Here, Wang et al. investigated the role of shelterin in telomeric heterochromatin assembly. The authors demonstrated that shelterin is first recruited to chromosome ends through direct association with CLRC through the Ccq1 subunit and that the proper connection of shelterin components allows CLRC to skip telomeric repeats to internal regions.
Keywords: shelterin, telomere, heterochromatin, spreading
Abstract
Telomeric regions contain prominent sites of heterochromatin, which is associated with unique histone modification profiles such as the methylation of histone H3 at Lys9 (H3K9me). In fission yeast, the conserved telomeric shelterin complex recruits the histone H3K9 methyltransferase complex CLRC to establish subtelomeric heterochromatin. Although many shelterin mutations affect subtelomeric heterochromatin assembly, the mechanism remains elusive due to the diverse functions of shelterin. Through affinity purification, we found that shelterin directly associates with CLRC through the Ccq1 subunit. Surprisingly, mutations that disrupt interactions between shelterin subunits compromise subtelomeric heterochromatin without affecting CLRC interaction with shelterin component Pot1, located at chromosome ends. We further discovered that telomeric repeats are refractory to heterochromatin spreading and that artificial restoration of shelterin connections or increased heterochromatin spreading rescued heterochromatin defects in these shelterin mutants. Thus, subtelomeric heterochromatin assembly requires both the recruitment of CLRC by shelterin to chromosome ends and the proper connection of shelterin components, which allows CLRC to skip telomeric repeats to internal regions.
Eukaryotic genomes contain large amounts of repetitive DNA elements at centromeres and telomeres that assemble constitutive heterochromatin, which is critical for preventing homologous recombination between repeats to maintain genome integrity (Blasco 2007; Grewal and Jia 2007). These heterochromatic regions are associated with distinct histone modification profiles and chromatin proteins, including low levels of acetylation, high levels of methylation at histone H3 Lys9 (H3K9me), and heterochromatin protein 1 (HP1) family proteins (Grewal and Jia 2007). Heterochromatin formation requires concerted actions of diverse histone-modifying activities, which are classified into three distinct steps: initiation, spreading, and maintenance (Wang et al. 2014a). Initiation of heterochromatin assembly takes place at nucleation centers via targeting of histone H3K9 methyltransferases by sequence-specific DNA-binding proteins or noncoding RNAs, leading to H3K9me (Castel and Martienssen 2013; Cohen and Jia 2014). Heterochromatin then spreads from these nucleation centers through a positive feedback loop involving H3K9me-mediated recruitment of additional histone H3K9 methyltransferases that methylate adjacent nucleosomes, resulting in an “inch worm”-like spreading (Talbert and Henikoff 2006; Wang et al. 2014a). Similarly, the recruitment of H3K9 methyltransferases by pre-existing H3K9me on old histones allows the newly incorporated histones during DNA replication to be methylated as well, resulting in stable maintenance of heterochromatin domains through cell divisions even without the initiation signal (Audergon et al. 2015; Ragunathan et al. 2015). Although the mechanisms of heterochromatin assembly at centromeres have been extensively studied, those that regulate initiation and spreading of heterochromatin at telomeres are not well understood.
The fission yeast Schizosaccharomyces pombe serves as a perfect model organism for the study of heterochromatin assembly and telomere biology. The heterochromatin assembly machinery in fission yeast is highly conserved compared with those of higher eukaryotes (Grewal and Jia 2007). The SUV39 family SET domain protein Clr4, the sole H3K9 methyltransferase in this organism, is critical for heterochromatin assembly (Rea et al. 2000; Nakayama et al. 2001). Clr4 associates with an E3 ubiquitin ligase (composed of Cul4, Rik1, Raf1, and Raf2) to form the CLRC complex, which is required for H3K9me in vivo (Rea et al. 2000; Nakayama et al. 2001; Hong et al. 2005; Horn et al. 2005; Jia et al. 2005). H3K9me recruits HP1 family proteins Swi6 and Chp2 to further compact chromatin (Nakayama et al. 2001; Sadaie et al. 2004). These HP1 proteins in turn recruit additional chromatin-modifying activities, such as the SHREC complex (composed of the histone deacetylase Clr3, chromatin remodeling protein Mit1, and additional proteins Clr1, Clr2, and Ccq1) to regulate histone acetylation and nucleosome positioning at heterochromatin (Sugiyama et al. 2007; Motamedi et al. 2008). Heterochromatin in fission yeast is mainly present at pericentric regions, subtelomeres, and the silent mating-type locus, all of which contain a similar repeat element that initiates heterochromatin through an RNAi-mediated pathway (Grewal and Jia 2007). In addition, DNA-binding proteins such as stress-activated ATF/CREB proteins Atf1/Pcr1 initiate heterochromatin at the silent mating type region, and the telomere-binding protein complex shelterin initiates heterochromatin at telomeres (Jia et al. 2004; Kim et al. 2004; Kanoh et al. 2005).
Telomeres and the telomere-binding shelterin complex in fission yeast are also highly conserved with respect to those of mammals (Jain and Cooper 2010). Telomeric DNA is composed of dsDNA repeats followed by a single-stranded overhang. Shelterin contains six subunits: Taz1, Rap1, Poz1, Tpz1, Pot1, and Ccq1 (Miyoshi et al. 2008). Taz1 (homolog of mammalian TRF1/TRF2) binds to the double-stranded portion of telomeric DNA (Cooper et al. 1997). The Tpz1–Pot1 subcomplex (homologous to the mammalian TTP1–POT1 complex) binds to the ssDNA overhang (Baumann and Cech 2001; Miyoshi et al. 2008). Rap1 and Poz1 (similar to mammalian RAP1 and TIN2, respectively) bridge Taz1 with Tpz1–Pot1 (Miyoshi et al. 2008). Tpz1 also associates with Ccq1, which is involved in telomerase recruitment and activation (Miyoshi et al. 2008; Tomita and Cooper 2008; Moser et al. 2011; Yamazaki et al. 2012; Jun et al. 2013; Armstrong et al. 2014; Harland et al. 2014).
Shelterin is required for heterochromatin assembly near native telomeres. For example, mutations of Taz1, Rap1, Poz1, Tpz1, and Ccq1 all resulted in silencing defects of reporter genes inserted near telomeric repeats (Cooper et al. 1997; Nimmo et al. 1998; Kanoh and Ishikawa 2001; Kanoh et al. 2005; Sugiyama et al. 2007; Fujita et al. 2012; Tadeo et al. 2013; Kallgren et al. 2014). In addition, insertion of telomeric repeats at an internal chromosome location or artificial recruitment of shelterin component Rap1 to DNA induces ectopic heterochromatin in a shelterin- and CLRC-dependent manner, suggesting that shelterin recruits CLRC to telomeric DNA (Kanoh et al. 2005; Tadeo et al. 2013). However, mechanistic dissection of subtelomeric heterochromatin assembly has been difficult due to the multiple functions that shelterin performs. For instance, the Tpz1–Pot1 subcomplex is required for chromosome end protection, and their null mutations result in the loss of telomeric DNA and chromosome circularization (Baumann and Cech 2001; Miyoshi et al. 2008). In addition, shelterin is required for telomere length control. Mutations such as taz1Δ, rap1Δ, or poz1Δ result in massive elongation of telomeres (Cooper et al. 1997; Chikashige and Hiraoka 2001; Kanoh and Ishikawa 2001; Fujita et al. 2012; Jun et al. 2013; Harland et al. 2014). In contrast, ccq1Δ results in defective telomerase recruitment and activation, leading to telomere shortening (Miyoshi et al. 2008; Tomita and Cooper 2008; Moser et al. 2011; Yamazaki et al. 2012; Jun et al. 2013; Armstrong et al. 2014; Harland et al. 2014). In addition, shelterin components are recruited to telomeres through both dsDNA and ssDNA, and the deletion of a single component of shelterin generally does not abolish the recruitment of other shelterin subunits to telomeres (Miyoshi et al. 2008). Therefore, it is unlikely that all of the known shelterin mutations block CLRC recruitment, and shelterin may play additional roles in subtelomeric heterochromatin assembly in addition to recruiting CLRC.
Here we show that shelterin physically associates with CLRC, mediated by the Ccq1 subunit. Interestingly, aside from ccq1Δ, mutations in shelterin components that affect subtelomeric heterochromatin still maintain CLRC interaction with shelterin subunit Pot1, which binds to the single-stranded end of telomeric DNA. A common feature of these mutations is that they affect the connections between the CLRC and double-stranded telomeric DNA, and forced restoration of their proper connections rescues subtelomeric heterochromatin defects. We further showed that insertion of telomeric repeats into an ectopic heterochromatin domain blocks heterochromatin spreading when these repeats do not recruit Clr4. Moreover, enhancing heterochromatin spreading alleviates subtelomeric heterochromatin defects in shelterin connection mutants. These results suggest that the recruitment of CLRC to the ssDNA portion of telomeres is not sufficient for heterochromatin assembly at subtelomeric regions due to the effects of telomeric repeats in blocking CLRC spreading and that the proper connections between shelterin components play a vital role in promoting CLRC spreading over telomeric repeats into subtelomeric regions.
Results
Shelterin associates with CLRC
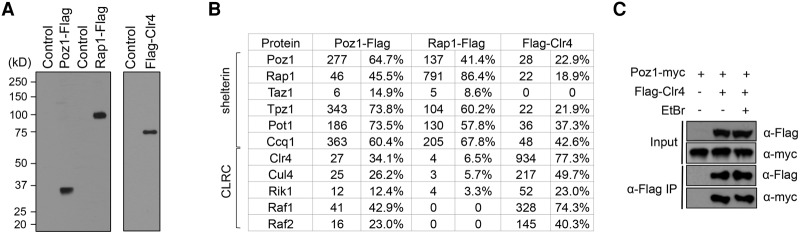
To understand the mechanism by which shelterin recruits heterochromatin assembly factors to telomeres, we generated a Flag-tagged version of shelterin component Poz1 at its endogenous chromosome location (Tadeo et al. 2013). Affinity purification of Poz1-Flag followed by mass spectrometry identified high levels of peptides corresponding to shelterin components such as Rap1, Tpz1, Pot1, Ccq1, and, to a lesser extent, Taz1 (Fig. 1A,B). Interestingly, members of CLRC, such as Clr4, Cul4, Rik1, Raf1, and Raf2, were also identified. Similarly, affinity purification of another shelterin component, Rap1-Flag, also identified both shelterin and CLRC components (Fig. 1A,B). Likewise, affinity purification of Flag-Clr4 identified CLRC and most of the shelterin subunits as well (Fig. 1A,B).
Figure 1.
Shelterin associates with CLRC in vivo. (A) Western blot analyses of affinity-purified protein complexes with a Flag antibody. (B) Mass spectrometry analyses of purified protein complexes. The spectral count numbers (left), which are normalized to the amino acid number of each protein, and the sequence coverage of each protein (right) are indicated. (C) Coimmunoprecipitation analyses of Clr4 and Poz1. Immunoprecipitation was performed with Flag-agarose beads, and Western blot analyses were performed with Flag and myc antibodies.
To confirm the shelterin–CLRC interaction, we performed coimmunoprecipitation (co-IP) analyses using extracts from a strain expressing both Flag-Clr4 and Poz1-myc at their endogenous chromosomal loci. When immunoprecipitation was performed with a Flag antibody, the specific association of Poz1-myc was also detected. Moreover, the interaction was resistant to ethidium bromide treatment, indicating that their interaction is independent of DNA (Fig. 1C). These results demonstrate that shelterin and CLRC physically associate with each other in vivo.
Ccq1 mediates interaction between shelterin and CLRC
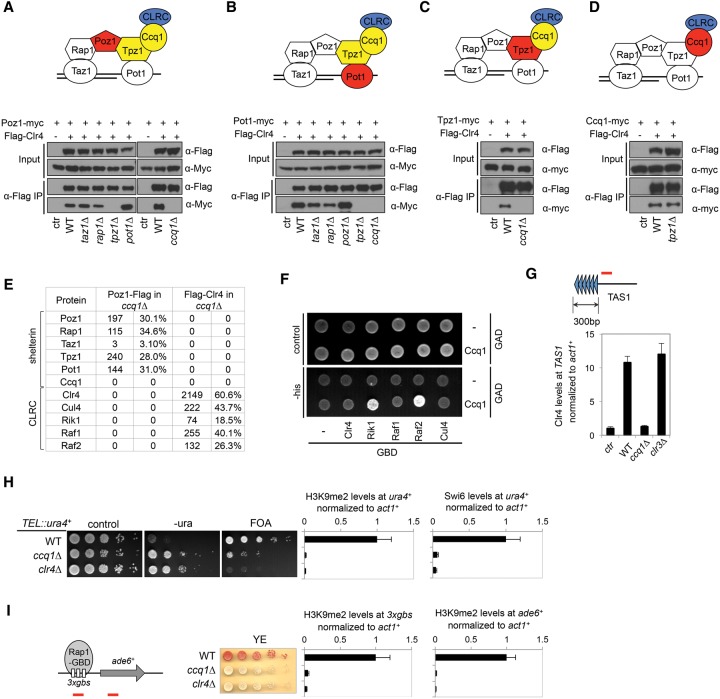
We then used co-IP analyses to determine which subunit of shelterin mediates interaction with CLRC. We found that Clr4–Poz1 interaction remained intact in taz1Δ, rap1Δ, and pot1Δ cells but was lost in tpz1Δ and ccq1Δ cells (Fig. 2A). In addition, the Clr4–Pot1 interaction was not affected by taz1Δ, rap1Δ, or poz1Δ but was abolished by tpz1Δ and ccq1Δ (Fig. 2B). Furthermore, the Clr4–Tpz1 interaction was disrupted in ccq1Δ cells, but the Clr4–Ccq1 interaction was unaffected in tpz1Δ cells (Fig. 2C,D), indicating that Ccq1 mediates the interaction between CLRC and shelterin. To confirm this idea, we performed affinity purifications of Flag-Clr4 and Poz1-Flag in ccq1Δ followed by mass spectrometry to identify associated proteins. We found that, in the absence of Ccq1, none of the shelterin subunits was identified in Flag-Clr4 purification, and none of the CLRC subunits was present in Poz1-Flag purification (Fig. 2E).
Figure 2.
Ccq1 mediates shelterin–CLRC interaction. (A–D) Co-IP analyses of shelterin–CLRC interaction. Flag-Clr4 (blue) was used for immunoprecipitation (IP) with Flag-agarose beads, and myc-tagged shelterin subunits (red) were used for co-IP detection. The shelterin subunits required for interaction are labeled in yellow. (E) Mass spectrometry analyses of purified protein complexes. The spectral counts (left), which were normalized to the amino acid number of each protein, and the sequence coverage of each protein (right) are indicated. (F) Yeast two-hybrid assay to measure the interaction between the Ccq1 and CLRC subunits. Each individual CLRC subunit was fused with the Gal4 DNA-binding domain, and Ccq1 was fused with the Gal4 DNA activation domain. Interaction between CLRC and Ccq1 resulted in the activation of a HIS3 reporter gene, allowing cells to grow in the absence of histidine. (G) Chromatin immunoprecipitation (ChIP) analyses of Flag-Clr4 levels at TAS1 (indicated by a red bar), which is immediately adjacent to telomeric repeats, normalized to act1+. (H, left) Tenfold serial dilution analyses of the indicated yeast strains grown on the indicated media to measure the expression of TEL::ura4+. (Right) ChIP analyses of H3K9me2 and Swi6 levels at TEL::ura4+, normalized to act1+. (I, left) Tenfold serial dilution analyses of indicated yeast strains grown on low-adenine medium (YE) to measure the expression of 3gbs-ade6+. (Right) ChIP analyses of H3K9me2 levels at 3gbs and ade6+(indicated by red bars at the left), normalized to act1+.
We also attempted to identify the subunit of CLRC that mediates interaction with shelterin through co-IP analyses. However, Clr4–Ccq1 interaction was significantly reduced in raf1Δ, raf2Δ, or rik1Δ cells (data not shown), suggesting that each of these CLRC subunits is likely required for the assembly of CLRC or that multiple CLRC subunits make independent contributions to shelterin–CLRC interaction. We then performed a yeast two-hybrid assay by fusing Ccq1 with the Gal4-activating domain (GAD) and each CLRC subunit with the Gal4-binding domain (GBD). We found that Rik1 and Raf2, but not any other CLRC subunit, interact with Ccq1 in this assay (Fig. 2F).
In addition to being a component of shelterin, Ccq1 copurifies with the SHREC histone deacetylase complex (Sugiyama et al. 2007; Motamedi et al. 2008). Although other members of SHREC are required for heterochromatin assembly at various chromosomal locations, Ccq1 is only required for telomeric heterochromatin assembly and is proposed to recruit SHREC to telomeric regions (Sugiyama et al. 2007). We consistently detected the SHREC subunit Clr3 in our mass spectrometry analyses of Poz1-Flag, Rap1-Flag, and Flag-Clr4 purifications but not when Poz1-Flag was purified from ccq1Δ cells (Supplemental Fig. S1A). Interestingly, other SHREC components were not detected or were present only at very low levels. Chromatin immunoprecipitation (ChIP) analyses showed that both Clr4 and Clr3 were delocalized from sequences immediately next to native telomeres (telomere-associated sequences [TAS1]) in ccq1Δ cells (Fig. 2G; Supplemental Fig. S1C). Furthermore, the localization of Clr4 to telomeres was unaffected by clr3Δ. However, Clr3 was reduced at, although not abolished from, TAS1 in clr4Δ cells (Fig. 2G; Supplemental Fig. S1D), consistent with previous studies showing that the localization of Clr3 to heterochromatin requires HP1 proteins (Yamada et al. 2005; Motamedi et al. 2008; Sadaie et al. 2008). Therefore, the effect of ccq1Δ on Clr4 localization to telomeres directly results from the loss of interaction between shelterin and Clr4 rather than an indirect effect on Clr3 localization.
We next examined silencing of TEL::ura4+, in which the ura4+ reporter is inserted immediately adjacent to telomeric repeats on a minichromosome (Nimmo et al. 1994). There is no subtelomeric DNA near this reporter, thus making it highly sensitive to shelterin mutations and avoiding complications of redundant heterochromatin initiation pathways within subtelomeric DNA. Consistent with the requirement of Ccq1 for the localization of Clr4 to telomeres, we found that silencing of TEL::ura4+ was defective in ccq1Δ cells, accompanied by the complete loss of H3K9me2 and Swi6 at the reporter and TAS1, similar to clr4Δ cells (Fig. 2H; Supplemental Fig. S1B). The residual growth on FOA medium is mainly due to increased telomere recombination in ccq1Δ cells, which resulted in the loss of the ura4+ reporter gene (confirmed by PCR analysis of individual FOA-resistant colonies) (data not shown).
Ectopic recruitment of the Rap1-GBD fusion protein to Gal4-binding sites silences an adjacent ade6+ reporter gene (3xgbs-ade6+) in a CLRC-dependent manner, resulting in the formation of red colonies when cells are grown on low-adenine (YE) medium (Tadeo et al. 2013). We found that Rap1-GBD failed to silence 3xgbs-ade6+ in ccq1Δ cells, as indicated by the formation of white colonies on YE medium, with the complete loss of H3K9me2 at the body of the ade6+ gene as well as the Gal4-binding sites (Fig. 2I). Moreover, H3K9me2 depends on Poz1 and Tpz1, which are required to link Ccq1 with Rap1, but independent of Taz1 or Pot1, which are not expected to affect Ccq1–Rap1 interaction (Supplemental Fig. S2; Tadeo et al. 2013). These results further support the role of Ccq1 in mediating CLRC–shelterin interaction.
The interaction between shelterin components is required for subtelomeric heterochromatin assembly
We previously isolated a poz1-W209A mutant that affects subtelomeric heterochromatin assembly without obvious impact on telomere length control (Tadeo et al. 2013). Co-IP analyses showed that Poz1-W209A still interacts with CLRC to a degree comparable with wild-type Poz1 (Fig. 3A). Moreover, Poz1-W209A had no effect on Poz1–Tpz1 interaction but significantly reduced Poz1–Rap1 interaction (Fig. 3B,C). Therefore, the Poz1-W209A mutation specifically affects the interaction between Rap1 and Poz1 (Fig. 3D). While a strong interaction of Rap1–Poz1 is required for telomere silencing, the residual Rap1–Poz1 interaction seems strong enough for telomere length maintenance. These results raise the possibility that the proper connection of shelterin components is essential for heterochromatin assembly at subtelomeres. To test this idea, we investigated other mutations that selectively disrupt interactions between shelterin subunits. A previous study showed that Tpz1-I501R specifically ablates Poz1–Tpz1 interaction and that Tpz1-I200R specifically disrupts Tpz1–Pot1 interaction (Fig. 3D; Jun et al. 2013). Like poz1-W209A, tpz1-I501R resulted in loss of silencing of TEL::ura4+ and loss of H3K9me2 and Swi6 at the reporter (Fig. 3E). However, tpz1-I200R has no silencing defects at TEL::ura4+, and ChIP analyses showed normal H3K9me2 and Swi6 levels at the ura4+ reporter gene (Fig. 3E). These results suggest that the proper connection from Rap1 to Tpz1, but not the Tpz1–Pot1 connection, is required for subtelomeric heterochromatin formation. To further confirm this idea, we examined the requirement of shelterin connections in Rap1-GBD-mediated ectopic heterochromatin assembly. Supporting our hypothesis, H3K9me2 levels were abolished in all mutants that are defective in subtelomeric heterochromatin formation (poz1Δ, poz1-W209A, tpz1Δ, and tpz1-I501R) but remained unchanged in tpz1-I200R cells (Supplemental Fig. S2B).
Figure 3.
Proper connection between shelterin subunits is critical for telomeric heterochromatin assembly. (A,B,C,F) Co-IP analyses to examine the effect of poz1-W209A on shelterin–CLRC or shelterin subunit interactions. (D) Schematic diagram of the shelterin–CLRC interaction in each mutant background. (E, left) Tenfold serial dilution analyses of the indicated yeast strains grown on indicated the media to measure the expression of TEL::ura4+. (Right) ChIP analysis of H3K9me2 and Swi6 levels at TEL::ura4+, normalized to act1+. (G) ChIP analyses of Flag-Clr4 levels at TAS1, normalized to act1+.
Recruitment of CLRC to double-stranded telomeric DNA is required for subtelomeric heterochromatin
The differences in the ability of poz1-W209A, tpz1-I501R, and tpz1-I200R mutants to assemble heterochromatin raise the possibility that interaction between CLRC and dsDNA-binding proteins, but not ssDNA-binding protein Pot1, is required for subtelomeric heterochromatin. To test this idea, we performed co-IP analyses and found that poz1-W209A, tpz1-I501R, and poz1Δ, which showed loss of silencing at TEL::ura4+, all disrupted the interaction between Clr4 and Rap1 but still maintained the interaction between Clr4 and Pot1 (Fig. 3F). On the other hand, the Clr4–Pot1 interaction, but not the Clr4–Rap1 interaction, was disrupted in tpz1-I200R cells, which had no silencing defects at TEL::ura4+ (Fig. 3F). Furthermore, ChIP analysis showed that Flag-Clr4 enrichment at TAS1 and TEL::ura4+ was abolished in poz1-W209A, tpz1-I501R, and poz1Δ cells but remained intact in tpz1-I200R cells (Fig. 3G; Supplemental Fig. S3A). Clr4 levels at more telomere-distal regions were not affected due to the presence of an RNAi-dependent pathway to recruit CLRC (Supplemental Fig. S3C; Kanoh et al. 2005). These results suggest that the association of CLRC with the dsDNA-binding part of shelterin is essential for subtelomeric heterochromatin assembly. In addition, Clr3 enrichment at subtelomeres was also reduced in all shelterin mutants that affected Clr4 recruitment but remained at wild-type levels in the tpz1-I200R mutant, which has no effects on Clr4 recruitment to telomeres (Supplemental Fig. S1C,D).
Restoring the proper association between shelterin components rescues subtelomeric heterochromatin assembly defects of shelterin mutants
To examine whether a compromised Poz1–Rap1 interaction is responsible for heterochromatin assembly defects in poz1-W209A cells, we generated a strain expressing Poz1-W209A with a GBP (GFP-binding protein) tag (Rothbauer et al. 2008) and crossed it into a strain expressing Rap1-GFP. The interaction between GBP and GFP is expected to restore the interaction between Rap1 and Poz1-W209A. Indeed, the TEL::ura4+ reporter was efficiently silenced in poz1-W209A-GBP rap1-GFP cells, accompanied by the restoration of heterochromatin hallmarks such as H3K9me2 and Swi6 (Fig. 4A). Similarly, tpz1-I501R-GBP poz1-GFP cells, which are expected to restore interaction between Tpz1 and Poz1, rescued silencing at TEL::ura4+ as well as H3K9me2 and Swi6 levels (Fig. 4B). In addition to the restoration of subtelomeric heterochromatin, tpz1-I501R-GBP poz1-GFP cells also alleviated telomere length regulation defects associated with tpz1-I501R, further indicative of the restoration of interaction between Tpz1 and Poz1 (Supplemental Fig. S4). Interestingly, forced Tpz1–Rap1 interaction through Tpz1-GBP and Rap1-GFP in poz1Δ cells was able to silence TEL::ura4+ and restored H3K9me2 and Swi6 levels at the reporter gene (Fig. 4C). Moreover, the telomere length was gradually reduced to wild-type level in tpz1-GBP rap1-GFP poz1Δ cells compared with poz1Δ cells (Supplemental Fig. S4). The fact that restoration of the linkage between the dsDNA and ssDNA telomere-binding complexes successfully rescues silencing defects caused by point mutants that specifically disrupt these interactions suggests that the overall architecture of the shelterin complex rather than the identity or specialized structure of the interaction is crucial for subtelomeric heterochromatin assembly. A recent study showing minishelterin, in which Taz1 is artificially fused to Tpz1, can maintain telomere length homeostasis and subtelomeric silencing even in the absence of Rap1 and Poz1 further supports this view (Pan et al. 2015).
Figure 4.
Proper connection between shelterin subunits is critical for subtelomeric heterochromatin assembly. (Left) Schematic diagram of the constructs. (Middle) Tenfold serial dilution analyses of indicated yeast strains grown on the indicated media to measure the expression of TEL::ura4+. (Right) ChIP analyses of H3K9me2 and Swi6 levels at TEL::ura4+, normalized to act1+.
Telomeric repeats block heterochromatin spreading
The fact that CLRC maintains its interaction with Pot1 in poz1Δ, tpz1-I501R, and poz1-W209A cells suggests that the subtelomeric silencing defect is not a result of impaired CLRC recruitment but is instead due to a failure of CLRC to spread from the chromosome ends over telomeric repeats. Telomere elongation increases the distance between Pot1-bound Clr4 and TEL::ura4+ in poz1Δ and tpz1-I501R cells, which might contribute to the inability of Clr4 spreading to silence the reporter. However, poz1-W209A has very little effect on telomere length (Tadeo et al. 2013). The fact that poz1-W209A lost telomere silencing similar to poz1Δ and tpz1-I501R indicates that even the normal length of telomeric repeats is sufficient to block the spreading of Clr4 from the chromosome ends to internal regions when the connection between CLRC and the dsDNA region of the telomeric repeat is compromised.
We then directly examined whether telomeric repeats block heterochromatin spreading at an ectopic heterochromatin domain. Artificially targeting a GBD-Clr4-ΔCD fusion protein to three copies of Gal4-binding sites (3xgbs) silences an adjacent ade6+ reporter gene (3xgbs-ade6+), as indicated by red colony color when cells were grown on YE medium (Supplemental Fig. S5A; Kagansky et al. 2009). We inserted between 3xgbs and ade6+ either 257 base pairs (bp) of telomeric repeat DNA (3xgbs-tel-ade6+) or 300 bp of control DNA from the leu1 locus (3xgbs-control-ade6+) (Fig. 5A). The control sequence had no effects on silencing of the ade6+ reporter by GBD-Clr4-ΔCD, indicating efficient CLRC spreading (Supplemental Fig. S5A). In the absence of GBD-Clr4-ΔCD, the telomeric repeats silenced the ade6+ reporter, and silencing was lost in taz1Δ, rap1Δ, poz1Δ, and ccq1Δ cells (Supplemental Fig. S5B). Such a result is consistent with shelterin-mediated recruitment of CLRC to telomeric repeats. Interestingly, in ccq1Δ cells, which abolished shelterin–CLRC interaction, GBD-Clr4-ΔCD failed to silence 3xgbs-tel-ade6+ (Fig. 5B). Moreover, ChIP analyses showed that H3K9me2 and Swi6 were enriched at 3xgbs, but not ade6+, in ccq1Δ cells (Fig. 5B), suggesting that CLRC failed to spread over the telomeric repeats. In contrast, GBD-Clr4-ΔCD efficiently silences 3xgbs-control-ade6+ in ccq1Δ cells, and H3K9me2 and Swi6 spread to the ade6+ reporter (Fig. 5B). These results suggest that shelterin-bound telomeric repeats are refractory to heterochromatin spreading.
Figure 5.
Telomeric repeats block heterochromatin spreading. (A) Schematic diagrams of the reporter genes used. Red bars indicate the positions of the PCR fragments used in ChIP analyses. (B, left) Tenfold serial dilution analyses of the indicated yeast strains grown on YE medium to measure the expression of ade6+. (Right) ChIP analyses of H3K9me2 and Swi6 levels at 3xgbs and ade6+, normalized to act1+.
In both budding and fission yeasts, telomeric DNA has been suggested to form nonnucleosomal structures (Chikashige et al. 1989; Wright et al. 1992). We performed MNase digestion of chromatin followed by Southern blot analyses with probes derived from pericentric and telomeric repeats. Pericentric chromatin showed a characteristic nucleosomal ladder pattern, indicative of well-positioned nucleosomes (Supplemental Fig. S6). In contrast, telomeric chromatin showed a diffuse pattern, indicative of low nucleosome density or alternative nucleosome structures (Supplemental Fig. S6). Interestingly, GBD-Clr4-ΔCD silenced 3xgbs-tel-ade6+ in taz1Δ cells (Supplemental Fig. S5C). Given that Taz1 is responsible for recruiting other shelterin components in this setting, the results suggest that telomeric repeats and its associated chromatin structure, at least when they are only 257 bp long, are insufficient to block CLRC spreading in the absence of bound shelterin. Telomeres associate with the nuclear periphery through the interaction between Rap1 and inner nuclear membrane proteins Bqt3–Bqt4 (Chikashige et al. 2009). GBD-Clr4-ΔCD failed to silence 3xgbs-tel-ade6+ in ccq1Δ bqt3Δ cells (Supplemental Fig. S5C), suggesting that the ability of shelterin-bound telomeric repeats to block CLRC spreading is not due to the tethering of these repeats to the nuclear periphery but rather that the bound shelterin creates a physical barrier.
We also found that poz1Δ had stronger silencing defects at TEL::ura4+ compared with taz1Δ (Supplemental Fig. S5D). Given that taz1Δ and poz1Δ have similar effects on telomere length, such a difference might be attributed to the binding of Taz1 to telomeric repeats in poz1Δ cells, which more efficiently blocks CLRC spreading. Consistent with this idea, poz1Δ taz1Δ partially rescued silencing defects of TEL::ura4+ compared with poz1Δ (Supplemental Fig. S5D).
Increased heterochromatin spreading restores silencing at subtelomeres in shelterin mutants
If the heterochromatin defects of shelterin connection mutations are due to the failure of CLRC spreading over telomeric repeats, we expect that a genetic background that is more permissive to heterochromatin spreading might overcome this barrier and rescue silencing defects in shelterin mutants that disrupt shelterin interactions. Our previous work showed that the Mst2 histone H3K14 acetyltransferase complex promotes histone turnover at heterochromatin regions, and mst2Δ results in increased heterochromatin spreading into subtelomeric regions (Reddy et al. 2011; Wang et al. 2012, 2015). We found that silencing of TEL::ura4+ was significantly restored in mst2Δ poz1Δ, mst2Δ poz1-W209A, and mst2Δ tpz1-I501R double mutants (Fig. 6A; Supplemental Fig. S7A). In addition, H3K9me2 and Swi6 levels at TEL::ura4+ were restored to near wild-type levels (Fig. 6A). Moreover, mutation of the active site of Mst2 (mst2-G250E) or other components of the Mst2 complex required for its acetyltransferase activity (nto1Δ and ptf2Δ) (Wang et al. 2012) also rescued poz1Δ in silencing of TEL::ura4+, consistent with the fact that the enzymatic activity of the Mst2 complex counteracts heterochromatin spreading (Supplemental Fig. S7B).
Figure 6.
Increased heterochromatin spreading alleviates telomeric silencing defects caused by compromised shelterin connections. (A,E, left) Tenfold serial dilution analyses of the indicated yeast strains grown on the indicated media to measure the expression of TEL::ura4+. (Right) ChIP analyses of H3K9me2 and Swi6 levels at TEL::ura4+, normalized to act1+. (B) Co-IP analysis of Clr4–Rap1 interaction in different mutant backgrounds. (C) Southern blot analysis to measure telomere length. (D) ChIP analyses of Flag-Clr4 levels at TAS1, normalized to act+1.
Co-IP analysis showed that Clr4 failed to interact with Rap1 in mst2Δ poz1Δ cells, similar to poz1Δ cells, suggesting that mst2Δ does not rescue CLRC–shelterin interaction in poz1Δ cells (Fig. 6B). Moreover, telomere length remains the same in poz1Δ and mst2Δ poz1Δ cells, suggesting that shelterin connections are not restored (Fig. 6C). However, ChIP analysis showed increased levels of Clr4 at TAS1 and TEL::ura4+ in mst2Δ poz1Δ cells (Fig. 6D; Supplemental Fig. S3B), consistent with the idea that the rescue of TEL::ura4+ silencing in mst2Δ poz1Δ cells is due to increased spreading of CLRC from the telomere end to the reporter. We also observed restored levels of Clr3 at TAS1 in mst2Δ poz1Δ cells (Supplemental Fig. S1E), which is in line with the idea that Clr3 is recruited to the telomeres by CLRC-mediated heterochromatin formation. In addition, overexpression of Swi6 or epe1Δ, which promote heterochromatin spreading (Ayoub et al. 2003; Wang et al. 2013), also rescued telomeric heterochromatin defects in poz1Δ cells (Supplemental Fig. S7C). Furthermore, introducing a clr4-W31G mutation, which abolishes heterochromatin spreading from nucleation centers (Zhang et al. 2008), resulted in loss of silencing at TEL::ura4+ in mst2Δ poz1Δ or epe1Δ poz1Δ cells (Fig. 6E; Supplemental S7D).
To rule out the possibility that the restoration of silencing at TEL::ura4+ in mst2Δ poz1Δ cells is due to heterochromatin maintenance, we performed a heterochromatin re-establishment assay. We crossed mst2Δ poz1Δ cells with mst2Δ poz1Δ clr4Δ cells containing TEL::ura4+ to obtain mst2Δ poz1Δ TEL::ura4+ cells, in which the TEL::ura4+ was inherited without pre-existing H3K9me. Dilution analyses showed that the resulting mst2Δ poz1Δ cells were able to establish heterochromatin at the reporter gene de novo, accompanied by the enrichment of H3K9me2 and Swi6 at the reporter (Supplemental Fig. S8).
Although we expect that Clr4 is recruited to ssDNA in poz1Δ cells through its interaction with Pot1 (Fig. 3F), we cannot directly measure the enrichment of Clr4 at the ssDNA given the identical sequences of the dsDNA and ssDNA portions of telomeric repeats. Nonetheless, given that ccq1Δ resulted in complete loss of Clr4 from telomeres (Fig. 2G, since the fragment used for PCR amplification in ChIP analysis is <300 bp from the chromosome end, the result also reflects the binding of CLRC to telomeric repeats), the fact that mst2Δ ccq1Δ failed to silence TEL::ura4+ suggests that Mst2 antagonizes telomeric silencing downstream from CLRC recruitment (Fig. 6E). Therefore, recruitment of CLRC to telomeric ssDNA is a prerequisite of mst2Δ to rescue the silencing defects associated with poz1Δ, although it is also possible that the failure of mst2Δ ccq1Δ cells to assemble telomeric heterochromatin is the result of the loss of both CLRC and SHREC from telomeres.
Discussion
In eukaryotic cells, telomeres are prominent sites of constitutive heterochromatin. However, the mechanism of telomeric heterochromatin assembly is poorly understood, making it difficult to dissect its biological functions. In fission yeast, shelterin is required for the establishment of subtelomeric heterochromatin, and it has been suggested that shelterin is responsible for the recruitment of the histone H3K9 methyltransferase CLRC to telomeric repeats (Kanoh et al. 2005; Tadeo et al. 2013). However, mutations of individual shelterin components do not significantly affect the association of other shelterin components with telomeres due to independent shelterin recruitment pathways through both the ssDNA and the dsDNA portions of telomeric repeats (Miyoshi et al. 2008). Therefore, it is difficult to explain why many different shelterin mutants lead to subtelomeric heterochromatin assembly defects if the only function of shelterin is to recruit CLRC. Our results show that shelterin recruits the histone H3K9 methyltransferase CLRC through protein–protein interactions mediated by the Ccq1 subunit. Consistently, ccq1Δ results in defective recruitment of CLRC to telomeres. Interestingly, mutations other than ccq1Δ that affect subtelomeric heterochromatin still maintain the CLRC–Pot1 interaction, suggesting that shelterin plays other crucial roles in subtelomeric heterochromatin assembly aside from recruiting CLRC to chromosome ends. A common theme is that these mutations affect the association between CLRC and the dsDNA-binding portion of shelterin, and restoring such connections can rescue heterochromatin defects.
Heterochromatin has the propensity to spread from the site of initiation to neighboring regions generally in a sequence-independent manner (Wang et al. 2014a). However, the failure of CLRC to spread from the site of recruitment (the Tpz1–Pot1 subcomplex located at the ssDNA of telomeres) over telomeric repeats to more internal regions in mutations that disrupt interactions between shelterin subunits suggests that telomeric repeats are refractory to heterochromatin spreading. The proper connection of shelterin components overcomes this barrier by promoting direct interaction between CLRC and chromosomal regions internal to telomeres, allowing heterochromatin assembly (Fig. 7). Therefore, the ability of CLRC to regulate heterochromatin assembly depends on not only its recruitment but also the local chromatin context.
Figure 7.

A model for telomeric heterochromatin assembly. In wild-type cells, the connection between shelterin allows CLRC to skip telomeric DNA to reach nucleosomes at subtelomeric regions to initiate histone H3K9me. In poz1Δ cells, although CLRC is still recruited to chromosome ends through interaction with Pot1–Tpz1–Ccq1, the paucity of nucleosomes on double-stranded telomeric DNA and the presence of shelterin components on these repeats prevent heterochromatin from spreading into internal chromosomal regions. In mst2Δ poz1Δ cells, the reduction of histone turnover allows H3K9me of the residual nucleosomes on telomeric DNA, which provides a platform for heterochromatin to spread.
It is interesting that, without CLRC recruitment, a short stretch of repeats only a little more than one nucleosome length can function as a barrier to heterochromatin spreading at an ectopic site. Such a barrier function requires the binding of shelterin to form a physical block to CLRC spreading (Fig. 5). At telomeres, CLRC spreading from the chromosome ends to subtelomeric regions is also affected by taz1Δ, which compromises the binding of shelterin to double-stranded telomeric DNA and increases the distances between chromosome end-bound CLRC and subtelomeres. Therefore, telomeric DNA also contribute to barrier activity when they are of sufficient length. It has been shown that telomeric DNA sequences from diverse species disfavor nucleosome assembly in vitro and in vivo (Chikashige et al. 1989; Wright et al. 1992; Cacchione et al. 1997; Ichikawa et al. 2014). The wrapping of DNA around a histone octamer requires sharp bending of DNA, which is facilitated by a 10-bp periodicity of AA/TT/TA dinucleotides (Segal et al. 2006). The 6- to 8-bp periodicity of telomeric repeats makes it difficult to maintain such phasing. Moreover, telomeric DNA can assemble G quadruplexes, which might also contribute to the formation of alternative chromatin structures at telomeres (Lipps and Rhodes 2009). Regardless of the mechanism, the nucleosome density at telomeres might be low enough to prevent the proximity-mediated positive feedback mechanism that facilitates spreading of heterochromatin, and the connections between shelterin components allow the heterochromatin machinery to skip telomeric repeats (Fig. 7). Although the telomeric chromatin structure, indicated by the MNase digestion pattern, is different from that of the chromosome arms, nucleosomes might still be present at fission yeast telomeres, as indicated by the detection of histone post-translational modifications (Carneiro et al. 2010). The fact that increased heterochromatin spreading overcomes silencing defects in the absence of shelterin connections further supports the existence of nucleosomes, most likely at lower levels, at telomeric DNA. However, these nucleosomes may be rapidly turned over or poorly positioned or possess post-translational modifications that prohibit heterochromatin assembly. The binding of shelterin to telomeric repeat DNA might further strengthen the barrier activity, as shelterin binding can directly impact the formation of nucleosomes over telomeric repeats. In addition, the binding of shelterin also creates a physical barrier for the spread of CLRC. Therefore, the effects of telomeric repeats in blocking CLRC spreading at native telomeres might be the combined action of telomeric chromatin structure and the binding of shelterin.
In budding yeast, the looping back of telomeres is thought to strengthen telomere position effect, a measure of heterochromatin integrity (Strahl-Bolsinger et al. 1997; de Bruin et al. 2000). However, such a telomeric chromatin organization differs from what we discovered in fission yeast. Budding yeast heterochromatin machinery is significantly divergent from that of fission yeast and is established at telomeres by RAP1, which directly binds to the dsDNA portion of telomere repeats and recruits SIR proteins (Rusche et al. 2003). Therefore, even though telomeres lack nucleosome organization, looping at telomeres is not required to bypass the telomeric DNA, and it is likely that such a structure is required for other aspects of gene silencing.
In fission yeast, although loss of heterochromatin factors has no effect on telomere length control or chromosome end protection, heterochromatin does affect the mode of survival of cells without telomerase (Ekwall et al. 1996; Tuzon et al. 2004; Jain et al. 2010). However, as heterochromatin assembly factors are involved in heterochromatin formation genome-wide, it is unknown whether such phenotypes are the direct result of loss of telomeric heterochromatin or an indirect effect of losing heterochromatin at nontelomeric locations. Our studies showing that most mutations of shelterin affect connections between shelterin subunits rather than shelterin–CLRC interactions suggest that these mutations are unlikely to be definitive in addressing the functions of telomeric heterochromatin. Nonetheless, our results do point out that Ccq1 is the critical subunit that mediates interaction between CLRC and shelterin. It is important to note that Ccq1 is a multifunctional protein involved in telomerase recruitment and DNA damage checkpoint suppression (Tomita and Cooper 2008). It is also part of the SHREC complex, in addition to its role in CLRC recruitment to telomeres (Sugiyama et al. 2007; Motamedi et al. 2008). Further isolation of separation-of-function mutations of Ccq1 holds great potential for genetically dissecting the function of telomeric heterochromatin in telomere length regulation and other aspects of chromosome biology.
It has long been known that telomeres form heterochromatin structures in diverse organisms (Blasco 2007). However, the function of telomeric heterochromatin is not well understood. In Drosophila, retrotransposons that form heterochromatin replace telomeric repeats, and heterochromatin proteins are directly involved in the recruitment of chromosome end protection factors (Mason et al. 2008). In fission yeast, the emergence of HAATI (heterochromatin amplification-mediated and telomerase-independent) survivors in response to telomerase loss also suggests a direct role of telomeric heterochromatin in the recruitment of telomere proteins (Jain et al. 2010), possibly mediated by direct protein–protein interactions between shelterin and CLRC. Moreover, heterochromatin is also involved in regulating transcription of telomeric noncoding RNA TERRA, which has been increasingly recognized to play important roles in telomere functions (Azzalin and Lingner 2015). Thus, a more complete understanding of the mechanism of telomeric heterochromatin assembly will allow a more definitive dissection of the functions of telomeric heterochromatin.
Materials and methods
Fission yeast strains and genetic analyses
Yeast strains containing Poz1-myc, Poz1-GBP, Poz1-W209A-GBP, Tpz1-GBP, Tpz1-myc, Tpz1-I501R-GBP, and Pot1-myc were generated by a PCR-based module method (Bahler et al. 1998). Due to the effect of C-terminal tags on the functionality of Tpz1 (Kallgren et al. 2014), the Tpz1-I501R and Tpz1-I200R used in this study were constructed without any tags unless otherwise noted. All strains containing tpz1Δ and pot1Δ were first constructed in diploid cells, followed by tetrad dissection to obtain desired haploid colonies. The 3xgbs-tel-ade6+ reporter was constructed by inserting 257-bp telomeric DNA sequences between 3xgbs and the ade6+ reporter gene, and the 3xgbs-control-ade6+ reporter was constructed by inserting 306-bp DNA from the leu1+ locus (corresponding to amino acids 230–331) between 3xgbs and the ade6+ reporter gene. A list of the yeast strains used is in Supplemental Table 1. For serial dilution plating assays, 10-fold dilutions of a mid-log-phase culture were plated on the indicated media and grown for 3 d at 30°C.
Protein purification, co-IP, and mass spectrometry analyses
Exponentially growing yeast cells were harvested, washed with 2× HC buffer (300 mM HEPES-KOH at pH 7.6, 2 mM EDTA, 100 mM KCl, 20% glycerol, 2 mM DTT, protease inhibitor cocktail [Roche]), and frozen in liquid nitrogen. Crude cell extracts were prepared by vigorously blending frozen yeast cells with dry ice using a household blender, followed by incubation with 30 mL of 1× HC buffer containing 250 mM KCl for 30 min. The lysate was cleared by centrifugation at 82,700g for 3 h. The supernatants were precleared with protein A agarose, incubated with 200 μL of Flag-agarose overnight, and washed eight times with 1× HC containing 250 mM KCl. For mass spectrometry analysis, bound proteins were eluted with 200 μg/mL 3xFlag peptides followed by TCA precipitation. MudPIT (multidimensional protein identification technology) mass spectrometry analysis was performed as described previously (Wang et al. 2014b). For co-IP analysis, bound proteins were resolved by SDS-PAGE followed by Western blot analyses with Myc (Santa Cruz Biotechnology), Flag (Sigma), and HA (3F10; Roche) antibodies.
ChIP analyses
ChIP analyses were performed as described previously (Wang et al. 2015). The antibodies used were H3K9me2 (Abcam), Swi6 (Reddy et al. 2011), and Flag (Sigma). Quantitative real-time PCR (qPCR) was performed with Maxima SYBR Green qPCR master mix (Fermentas) in a StepOne Plus real-time PCR system (Applied Biosystems). DNA serial dilutions were used as templates to generate a standard curve of amplification for each pair of primers, and the relative concentration of target sequence was calculated accordingly. An act1 fragment was used as reference to calculate the enrichment of ChIP over WCE for each target sequence. The numbers are the averages of three experiments, and error bars represent standard deviation. A list of the DNA oligos used is in Supplemental Table 2.
Yeast two-hybrid assay
Full-length individual CLRC subunits were cloned into the XmaI/BamHI site of pGBT9 (Clontech) to generate fusion proteins with the GAL4 DNA-binding domain. Full-length Ccq1 was cloned into the XmaI/BamHI site of pGAD424 (Clontech) to generate fusion proteins with the GAL4 activation domain. Both plasmids were transformed into the budding yeast strain pJ69-4A, and transformants were selected on medium lacking tryptophan and leucine to maintain both plasmids and were confirmed by PCR analyses. The interaction of the two proteins was indicated by the activation of a HIS3 reporter, allowing growth on medium lacking histidine.
Telomere length analysis
Genomic DNA was isolated, digested with EcoRI, and separated on 1% agarose gel. Hybridization with a radiolabeled telomeric DNA probe was performed as previously described (Moser et al. 2011).
Supplementary Material
Acknowledgments
We thank Junichi Nakayama for the Clr3-Flag strain, Fred Chang and Jamie Moseley for the GBP plasmid, and Ruben Dorador for technical assistance. This work was supported by National Institutes of Health grants (R01-GM085145 to S.J., and R01-GM098943 to F.Q.), the National Center for Research Resources (P41-RR011823), and the National Institute of General Medical Sciences (P41-GM103533). X.T. was a Fulbright Scholar. F.Q. was supported by a Basil O'Connor Starter Scholar Research Award from the March of Dimes and a Beginning Grant-in-aid from the American Heart Association.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.266718.115.
References
- Armstrong CA, Pearson SR, Amelina H, Moiseeva V, Tomita K. 2014. Telomerase activation after recruitment in fission yeast. Curr Biol 24: 2006–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audergon PN, Catania S, Kagansky A, Tong P, Shukla M, Pidoux AL, Allshire RC. 2015. Epigenetics. Restricted epigenetic inheritance of H3K9 methylation. Science 348: 132–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayoub N, Noma K, Isaac S, Kahan T, Grewal SI, Cohen A. 2003. A novel jmjC domain protein modulates heterochromatization in fission yeast. Mol Cell Biol 23: 4356–4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzalin CM, Lingner J. 2015. Telomere functions grounding on TERRA firma. Trends Cell Biol 25: 29–36. [DOI] [PubMed] [Google Scholar]
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A III, Steever AB, Wach A, Philippsen P, Pringle JR. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14: 943–951. [DOI] [PubMed] [Google Scholar]
- Baumann P, Cech TR. 2001. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 292: 1171–1175. [DOI] [PubMed] [Google Scholar]
- Blasco MA. 2007. The epigenetic regulation of mammalian telomeres. Nat Rev Genet 8: 299–309. [DOI] [PubMed] [Google Scholar]
- Cacchione S, Cerone MA, Savino M. 1997. In vitro low propensity to form nucleosomes of four telomeric sequences. FEBS Lett 400: 37–41. [DOI] [PubMed] [Google Scholar]
- Carneiro T, Khair L, Reis CC, Borges V, Moser BA, Nakamura TM, Ferreira MG. 2010. Telomeres avoid end detection by severing the checkpoint signal transduction pathway. Nature 467: 228–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castel SE, Martienssen RA. 2013. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat Rev Genet 14: 100–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikashige Y, Hiraoka Y. 2001. Telomere binding of the Rap1 protein is required for meiosis in fission yeast. Curr Biol 11: 1618–1623. [DOI] [PubMed] [Google Scholar]
- Chikashige Y, Kinoshita N, Nakaseko Y, Matsumoto T, Murakami S, Niwa O, Yanagida M. 1989. Composite motifs and repeat symmetry in S. pombe centromeres: direct analysis by integration of NotI restriction sites. Cell 57: 739–751. [DOI] [PubMed] [Google Scholar]
- Chikashige Y, Yamane M, Okamasa K, Tsutsumi C, Kojidani T, Sato M, Haraguchi T, Hiraoka Y. 2009. Membrane proteins Bqt3 and -4 anchor telomeres to the nuclear envelope to ensure chromosomal bouquet formation. J Cell Biol 187: 413–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen AL, Jia S. 2014. Noncoding RNAs and the borders of heterochromatin. Wiley Interdiscip Rev RNA 5: 835–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JP, Nimmo ER, Allshire RC, Cech TR. 1997. Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature 385: 744–747. [DOI] [PubMed] [Google Scholar]
- de Bruin D, Kantrow SM, Liberatore RA, Zakian VA. 2000. Telomere folding is required for the stable maintenance of telomere position effects in yeast. Mol Cell Biol 20: 7991–8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekwall K, Nimmo ER, Javerzat JP, Borgstrom B, Egel R, Cranston G, Allshire R. 1996. Mutations in the fission yeast silencing factors clr4+ and rik1+ disrupt the localisation of the chromo domain protein Swi6p and impair centromere function. J Cell Sci 109 (Pt 11): 2637–2648. [DOI] [PubMed] [Google Scholar]
- Fujita I, Tanaka M, Kanoh J. 2012. Identification of the functional domains of the telomere protein Rap1 in Schizosaccharomyces pombe. PLoS One 7: e49151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI, Jia S. 2007. Heterochromatin revisited. Nat Rev Genet 8: 35–46. [DOI] [PubMed] [Google Scholar]
- Harland JL, Chang YT, Moser BA, Nakamura TM. 2014. Tpz1–Ccq1 and Tpz1–Poz1 interactions within fission yeast shelterin modulate Ccq1 Thr93 phosphorylation and telomerase recruitment. PLoS Genet 10: e1004708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong EJ, Villen J, Gerace EL, Gygi SP, Moazed D. 2005. A cullin E3 ubiquitin ligase complex associates with Rik1 and the Clr4 histone H3-K9 methyltransferase and is required for RNAi-mediated heterochromatin formation. RNA Biol 2: 106–111. [DOI] [PubMed] [Google Scholar]
- Horn PJ, Bastie JN, Peterson CL. 2005. A Rik1-associated, cullin-dependent E3 ubiquitin ligase is essential for heterochromatin formation. Genes Dev 19: 1705–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa Y, Morohashi N, Nishimura Y, Kurumizaka H, Shimizu M. 2014. Telomeric repeats act as nucleosome-disfavouring sequences in vivo. Nucleic Acids Res 42: 1541–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain D, Cooper JP. 2010. Telomeric strategies: means to an end. Annu Rev Genet 44: 243–269. [DOI] [PubMed] [Google Scholar]
- Jain D, Hebden AK, Nakamura TM, Miller KM, Cooper JP. 2010. HAATI survivors replace canonical telomeres with blocks of generic heterochromatin. Nature 467: 223–227. [DOI] [PubMed] [Google Scholar]
- Jia S, Noma K, Grewal SI. 2004. RNAi-independent heterochromatin nucleation by the stress-activated ATF/CREB family proteins. Science 304: 1971–1976. [DOI] [PubMed] [Google Scholar]
- Jia S, Kobayashi R, Grewal SI. 2005. Ubiquitin ligase component Cul4 associates with Clr4 histone methyltransferase to assemble heterochromatin. Nat Cell Biol 7: 1007–1013. [DOI] [PubMed] [Google Scholar]
- Jun HI, Liu J, Jeong H, Kim JK, Qiao F. 2013. Tpz1 controls a telomerase-nonextendible telomeric state and coordinates switching to an extendible state via Ccq1. Genes Dev 27: 1917–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagansky A, Folco HD, Almeida R, Pidoux AL, Boukaba A, Simmer F, Urano T, Hamilton GL, Allshire RC. 2009. Synthetic heterochromatin bypasses RNAi and centromeric repeats to establish functional centromeres. Science 324: 1716–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallgren SP, Andrews S, Tadeo X, Hou H, Moresco JJ, Tu PG, Yates JR III, Nagy PL, Jia S. 2014. The proper splicing of RNAi factors is critical for pericentric heterochromatin assembly in fission yeast. PLoS Genet 10: e1004334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanoh J, Ishikawa F. 2001. spRap1 and spRif1, recruited to telomeres by Taz1, are essential for telomere function in fission yeast. Curr Biol 11: 1624–1630. [DOI] [PubMed] [Google Scholar]
- Kanoh J, Sadaie M, Urano T, Ishikawa F. 2005. Telomere binding protein Taz1 establishes Swi6 heterochromatin independently of RNAi at telomeres. Curr Biol 15: 1808–1819. [DOI] [PubMed] [Google Scholar]
- Kim HS, Choi ES, Shin JA, Jang YK, Park SD. 2004. Regulation of Swi6/HP1-dependent heterochromatin assembly by cooperation of components of the mitogen-activated protein kinase pathway and a histone deacetylase Clr6. J Biol Chem 279: 42850–42859. [DOI] [PubMed] [Google Scholar]
- Lipps HJ, Rhodes D. 2009. G-quadruplex structures: in vivo evidence and function. Trends Cell Biol 19: 414–422. [DOI] [PubMed] [Google Scholar]
- Mason JM, Frydrychova RC, Biessmann H. 2008. Drosophila telomeres: an exception providing new insights. Bioessays 30: 25–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi T, Kanoh J, Saito M, Ishikawa F. 2008. Fission yeast Pot1-Tpp1 protects telomeres and regulates telomere length. Science 320: 1341–1344. [DOI] [PubMed] [Google Scholar]
- Moser BA, Chang YT, Kosti J, Nakamura TM. 2011. Tel1ATM and Rad3ATR kinases promote Ccq1-Est1 interaction to maintain telomeres in fission yeast. Nat Struct Mol Biol 18: 1408–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motamedi MR, Hong EJ, Li X, Gerber S, Denison C, Gygi S, Moazed D. 2008. HP1 proteins form distinct complexes and mediate heterochromatic gene silencing by nonoverlapping mechanisms. Mol Cell 32: 778–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. 2001. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292: 110–113. [DOI] [PubMed] [Google Scholar]
- Nimmo ER, Cranston G, Allshire RC. 1994. Telomere-associated chromosome breakage in fission yeast results in variegated expression of adjacent genes. EMBO J 13: 3801–3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmo ER, Pidoux AL, Perry PE, Allshire RC. 1998. Defective meiosis in telomere-silencing mutants of Schizosaccharomyces pombe. Nature 392: 825–828. [DOI] [PubMed] [Google Scholar]
- Pan L, Hildebrand K, Stutz C, Thoma N, Baumann P. 2015. Minishelterins separate telomere length regulation and end protection in fission yeast. Genes Dev 29: 1164–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragunathan K, Jih G, Moazed D. 2015. Epigenetics. Epigenetic inheritance uncoupled from sequence-specific recruitment. Science 348: 1258699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, et al. 2000. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406: 593–599. [DOI] [PubMed] [Google Scholar]
- Reddy BD, Wang Y, Niu L, Higuchi EC, Marguerat SB, Bahler J, Smith GR, Jia S. 2011. Elimination of a specific histone H3K14 acetyltransferase complex bypasses the RNAi pathway to regulate pericentric heterochromatin functions. Genes Dev 25: 214–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbauer U, Zolghadr K, Muyldermans S, Schepers A, Cardoso MC, Leonhardt H. 2008. A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol Cell Proteomics 7: 282–289. [DOI] [PubMed] [Google Scholar]
- Rusche LN, Kirchmaier AL, Rine J. 2003. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu Rev Biochem 72: 481–516. [DOI] [PubMed] [Google Scholar]
- Sadaie M, Iida T, Urano T, Nakayama J. 2004. A chromodomain protein, Chp1, is required for the establishment of heterochromatin in fission yeast. EMBO J 23: 3825–3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadaie M, Kawaguchi R, Ohtani Y, Arisaka F, Tanaka K, Shirahige K, Nakayama J. 2008. Balance between distinct HP1 family proteins controls heterochromatin assembly in fission yeast. Mol Cell Biol 28: 6973–6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Fondufe-Mittendorf Y, Chen L, Thastrom A, Field Y, Moore IK, Wang JP, Widom J. 2006. A genomic code for nucleosome positioning. Nature 442: 772–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M. 1997. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev 11: 83–93. [DOI] [PubMed] [Google Scholar]
- Sugiyama T, Cam HP, Sugiyama R, Noma K, Zofall M, Kobayashi R, Grewal SI. 2007. SHREC, an effector complex for heterochromatic transcriptional silencing. Cell 128: 491–504. [DOI] [PubMed] [Google Scholar]
- Tadeo X, Wang J, Kallgren SP, Liu J, Reddy BD, Qiao F, Jia S. 2013. Elimination of shelterin components bypasses RNAi for pericentric heterochromatin assembly. Genes Dev 27: 2489–2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert PB, Henikoff S. 2006. Spreading of silent chromatin: inaction at a distance. Nat Rev Genet 7: 793–803. [DOI] [PubMed] [Google Scholar]
- Tomita K, Cooper JP. 2008. Fission yeast Ccq1 is telomerase recruiter and local checkpoint controller. Genes Dev 22: 3461–3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuzon CT, Borgstrom B, Weilguny D, Egel R, Cooper JP, Nielsen O. 2004. The fission yeast heterochromatin protein Rik1 is required for telomere clustering during meiosis. J Cell Biol 165: 759–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Kallgren SP, Reddy BD, Kuntz K, Lopez-Maury L, Thompson J, Watt S, Ma C, Hou H, Shi Y, et al. 2012. Histone H3 lysine 14 acetylation is required for activation of a DNA damage checkpoint in fission yeast. J Biol Chem 287: 4386–4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Tadeo X, Hou H, Tu PG, Thompson J, Yates JR III, Jia S. 2013. Epe1 recruits BET family bromodomain protein Bdf2 to establish heterochromatin boundaries. Genes Dev 27: 1886–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Lawry ST, Cohen AL, Jia S. 2014a. Chromosome boundary elements and regulation of heterochromatin spreading. Cell Mol Life Sci 71: 4841–4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Tadeo X, Hou H, Andrews S, Moresco JJ, Yates JR III, Nagy PL, Jia S. 2014b. Tls1 regulates splicing of shelterin components to control telomeric heterochromatin assembly and telomere length. Nucleic Acids Res 42: 11419–11432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Reddy BD, Jia S. 2015. Rapid epigenetic adaptation to uncontrolled heterochromatin spreading. Elife 4: e06179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright JH, Gottschling DE, Zakian VA. 1992. Saccharomyces telomeres assume a non-nucleosomal chromatin structure. Genes Dev 6: 197–210. [DOI] [PubMed] [Google Scholar]
- Yamada T, Fischle W, Sugiyama T, Allis CD, Grewal SI. 2005. The nucleation and maintenance of heterochromatin by a histone deacetylase in fission yeast. Mol Cell 20: 173–185. [DOI] [PubMed] [Google Scholar]
- Yamazaki H, Tarumoto Y, Ishikawa F. 2012. Tel1(ATM) and Rad3(ATR) phosphorylate the telomere protein Ccq1 to recruit telomerase and elongate telomeres in fission yeast. Genes Dev 26: 241–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Mosch K, Fischle W, Grewal SI. 2008. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat Struct Mol Biol 15: 381–388. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.