. Somatic RHOA mutation in angioimmunoblastic T cell lymphoma. Nat Genet 46: 171–175. [DOI] [PubMed] [Google Scholar]
- Scourzic L, Mouly E, Bernard OA. 2015. TET proteins and the control of cytosine demethylation in cancer. Genome Med 7: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seisenberger S, Peat JR, Reik W. 2013. Conceptual links between DNA methylation reprogramming in the early embryo and primordial germ cells. Curr Opin Cell Biol 25: 281–288. [DOI] [PubMed] [Google Scholar]
- Sérandour AA, Avner S, Oger F, Bizot M, Percevault F, Lucchetti-Miganeh C, Palierne G, Gheeraert C, Barloy-Hubler F, Péron CL, et al. 2012. Dynamic hydroxymethylation of deoxyribonucleic acid marks differentiation-associated enhancers. Nucleic Acids Res 40: 8255–8265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Wu H, Diep D, Yamaguchi S, D'Alessio AC, Fung H-L, Zhang K, Zhang Y. 2013. Genome-wide analysis reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics. Cell 153: 692–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shide K, Kameda T, Shimoda H, Yamaji T, Abe H, Kamiunten A, Sekine M, Hidaka T, Katayose K, Kubuki Y, et al. 2012. TET2 is essential for survival and hematopoietic stem cell homeostasis. Leukemia 26: 2216–2223. [DOI] [PubMed] [Google Scholar]
- Shih AH, Jiang Y, Meydan C, Shank K, Pandey S, Barreyro L, Antony-Debre I, Viale A, Socci N, Sun Y, et al. 2015. Mutational cooperativity linked to combinatorial epigenetic gain of function in acute myeloid leukemia. Cancer Cell 27: 502–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Rechkoblit O, Bestor TH, Patel DJ. 2011. Structure of DNMT1–DNA complex reveals a role for autoinhibition in maintenance DNA methylation. Science 331: 1036–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C-X, Szulwach KE, Dai Q, Fu Y, Mao S-Q, Lin L, Street C, Li Y, Poidevin M, Wu H, et al. 2013a. Genome-wide profiling of 5-formylcytosine reveals its roles in epigenetic priming. Cell 153: 678–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song SJ, Poliseno L, Song MS, Ala U, Webster K, Ng C, Beringer G, Brikbak NJ, Yuan X, Cantley LC, et al. 2013b. MicroRNA-antagonism regulates breast cancer stemness and metastasis via TET-family-dependent chromatin remodeling. Cell 154: 311–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucie E, Hanssens K, Mercher T, Georgin-Lavialle S, Damaj G, Livideanu C, Chandesris MO, Acin Y, Létard S, de Sepulveda P, et al. 2012. In aggressive forms of mastocytosis, TET2 loss cooperates with c-KITD816V to transform mast cells. Blood 120: 4846–4849. [DOI] [PubMed] [Google Scholar]
- Spruijt CG, Gnerlich F, Smits AH, Pfaffeneder T, Jansen PWTC, Bauer C, Münzel M, Wagner M, Müller M, Khan F, et al. 2013. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell 152: 1146–1159. [DOI] [PubMed] [Google Scholar]
- Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Schöler A, van Nimwegen E, Wirbelauer C, Oakeley EJ, Gaidatzis D, et al. 2011. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature 480: 490–495. [DOI] [PubMed] [Google Scholar]
- Stroud H, Feng S, Morey Kinney S, Pradhan S, Jacobsen SE. 2011. 5-hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cells. Genome Biol 12: R54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szulwach KE, Li X, Li Y, Song C-X, Han JW, Kim S, Namburi S, Hermetz K, Kim JJ, Rudd MK, et al. 2011. Integrating 5-hydroxymethylcytosine into the epigenomic landscape of human embryonic stem cells. PLoS Genet 7: e1002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szwagierczak A, Bultmann S, Schmidt CS, Spada F, Leonhardt H. 2010. Sensitive enzymatic quantification of 5-hydroxymethylcytosine in genomic DNA. Nucleic Acids Res 38: e181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taberlay PC, Statham AL, Kelly TK, Clark SJ, Jones PA. 2014. Reconfiguration of nucleosome-depleted regions at distal regulatory elements accompanies DNA methylation of enhancers and insulators in cancer. Genome Res 24: 1421–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. 2009. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324: 930–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, Campos C, Fabius AWM, Lu C, Ward PS, et al. 2012. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature 483: 479–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valinluck V, Sowers LC. 2007. Endogenous cytosine damage products alter the site selectivity of human DNA maintenance methyltransferase DNMT1. Cancer Res 67: 946–950. [DOI] [PubMed] [Google Scholar]
- Wang L, Zhang J, Duan J, Gao X, Zhu W, Lu X, Yang L, Zhang J, Li G, Ci W, et al. 2014. Programming and inheritance of parental DNA methylomes in mammals. Cell 157: 979–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Zhou Y, Xu L, Xiao R, Lu X, Chen L, Chong J, Li H, He C, Fu X-D, et al. 2015a. Molecular basis for 5-carboxycytosine recognition by RNA polymerase II elongation complex. Nature 523: 621–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Xiao M, Chen X, Chen L, Xu Y, Lv L, Wang P, Yang H, Ma S, Lin H, et al. 2015b. WT1 recruits TET2 to regulate its target gene expression and suppress leukemia cell proliferation. Mol Cell 57: 662–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissmann S, Alpermann T, Grossmann V, Kowarsch A, Nadarajah N, Eder C, Dicker F, Fasan A, Haferlach C, Haferlach T, et al. 2012. Landscape of TET2 mutations in acute myeloid leukemia. Leukemia 26: 934–942. [DOI] [PubMed] [Google Scholar]
- Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PAC, Rappsilber J, Helin K. 2011. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature 473: 343–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K, Christensen J, Helin K. 2012. DNA methylation: TET proteins—guardians of CpG islands? EMBO Rep 13: 28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Zhang Y. 2014. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell 156: 45–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, D'Alessio AC, Ito S, Wang Z, Cui K, Zhao K, Sun YE, Zhang Y. 2011a. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev 25: 679–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, D'Alessio AC, Ito S, Xia K, Wang Z, Cui K, Zhao K, Sun YE, Zhang Y. 2011b. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature 473: 389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Wu X, Shen L, Zhang Y. 2014. Single-base resolution analysis of active DNA demethylation using methylase-assisted bisulfite sequencing. Nat Biotechnol 32: 1231–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao M, Yang H, Xu W, Ma S, Lin H, Zhu H, Liu L, Liu Y, Yang C, Xu Y, et al. 2012. Inhibition of α-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes Dev 26: 1326–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M, Lu C, Wang J, McLellan MD, Johnson KJ, Wendl MC, McMichael JF, Schmidt HK, Yellapantula V, Miller CA, et al. 2014. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med 20: 1472–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Bian C, Lam R, Dong A, Min J. 2011a. The structural basis for selective binding of non-methylated CpG islands by the CFP1 CXXC domain. Nat Commun 2: 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim S-H, Ito S, Yang C, Wang P, Xiao M-T, et al. 2011b. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell 19: 17–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Wu F, Tan L, Kong L, Xiong L, Deng J, Barbera AJ, Zheng L, Zhang H, Huang S, et al. 2011c. Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol Cell 42: 451–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Xu C, Kato A, Tempel W, Abreu JG, Bian C, Hu Y, Hu D, Zhao B, Cerovina T, et al. 2012. Tet3 CXXC domain and dioxygenase activity cooperatively regulate key genes for Xenopus eye and neural development. Cell 151: 1200–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki J, Taby R, Vasanthakumar A, Macrae T, Ostler KR, Shen L, Kantarjian HM, Estecio MR, Jelinek J, Godley LA, et al. 2012. Effects of TET2 mutations on DNA methylation in chronic myelomonocytic leukemia. Epigenetics 7: 201–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki J, Jelinek J, Lu Y, Cesaroni M, Madzo J, Neumann F, He R, Taby R, Vasanthakumar A, Macrae T, et al. 2015. TET2 mutations affect non-CpG Island DNA methylation at enhancers and transcription factor-binding sites in chronic myelomonocytic leukemia. Cancer Res 75: 2833–2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Liu Y, Bai F, Zhang J-Y, Ma S-H, Liu J, Xu Z-D, Zhu H-G, Ling Z-Q, Ye D, et al. 2013. Tumor development is associated with decrease of TET gene expression and 5-methylcytosine hydroxylation. Oncogene 32: 663–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yardimci H, Zhang Y. 2015. Charting oxidized methylcytosines at base resolution. Nat Struct Mol Biol 22: 656–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin R, Mao S-Q, Zhao B, Chong Z, Yang Y, Zhao C, Zhang D, Huang H, Gao J, Li Z, et al. 2013. Ascorbic acid enhances Tet-mediated 5-methylcytosine oxidation and promotes DNA demethylation in mammals. J Am Chem Soc 135: 10396–10403. [DOI] [PubMed] [Google Scholar]
- Yu M, Hon GC, Szulwach KE, Song C-X, Zhang L, Kim A, Li X, Dai Q, Shen Y, Park B, et al. 2012. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell 149: 1368–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Zhang Y-L, Pan W-W, Li X-M, Wang Z-W, Ge Z-J, Zhou J-J, Cang Y, Tong C, Sun Q-Y, et al. 2013. CRL4 complex regulates mammalian oocyte survival and reprogramming by activation of TET proteins. Science 342: 1518–1521. [DOI] [PubMed] [Google Scholar]
- Zhang H, Zhang X, Clark E, Mulcahey M, Huang S, Shi YG. 2010. TET1 is a DNA-binding protein that modulates DNA methylation and gene transcription via hydroxylation of 5-methylcytosine. Cell Res 20: 1390–1393. [DOI] [PubMed] [Google Scholar]
- Zhang L, Lu X, Lu J, Liang H, Dai Q, Xu G-L, Luo C, Jiang H, He C. 2012. Thymine DNA glycosylase specifically recognizes 5-carboxylcytosine-modified DNA. Nat Chem Biol 8: 328–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Zhao K, Shen Q, Han Y, Gu Y, Li X, Zhao D, Liu Y, Wang C, Zhang X, et al. 2015. Tet2 is required to resolve inflammation by recruiting Hdac2 to specifically repress IL-6. Nature 525: 389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Chen L, Dawlaty MM, Pan F, Weeks O, Zhou Y, Cao Z, Shi H, Wang J, Lin L, et al. 2015. Combined loss of Tet1 and Tet2 promotes B cell, but not myeloid malignancies, in mice. Cell Rep 13: 1692–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Xiong J, Wang M, Yang N, Wong J, Zhu B, Xu R-M. 2014. Structural basis for hydroxymethylcytosine recognition by the SRA domain of UHRF2. Mol Cell 54: 879–886. [DOI] [PubMed] [Google Scholar]
- Ziller MJ, Gu H, Müller F, Donaghey J, Tsai LT-Y, Kohlbacher O, De Jager PL, Rosen ED, Bennett DA, Bernstein BE, et al. 2013. Charting a dynamic DNA methylation landscape of the human genome. Nature 500: 477–481. [DOI] [PMC free article] [PubMed] [Google Scholar]