Chen et al. generated a FIP200-4A mutant knock-in mouse model and found that specifically blocking FIP200 interaction with Atg13 abolishes autophagy in vivo. Analysis of the new mouse model showed that nonautophagic functions of FIP200 are sufficient to fully support embryogenesis by maintaining a protective role in TNFα-induced apoptosis. However, FIP200-mediated canonical autophagy is required to support neonatal survival and tumor cell growth.
Keywords: autophagy, FIP200, knock-in mutant mouse, embryogenesis, tumor growth
Abstract
Autophagy is an evolutionarily conserved cellular process controlled through a set of essential autophagy genes (Atgs). However, there is increasing evidence that most, if not all, Atgs also possess functions independent of their requirement in canonical autophagy, making it difficult to distinguish the contributions of autophagy-dependent or -independent functions of a particular Atg to various biological processes. To distinguish these functions for FIP200 (FAK family-interacting protein of 200 kDa), an Atg in autophagy induction, we examined FIP200 interaction with its autophagy partner, Atg13. We found that residues 582–585 (LQFL) in FIP200 are required for interaction with Atg13, and mutation of these residues to AAAA (designated the FIP200-4A mutant) abolished its canonical autophagy function in vitro. Furthermore, we created a FIP200-4A mutant knock-in mouse model and found that specifically blocking FIP200 interaction with Atg13 abolishes autophagy in vivo, providing direct support for the essential role of the ULK1/Atg13/FIP200/Atg101 complex in the process beyond previous studies relying on the complete knockout of individual components. Analysis of the new mouse model showed that nonautophagic functions of FIP200 are sufficient to fully support embryogenesis by maintaining a protective role in TNFα-induced apoptosis. However, FIP200-mediated canonical autophagy is required to support neonatal survival and tumor cell growth. These studies provide the first genetic evidence linking an Atg's autophagy and nonautophagic functions to different biological processes in vivo.
Autophagy is a highly conserved cellular process characterized by the formation of double-membraned autophagosomes followed by their fusion with lysosomes for the degradation of bulk cytoplasmic materials. Pioneering studies in yeast revealed a series of essential autophagy genes (Atgs) whose products act in several protein complexes required for the induction, initiation, and maturation of autophagosomes (He and Klionsky 2009; Nakatogawa et al. 2009). The corresponding mammalian homologs for many yeast Atgs have been identified. Knockout studies of these genes in mice have revealed important roles of autophagy in a variety of developmental, physiological, and disease processes (Levine and Kroemer 2008; Mizushima and Levine 2010; Mizushima and Komatsu 2011; Rubinsztein et al. 2011; White 2012; Jiang et al. 2015). While many studies showed that disruption of different Atgs led to similar phenotypes as a consequence of autophagy inhibition, other studies indicated differential roles of Atgs in particular biological functions. For example, deletion of beclin1 (mammalian homolog of yeast Atg6 gene, required for autophagy initiation) led to early embryonic lethality (Yue et al. 2003), whereas knockout of mammalian Atg5 or Atg7 (involved in autophagosome maturation) did not impair embryogenesis; Atg5- and Atg7-null mice survived until shortly after birth (Kuma et al. 2004a; Komatsu et al. 2005). The underlying mechanisms for such differential roles of Atgs in embryonic development and other biological and disease processes are largely unknown at present.
FIP200 (FAK family-interacting protein of 200 kDa) is the mammalian counterpart of the yeast Atg17 gene and functions as a component of the ULK1/Atg13/FIP200/Atg101 complex essential for autophagy induction (Hara et al. 2008; Ganley et al. 2009; Hosokawa et al. 2009a,b; Jung et al. 2009; Mercer et al. 2009). Extensive studies in various cell lines and mouse models demonstrated that FIP200 is indispensable for autophagy in all mammalian systems examined so far. Global knockout of FIP200 leads to late embryonic lethality caused by severe heart and liver degeneration (Gan et al. 2006). Conditional knockout of FIP200 in different tissues showed its diverse functions, such as promotion of mammary tumor development and progression and maintenance of hematopoietic and neural stem cells (Liang et al. 2010; Liu et al. 2010, 2013; Wei et al. 2011; Ma et al. 2013; Wang et al. 2013). Whereas some of the defects upon FIP200 deletion were also observed in mice with knockout of other Atgs, other phenotypes, such as embryonic lethality, were not shared by the loss of Atg5 or Atg7 as discussed above. These observations indicate that, despite being essential components of the canonical autophagy pathway, Atgs may have distinct nonautophagic roles. Indeed, FIP200 and beclin1 have been shown to interact with other proteins to regulate diverse cellular functions independently of or in addition to their roles in autophagy (Gan and Guan 2008; He and Levine 2010; Liu et al. 2011; Boya et al. 2013). Moreover, Atg7, which was thought to be exclusively involved in autophagy, has recently been shown to regulate cell cycle and cell death pathways independently of its E1-like enzymatic activity, which is essential for autophagy (Lee et al. 2012). Nevertheless, the potential nonautophagic functions of various Atgs with regard to their different roles in biological and disease processes in vivo (either alone or in combination with their function in canonical autophagy) are yet to be examined.
In this study, we set out to address these important questions using FIP200 as a model, taking advantage of its several well-characterized additional interactions with other cellular proteins besides its role in canonical autophagy (Gan and Guan 2008). We identified residues 582–585 (LQFL) in FIP200, required for its interaction with Atg13 and autophagy function in vitro. By generation and analysis of a FIP200 knock-in mouse model that mutates LQFL to AAAA in the endogenous FIP200 gene, we demonstrated that nonautophagic functions of FIP200 are sufficient to fully support embryogenesis by maintaining its protective function in TNFα-induced apoptosis, but the autophagy function of FIP200 is required to support neonatal survival and tumor cell growth. These studies provide significant mechanistic insights into the integration of canonical autophagy and nonautophagic functions in the regulation of developmental and disease processes.
Results
Identification of residues 582–585 LQFL in FIP200, required for its interaction with Atg13 and autophagy induction
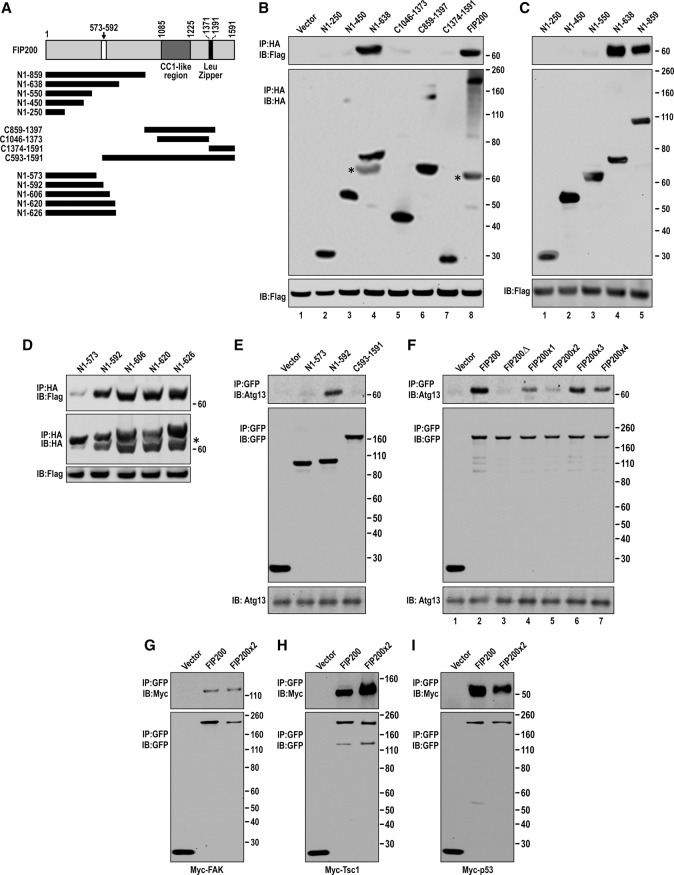
To study the respective contributions of FIP200-mediated autophagy and its potential autophagy-independent functions to various biological processes, we were interested in creating a specific FIP200 mutation that would abolish its role in canonical autophagy without affecting its interaction with other proteins. Given its involvement in the ULK1/Atg13/FIP200/Atg101 complex for autophagy induction, we first generated a series of overlapping FIP200 segments from both the N-terminal and C-terminal regions and tested them for binding to overexpressed Flag-Atg13 by coimmunoprecipitation (Fig. 1A–C). We found that the FIP200 N-terminal 638 residues (N1–638), but not N1–250 or N1–450, associated with Flag-Atg13 (Fig. 1B, lanes 1–4). In contrast, residues in the C-terminal region containing C1046–1373, C859–1397, or C1374–1591 did not bind to Flag-Atg13 (Fig. 1B, lanes 5–7). Further analysis with additional fragments showed that N1–638 as well as N1–859, but not N1–550, bound to Flag-Atg13 (Fig. 1C). This result suggested that the region between N551 and N638 is required for the interaction of FIP200 with Atg13. To further narrow down binding sites, we next analyzed another series of HA-tagged FIP200 fragments spanning the N terminus of FIP200 (see Fig. 1A). As shown in Figure 1D, the N1–573 segment, but not N1–592 or larger segments, exhibited significantly diminished binding to Flag-Atg13. A select set of the fragments was then analyzed as GFP fusion proteins for their binding to endogenous Atg13. Consistent with results using HA-tagged fragments, N1–592 bound to Atg13, whereas the smaller N1–573 or C593–1591 did not (Fig. 1E). We next created vectors encoding GFP-tagged FIP200 or the FIP200Δ mutant lacking residues between N573 and N592 and verified that this deletion mutant failed to bind to endogenous Atg13 in the full-length FIP200 context (Fig. 1F, lanes 2,3). Together, these data provide further support that residues between amino acids 573 and 592 (marked in Fig. 1A) are essential for FIP200 interaction with Atg13.
Figure 1.
Identification of residues 582–585 LQFL in FIP200, required for its interaction with Atg13 and autophagy induction. (A) Schematic of the FIP200 fragments used in B–E. A white box marks residues 573–592, required for Atg13 binding. (B–D) HEK293T cells were cotransfected with expression vectors encoding Flag-tagged Atg13 and HA-tagged FIP200 or its segments or vector alone as a control as indicated. Lysates were prepared and immunoprecipitated by anti-HA agarose beads followed by immunoblotting with antibodies as indicated. (E) The GFP-tagged FIP200 N-terminal or C-terminal segments indicated in A were transfected into HEK293T cells. The lysates were prepared and immunoprecipitated by anti-GFP antibody followed by immunoblotting with antibodies as indicated. (F) HEK293T cells were transfected with expression vectors encoding GFP-tagged-FIP200 or its mutants as indicated. Lysates were prepared and immunoprecipitated by anti-GFP followed by immunoblotting with antibodies as indicated. (G–I) GFP-tagged FIP200 or FIP200-4A mutant was cotransfected with Myc-FAK (G), Myc-Tsc1 (H), or Myc-P53 (I), respectively, into HEK293T cells. Lysates were prepared and immunoprecipitated with anti-GFP antibody followed by immunoblotting using antibodies as indicated. Please note that the bands marked by asterisks in the bottom panels of B and D are Flag-tagged Atg13 that was not stripped after immunoblotting using anti-Flag. Molecular weight markers (kilodaltons) are shown at the right.
To determine the specific residues within this region for Atg13 binding, we generated an additional set of point mutations in the full-length FIP200 context by alanine-scanning mutagenesis, each converting four residues from 578 and 593 to Ala (i.e., 578–581, 582–585, 586–589, and 590–593, designated as FIP200x1–x4, respectively). Expression vectors encoding this set of mutants with GFP tags were transfected into HEK293T cells to determine the interaction between FIP200 and Atg13 by coimmunoprecipitation. We found that the FIP200x2 mutation (LQFL to AAAA) showed significantly reduced binding to Atg13 to the level of the FIP200Δ mutant; the others had little or only moderate inhibition of the association (Fig. 1F). Importantly, the FIP200x2 mutant maintained binding to FAK (Fig. 1G), Tsc1 (Fig. 1H), and p53 (Fig. 1I), suggesting that mutation of residues 582–585 did not cause major structural alteration, a possible cause for disrupted Atg13 binding. These results identified the highly conserved residues 582–585 LQFL (Supplemental Fig. S1) of FIP200 as critical determinants of Atg13 binding. For simplicity, the FIP200x2 mutant disrupting binding to Atg13 was redesignated as the FIP200-4A mutant for the following studies.
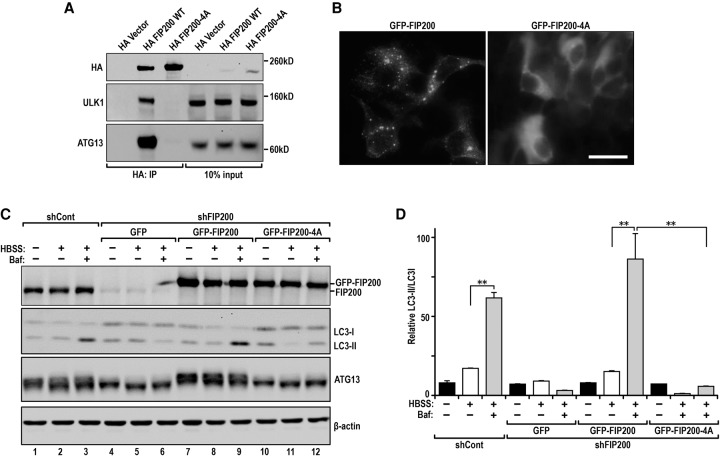
To evaluate the effect of disrupting the interaction between FIP200 and Atg13 on autophagy, we examined the interaction of the FIP200-4A mutant with another partner, ULK1, and its subcellular localization. Previous studies showed that Atg13 knockdown reduced association of ULK1 with FIP200, suggesting that Atg13 serves as a link in the ULK1/Atg13/FIP200/Atg101 complex to mediate ULK1 binding to FIP200 (Hosokawa et al. 2009a). Consistent with this finding, we found that the FIP200-4A mutant lost association with ULK1 (Fig. 2A). Furthermore, while wild-type GFP-FIP200 was detected in puncta (Hara et al. 2008), the GFP-FIP200-4A mutant only showed diffuse cytoplasmic localization in starved HEK293T cells (Fig. 2B). These results indicated that disruption of FIP200 binding to Atg13 could abolish the formation of the ULK1/Atg13/FIP200/Atg101 complex. To test whether the FIP200-4A mutation also inhibits autophagy, we analyzed LC3 conversion in FIP200 knockdown HEK293T cells with or without overexpression of the FIP200-4A mutant. As expected from previous findings in FIP200 knockout mouse embryonic fibroblasts (MEFs) (Hara et al. 2008), knockdown of FIP200 in HEK293T cells inhibited starvation-induced autophagy, as measured by reduced LC3 lipidation in the presence of bafilomycin A1 (BafA1) to inhibit degradation of LC3-II (Fig. 2C,D, cf. lanes 3 and 6). Re-expression of wild-type GFP-FIP200 restored the reduced LC3 lipidation in these cells (see Fig. 2C,D, lane 9). In contrast, re-expression of the GFP-FIP200-4A mutant did not rescue the defective autophagy in these cells (see Fig. 2C,D, lane 12). These results suggest that the FIP200-4A mutation disrupting FIP200 interaction with Atg13 abolishes autophagy.
Figure 2.
The FIP200-4A mutant unable to bind ATG13 is defective in amino acid starvation-induced autophagy. (A) HEK293T cells were transfected with expression vectors encoding HA-tagged FIP200 and FIP200-4A mutant or vector alone as a control as indicated. Lysates were prepared and immunoprecipitated by anti-HA followed by immunoblotting using anti-HA (top), anti-ULK1 (middle), or anti-Atg13 (bottom). (B) GFP-FIP200 (left) and GFP-FIP200-4A mutant puncta formation in starved HEK293T cells. (C,D) shRNA FIP200 knockdown HEK293T cells reconstituted with shRNA-resistant GFP-FIP200 or the GFP-FIP200-4A mutant were either left untreated or starved (Hank's balanced salt solution [HBSS]) for 2 h and then analyzed by immunoblotting with the indicated antibodies. (C) BafA1 (100 nM), an inhibitor of lysosome degradation, was included for 2 h in samples as indicated. (D) Quantification of LC3I-to-LC3II conversion in FIP200 knockdown HEK293T cells reconstituted with GFP-FIP200 and GFP-FIP200-4A upon starvation as shown in C. (**) P < 0.01.
Disruption of FIP200 interaction with Atg13 abolishes autophagy in vivo
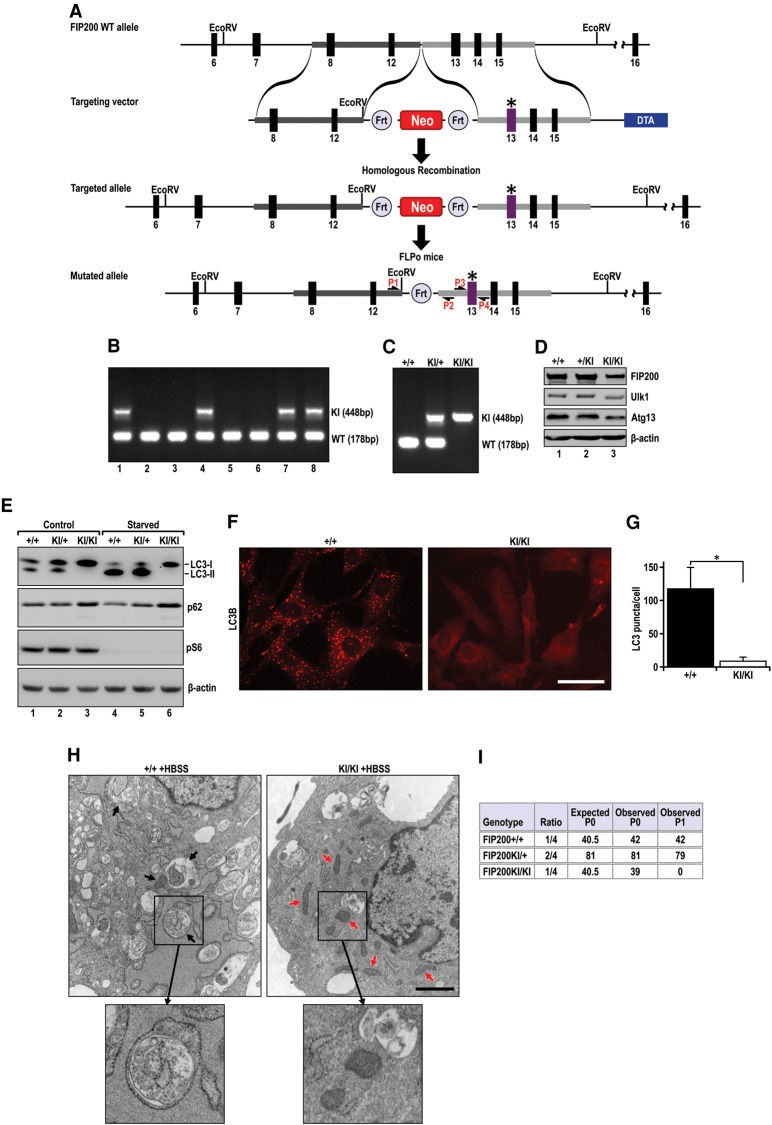
To assess the role of FIP200 interaction with Atg13 in the regulation of autophagy in vivo, we generated a FIP200 knock-in mutant mouse (designated FIP200+/KI) containing the FIP200-4A allele with the highly conserved residues 582–585 LQFL mutated to AAAA in the endogenous FIP200 gene using a homologous recombination-based gene knock-in approach as described previously (Fig. 3A; Zhao et al. 2010; Fan et al. 2013). Consistent with the results that both FIP200+/− and FIP200f/− mice are viable, fertile, and indistinguishable from wild-type mice (i.e., one functional FIP200 allele is sufficient for normal mouse development and function) (Gan et al. 2006; Wei et al. 2009), viable and apparently normal FIP200+/KI mice were obtained at Mendelian ratios (Fig. 3B). FIP200+/KI mice did not exhibit any apparent phenotypes under observation for up to 14 mo (data not shown).
Figure 3.
Disruption of FIP200 interaction with Atg13 blocks autophagy in MEFs. (A) Schematic representation of the FIP200-4A mutant knock-in targeting vector and the targeted allele of the FIP200-4A mutant. Solid boxes represent regions of vector homology with the target locus; thick, numbered boxes represent the exons flanking homology regions. Site-directed mutagenesis was used to introduce the LQFL-to-AAAA mutation at the 582–585 amino acid residues. The asterisk denotes the mutated residues on exon 13. (EcoRV) EcoRV sites; (neo) neomycin-resistant gene cassette; (DTA) diphtheria toxin gene. P1 and P2 are a pair of primers for genotyping. P3 and P4 are a pair of primers for sequencing amplified genomic DNA with mutations. (B) Genomic DNA was extracted from mouse tails and analyzed by PCR using P1 and P2 to detect the FIP200 wild-type allele (178 base pairs [bp]) or knock-in allele (448 bp). Representative genotypes of a litter of mice from crosses of FIP200+/KI mice with wild-type B6 mice. (C) Representative genotypes of MEF cells prepared from embryonic day 13.5 (E13.5) embryos of intercrosses of FIP200+/KI mice. (D) Lysates from different MEFs were analyzed by immunoblotting using antibodies against various proteins as indicated. (E) MEFs were incubated in fresh growth medium (control) or HBSS (starvation) in the presence of 100 nM BafA1 for 2 h. Lysates were then prepared and analyzed by immunoblotting using antibodies against proteins as indicated. (F) FIP200+/+ and FIP200KI/KI MEFs were starved in HBSS for 2 h, fixed, and stained for endogenous LC3B. Representative images are shown. Bar, 10 μm. (G) Quantification of LC3B puncta number in starved MEF cells. At least 50 cells per experiment were counted. (**) P < 0.01. (H) Representative transmission electron microscopy images of starved MEF FIP200+/+ and MEF FIP200KI/KI cells. Arrows indicate autophagosomal structures (left panel) and mitochondria (right panel), respectively. Bars, 500 nm. (I) Genotypes of progeny from FIP200+/KI intercrosses.
To examine whether the FIP200-4A mutation blocks autophagy in vivo, we first obtained and analyzed FIP200KI/KI MEFs as well as FIP200+/KI and FIP200+/+ MEFs as controls from embryos of intercrosses of FIP200+/KI mice (Fig. 3C). We noted slightly reduced expression levels of ULK1 and Atg13 as well as a small shift of mobility of these bands in FIP200KI/KI MEFs (Fig. 3D), possibly caused by their decreased phosphorylation and stability due to impaired interaction of FIP200-4A and Atg13 in mutant MEFs. Under normal growth conditions, in contrast to the presence of both LC3-I and LC3-II in wild-type and heterozygous knock-in MEFs, LC3-II was almost undetectable in homozygous knock-in MEFs (Fig. 3E, lanes 1–3), suggesting a significant decrease in basal autophagy upon disruption of FIP200 interaction with Atg13. Comparable mTORC1 activation was found in homozygous knock-in MEFs and controls, and starvation significantly inhibited its activation in all MEFs, as measured by pS6 levels in these cells (Fig. 3E, lanes 1–6). Upon starvation in the presence of lysosomal inhibitor BafA1, increased LC3-II was found compared with normal conditions in wild-type and heterozygous knock-in MEFs (Fig. 3E, cf. lanes 4,5 and lanes 1,2). However, LC3-II was still undetectable in homozygous knock-in MEFs, indicating a blockage of starvation-induced autophagy in these cells. We also found a slightly increased accumulation of p62 in homozygous knock-in MEFs compared with wild-type and heterozygous knock-in MEFs under both basal and starvation conditions (Fig. 3E). Moreover, immunofluorescent staining showed that the number of endogenous LC3 puncta was significantly decreased in starved homozygous knock-in MEFs compared with wild-type MEFs (Fig. 3F,G). Last, autophagosomes with double membranes (Fig. 3H, left panel, arrows) were observed in starved wild-type MEFs but not in homozygous knock-in MEFs under transmission electron microscopy. We also detected an increased number of mitochondria with heterogeneous sizes (Fig. 3H, right panel, red arrows) in homozygous knock-in MEFs, similar to our previous findings in FIP200-null postnatal neural stem cells (Wang et al. 2013).
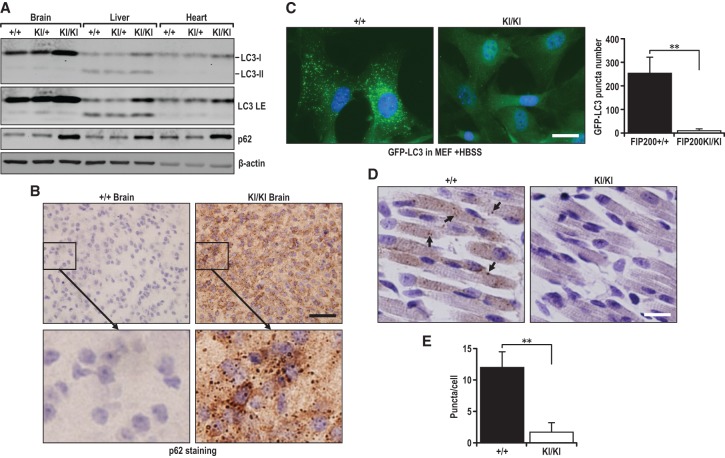
Intercrosses between FIP200+/KI mice also produced viable homozygous FIP200KI/KI offspring at the expected Mendelian ratios at postnatal day 0 (P0) (Fig. 3I). Genotyping of tail DNA verified the presence of both knock-in alleles in the P0 FIP200KI/KI pups (Supplemental Fig. S2A). Direct sequencing of the PCR-amplified DNA further confirmed the 582–585 LQFL-to-AAAA mutation of the knock-in allele (Supplemental Fig. S2B). We then examined autophagy by measuring LC3-I-to-LC3-II conversion and p62 accumulation in a number of tissues of neonatal mice. Analysis of lysates from the brain, liver, and heart showed increased amounts of LC3-I relative to LC3-II in homozygous knock-in mice compared with that in wild-type and heterozygous knock-in mice for all three tissues (Fig. 4A), suggesting decreased autophagy activity for LC3-I-to-LC3-II conversion. Consistent with these observations, increased p62 accumulation was found in these tissues of homozygous knock-in mice. Moreover, the accumulation of p62 aggregates was also readily detected in the brains of homozygous knock-in mice but not wild-type mice (Fig. 4B). To further assess the inhibition of autophagy by FIP200-4A mutation, we crossed FIP200+/KI mice with the GFP-LC3 reporter transgenic mice to generate FIP200KI/KI;GFP-LC3 mice for monitoring autophagy in live cells and in vivo directly, as described previously (Mizushima 2009). Analysis of MEFs from FIP200KI/KI;GFP-LC3 mice showed a lack of LC3 puncta compared with MEFs from control GFP-LC3 mice under starvation conditions (Fig. 4C). Furthermore, autophagosome puncta were greatly decreased in the muscles of neonate FIP200KI/KI;GFP-LC3 mice compared with control GFP-LC3 mice (Fig. 4D,E). Taken together, these results demonstrate that the FIP200-4A mutation that disrupts FIP200 interaction with Atg13 in the ULK1/Atg13/FIP200/Atg101 complex abrogates autophagy in vivo.
Figure 4.
FIP200KI/KI homozygous mice are defective in autophagy. (A) Various FIP200KI/KI neonatal brain, liver, and heart tissue lysates were analyzed by immunoblotting with the indicated antibodies. (B) Immunohistochemistry (IHC) staining of p62 of wild-type and homozygous FIP200 knock-in neonatal brains. (C) GFP-LC3 puncta in FIP200 wild-type and FIP200KI/KI MEFs after starvation with HBSS for 2 h. (D) Neonatal muscle sections from mice expressing GFP-LC3 were stained with anti-GFP antibody. (E) The puncta dot structures were quantified. (**) P < 0.01.
Nonautophagic functions of FIP200 are sufficient to fully support embryogenesis by maintaining its protective function in TNFα-induced apoptosis
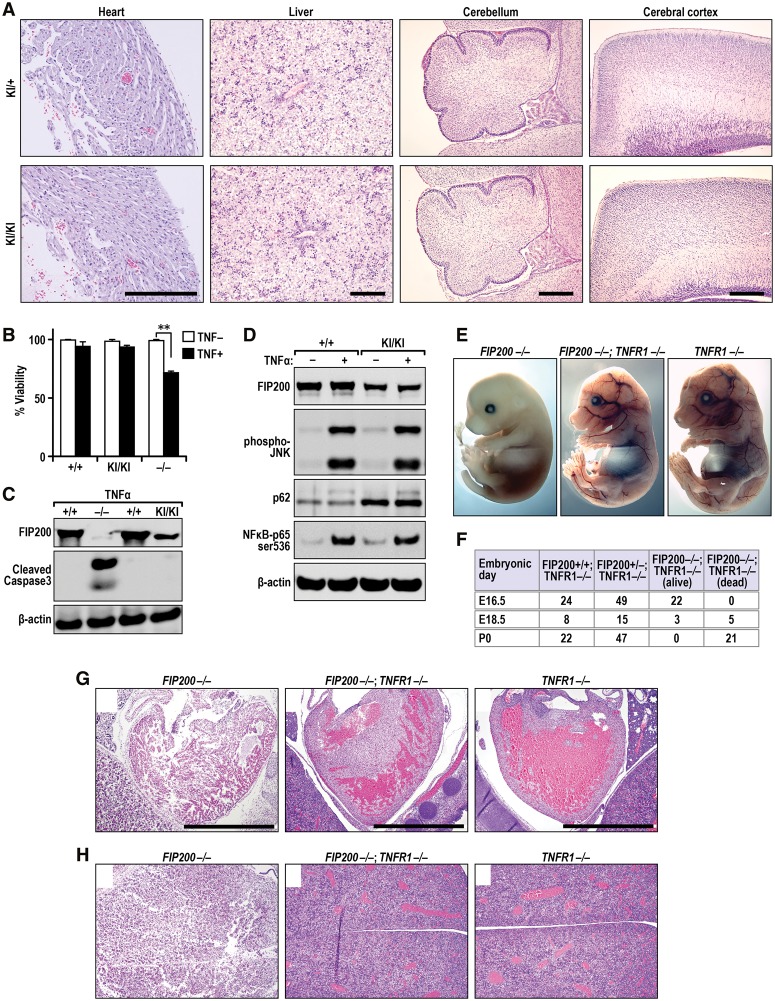
Previous studies have shown that FIP200 knockout results in embryonic lethality by embryonic day 16.5 (E16.5) (Gan et al. 2006). Therefore, our findings of the viable FIP200KI/KI pups born at Mendelian ratios (see Fig. 3I) suggest that nonautophagic functions of the FIP200-4A mutant were able to rescue the embryonic lethality of FIP200 knockout mice. Indeed, in contrast to the extensive degeneration of liver and heart tissues observed in FIP200 knockout embryos (Gan et al. 2006), H&E staining of heart, liver, and brain tissues of homozygous knock-in pups at P0 were indiscernible from FIP200+/KI controls (Fig. 5A) despite deficient autophagy in these tissues (see Fig. 4A,B). In addition to functioning in the ULK1/Atg13/FIP200/Atg101 complex for autophagy induction, FIP200 has been shown to interact with other proteins to regulate various cellular functions (Gan and Guan 2008). Previous studies showed that FIP200 knockout MEFs were sensitized to TNFα-induced cell death, which may account for the extensive apoptosis in the liver and heart, leading to embryonic lethality of FIP200 knockout mice by E16.5 (Gan et al. 2006). However, we did not detect any significant apoptotic cells in the livers and hearts of homozygous knock-in mice (data not shown), raising the interesting possibility that the FIP200-4A mutant may rescue the embryonic-lethal phenotype by maintaining its protective function in TNFα-induced apoptosis. To test such a possibility, we examined the responses of homozygous knock-in MEFs to TNFα stimulation. We found that, in contrast to the increased sensitivity to TNFα-induced apoptosis in knockout MEFs (Gan et al. 2006), treatment with TNFα did not cause significant cell death of homozygous knock-in MEFs as measured by either cell viability or cleaved caspase 3 levels (Fig. 5B,C). Previous studies showed that reduced JNK activation in knockout MEFs in response to TNFα stimulation was responsible for their decreased survival (Gan et al. 2006). However, homozygous knock-in MEFs showed a level of JNK activation similar to that of wild-type MEFs upon TNFα treatment (Fig. 5D), providing further support that nonautophagic functions of FIP200 were able to protect the cells from TNFα-induced apoptosis.
Figure 5.
Normal morphology of FIP200KI/KI pups at P0 and normal heart and liver development in FIP200−/−;TNFR1−/− embryos at E16.5. (A) Histological sections from the hearts, livers, cerebella, and cerebral cortexes of FIP200+/KI (top panels) and FIP200KI/KI (bottom panels) pups at P0 were analyzed by H/E staining. Bars, 500 µm. (B,C) MEFs from FIP200+/+, FIP200−/−, and FIP200KI/KI (knock-in) embryos were treated for 1 d with or without 50 ng/mL TNFα as indicated. They were analyzed for viability by Trypan blue assay (B) or immunoblotting using antibodies as indicated (C). (D) MEFs from FIP200+/+ (wild-type) and FIP200KI/KI (knock-in) embryos were treated for 10 min with or without 50 ng/mL TNFα as indicated. They were analyzed by immunoblotting using antibodies as indicated. (E) Gross examination of whole-mount FIP200−/−, FIP200−/−;TNFR1−/−, and TNFR1−/− embryos at E16.5. (F) Genotypes of progeny from FIP200+/−,TNFR1−/− intercrosses. (G,H) Histological sections from the hearts (G) and livesr (H) of FIP200−/−, FIP200−/−;TNFR1−/−, and TNFR1−/− embryos at E16.5. Bar, 1 mm.
Previous studies suggested that the TNFα pathway plays a crucial role during embryogenesis and is involved in embryonic lethality caused by a variety of abnormalities in different knockout mice (Dillon et al. 2014; Peltzer et al. 2014). To investigate directly whether increased sensitivity to TNFα-induced cell death is responsible for the embryonic lethality of FIP200 knockout mice, we generated FIP200−/−;TNFR1−/− embryos by crossing FIP200+/− (Gan et al. 2006) and TNFR1−/− (Peschon et al. 1998) mice and examined the survival probability of the embryos. In contrast to the complete lethality of FIP200−/− embryos (Gan et al. 2006), FIP200−/−;TNFR1−/− embryos were viable at E16.5 with near-Mendelian ratios (Fig. 5E,F). A fraction of viable FIP200−/−;TNFR1−/− embryos was even found at E18.5, but no FIP200−/−;TNFR1−/− pups were born at P0 (data not shown). At E16.5, the hearts and livers of FIP200−/− embryos were irregular and degenerated. The myocardiocytes and hepatocytes were undergoing apoptosis, as suggested by condensed or fragmented nuclei (Fig. 5G,H). This is consistent with our previous study that showed that the significant defects in the formation and development of the myocardium and liver were the most likely causes of the embryonic lethality in FIP200 knockout embryos (Gan et al. 2006). The hearts and livers of TNFR1−/− and FIP200−/−;TNFR1−/− embryos were normal at E16.5 (Fig. 5G,H). Moreover, FIP200−/−;TNFR1−/−, TNFR1−/−, and control FIP200+/+ embryos showed comparable levels of apoptosis (Supplemental Fig. S3), suggesting that TNFR1 deletion rescued the increased apoptosis caused by FIP200 inactivation (Gan et al. 2006).
Together, these results demonstrated that FIP200 knockout caused embryonic lethality by E16.5 due to abnormalities in the TNFα pathway (Gan et al. 2006), which could be rescued by blockade of TNFα signaling through TNFR1 ablation. The results also suggested that nonautophagic functions of the FIP200-4A mutant allowed passage of this stage controlled by TNFα signaling and fully supported the completion of embryogenesis until birth at least partially by maintaining the FIP200 protective function in TNFα-induced apoptosis.
The autophagy function of FIP200 is required to support neonatal survival and tumor cell growth in vivo
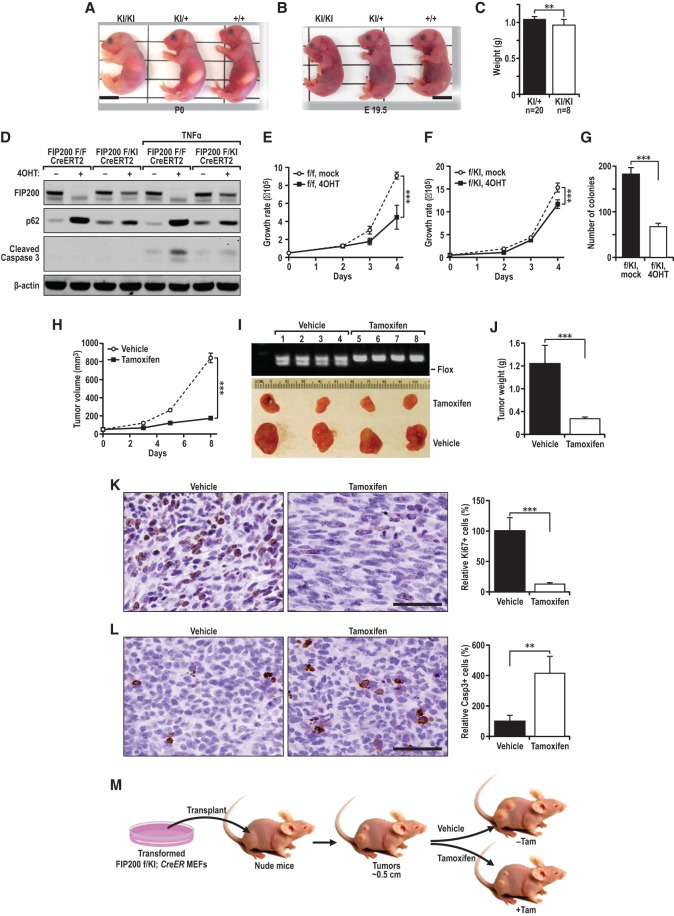
Although they survived at birth, all homozygous knock-in pups died within 24 h after birth (see Fig. 3I), suggesting that the autophagy function of FIP200 is still required for the survival of neonates. We noted that homozygous knock-in neonates lacked milk in their stomachs and were slightly smaller than the FIP200+/KI and FIP200+/+ control littermates (Fig. 6A), which were similar to the phenotypes of Atg5 knockout mice described previously (Kuma et al. 2004b). Analysis of E19.5 embryos obtained by cesarean section also revealed a slightly smaller size as well as reduced weight of homozygous knock-in embryos compared with the FIP200+/KI and FIP200+/+ control littermates (Fig. 6B,C). These results suggest that specific blockage of FIP200 autophagy function leads to neonatal lethality due to insufficient sucking as well as potentially other nutritional deficiencies, as proposed previously for the Atg5 knockout mice (Kuma et al. 2004b).
Figure 6.
The autophagy function of FIP200 is required to support neonatal survival and tumor cell growth in vivo. (A) Representative morphology of a FIP200KI/KI homozygous neonate compared with its wild-type littermates at P0. (B) Representative neonates of the indicated genotypes delivered by cesarean section at E19.5. (C) The mean body weight of FIP200+/KI and FIP200KI/KI mice. (D) MEF FIP200flox/flox or FIP200flox/KI CreERT2 cells (transformed by E1A/Ras) were treated with 10 nM 4-OHT or control for 72 h followed by addition of 50 ng/mL TNFα for 12 h. Cell lysates were then prepared and analyzed by immunoblotting using various antibodies as indicated. (E–G). The cells treated with 4-OHT as in D were used for proliferation assay (E,F) and anchorage-independent growth assay in soft agar (G). (H–M).Transformed FIP200f/KI;CreERT2 MEFs were injected subcutaneously into athymic nude mice. (H) Animals (n = 4 each) were treated with vehicle control (−TAM [tamoxifen]; dotted line) or TAM (+TAM; solid line) by intraperitoneal (i.p.) injection, and single tumor growth was measured at the indicated time points. Data points represent means of tumor volume. Tumor burden images (I) and average tumor weight (J) at the final time point are shown. Tumor sections were harvested from the recipient mice at the final time point and analyzed by IHC using anti-Ki67 antibody (K) or anti-cleaved caspase 3 antibody (L). (M) Schematics for the deletion of floxed FIP200 induced by TAM after transplantation of transformed FIP200f/KI;CreERT2 MEFs as described in the Materials and Methods. Bars, 100 μm. (**) P < 0.01; (***) P < 0.001.
Previous studies have suggested that autophagy could have either tumor-suppressive or tumor-promoting functions (White 2012; Kenific and Debnath 2015). Although different models and contexts may explain the differential outcomes of deleting different Atgs, an alternative explanation is that some of the effects were due to the loss of nonautophagic (thus different) functions of these genes. Taking advantage of the specific blockage of its autophagy function by the FIP200-4A mutation, we examined whether the loss of FIP200 autophagy function alone is sufficient to inhibit tumor growth of transformed MEFs, as found in FIP200-null transformed MEFs in our recent report (Wei et al. 2014). MEFs were isolated from embryos with the FIP200KI/f genotype from intercrosses between FIP200+/KI and FIP200f/f mice and then transformed by E1A/H-RasV12 and infected with MSCV.CreERT2, as described previously (Wei et al. 2014). The resulting transformed FIP200KI/f CreER MEFs were first examined for their proliferation in vitro compared with transformed FIP200f/f CreER MEFs (Wei et al. 2014). As expected, treatment of transformed FIP200f/f CreER MEFs with 4-OHT to induce FIP200 deletion inhibited autophagy, as measured by increased p62 accumulation (Fig. 6D). Similar treatment of transformed FIP200KI/f CreER MEFs induced deletion of the floxed FIP200 allele, leaving only the knock-in allele encoding FIP200-4A, and also reduced autophagy as seen by increased p62, although to a lesser extent than FIP200 knockout cells. In addition, similar to our observations in nontransformed MEFs, TNFα-induced cell death was increased in FIP200-null transformed MEFs (i.e., transformed FIP200f/f CreER MEFs after 4-OHT treatment) but not appreciably in transformed MEFs with the FIP200-4A mutation (i.e., transformed FIP200KI/f CreER MEFs after 4-OHT treatment). Interestingly, both FIP200 deletion and FIP200-4A mutation (although to a lesser extent) reduced tumor cell proliferation (Fig. 6E,F). FIP200-4A mutation also reduced the colony-forming efficiency of the transformed MEFs in vitro (Fig. 6G).
To examine specifically the role of FIP200 autophagy function in tumor growth, transformed FIP200KI/f CreER MEFs were transplanted into nude mice to allow tumor growth until ∼0.5 cm in diameter followed by tamoxifen (TAM) treatment to induce deletion of the floxed FIP200 allele, as described previously (Wei et al. 2014). We found that conversion to the FIP200-4A mutation (encoded by the knock-in allele after the deletion of the floxed allele) significantly inhibited tumor growth in vivo (Fig. 6H–J). Staining of tumor sections for Ki67 and cleaved caspase 3 showed reduced numbers of proliferative cells and increased apoptotic cells following TAM treatment compared with vehicle treatment (Fig. 6K,L). These results demonstrated that the autophagy function of FIP200 is required to promote tumor growth in vivo (Fig. 6M).
Discussion
Autophagy is orchestrated by a set of essential Atgs in several protein complexes, including the ULK1/Atg13/FIP200/Atg101 complex for autophagy induction in mammalian cells (Hara et al. 2008; Ganley et al. 2009; Hosokawa et al. 2009a,b; Jung et al. 2009; Mercer et al. 2009). Building on the elegant mutational analyses in yeast demonstrating the requirement for each gene in autophagy, knockout studies in mice showed essential roles of these genes in autophagy in mammals, revealing a multitude of functions of autophagy in mammalian development as well as various physiological and disease processes (Levine and Kroemer 2008; Mizushima and Levine 2010; Mizushima and Komatsu 2011; Rubinsztein et al. 2011; White 2012; Jiang et al. 2015). However, the interpretation of the observations from knockout studies to link autophagy (or particular Atgs) to various phenotypes is confounded by the increasing evidence that many, if not all, of these genes perform other functions besides their role in the canonical autophagy pathway (Subramani and Malhotra 2013). Moreover, observations that interactions among the components of the ULK1/Atg13/FIP200/Atg101 complex were not affected during starvation-induced autophagy raises additional questions about whether these components function as a complex or separately (alone or possibly through interaction with other proteins). In this study, by determining the critical residues in FIP200 for binding to Atg13 and creating a FIP200 knock-in mutant allele in mice, we identified distinct roles of the FIP200 canonical autophagy function and its autophagy-independent function in embryogenesis, cell survival, and tumor growth in vivo. These results also demonstrate that while FIP200 possesses nonautophagic functions, its canonical autophagy function depends on its interaction with Atg13 and therefore provide direct support for the essential role of the ULK1/Atg13/FIP200/Atg101 complex formation in vivo.
We found that, unlike the embryonic lethality in the FIP200 knockout mice, the FIP200-4A mutation allowed the completion of embryogenesis until birth. This clearly established a biological role for an autophagy-independent function of FIP200. These results shed significant light on the previous puzzling observations that knockout of some Atgs, like FIP200 and beclin1, caused embryonic lethality, but ablation of Atg5 and Atg7 did not (Yue et al. 2003; Kuma et al. 2004a; Komatsu et al. 2005; Gan et al. 2006; Mizushima et al. 2010). They implicate that the embryonic-lethal phenotype of beclin1 knockout and perhaps knockout of some other Atgs could be caused by the loss of functions other than their essential role in canonical autophagy. Future studies will be necessary to examine these possibilities that would support a broader conclusion that canonical autophagy is not required for embryogenesis.
Although FIP200 interacts with several other proteins and potentially regulates different intracellular signaling pathways (Gan and Guan 2008), our results suggest that nonautophagic functions of FIP200 (i.e., retained in the FIP200-4A mutation) supported embryogenesis by maintaining its protective function from TNFα-induced cell death. Like FIP200-4A homozygous knock-in mice, ablation of TNFR1 in FIP200 knockout mice prevented the lethality by E16.5 observed previously (Gan et al. 2006). These results are consistent with recent reports suggesting a critical role of the TNFα pathway as a “checkpoint” during embryogenesis that triggers embryonic lethality in response to various defects of knockout animals (Dillon et al. 2014; Peltzer et al. 2014). On the other hand, whereas FIP200-4A homozygous knock-in mice survived until birth, TNFR1 knockout did not allow completion of embryogenesis of FIP200 knockout mice. Therefore, other nonautophagic functions of FIP200 may also contribute to its role in supporting embryogenesis. The current mouse model and MEFs derived from these mice can be used to further characterize additional nonautophagic functions and the respective contributions of canonical autophagy and nonautophagic functions to roles of FIP200 in vivo. It will also be interesting to use this model to examine the premise that FIP200 serves as an intracellular node to coordinate the induction of autophagy and other signaling pathways. The FIP200-4A mutant knock-in mice in which FIP200 interactions with FAK, Tsc1, and p53 are maintained will be a valuable resource to pursue potential cross-talk of signaling pathways with autophagy in future studies.
The perinatal lethality of the FIP200-4A mutant knock-in mice is consistent with previous observations for Atg5 knockout mice (Kuma et al. 2004a) and further validates a role for autophagy in postnatal survival and growth. Our analysis of the FIP200-4A mutant in transformed MEFs provides direct support for a role of FIP200-mediated autophagy in tumor cell growth. Because previous studies using conditional knockout of various Atgs, including FIP200, have shown a role for autophagy to promote or suppress tumorigenesis in different systems (Kenific and Debnath 2015; White 2015), the availability of the FIP200-4A mutant specifically disrupting its autophagy function will allow us to clarify whether some of the conflicting results could be caused by the potential loss of nonautophagic functions associated with FIP200 and/or other Atgs. In the context of breast cancer, we found previously that FIP200 ablation inhibited breast cancer development and progression through both decreasing growth of tumor cells and increased immune surveillance (Wei et al. 2011). However, it should be noted that the current study evaluating tumor growth in vivo was performed in immunocompromised nude mice. Therefore, it will be particularly interesting to determine whether FIP200-mediated autophagy and its nonautophagic functions differentially contribute to promoting tumor cell growth and preventing the increased immune surveillance by analyzing the effect of the FIP200-4A mutation in breast cancer models in future studies. Likewise, the availability of the FIP200-4A knock-in mice and cells derived from these mice will help to clarify the role of FIP200 in other systems, such as its role in the maintenance of neural stem cells (Wang et al. 2013).
In addition to significant insights into the physiological role of both autophagy-dependent and -independent functions of FIP200 in vivo, our studies also shed light on the interaction of various components in the ULK1/Atg13/FIP200/Atg101 complex. Our studies suggested the possibility of Atg13 serving as a link for association of ULK1 and FIP200. Interestingly, a recent study showed that an Atg13 mutant lacking binding to ULK1/2 was capable of partially restoring autophagy in Atg13-deficient cells (Hieke et al. 2015), suggesting that this interaction is not essential for autophagy induction. On the other hand, Atg101 interaction with Atg13 has been shown to play an essential role in autophagy through analysis of an Atg101 mutant lacking binding to Atg13 (Suzuki et al. 2015). These data are consistent with the idea that Atg13 acts as a central hub for the complex, whereas the other components may play a regulatory role within the complex (e.g., ULK1/2 as a kinase that phosphorylates other components in the complex) (Hosokawa et al. 2009a; Jung et al. 2009) or connecting to other components of the autophagy machinery (e.g., Atg101 for downstream factors) (Suzuki et al. 2015) or to intracellular signaling pathways (e.g., FIP200 to TNFα signaling, as suggested here). Interestingly, a very recent study reported that Atg13 knockout mice showed some phenotypes similar to that of FIP200 knockout mice (i.e., embryonic lethality and hypersensitivity to TNFa) (Kaizuka and Mizushima 2015). Together with the present study, these data raise the interesting possibility that nonautophagic functions of both Atg13 and FIP200 are important for embryonic survival and suppression of TNFα sensitivity even though their direct binding is not essential. However, we cannot exclude an alternative explanation that a possible weak interaction of the FIP200-4A mutant with Atg13 (see Fig. 1F) may be sufficient to exert the nonautophagic functions but not the autophagic functions. Future studies with Atg13 and/or other FIP200 mutants that completely block their interactions will be interesting to explore these possibilities. Likewise, similar approaches for components of the other autophagy complexes will significantly enhance our overall understanding of autophagy processes and biological functions in vivo. For example, analysis of beclin1 mutants disrupting its interaction with other proteins in the beclin1 complex specifically for autophagy function will help to delineate the relative contribution of autophagy-dependent and -independent functions to its tumor-suppressive roles in many cancers.
Materials and methods
Generation of FIP200 knock-in mice
Genomic regions of mouse FIP200 were PCR-amplified from C57BL/6J genomic DNA (Jackson Laboratory, 000664) by high-fidelity PCR (Invitrogen high-fidelity PCR system) and cloned into PGKneoF2L2DTA (Addgene, plasmid no. 13445) (Hoch and Soriano 2006). A 3.3-kb genomic sequence (3′ targeting arm) was cloned into the pENTR-SD vector (Invitrogen), and site-directed mutagenesis was used to produce the 582–585 LQFL-to-AAAA substitution in the exon 13 coding region of mouse genomic FIP200. The 3′ arm with mutations was PCR-amplified, digested with NheI and SalI, and cloned downstream from the 3′ loxP sequence in pGKneoF2L2DTA. The 3.2-kb genomic sequence (5′ targeting arm) was amplified by PCR, digested with SacII and NotI, and cloned upstream of the 5′ loxP sequence. The resulting pGKneo-5arm-F2L2-3armKI-DTA vector was verified by restriction mapping and DNA sequencing. Subsequently, the targeting vector was linearized and electroporated into C57BL/6 embryonic stem (ES) cells in the University of Michigan Transgenic Animal Model Core. The neomycin-resistant ES clones were first selected by G418 and screened for correct gene targeting by PCR and Southern blot. Three positive ES clones were then microinjected into C57BL/6 blastocysts to produce chimeras. Following mating of chimeric mice with albino C57BL/6 mice, PCR and sequencing of mouse genomic DNA were used to confirm germline transmission of the FIP200 knock-in mutation. The neomycin cassette was removed from the neo-FIP200+/KI mice by mating with FLPo transgenic mice on a C57BJ/6 background.
Mice, genotyping, and cell culture
FIP200f/f mice (Gan et al. 2006) were used to cross with FIP200+/KI mice to produce FIP200f/KI mice. FIP200−/− mice were generated as described previously (Gan et al. 2006). TNFR1−/− mice were obtained from The Jackson Laboratory. To generate FIP200KI/KI mice with GFP-LC3, we crossed FIP200+/KI mice with GFP-LC3 transgenic mice (Mizushima 2004). PCR was used to identify the FIP200 knock-in allele using primers P1 (5′-GCCGCTTCATTATTTGAGTCAGAGTAGCTTG-3′) and P2 (5′-GGTGGAAGGACATACCATTTCCTAAATTTCTTTG-3′). Mice were housed and handled according to local, state, and federal regulations, and all experimental procedures were carried out according to the guidelines of the Institutional Animal Care and Use Committee at University of Michigan and University of Cincinnati. Genotyping was performed by PCR analysis of ear or tail DNA.
HEK293T and MEF cells were maintained in DMEM supplemented with 10% FBS. Cell viability was measured by Trypan blue exclusion.
Preparation of primary and transformed MEFs capable of inducible deletion of floxed FIP200
E13.5 embryos were isolated from the timed intercrosses of FIP200f/f mice with FIP200f/KI mice and used for the preparation of MEFs with FIP200f/f and FIP200f/KI genotypes essentially as described previously (Wei et al. 2011). E1A/H-RasV12 transformed MEF FIP200flox/KI cells were generated as previously described (Wei et al. 2011) and subsequently infected by MSCV.CreERT2.puro or MSCV vector alone (Kumar et al. 2009) to produce transformed FIP200f/KI;CreERT2 MEFs, as described previously (Ventura et al. 2007). In some experiments, the transformed MEFs were treated with 4-OHT (Sigma) to delete the floxed FIP200 allele via Cre induction, as described (Ventura et al. 2007). In other experiments, these cells were transplanted into recipient nude mice followed by treatment with TAM to delete the floxed FIP200 allele via Cre induction in vivo (see the details below).
Tumor cell transplantation, treatment with TAM, and tumor size monitoring
Transformed MEFs cells were harvested, washed twice in DMEM, and then injected (5 × 105 cells in 1:1 PBS:Matrigel [BD Bioscience]) into the flank region of 7-wk-old female athymic nude mice. After tumors appeared, tumor size was monitored, as described previously. Once the tumors reached ∼0.5 cm in diameter, mice were randomly assigned and continuously treated for 3 d with 100 µL of 10 mg/mL TAM in corn oil or with an equal volume of vehicle. Tumor size was then measured at the indicated time points. Tumor volume was calculated using the formula volume = (width2 × length)/2, as described previously (Wei et al. 2011).
DNA expression and knockdown
The expression vectors pKH3-HA-FIP200, pKH3-HA-FIP-NT, pKH3-HA-FIP-MD, pKH3-HA-FIP-CT, and pHAN-Myc-FAK (Abbi and Guan 2002); pHAN-myc-p53 and pEGFP-FIP200 (Melkoumian et al. 2005); and pHAN-TSC1 (Gan et al. 2005) have been described previously. All other HA-FIP200 and GFP-FIP200 mutants were generated by PCR-based site-directed mutagenesis. p3xFlag-CMV10-hAtg13 was a gift from Noboru Mizushima (Addgene, plasmid no. 22872). Human FIP200 shRNA(V2LHS_229754) was obtained from Open Biosystems. Transient transfection of HEK293T cells was performed using Lipofectamine 2000 (Life Technologies, 11668019). For the production of recombinant retroviruses, HEK293T cells were transfected with pMSCVpuro retroviral vectors with packaging vector using Lipofectamine 2000 (Life Technologies, 11668019). MEF cells were incubated with retroviral supernatant fractions containing 6 µg/mL polybrene (Sigma-Aldrich, H9268-106) and selected in medium containing 1 µg/mL puromycin.
Immunoprecipitation and immunoblotting
To test the interaction of FIP200 fragments and Atg13, HEK293T cells were cotransfected with expression vectors encoding Flag-tagged Atg13 and HA-tagged FIP200 or its segments. Lysates were prepared with immunoprecipitation lysis buffer (25 mM Tris-HCl at pH 7.5, 150 mM NaCl, 1% NP-40 with Halt protease inhibitor cocktail) for immunoprecipitation followed by immunoblotting, as described previously (Wei et al. 2011). In other experiments, total neonatal brain, liver, and heart lysates were prepared by homogenizing frozen tissue in RIPA buffer (50 mM Tris, 150 mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate, 1% Nonidet P-40, protease inhibitor mixture; Roche). Lysates were then resolved by SDS-PAGE and analyzed to detect proteins of interest. The following antibodies were used: monoclonal anti-HA-agarose (Sigma, A2095), rabbit anti-HA (Santa Cruz Biotechnology, sc-805), rabbit anti-Flag (SAB) (Signalwayantibody, T503), rabbit anti-GFP (Invitrogen, A11222), mouse anti-Myc (Santa Cruz Biotechnology, sc-40), rabbit anti-ULK1 (Cell Signaling, 8054), rabbit anti-Atg13 (Cell Signaling, 6940), rabbit anti-FIP200 (Ptglab, 17250), rabbit anti-LC3B (Cell Signaling, 2775), rabbit anti-P62 (Cell Signaling, 5114), rabbit anti-Ser240/244 phospho-S6 (Cell Signaling, 5364), rabbit anti-cleaved caspase 3 (Cell Signaling, 9661), rabbit anti-phospho-JNK (Thr183/Tyr185) (Cell Signaling,4668), and rabbit anti-phospho-NF-κB p65 (Ser536) (Cell Signaling, 3033). Goat anti-mouse IgG-HRP and goat anti-rabbit IgG-HRP (Jackson Immunology) were used as secondary antibodies. Bands were visualized with an ECL detection kit (Advansta).
Immunohistochemistry (IHC), immunofluorescence (IF), TUNEL assay, and transmission electron microscopy
Tumor samples were fixed overnight in 10% phosphate-buffered formalin (Fisher Scientific) and embedded in paraffin. They were then sectioned at a 5-μm depth and subjected to IHC or IF as previously described (Wei et al. 2009, 2011). The primary antibodies used included p62 (Enzo Life Sciences), anti-Ki67 (Spring Bioscience), and anti-cleaved caspase 3 (Cell Signaling, 9661). Sections were counterstained with hematoxylin for IHC. For indirect IF, MEF cells were grown on eight-well Lab-Tek II chamber slides (Nalge Nunc International), fixed with 100% cold methanol, and blocked in PBS containing 2% BSA. Primary antibodies against LC3B (1:200; Cell Signaling, 3868) were used. Alexa fluor 594-conjugated chicken anti-rabbit IgG (H+L) antibody from Life Technologies (A-21442) was used for detection. Apoptotic cells in E16.5 liver and heart sections were detected by TUNEL assay according to the protocol provided by the manufacturer with the in situ cell death detection kit TMR Red (Roche). For transmission electron microscopy, MEF cells were prepared and analyzed as described previously (Liang et al. 2010).
Statistical analysis
Statistical significance was evaluated by unpaired t-test using P < 0.05 as indicative of statistical significance.
Supplementary Material
Acknowledgments
We are grateful to Dr. Thom Saunders, Dr. Zilin Nie, Dr. Dongping Wei, and Dr. Kaiyu Ma at the University of Michigan Transgenic Core for help in the generation of FIP200-4A mutant knock-in mice, and Dr. Noboru Mizushima at the University of Tokyo for providing the GFP-LC3 reporter transgenic mice. We thank our colleagues Dr. Maria Czyzyk-Krzeska and Dr. Ana Mesquite for their critical reading and helpful comments of the manuscript. We thank Glenn Doerman for figure preparation, and Dr. Belinda Peace for editing the manuscript. We also thank Nigel Muhammad and Mike Haas for their assistance. This research was supported by National Institutes of Health grants R01CA163493, R01HL073394, and R01NS094144 to J.-L.G.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.276428.115.
References
- Abbi S, Guan JL. 2002. Focal adhesion kinase: protein interactions and cellular functions. Histol Histopathol 17: 1163–1171. [DOI] [PubMed] [Google Scholar]
- Boya P, Reggiori F, Codogno P. 2013. Emerging regulation and functions of autophagy. Nat Cell Biol 15: 713–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon CP, Weinlich R, Rodriguez DA, Cripps JG, Quarato G, Gurung P, Verbist KC, Brewer TL, Llambi F, Gong YN, et al. 2014. RIPK1 blocks early postnatal lethality mediated by caspase-8 and RIPK3. Cell 157: 1189–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H, Zhao X, Sun S, Luo M, Guan JL. 2013. Function of focal adhesion kinase scaffolding to mediate endophilin A2 phosphorylation promotes epithelial–mesenchymal transition and mammary cancer stem cell activities in vivo. J Biol Chem 288: 3322–3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan B, Guan JL. 2008. FIP200, a key signaling node to coordinately regulate various cellular processes. Cell Signal 20: 787–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan B, Melkoumian ZK, Wu X, Guan KL, Guan JL. 2005. Identification of FIP200 interaction with the TSC1–TSC2 complex and its role in regulation of cell size control. J Cell Biol 170: 379–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan B, Peng X, Nagy T, Alcaraz A, Gu H, Guan JL. 2006. Role of FIP200 in cardiac and liver development and its regulation of TNFα and TSC–mTOR signaling pathways. J Cell Biol 175: 121–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganley IG, Lam du H, Wang J, Ding X, Chen S, Jiang X. 2009. ULK1⋅ATG13⋅FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem 284: 12297–12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T, Takamura A, Kishi C, Iemura S, Natsume T, Guan JL, Mizushima N. 2008. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol 181: 497–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Klionsky DJ. 2009. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet 43: 67–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Levine B. 2010. The Beclin 1 interactome. Curr Opin Cell Biol 22: 140–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieke N, Loffler AS, Kaizuka T, Berleth N, Bohler P, Driessen S, Stuhldreier F, Friesen O, Assani K, Schmitz K, et al. 2015. Expression of a ULK1/2 binding-deficient ATG13 variant can partially restore autophagic activity in ATG13-deficient cells. Autophagy 11: 1471–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch RV, Soriano P. 2006. Context-specific requirements for Fgfr1 signaling through Frs2 and Frs3 during mouse development. Development 133: 663–673. [DOI] [PubMed] [Google Scholar]
- Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, et al. 2009a. Nutrient-dependent mTORC1 association with the ULK1–Atg13–FIP200 complex required for autophagy. Mol Biol Cell 20: 1981–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N, Sasaki T, Iemura S, Natsume T, Hara T, Mizushima N. 2009b. Atg101, a novel mammalian autophagy protein interacting with Atg13. Autophagy 5: 973–979. [DOI] [PubMed] [Google Scholar]
- Jiang X, Overholtzer M, Thompson CB. 2015. Autophagy in cellular metabolism and cancer. J Clin Invest 125: 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, Kundu M, Kim DH. 2009. ULK–Atg13–FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell 20: 1992–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaizuka T, Mizushima N. 2015. Atg13 is essential for autophagy and cardiac development in mice. Mol Cell Biol 36: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenific CM, Debnath J. 2015. Cellular and metabolic functions for autophagy in cancer cells. Trends Cell Biol 25: 37–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, et al. 2005. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol 169: 425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. 2004a. The role of autophagy during the early neonatal starvation period. Nature 432: 1032–1036. [DOI] [PubMed] [Google Scholar]
- Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. 2004b. The role of autophagy during the early neonatal starvation period. Nature 432: 1032–1036. [DOI] [PubMed] [Google Scholar]
- Kumar MS, Pester RE, Chen CY, Lane K, Chin C, Lu J, Kirsch DG, Golub TR, Jacks T. 2009. Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev 23: 2700–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee IH, Kawai Y, Fergusson MM, Rovira II, Bishop AJ, Motoyama N, Cao L, Finkel T. 2012. Atg7 modulates p53 activity to regulate cell cycle and survival during metabolic stress. Science 336: 225–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Kroemer G. 2008. Autophagy in the pathogenesis of disease. Cell 132: 27–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang CC, Wang C, Peng X, Gan B, Guan JL. 2010. Neural-specific deletion of FIP200 leads to cerebellar degeneration caused by increased neuronal death and axon degeneration. J Biol Chem 285: 3499–3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Lee JY, Wei H, Tanabe O, Engel JD, Morrison SJ, Guan JL. 2010. FIP200 is required for the cell-autonomous maintenance of fetal hematopoietic stem cells. Blood 116: 4806–4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Xia H, Kim M, Xu L, Li Y, Zhang L, Cai Y, Norberg HV, Zhang T, Furuya T, et al. 2011. Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell 147: 223–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Fang F, Yuan H, Yang D, Chen Y, Williams L, Goldstein SA, Krebsbach PH, Guan JL. 2013. Suppression of autophagy by FIP200 deletion leads to osteopenia in mice through the inhibition of osteoblast terminal differentiation. J Bone Miner Res 28: 2414–2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D, Molusky MM, Song J, Hu CR, Fang F, Rui C, Mathew AV, Pennathur S, Liu F, Cheng JX, et al. 2013. Autophagy deficiency by hepatic FIP200 deletion uncouples steatosis from liver injury in NAFLD. Mol Endocrinol 27: 1643–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melkoumian ZK, Peng X, Gan B, Wu X, Guan JL. 2005. Mechanism of cell cycle regulation by FIP200 in human breast cancer cells. Cancer Res 65: 6676–6684. [DOI] [PubMed] [Google Scholar]
- Mercer CA, Kaliappan A, Dennis PB. 2009. A novel, human Atg13 binding protein, Atg101, interacts with ULK1 and is essential for macroautophagy. Autophagy 5: 649–662. [DOI] [PubMed] [Google Scholar]
- Mizushima N. 2004. Methods for monitoring autophagy. Int J Biochem Cell Biol 36: 2491–2502. [DOI] [PubMed] [Google Scholar]
- Mizushima N. 2009. Methods for monitoring autophagy using GFP-LC3 transgenic mice. Methods Enzymol 452: 13–23. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Komatsu M. 2011. Autophagy: renovation of cells and tissues. Cell 147: 728–741. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Levine B. 2010. Autophagy in mammalian development and differentiation. Nat Cell Biol 12: 823–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Levine B. 2010. Methods in mammalian autophagy research. Cell 140: 313–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. 2009. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol 10: 458–467. [DOI] [PubMed] [Google Scholar]
- Peltzer N, Rieser E, Taraborrelli L, Draber P, Darding M, Pernaute B, Shimizu Y, Sarr A, Draberova H, Montinaro A, et al. 2014. HOIP deficiency causes embryonic lethality by aberrant TNFR1-mediated endothelial cell death. Cell Rep 9: 153–165. [DOI] [PubMed] [Google Scholar]
- Peschon JJ, Torrance DS, Stocking KL, Glaccum MB, Otten C, Willis CR, Charrier K, Morrissey PJ, Ware CB, Mohler KM. 1998. TNF receptor-deficient mice reveal divergent roles for p55 and p75 in several models of inflammation. J Immunol 160: 943–952. [PubMed] [Google Scholar]
- Rubinsztein DC, Marino G, Kroemer G. 2011. Autophagy and aging. Cell 146: 682–695. [DOI] [PubMed] [Google Scholar]
- Subramani S, Malhotra V. 2013. Non-autophagic roles of autophagy-related proteins. EMBO Rep 14: 143–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Kaizuka T, Mizushima N, Noda NN. 2015. Structure of the Atg101–Atg13 complex reveals essential roles of Atg101 in autophagy initiation. Nat Struct Mol Biol 22: 572–580. [DOI] [PubMed] [Google Scholar]
- Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, Newman J, Reczek EE, Weissleder R, Jacks T. 2007. Restoration of p53 function leads to tumour regression in vivo. Nature 445: 661–665. [DOI] [PubMed] [Google Scholar]
- Wang C, Liang CC, Bian ZC, Zhu Y, Guan JL. 2013. FIP200 is required for maintenance and differentiation of postnatal neural stem cells. Nat Neurosci 16: 532–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H, Gan B, Wu X, Guan JL. 2009. Inactivation of FIP200 leads to inflammatory skin disorder, but not tumorigenesis, in conditional knock-out mouse models. J Biol Chem 284: 6004–6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H, Wei S, Gan B, Peng X, Zou W, Guan JL. 2011. Suppression of autophagy by FIP200 deletion inhibits mammary tumorigenesis. Genes Dev 25: 1510–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H, Wang C, Croce CM, Guan JL. 2014. p62/SQSTM1 synergizes with autophagy for tumor growth in vivo. Genes Dev 28: 1204–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E. 2012. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer 12: 401–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E. 2015. The role for autophagy in cancer. J Clin Invest 125: 42–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Z, Jin S, Yang C, Levine AJ, Heintz N. 2003. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci 100: 15077–15082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Peng X, Sun S, Park AY, Guan JL. 2010. Role of kinase-independent and -dependent functions of FAK in endothelial cell survival and barrier function during embryonic development. J Cell Biol 189: 955–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.