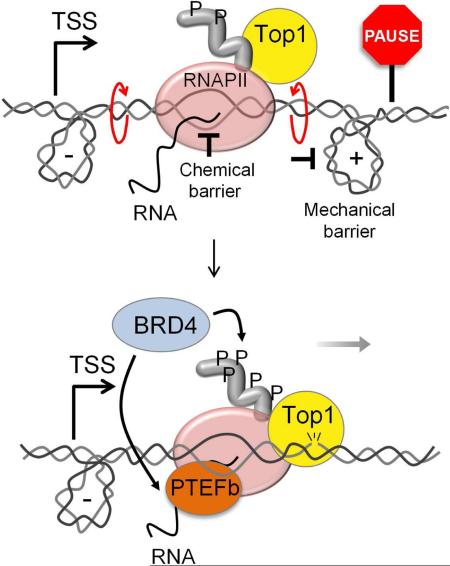
SUMMARY
We report a mechanism through which the transcription machinery directly controls topoisomerase 1 (TOP1) activity to adjust DNA topology throughout the transcription cycle. By comparing TOP1 occupancy using ChIP-Seq, versus TOP1 activity using TOP1-Seq, a method reported here to map catalytically engaged TOP1, TOP1 bound at promoters was discovered to become fully active only after pause-release. This transition coupled the phosphorylation of the carboxyl-terminal-domain (CTD) of RNA polymerase II (RNAPII) with stimulation of TOP1 above its basal rate, enhancing its processivity. TOP1 stimulation is strongly dependent on the kinase activity of BRD4, a protein that phosphorylates Ser2-CTD and regulates RNAPII pause-release. Thus the coordinated action of BRD4 and TOP1 overcame the torsional stress opposing transcription as RNAPII commenced elongation, but preserved negative supercoiling that assists promoter melting at start sites. This nexus between transcription and DNA topology promises to elicit new strategies to intercept pathological gene expression.
Graphical Abstract
INTRODUCTION
The generation of DNA topological stress is intrinsic to transcription. Our knowledge of transcriptional mechanisms has been gleaned mainly by study centered on transcription factors and chromatin regulation, while the mechanical and topological properties of the DNA during transcription have been less investigated. Accruing functional and structural data describing the dynamics of DNA and DNA-binding factors during transcription promise new insights into the mechanics of gene expression (Kouzine et al., 2013; Sainsbury et al., 2013). The main steps of transcription are coupled with the concomitant reorganization of chromatin and the generation of torsional stress, also called supercoiling (Cheung and Cramer, 2012; Wang et al., 1998). Unless dissipated or enzymatically disposed of, this torsional stress may interfere with the regulatory steps of transcription (Ma et al., 2013; Roca, 2011). TOP1 catalyzes changes in the linkage between DNA strands by transiently breaking one strand, swiveling about the unbroken strand and resealing the nick (Champoux, 2001). Early studies demonstrated the need for TOP1 as a DNA swivel during transcription (Brill et al., 1987). Transcriptional activation of the Hsp70 genes in Drosophila results in rapid recruitment of TOP1, which behaves as an elongation factor associated with active genes (Zobeck et al., 2010). TOP1 is dispensable for cell viability in yeast and its loss can be compensated by other topoisomerases (Uemura and Yanagida, 1984), but it is essential for early development in multicellular eukaryotes, suggesting specific functions not shared with other relaxases.
The ability of TOP1 to facilitate transcription relates to two roles whose interrelationship remains largely unexplored. First, TOP1 relaxes torsional stress that would otherwise accumulate and interfere with transcription elongation. In the “twin domain model” (Liu and Wang, 1987) as DNA is screwed through the transcription machinery, positive supercoils are driven ahead and negative supercoils trail RNAPII. Positive supercoils can impede elongation (Gartenberg and Wang, 1992; Joshi et al., 2010) whereas negative supercoils support DNA melting and favor initiation (Dunaway and Ostrander, 1993; Parvin and Sharp, 1993). At regulatory sequences negative supercoils can also drive duplex B-DNA into single-stranded or other non-B DNA conformations changing the transcriptional output of the gene (Kouzine et al., 2008; Liu et al., 2006). Because TOP1 drains supercoils of both signs, it is unknown if and how its activity might be differentially programmed to promote both initiation and elongation or to regulate other genetic activities (Baranello et al., 2012). Neither TOP1 relaxation nor transcription is instantaneous and so a dynamic balance must couple these processes (Kouzine et al., 2008).
During pre-initiation complex (PIC) assembly, TOP1 may interact with TFIID and TFIIA to potentiate or repress transcription in vitro non-catalytically (Kretzschmar et al., 1993; Merino et al., 1993; Shykind et al., 1997). TOP1 may assist nucleosome disassembly at promoters prior to transcription initiation (Durand-Dubief et al., 2010) and help orchestrate the program of induction or repression of gene expression (Pedersen et al., 2012). Together these studies suggest that TOP1 participates in early phases of transcription, but do not reveal its scope and mechanism in vivo. Recently, the genomic mapping of transcription-generated supercoils in cell lines has provided insight into DNA dynamics at transcribed loci and into TOP1 function at transcription start sites (TSSs) (Kouzine et al., 2013; Teves and Henikoff, 2014). Dynamic supercoils spread ~1.5 kilobases from the TSSs of active genes, and TOP1 activity predominated at medium output promoters bearing paused RNAPII. Some paused promoters are extremely sensitive to camptothecin (CPT), a TOP1-selective inhibitor (Khobta et al., 2006). TOP1 therefore may participate in RNAPII promoter-proximal pausing, one of the most pervasive and highly regulated events in the transcription cycle (Adelman and Lis, 2012). However few experiments have probed a regulatory role for TOP1 in pausing.
In this study, the genome-wide distributions of TOP1's sites of binding versus catalytic engagement were determined. Though concordant in gene bodies, the two maps were discordant at the promoters of active genes where the bound TOP1 was relatively inactive indicating that TOP1 action is coordinated with the progression through the transcription cycle. Supercoiled DNA-relaxation assays using components of the transcription machinery, revealed the phosphorylated CTD of the largest subunit of RNAPII to be a potent activator of TOP1. These results suggest that the level of supercoiling is actively managed by the transcription machinery to melt DNA at TSSs, hold-back RNAPII at pause sites or to accelerate elongation and help control the transcriptional output of virtually any gene.
RESULTS
TOP1 Functionally and Physically Associates with the Transcriptional Machinery
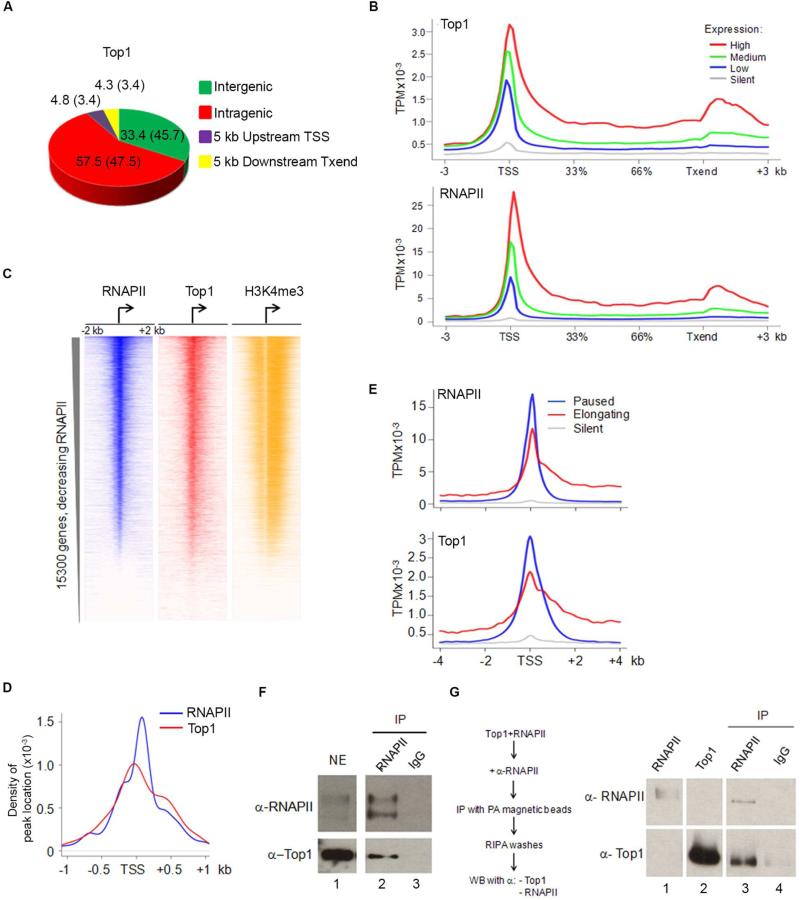
Specific predictions were formulated to discriminate between architectural/non-catalytic versus predominantly catalytic roles for TOP1 in the regulation of genes transcribed by RNAPII. If TOP1 operates principally to relax transcription-generated supercoils, then high levels of enzyme should populate the bodies of active genes. Alternatively, if TOP1 is a major promoter factor regulating PIC assembly, it should accumulate mainly at TSSs. These predictions were evaluated using Chromatin Immuno-Precipiation-Sequencing (ChIP-Seq) of native TOP1 in HCT116 WT, a colorectal cancer cell line previously used for studies of TOP1 (Miao et al., 2007).
67% of DNA bound TOP1 was localized from 5 kb upstream of TSSs to 5 kb downstream of transcription termination site (Txend) (Figure 1A), indicating that TOP1 disposition is related to transcription (Figure S1A). From first principles (Liu and Wang, 1987), increased gene transcription should increase dynamic supercoiling and demand more TOP1 recruitment. At genes with high (100-95%), medium (95-50%) low (bottom 50% of genes with significant output) or silent (genes with non-significant output) expression as measured by RNA-Sequencing (RNA-Seq) (Figure 1B, top panel), TOP1 recruitment was indeed associated with expression level. TOP1 enrichment on high output genes extended from TSSs through to Txends. The distribution of TOP1 at promoters qualitatively matched the distribution of negative supercoiling as measured with psoralen (Kouzine et al., 2013; Teves and Henikoff, 2014).
Figure 1. TOP1 Functionally and Physically Associates With the Transcriptional Machinery.
A) TOP1 occupancy in HCT116 cells. +5Kb and −5Kb indicate tags within 5Kb upstream of TSS or downstream of Txend respectively; numbers in parenthesis are values expected if randomly distributed. B) TOP1 and RNAPII occupancy (as sequence tags per million, TPM) across genes classified by expression. C) Heat map of RNAPII, TOP1 and H3k4me3 near TSSs of human protein coding genes ranked from highest to lowest RNAPII level. D) Density distribution of RNAPII and TOP1 peaks around TSSs identified by QuEST. E) RNAPII and TOP1 distribution around TSSs for elongating, paused and silent genes. F) Nuclear extracts and G) recombinant proteins immunoprecipitated with anti-RNAPII or non-immune IgG and probed for TOP1 and RNAPII. See also Figure S1
TOP1 promoter binding was compared with RNAPII loading (Figure 1B, bottom panel). RNAPII ChIP-Seq data were used to generate heat maps (Figure 1C) where promoters were first sorted according to RNAPII occupancy, then inspected for H3K4me3 modification to verify promoter activity and finally the examination of TOP1 revealed that its levels and localization paralleled RNAPII (Spearman's correlation: 0.57). Two populations of TOP1 binding were noted (Figure 1D); the first was slightly upstream of the major peak of RNAPII whereas the second was coincident with early elongating RNAPII. This pattern seemed indicative of a role for TOP1 in transcription beyond its recruitment directly to torsionally strained DNA to dissipate supercoils.
To see if the TOP1 distribution was related with RNAPII pausing, genes were classified by pausing index (PI) (Extended Experimental Procedures) as paused, elongating or silent and analyzed for TOP1 occupancy. TOP1 was abundant along gene bodies if RNAPII was elongating, but accumulated at TSSs where RNAPII was paused (Figure 1E). On highly active genes, TOP1 paralleled the RNAPII distribution from the TSS through the polyadenylation site where higher signals for both proteins were observed (Figure 1B, and S1A).
To test whether physical interaction between RNAPII and TOP1 contributed to their co-localization, pull-down assays were performed either from cellular extracts (Figure 1F) or from mixtures of purified recombinant proteins in vitro (Figure 1G). Purified TOP1 and RNAPII interacted directly in the absence of DNA (Figure 1G). TOP1-ChIP RNAPII-re-ChIP experiments revealed RNAPII and TOP1 co-associated at promoters and in the bodies of active, but not inactive genes (Figure S1B). Thus TOP1 seemed to be an integral part of the transcription machinery, from TSS to Txend.
TOP1 Activity is Stimulated in Gene Bodies
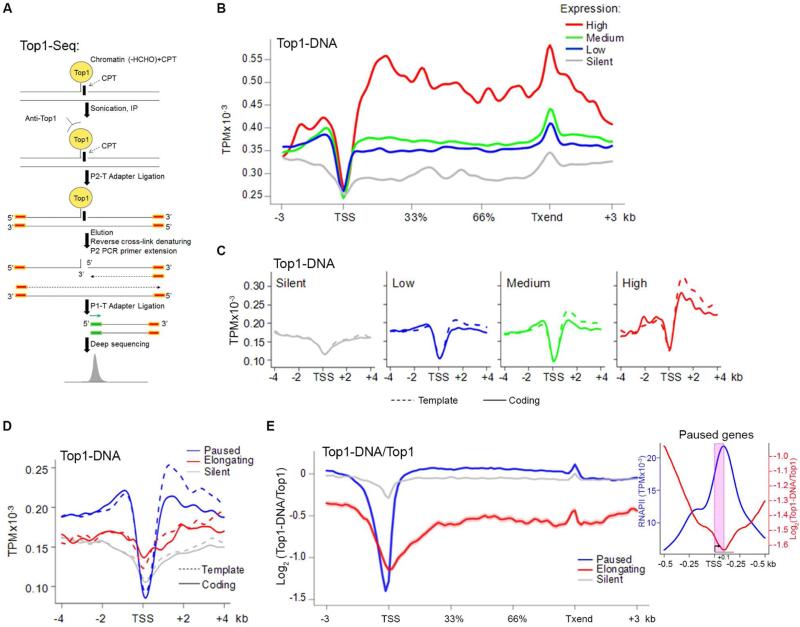
Co-localization with RNAPII might reflect either catalytic or non-catalytic roles for TOP1; to query whether TOP1 binding paralleled its activity we developed TOP1-Seq, a modified ChIP-Seq technique to immunoprecipitate only catalytically engaged TOP1. During the TOP1 catalytic cycle, the enzyme nicks DNA and forms a covalent intermediate that was trapped with a short (4 min) CPT treatment, immunoprecipitated with anti-TOP1 antibody and sequenced (Figure 2A) (Redinbo et al., 1998; Stewart et al., 1998). This brief CPT treatment neither altered chromatin structure (in bulk or at individual genes) nor elicited a DNA-damage response (Kouzine et al., 2013). Consequently TOP1-Seq enabled precise genomic mapping of catalytically engaged TOP1.
Figure 2. TOP1 Is Active Along Gene Bodies.
A) TOP1-Seq. B) TOP1-Seq profile at genes ranked by expression. C) Strand-specific TOP1-Seq around TSSs ranked by gene expression. D) Strand-specific TOP1-Seq at TSSs for paused, elongating and silent genes. E) Log-ratio of tags of TOP1-Seq and TOP1 ChIP-Seq across gene bodies. Shaded area indicates s.e.m. Inset. RNAPII density and TOP1 relative activity at paused promoters. TSS/pause region is shaded (pink). See also Figure S1.
Whereas promoter-bound TOP1-DNA complexes were sparse, the recovery of these complexes increased downstream and remained high through gene bodies. The magnitude of the increase was correlated with the level of gene expression (Spearman's correlation: 0.24) (Figure 2B, 2C, and S1A). Local maxima of TOP1-DNA complexes mapped to 3’ ends where positive supercoils accumulate (Joshi et al., 2010) and to ~1.5 kb downstream of the TSS (Figure 2B and 2C), perhaps reflecting the disposition of torsional stress (Kouzine et al., 2013). TOP1 complex formation downstream of the TSS was biased for the template strand (Figure 2C), suggesting that the movement of the transcriptional machinery must be coordinated with the activity of TOP1.
The TOP1-Seq patterns were compared at genes with paused versus elongating RNAPII (Figure 2D and S1C). Notably, while there was a gradual increase in the density of TOP1-associated cleavage sites along elongating genes, paused genes displayed a sharp increase in the abundance of TOP1 covalent complexes from promoters to gene bodies (Figure 2D). The ratio between TOP1 binding, as assessed by ChIP-Seq, and activity, as assessed by TOP1-Seq highlighted the transition to full activity (Figure 2E); strikingly the upswing in engaged, enzymatically active, TOP1 coincided with the zone of pause-release (Figure 2E, inset, and S1D). Because the promoters of paused genes are supercoiled to the same degree as elongating genes (Kouzine et al., 2013; Teves and Henikoff, 2014), this difference in TOP1 activity was not attributable simply to different levels of torsional stress residing at these types of promoters. Thus we hypothesized regulation of TOP1 activity at paused genes either by a factor(s) that blunts relaxation at the TSS or that stimulates the enzyme in gene bodies.
RNAPII Stimulates TOP1 Relaxation Above its Basal Activity
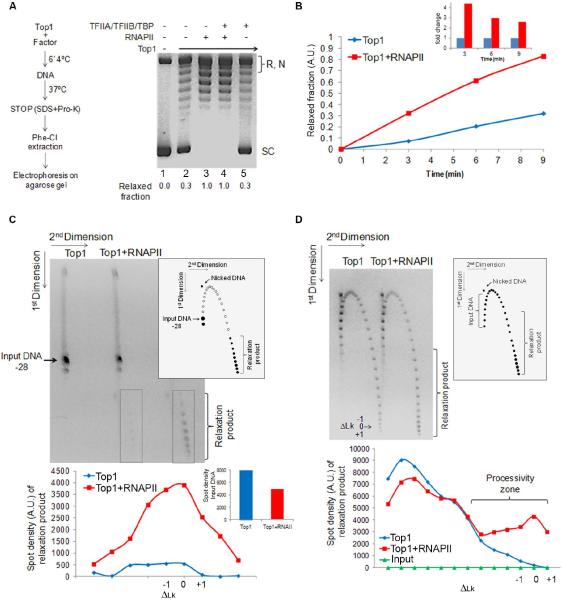
Studies showing that both catalytically active and inactive TOP1 interact with the general transcription machinery to repress basal or increase activated transcription in vitro (Carty and Greenleaf, 2002; Merino et al., 1993; Wu et al., 2010), did not ask if these interactions might reciprocally influence TOP1 activity. Therefore the main components of the transcription machinery were screened to see if they inhibited or stimulated plasmid relaxation by TOP1 (Figure 3A, left panel) (Extended Experimental Procedures). Surprisingly, RNAPII increased relaxation by TOP1 above its basal activity (Figure 3A). None of the other general transcription factors altered the basal relaxation rate of TOP1 (Figure 3A, and data not shown), so the stimulation seemed to be a specific property of RNAPII. Dose response (Figure S2A) and kinetic curves (Figure 3B) evidenced that RNAPII enhanced the initial velocity of TOP1 relaxation by at least 5-fold at approximately equimolar concentration. The activity of Escherichia coli topoisomerase I was not so enhanced indicating that the RNAPII stimulation of human TOP1 was due to a specific interaction (Figure S2B). Thus the transcription machinery seems equipped to coordinate both supercoil generation and removal.
Figure 3. RNAPII Stimulates TOP1 Activity Above Its Intrinsic Rate.
A) TOP1 alone or pre-incubated with RNAPII and/or general transcription factors TFIIA/TFIIB/TBP before plasmid DNA was added. Plasmid relaxation was checked by agarose gel electrophoresis. Supercoiled bands (SC) were quantified and compared to SC without TOP1 (1st lane). Numbers indicate relaxed fraction. R – relaxed DNA. N – nicked DNA. B) TOP1 was incubated on ice with or without RNAPII before addition of DNA. The graph shows the relaxed fraction. The inset shows the fold stimulation. C) The right diagram shows schematic trajectory of topoisomers in 2D gel (Extended Experimental Procedures). Filled circles indicate experimentally observed species. Unreacted −28 ΔLk topoisomer substrate and relaxed product topoisomers are indicated. Left. Distribution of product DNA topoisomers after incubation with TOP1 with or without RNAPII. Bottom panel quantifies spot distribution (relaxation product is indicated by grey boxes). Inset shows spot quantification of unreacted −28 ΔLk DNA topoisomer. D) Reactions performed as in C but the substrate was a population of supercoiled topoisomers in increased ionic strength buffer (Extended Experimental Procedures). See also Figure S2.
RNAPII Increases the Processivity of TOP1 Relaxation
To study how RNAPII influenced DNA relaxation by TOP1, two-dimensional gel electrophoresis was used to display the distribution of DNA topoisomers recovered from the reactions (Extended Experimental Procedures) (Figure 3C). The catalytic stimulation of TOP1 by RNAPII was dramatically visualized using limiting amounts of TOP1 acting on a purified topoisomer. Alone, TOP1 yielded a small amount of fully relaxed product whereas addition of RNAPII increased the recovery of relaxed species by ~8-fold (Figure 3C). Due to high enzymatic processivity, TOP1 yielded only products thermally distributed around the most fully relaxed topoisomer (ΔLk0) (Figure 3C) and so revealed no information concerning reaction intermediates. To visualize better the reaction pathway, experiments were repeated at physiological ionic strength (150mM NaCl) where TOP1 acts distributively (Extended Experimental Procedures). These experiments revealed that the population of products was distributed across the spectrum of Lk's with TOP1, and with RNAPII a subpopulation of DNA was processively driven to full relaxation (Figure 3D). To discriminate the influence of RNAPII on specific steps of the TOP1 reaction cycle (binding, cleavage, strand rotation, relegation) a DNA nicking assay was performed (Extended Experimental Procedures) revealing that RNAPII increased the population of cleavage intermediates within the TOP1 cycle (Figure S2C).
The CTD of RNAPII Stimulates DNA Relaxation via the N-Terminal Domain of TOP1
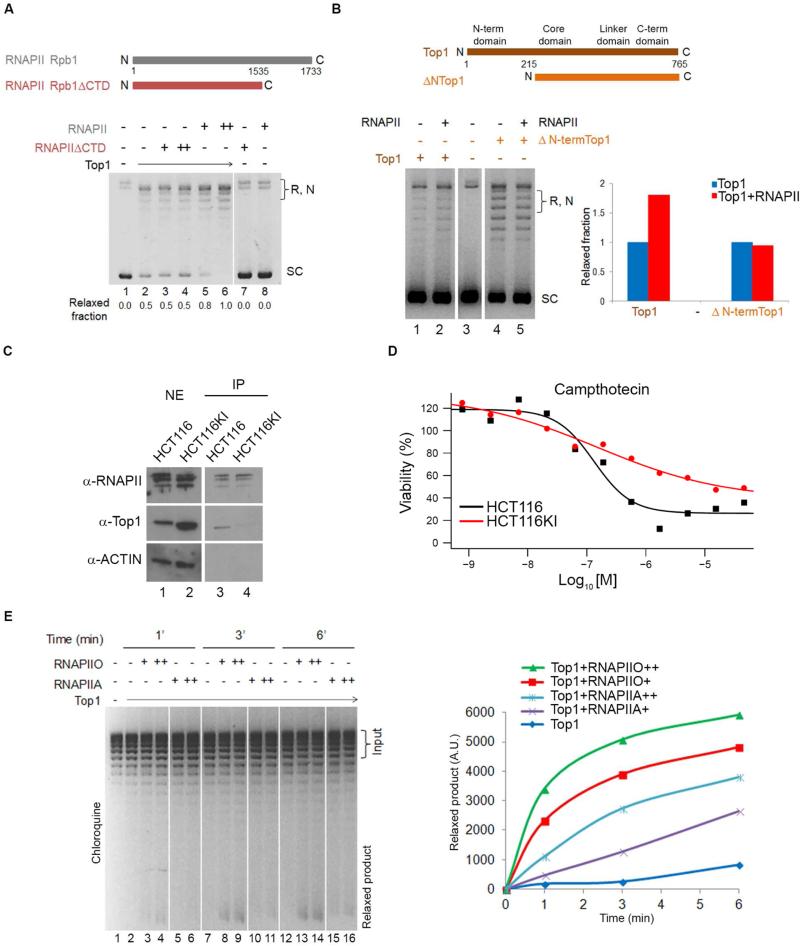
The N-terminal domain (NTD) of TOP1 binding with the CTD of RNAPII (Carty and Greenleaf, 2002; Wu et al., 2010) suggested that TOP1 stimulation was mediated through these domains. To evaluate the involvement of the RNAPII CTD in TOP1 activation, full length RNAPII and a truncated form lacking the CTD (Kim and Dahmus, 1988) (Figure 4A and S2D) were compared for stimulation of plasmid relaxation by TOP1; CTD-less RNAPII failed to stimulate DNA relaxation by TOP1 (Figure 4A). Full length and NTD-less (ΔNTOP1) TOP1 (Laco and Pommier, 2008) were tested for their capacity to be stimulated by RNAPII; only full-length TOP1 (Figure 4B) was efficiently stimulated.
Figure 4. Phosphorylated CTD of RNAPII Stimulates TOP1.
A) Upper panel. Representation of RPB1 subunit of RNAPII and a version lacking the CTD. Lower panel. TOP1 was pre-incubated alone or in combination with RNAPII or CTD-minus RNAPII. After relaxation, DNA was run under native conditions. Numbers quantify the relaxed fraction. B) Upper panel. Domain structure of the human TOP1 and of a truncated form lacking the NTD. Lower panel. Full length TOP1 and ΔN-term TOP1 were pre-incubated on ice with or without RNAPII. Relaxation assay and gel electrophoresis were performed as in A). Relevant lanes from the same gel were juxtaposed. The graph quantifies relaxation. C) Nuclear extracts from HCT116 and HCT116KI cells immunoprecipitated with anti-RNAPII and probed for TOP1 and RNAPII. ACTIN was used to assess non-specific binding. D) HCT116 and HCT116KI cells were treated with serial dilutions of TOP1 inhibitor CPT and viability was measured. E) RNAPIIA and RNAPIIO were pre-incubated with TOP1 then plasmid was added. Relaxation products were run in the presence of chloroquine and quantified as shown in graph. See also Figure S3 and S4.
To test whether the RNAPII CTD interacted with the TOP1 NTD in vivo, and if so, its impact on TOP1 activity, CRISPR/Cas9 was used to delete and replace exon 4 of TOP1 in its endogenous chromosomal location (Figure S3A) with tandem in-frame HA-tags. The lysine-rich exon 4 was judged to be a likely region to mediate interaction with phosphorylated residues of the RNAPII CTD. A homozygous knock-in (KI) clone derived from HCT116 WT cells in which both TOP1 alleles were mutated (HCT116KI) (Figure S3B) was selected for further studies. Co-immunoprecipitation of TOP1 with RNAPII was strikingly attenuated in the HCT116KI cells (Figure 4C) confirming that residues within exon 4 of the TOP1 facilitated TOP1 binding with RNAPII. Compared with WT, ex4KI cells expressed 3 times more TOP1 protein (Figure S3C). This phenotype did not reflect protein stabilization since exon 4 did not contain PEST-like sequence. Thus the loss of exon 4 seemed to drive a compensatory increase in TOP1 suggesting that the NTD, though dispensable for enzymatic activity in vitro (Redinbo et al., 1998), may be important to regulate TOP1 activity in vivo.
If RNAPII-CTD interacted with TOP1 NTD to increase processivity (Figure 3D) then RNAPII-stimulated TOP1 should spend more time in the DNA-cleaved state as the torsionally-stressed DNA spins through multiple rotations. Conversely, if the RNAPII/TOP1 association is impaired in the HCT116KI cells, the duration of the TOP1-DNA cleavage complex should be shortened. Consequently, the HCT116KI cells should be less sensitive to TOP1 inhibitors such as CPT that inhibits religation by intercalating at the TOP1-DNA interface (Strumberg et al., 2000). To assess the cellular response to the replacement of TOP1 exon 4, WT and HCT116KI cells were treated with TOP1 inhibitors and evaluated for growth and viability using an ATP-luminescence assay.
Cells without exon 4 of TOP1 were profoundly less sensitive to CPT; the 50% inhibition concentration (IC50) was shifted rightward (approximatively 10-100 fold) (Figure 4D) indicating lower toxicity. Notably this resistance to TOP1 inhibitors was restricted to those agents that stabilized the TOP1 covalent DNA-cleaved intermediate; β-lapachone, which acts pre-cleavage (Li et al., 1993), did not discriminate exon 4 plus or minus cells (Figure S3D). Taken together these observations suggested that the TOP1 NTD regulates the enzyme's activity, post-cleavage through interaction with RNAPII in vivo.
RNAPII Stimulates TOP1 Via the Phosphorylated CTD
Both RNAPII and TOP1 are abundant at TSSs (Figure 1B) and if RNAPII activates TOP1, why doesn't this happen at the TSS? One possibility would be that the stimulation of TOP1 is mediated by phosphorylated RNAPII (Figure S2E). With 52 consecutive repeats of the heptapeptide YSPTSPS, the CTD is a substrate for several protein kinases including the CDK7 subunit of TFIIH, CDK9 of PTEF-b and BRD4 (among others) (Kwak and Lis, 2013; Ramanathan et al., 2001). The pattern of CTD-phosphorylation constitutes a code (Egloff et al., 2012) that marks the transitions between different stages of the transcription cycle. RNAPII enters the PIC hypo-phosphorylated. During initiation, the CTD is phosphorylated on Ser5 primarily by CDK7 (Spangler et al., 2001; Tirode et al., 1999), and subsequently on Ser2 by BRD4 (Devaiah et al., 2012; Wu and Chiang, 2007), PTEF-b (Peterlin and Price, 2006) and other kinases during pause-release and elongation (Kwak and Lis, 2013). Therefore, we asked whether RNAPII phosphorylation affected TOP1 activity. Plasmid relaxation assays of TOP1 incubated with RNAPIIA (hypo-phosphorylated) or RNAPIIO (highly phosphorylated) (Figure 4E and S4A) strikingly demonstrated that the amount of activity paralleled the degree of RNAPII phosphorylation. Thus, TOP1 activity is controlled by the phosphorylation status of the RNAPII suggesting a mechanism to couple TOP1 catalysis with pause-release.
BRD4-phosphorylated CTD Stimulates TOP1
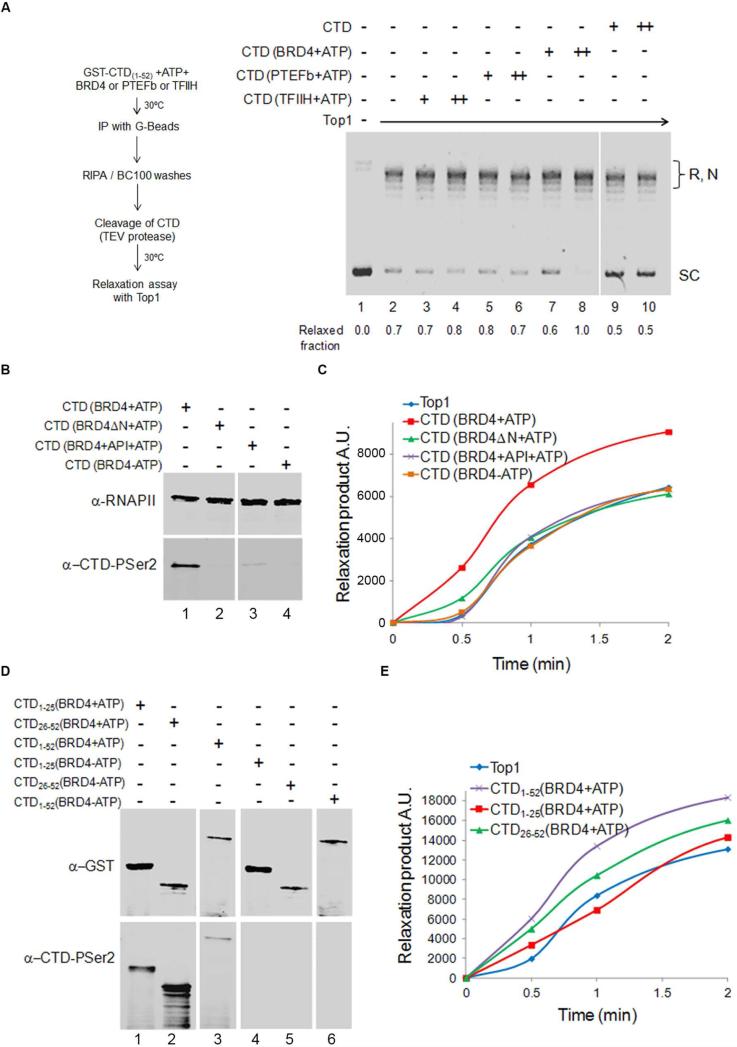
To examine whether TOP1 stimulation was associated with a specific CTD-kinase, GST-CTD was individually treated with the kinase components of TFIIH, PTEF-b or BRD4, in the presence or absence of ATP. The treated CTD was then purified from the reaction, cleaved from GST and incubated with TOP1 and plasmid DNA (Figure 5A and S4B). Unphosphorylated CTD was unable to stimulate TOP1. In contrast, ATP-dependent TOP1 stimulation was supported by BRD4-phosphorylated CTD (Figure 5A). This stimulation was not simply fostered by a physical interaction between TOP1 and BRD4 phopshorylated-CTD as evinced by pull down experiments that showed TOP1 forming protein-protein complexes with all the different CTDs, independent of their phosphorylation status (Figure S4C).
Figure 5. BRD4-phosphorylated RNAPII-CTD Stimulates TOP1 Relaxation.
A) GST-CTD(1-52) was phosphorylated with TFIIH, PTEF-b or BRD4, purified and co-incubated with TOP1 for plasmid relaxation. Numbers quantify relaxed fraction. B) GST-CTD(1-52) was phosphorylated with BRD4 plus (lane 1) or minus (lane 4) ATP, with kinase inhibitor apigenin (API, lane 3) or a BRD4 kinase mutant (BRD4ΔN, lane 2) was used. Reactions were immunoblotted for total RNAPII and phospho-Ser 2 CTD. After purification, CTDs were co-incubated with TOP1. Quantification of relaxation is graphed. D) GST-CTD(1-52), ,GST-CTD(1-25) and GST-CTD(26-52) were phosphorylated with BRD4 in presence (lanes 1, 2, 3) or absence (lanes 4, 5, 6) of ATP. Reactions were immunoblotted for GST and phospho-Ser2 CTD. After purification CTDs were co-incubated with TOP1 (E). Relaxation products were run and analyzed as in C). The graph shows quantification of relaxation. Relevant lanes from the same gel were juxtaposed. See also Figure S4 and S5.
Stimulation of TOP1 by CTD treated with BRD4 and ATP had the same structural requirements and sensitivities to inhibitors as BRD4-kinase activity (Devaiah et al., 2012). TOP1 stimulation was sensitive to apigenin that also suppressed BRD4 phosphorylation of Ser2 (Figure 5B lane 3, 5C purple line, and S4D). When a BRD4 mutant lacking the N-terminal domain (BRD4ΔN) was employed, CTD-phosphorylation failed (Figure 5B lane 2), as did stimulation of TOP1 relaxation (Figure 5C green line, and S4D).
The particular pattern of Ser2 phosphorylation within the CTD may dictate the degree of TOP1 activation. Truncated GST-CTDs bearing either the N-terminal repeats [GST-CTD(1-25)] or the C-terminal repeats [GST-CTD(26-52)] (Figure S4E upper panel), were BRD4-phosphorylated, purified (Figure S4B) and tested for stimulation of TOP1. Notably repeats 1-25 are comprised mainly of canonical YSPTSPS repeats whereas the majority of repeats 26-52 are non-canonical, if not unique (Eick and Geyer, 2013). Interestingly, BRD4 phosphorylated Ser2 to a much greater extent in GST-CTD(26-52) than in GST-CTD(1-25) (Figure 5D). Paralleling the extent of BRD4 phosphorylation, CTD(26-52) was a more potent activator of TOP1 than was CTD(1-25) (Figure 5E and S4E, lower panel). Because TOP1 bound all phosphorylated or unphosphorylated CTD forms to similar extents (Figure S4F), but only BRD4 phosphorylation supported stimulation of TOP1, it is likely that a particular conformation of the phosphorylated-CTD/TOP1 complex is required for maximal TOP1 activity.
TOP1 Activity is Inhibited when Ser2 CTD is Mutated
The characterization and the functional consequences of alternative patterns of heptad phosphorylation by CTD-kinases have been incompletely described. Ser2 of the CTD was an effective substrate for both the BRD4 and CDK9/PTEF-b kinases (Figure S4G) but only BRD4-phosphorylated CTD stimulated TOP1 (Figure 5C and S4H). To map more finely the TOP1-stimulating sites of BRD4 phosphorylation and to examine the relationships between BRD4 vs. PTEF-b CTD phosphorylation and TOP1 activity, a panel of GST-CTD(26-52) bearing Ser2 to Ala substitutions (Figure S5A) within blocks of heptads was generated. Phosphorylation of these mutants and WT CTD(26-52), by BRD4 versus PTEF-b showed distinct patterns of modification (Figure S5B lane 1). Diffuse migration of WT GST-CTD(26-52) treated with PTEF-b indicated heterogeneous phosphorylation, whereas tighter migration after BRD4 treatment suggested a more homogeneous and restricted pattern of phosphorylation (Figure S5B odd number lanes). No change in the band intensity or electrophoretic migration of any GST-CTD(26-52) mutants was apparent after treatment with PTEF-b, consistent with PTEF-b Ser2 targets being heterogeneously distributed throughout the CTD (Figure S5B even number lanes). In contrast Ser2 mutations within the first and the fifth blocks of GST-CTD(26-52) (mutant 1-M1 and mutant 5-M5) greatly reduced the level of BRD4-dependent Ser2 phosphorylation (Figure S5B and S5C); remarkably, TOP1 activity exactly paralleled the profile of phosphorylation. Surprisingly, TOP1 relaxation dropped below basal activity when combined with M1 or M5 (Figure S5D). Therefore specific patterns of BRD4-dependent CTD phosphorylation may be able to program multiple output states of TOP1 activity through direct interaction between TOP1 and phosphorylated-CTD (Figure S4F).
CPT Synergizes with Bromodomain and ExtraTerminal Domain (BET) Inhibitors to Kill HCT116 Cells
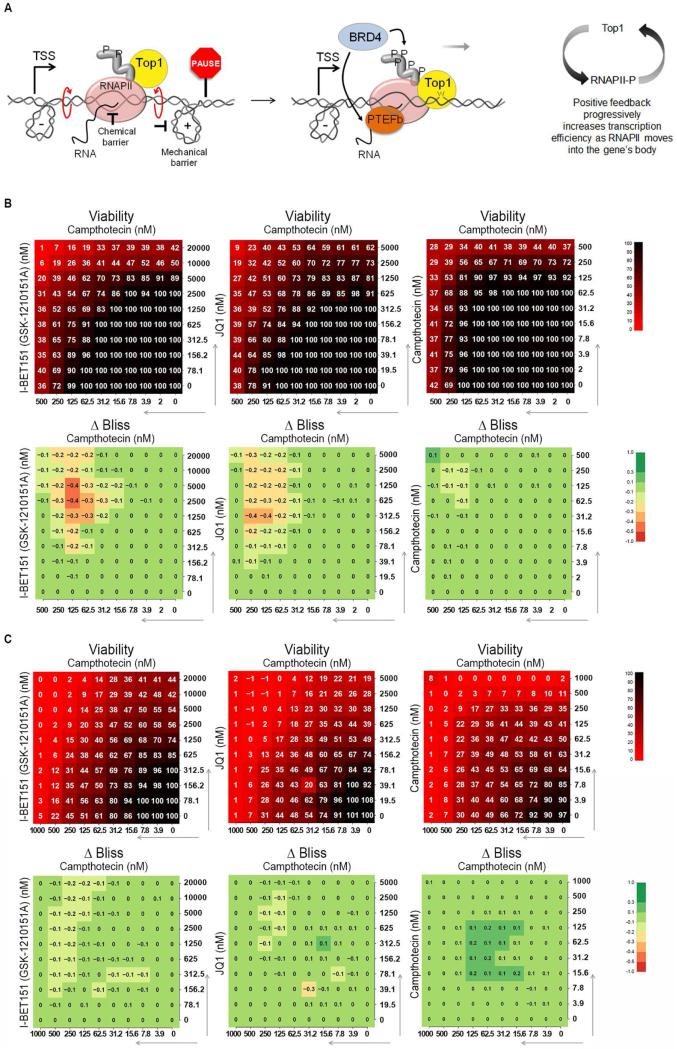
BRD4 belongs to the BET family, bromodomain proteins that interact with acetylated histones. Recent studies (Kanno et al., 2014) have also revealed bromodomain independent recruitment of BRD4 to TSSs. Accordingly, BRD4 stimulated TOP1 activity in vitro via RNAPII-CTD phosphorylation, independent of nucleosome acetylation. The coupling of TOP1 activity with BRD4 kinase suggested that the regulation of DNA topology may be linked to pause-release, paralleling BRD4's regulation of pause-release through PTEF-b (Liu et al., 2013). Though promoter pausing is central to the expression of most genes, much of the fundamental biochemistry of this regulatory process remains obscure. From first-principles, pausing must be imposed either by directly slowing the chemistry of nucleotide addition or by mechanically resisting RNAPII translocation along the template (Ma et al., 2013). Our data suggest that BRD4 may relieve both chemical and mechanical impediments to elongation via a chromatin-dependent recruitment of PTEF-b (sensitive to BET inhibitors such as I-BET151 or JQ1) (Filippakopoulos et al., 2010; Kanno et al., 2014; Yang et al., 2005) and chromatin-independent stimulation of TOP1, respectively (Figure 6A). If BRD4 acts through a BET-inhibitor sensitive arm to drive PTEF-b and through a BET-inhibitor insensitive, kinase-dependent arm to activate TOP1, then the simultaneous targeting of both arms with small molecules might synergistically interfere with transcription and so inhibit cell growth.
Figure 6. CPT Synergizes With BET Inhibitors in Killing HCT116 Cells.
A) Model. DNA supercoiling imposes a mechanical barrier to the progression of RNAPII and contributes to arrest at pause site. CTD phosphorylation stimulates TOP1 to relieve torsional stress and assist pause-release. Through a second arm, BRD4 activates the transcription machinery via PTEF-b. B) Combination response to BRD4 inhibitors I-BET151 or JQ1 with TOP1 inhibitor CPT and a self-cross of CPT. Negative values indicate synergy (Extended Experimental Procedures). C) As in B) but experiment performed on HCT116KI. See also Figure S6.
We interrogated a library of 1912 approved or investigational agents examined in cancer therapy. After drug activity was defined, specific agents of characterized mechanism including TOP1 and BET inhibitors were tested to identify synergistic combinations. Serial dilutions of each compound were combined and the effect on the viability of HCT116 WT cells was measured using an ATP-based assay (Extended Experimental Procedures). Synergistic toxicity between the TOP1 inhibitor CPT and the BRD4 inhibitors I-BET151 or JQ1 was prominent (Figure 6B). A self-cross of CPT that defines additivity is shown for reference, as are unrelated combinations that possessed no synergy (Figure 6B and S6). Remarkably, in the HCT116KI cells no synergistic toxicity between I-BET151/JQ1 and CPT was observed (Figure 6C). In the KI cells, constitutive TOP1 activity was uncoupled from RNAPII-stimulation and was independent of BET action suggesting a functional interaction between TOP1 and BRD4 that may prove susceptible to therapeutic intervention in cancer.
JQ1 Antagonizes BRD4 Binding and TOP1 Activity
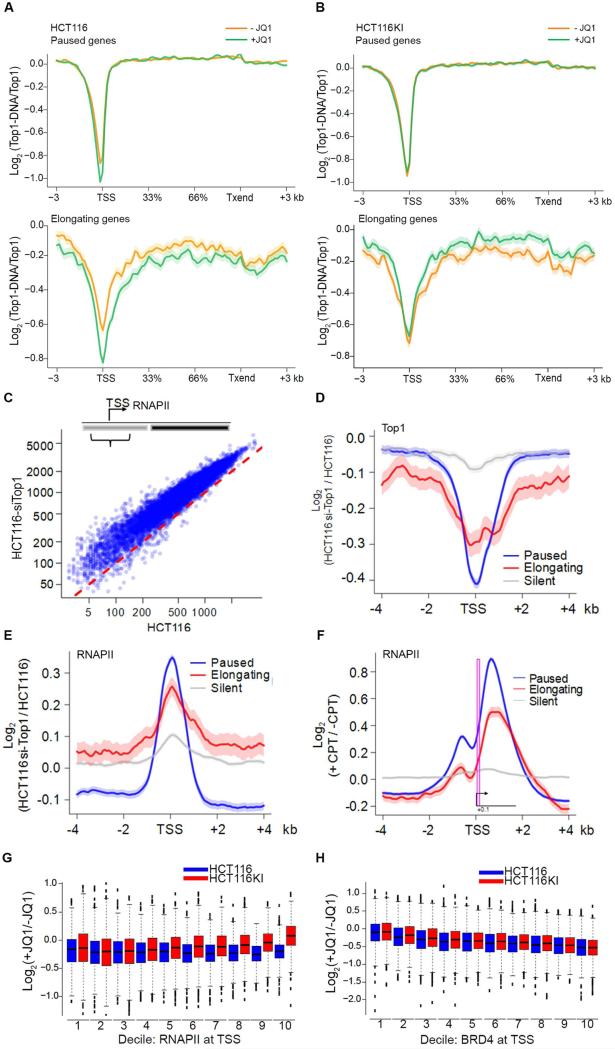
Our in vitro data clearly established a link between TOP1 relaxation and BRD4 kinase activity via phosphorylated CTD. To appraise the influence of BRD4 on the binding and activity of TOP1 at promoters, the occupancy of both proteins along genes, and TOP1 activity were monitored in HCT116 cells treated without and with JQ1. Without JQ1 77% of TOP1 promoter peaks co-localized with BRD4 (Figure S7A). Interestingly, at the TSSs of paused and elongating genes, JQ1 decreased both BRD4 binding (Figure S7B) and the fraction of active TOP1 (TOP1-Seq/TOP1 ChIP-Seq) (Figure 7A). In sharp contrast, in the ex4KI cells, both TOP1 binding and activity at promoters was unaffected (Figure 7B) by the JQ1-elicited decrement of BRD4 (Figure S7E), exactly as predicted if TOP1 activity remained at basal levels, uncoupled from stimulation by BRD4-phosphorylated CTD.
Figure 7. TOP1 Knockdown or Inhibition Elicit RNAPII Accumulation at TSSs.
A) HCT116 cells were treated with JQ1. The Log-ratio of tag profiles of TOP1-Seq and TOP1 ChIP-Seq across paused and elongating genes is shown. B) Same as A) but experiment performed on HCT116KI. C) Scatter plot showing change of RNAPII density at TSS between HCT116 and HCT116-siTOP1 cells (Wilcoxon rank-sum test, p-value < e-200). D) Relative TOP1 and E) RNAPII levels at TSSs. The curves show the average Log enrichment ratio of tag profiles in HCT116-siTOP1 versus HCT116 cells. F) Relative RNAPII levels at TSSs of elongating, paused and silent genes. The curves show average Log enrichment ratios of tag profiles in HCT116 treated or not with CPT. The TSS/pause region is boxed to show increased RNAPII there. G) Relative RNAPII levels at TSS. TSSs were sorted in deciles according to increasing levels of RNAPII. Log ratio of tag profiles in HCT116 and HCT116KI cells treated or not with JQ1 is shown. H) Same as G) but analysis was made with BRD4. See also Figure S7.
TOP1 Knockdown Promotes RNAPII Accumulation at TSS
A positive feedback loop develops between TOP1 and RNAPII during pause-release: as the CTD of RNAPII is progressively BRD4 phosphorylated, TOP1 activity is stimulated; as TOP1 activity is increased, removal of impeding torque in turn facilitates transcription (Figure 6A).
Because of the deletion of TOP1-exon4 in HCT116KI cells, TOP1 stimulation would be undermined by impaired interaction with the CTD, so accumulation of RNAPII at pause sites might be anticipated due to a buildup of torsional stress. That such a buildup of RNAPII was not noted (data not shown) might be rationalized if the excess supercoiling in these cells were mitigated by the compensatory increase in TOP1 expression, and/or by a more effective engagement of the BRD4/PTEF-b JQ1-sensitive arm of the pause-release pathway. These hypotheses were explored using two separate approaches.
If the increase in TOP1 expression (Figure S3C) compensates for pause-release then TOP1 knockdown or TOP1 poisoning with inhibitors would create an impediment to the progression of RNAPII downstream of the pause site. To test these predictions the genomic occupancy of RNAPII was evaluated in HCT116-siTOP1 cells in which TOP1 was knocked down by 80% (Miao et al., 2007) (Figure S7F), or in HCT116 cells after a short treatment with TOP1 inhibitor CPT.
Upon TOP1 knockdown, levels of RNAPII increased at TSSs (Figure 7C). Remarkably, at paused promoters, the reduction of TOP1 (Figure 7D) resulted in further accumulation of RNAPII (Figure 7E) suggestive of an inability of RNAPII to move into the gene bodies.
Poisoning of TOP1 with CPT impairs TOP1-mediated DNA uncoiling (Koster et al., 2007). After a short CPT treatment the RNAPII released from an average promoter was unable to elongate efficiently as RNAPII density increased in the vicinity and just downstream of the pause site compared to the untreated sample (Figure 7F and S7H). The increased CTD phosphorylation and the redeployment of stored PTEF-b consequent to CPT treatment reported by others (Amente et al., 2009; Khobta et al., 2006; Sordet et al., 2008) might represent a futile attempt of the system to compensate for TOP1 poisoning. Despite transcribing beyond the pause site, RNAPII was unable to spread beyond 1.5 kb downstream of the TSS due to the build-up of opposing torsional stress (Ma et al., 2013).
If the BRD4/PTEF-b JQ1-sensitive arm compensates for reduced TOP1 stimulation to ensure an efficient transition to elongation in HCT116KI, then treatment with JQ1 should impair pause-release. When HCT116KI cells were treated with JQ1, RNAPII occupancy increased at TSSs that were most laden with RNAPII (Figure 7G). This buildup of RNAPII was not observed in WT cells where the BRD4-CTD-TOP1 nexus was intact; notably, BRD4 was equivalently loaded at promoters in both cell lines (Figure 7H). Taken together these results reinforce the model that pause-release occurs in response to two parallel actions: RNAPII-stimulated TOP1 relaxation and BRD4 recruitment of PTEF-b (Jang et al., 2005; Patel et al., 2013).
DISCUSSION
Supercoiling is often seen as an unfortunate by-product of transcription that must be eliminated by topoisomerases to enable efficient RNA synthesis. Indeed, in prokaryotes, transcription rapidly drives supercoiling to levels high enough to arrest transcription intragenically in vivo and in vitro (Chong et al., 2014; Ma et al., 2013). The dyssynchronous action of TOP1A that removes negative supercoils trailing RNAP versus DNA gyrase that removes the positive supercoils pushed ahead of the RNAP, helps to explain the “bursting” pattern of promoter output in bacteria. Our data suggest that a different and finer mechanism of regulation might occur in eukaryotes: by directly putting the processes that generate and relieve torsional stress under the supervision of the transcription apparatus, the elastic properties of DNA might be harnessed for the regulation of genetic processes. This coupling might i) help to trap negative supercoiling at TSSs to promote DNA melting during PIC formation and re-initiation (Grunberg et al., 2012; Kouzine et al., 2013; Naughton et al., 2013), ii) apply torque that helps to hold-back RNAPII at pause sites and iii) drain positive supercoils from gene bodies that would otherwise impede elongation (Ma et al., 2013).
TOP1 Participates in Pausing Regulation
Pausing is a rate controlling step during the transcription of about 60% of genes. During transcription initiation RNAPII binds to promoters, melts the DNA, initiates RNA synthesis and pauses after transcribing ~50-100 nucleotides (Adelman and Lis, 2012). Pausing and pause-release seem not to result from a single molecular switch, but involve the interplay of many factors. The various stages of the transcription cycle preceding and following pausing are associated with covalent modifications of the RNAPII CTD; this CTD ‘code’ manages mRNA capping, mRNA splicing, histone methylation, and polyadenylation via a host of dynamic interactions with many partners (Eick and Geyer, 2013). This work places TOP1 among the factors that “read” the phosphorylation code by adjusting its activity to redistribute torsional stress along the gene. Ultimately pausing must be imposed by i) affecting the chemistry that adds nucleotides to nascent transcripts and/or ii) by the mechanical forces that physically block translocation and stall RNAPII. These forces may be applied by barriers, such as nucleosomes (Gilchrist et al., 2010), bound proteins or alternative DNA conformations, or when oppositional forces and torques transmitted through the DNA fiber exceed the stall force of RNAPII (Ma et al., 2013). During pausing the RNAPII remains stably associated with nascent RNA until further signals trigger abatement of chemical and the mechanical barriers and transition to productive elongation.
We propose that BRD4 facilitates pause-release in at least two independent ways: by recruiting/regulating the PTEF-b complex and by enhancing TOP1 relaxation especially within a zone ~1.5 kb downstream of the pause site (Figure 6A). Removal of opposing supercoils within this zone may suffice to reactivate a stalled RNAP; indeed Ma et al. demonstrated that an RNAP gradually resumes transcription after relaxation of opposing torque (Ma et al., 2013).
Tuning DNA Topology to Regulate Gene Expression
The NTD of TOP1 is required for the phosphorylated CTD to stimulate TOP1 activity (Figure 4). This disordered domain has often been removed for structural and mechanistic studies (Redinbo et al., 1998); yet it serves as a conduit for the stimulation of TOP1 by phosphorylated RNAPII CTD. Because the NTD is dispensable for TOP1 nicking-closing activity and is remote from the catalytic site (Redinbo et al., 1998), we infer that the phosphorylated RNAPII CTD stimulates TOP1 by an allosteric mechanism that increases the processivity of the enzyme. Catalysis by TOP1 is highly processive, relaxing all supercoils in one DNA molecule before moving on to the next under the usual low ionic strength in in vitro assay conditions (Dynan et al., 1981). Under physiological conditions (150mM NaCl) the reaction becomes more distributive, thus in the cell, TOP1 processivity supported by RNAPII likely contributes to maximal elongation rates.
TOP1 relaxation may be modulated by factors besides RNAPII. Earlier studies showed that TOP1 catalytic activity is augmented by the SV40 virus large T antigen of simian virus 40 (Simmons et al., 1998); the Werner protein (WRN), a DNA helicase required for the maintenance of genomic stability, interacts with and stimulates TOP1 (Laine et al., 2003). The transcription factor NKX3.1 stimulates TOP1 at hormone-responsive enhancers, to support eRNA transcription and expression of the associated gene (Bowen et al., 2007; Puc et al., 2015). All-in-all, these results belie the notion that TOP1 activity is constitutive and suggest that modulation of topoisomerase activity (Baranello et al., 2014) may be a regulatory modality, for example in neurons where topoisomerases regulate the expression of long genes, like those associated with synaptic function and autism (King et al., 2013).
New Combinatorial Treatment Involving TOP1 and BET Inhibitors
TOP1 is targeted by CPT and analogues, antineoplastic agents that stabilize the TOP1-DNA cleaved complex impairing replication and transcription (Pommier, 2006). Though the ternary complex TOP1-drug-DNA induces DNA damage that activates apoptosis in tumors with WT p53, these drugs often retain some efficacy even when p53 is mutated suggesting that secondary pathways also contribute to drug activity (Fedier et al., 2003). The recruitment and requirement for TOP1 at highly expressed genes is qualitatively similar to MYC-mediated transcriptome amplification (Nie et al., 2012). Therefore, TOP1 drugs might be expected to impair the expression of many MYC targets. Because JQ1 acts through BRD4 to depress MYC (Loven et al., 2013) and so indirectly (and directly) the transcriptional targets of MYC, and because JQ1 acts synergistically with CPT, we infer that transcriptional inhibition may contribute to the anti-neoplastic action of TOP1 poisons. Further illumination of the architectural features and the protein domains involved in RNAPII-mediated activation of TOP1 may seed new strategies to uncouple pharmacologically the TOP1/RNAPII positive feedback loop on transcription without causing the toxic DNA damage associated with most TOP1 inhibitors (Pommier, 2006, 2009).
EXPERIMENTAL PROCEDURES
CRISPR-Cas9 System
MIT CRISPR Design Tool selected small guide RNAs targeting TOP1 exon 4 that were cloned in pX330-PGK-puro-SV40PA (gift of R. Casellas; NIAID, NIH). The recombination donor was made by cloning a gBlock Gene Fragment in a ColE1 origin vector. The gBlock contained 3 HA tag sequences replacing most of exon 4, flanked by two homology arms. Following puromycin selection, clones were screened by genomic PCR, and verified by sequencing and immunoblot. Positive clones were grown from single cells prior to experiments.
Purification of Proteins
Human RNAPII and general transcription factors were purified according to (Maldonado et al., 1996). Human recombinant TOP1 and BRD4 were purified as described, respectively in (Zhelkovsky and Moore, 1994) and (Maruyama et al., 2002).
In Vitro Relaxation Assay and Gel Electrophoresis Topological Analysis
TOP1 was incubated with equimolar amount of recombinant factors for 3 or 6 min in ice in TOP1 buffer (10 mM Tris-HCl pH 7.5, 50mM KCl, 5 mM MgCl2, 0.1 mM EDTA, 15 ug/ml BSA). After the addition of plasmid DNA reactions were incubated at 37°C for different times and terminated with TE-SDS 1% proteinase K. Purified DNA products were analyzed on 1-D 1.4% (w/v) agarose gel in TAE buffer, pH 7.6, containing 15 μM chloroquine or 2-D agarose gel (Extended Experimental Procedures). In this case electrophoresis used 1.8% (w/v) agarose in TAE buffer containing 3.4 μM of chloroquine for the first and 15 μM of chloroquine for the second dimension. The distribution of topoisomers was visualized with Sybr Green. Quantification of the spots used Image Quant 5.2 software.
ChIP-Seq, RNA-Seq and TOP1-Seq
TOP1 and RNAPII ChIP-Seq were as described in (Barski et al., 2007) with minor changes. Total RNAs were purified; quality check used an Agilent Bioanalyzer. RNA-Seq library preparation and sequencing were as described in (Chepelev et al., 2009). TOP1-Seq did not include formaldehyde cross-linking and followed the ChIP-Seq protocol with minor modifications.
Supplementary Material
Article Highlights.
The DNA relaxation of TOP1 is coordinated with pause-release
TOP1 activity is stimulated by BRD4 dependent phosphorylation of RNAPII
The N-term domain of TOP1 mediates interaction and stimulation by RNAPII
BRD4 inhibitors and TOP1 inhibitors synergize in killing cells
ACKNOWLEGEMENTS
Our research is supported by the Intramural Research Program of the US National Institutes of Health, Center for Cancer Research of the National Cancer Institute, the National Heart, Lung and Blood Institute, and the National Library of Medicine. The ChIP-Seq and RNA-Seq libraries were sequenced by the NHLBI DNA Sequencing and Genomics Core Facility. Some Top1-Seq experiments were supported by NIH grant GM05905 to B.F. Pugh
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
D.L., L.B. F.K. and K.Z. conceived the project, designed the experiments and wrote the manuscript. L.B. performed most experiments. D.W. performed bioinformatics analyses. F.K. performed 2D gels. K.C. and K.Y.C. performed sequencing. B.N.D. prepared recombinant BRD4 and performed BRD4 kinase assays. H.C. performed CRISPR. B.A.L. and J.P. purified the transcription components. K.W., X.Z. and R.G. performed drug screening. Y.P provided human TOP1 and ΔNTOP1. C.T., D. S., Y.P., F.P., T.M.P., K.J. and B.A.L. gave input during the project and the writing.
REFERENCES
- Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nature reviews Genetics. 2012;13:720–731. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amente S, Gargano B, Napolitano G, Lania L, Majello B. Camptothecin releases P-TEFb from the inactive 7SK snRNP complex. Cell Cycle. 2009;8:1249–1255. doi: 10.4161/cc.8.8.8286. [DOI] [PubMed] [Google Scholar]
- Baranello L, Kouzine F, Wojtowicz D, Cui K, Przytycka TM, Zhao K, Levens D. DNA break mapping reveals topoisomerase II activity genome-wide. International journal of molecular sciences. 2014;15:13111–13122. doi: 10.3390/ijms150713111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranello L, Levens D, Gupta A, Kouzine F. The importance of being supercoiled: how DNA mechanics regulate dynamic processes. Biochimica et biophysica acta. 2012;1819:632–638. doi: 10.1016/j.bbagrm.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bowen C, Stuart A, Ju JH, Tuan J, Blonder J, Conrads TP, Veenstra TD, Gelmann EP. NKX3.1 homeodomain protein binds to topoisomerase I and enhances its activity. Cancer research. 2007;67:455–464. doi: 10.1158/0008-5472.CAN-06-1591. [DOI] [PubMed] [Google Scholar]
- Brill SJ, DiNardo S, Voelkel-Meiman K, Sternglanz R. Need for DNA topoisomerase activity as a swivel for DNA replication for transcription of ribosomal RNA. Nature. 1987;326:414–416. doi: 10.1038/326414a0. [DOI] [PubMed] [Google Scholar]
- Carty SM, Greenleaf AL. Hyperphosphorylated C-terminal repeat domain-associating proteins in the nuclear proteome link transcription to DNA/chromatin modification and RNA processing. Molecular & cellular proteomics : MCP. 2002;1:598–610. doi: 10.1074/mcp.m200029-mcp200. [DOI] [PubMed] [Google Scholar]
- Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annual review of biochemistry. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- Chepelev I, Wei G, Tang Q, Zhao K. Detection of single nucleotide variations in expressed exons of the human genome using RNA-Seq. Nucleic acids research. 2009;37:e106. doi: 10.1093/nar/gkp507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung AC, Cramer P. A movie of RNA polymerase II transcription. Cell. 2012;149:1431–1437. doi: 10.1016/j.cell.2012.06.006. [DOI] [PubMed] [Google Scholar]
- Chong S, Chen C, Ge H, Xie XS. Mechanism of transcriptional bursting in bacteria. Cell. 2014;158:314–326. doi: 10.1016/j.cell.2014.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah BN, Lewis BA, Cherman N, Hewitt MC, Albrecht BK, Robey PG, Ozato K, Sims RJ, 3rd, Singer DS. BRD4 is an atypical kinase that phosphorylates serine2 of the RNA polymerase II carboxy-terminal domain. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:6927–6932. doi: 10.1073/pnas.1120422109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunaway M, Ostrander EA. Local domains of supercoiling activate a eukaryotic promoter in vivo. Nature. 1993;361:746–748. doi: 10.1038/361746a0. [DOI] [PubMed] [Google Scholar]
- Durand-Dubief M, Persson J, Norman U, Hartsuiker E, Ekwall K. Topoisomerase I regulates open chromatin and controls gene expression in vivo. The EMBO journal. 2010;29:2126–2134. doi: 10.1038/emboj.2010.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynan WS, Jendrisak JJ, Hager DA, Burgess RR. Purification and characterization of wheat germ DNA topoisomerase I (nicking-closing enzyme). The Journal of biological chemistry. 1981;256:5860–5865. [PubMed] [Google Scholar]
- Egloff S, Dienstbier M, Murphy S. Updating the RNA polymerase CTD code: adding gene-specific layers. Trends in genetics : TIG. 2012;28:333–341. doi: 10.1016/j.tig.2012.03.007. [DOI] [PubMed] [Google Scholar]
- Eick D, Geyer M. The RNA polymerase II carboxy-terminal domain (CTD) code. Chemical reviews. 2013;113:8456–8490. doi: 10.1021/cr400071f. [DOI] [PubMed] [Google Scholar]
- Fedier A, Steiner RA, Schwarz VA, Lenherr L, Haller U, Fink D. The effect of loss of Brca1 on the sensitivity to anticancer agents in p53-deficient cells. Int J Oncol. 2003;22:1169–1173. [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartenberg MR, Wang JC. Positive supercoiling of DNA greatly diminishes mRNA synthesis in yeast. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:11461–11465. doi: 10.1073/pnas.89.23.11461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist DA, Dos Santos G, Fargo DC, Xie B, Gao Y, Li L, Adelman K. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell. 2010;143:540–551. doi: 10.1016/j.cell.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunberg S, Warfield L, Hahn S. Architecture of the RNA polymerase II preinitiation complex and mechanism of ATP-dependent promoter opening. Nature structural & molecular biology. 2012;19:788–796. doi: 10.1038/nsmb.2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Molecular cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- Joshi RS, Pina B, Roca J. Positional dependence of transcriptional inhibition by DNA torsional stress in yeast chromosomes. The EMBO journal. 2010;29:740–748. doi: 10.1038/emboj.2009.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno T, Kanno Y, LeRoy G, Campos E, Sun HW, Brooks SR, ahedi G, Heightman TD, Garcia BA, Reinberg D, et al. BRD4 assists elongation of both coding and enhancer RNAs by interacting with acetylated histones. Nature structural & molecular biology. 2014;21:1047–1057. doi: 10.1038/nsmb.2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khobta A, Ferri F, Lotito L, Montecucco A, Rossi R, Capranico G. Early effects of topoisomerase I inhibition on RNA polymerase II along transcribed genes in human cells. Journal of molecular biology. 2006;357:127–138. doi: 10.1016/j.jmb.2005.12.069. [DOI] [PubMed] [Google Scholar]
- Kim WY, Dahmus ME. Purification of RNA polymerase IIO from calf thymus. The Journal of biological chemistry. 1988;263:18880–18885. [PubMed] [Google Scholar]
- King IF, Yandava CN, Mabb AM, Hsiao JS, Huang HS, Pearson BL, Calabrese JM, Starmer J, Parker JS, Magnuson T, et al. Topoisomerases facilitate transcription of long genes linked to autism. Nature. 2013;501:58–62. doi: 10.1038/nature12504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster DA, Palle K, Bot ES, Bjornsti MA, Dekker NH. Antitumour drugs impede DNA uncoiling by topoisomerase I. Nature. 2007;448:213–217. doi: 10.1038/nature05938. [DOI] [PubMed] [Google Scholar]
- Kouzine F, Gupta A, Baranello L, Wojtowicz D, Ben-Aissa K, Liu J, Przytycka TM, Levens D. Transcription-dependent dynamic supercoiling is a short-range genomic force. Nature structural & molecular biology. 2013;20:396–403. doi: 10.1038/nsmb.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzine F, Sanford S, Elisha-Feil Z, Levens D. The functional response of upstream DNA to dynamic supercoiling in vivo. Nature structural & molecular biology. 2008;15:146–154. doi: 10.1038/nsmb.1372. [DOI] [PubMed] [Google Scholar]
- Kretzschmar M, Meisterernst M, Roeder RG. Identification of human DNA topoisomerase I as a cofactor for activator-dependent transcription by RNA polymerase II. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:11508–11512. doi: 10.1073/pnas.90.24.11508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak H, Lis JT. Control of transcriptional elongation. Annual review of genetics. 2013;47:483–508. doi: 10.1146/annurev-genet-110711-155440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laco GS, Pommier Y. Role of a tryptophan anchor in human topoisomerase I structure, function and inhibition. The Biochemical journal. 2008;411:523–530. doi: 10.1042/BJ20071436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laine JP, Opresko PL, Indig FE, Harrigan JA, von Kobbe C, Bohr VA. Werner protein stimulates topoisomerase I DNA relaxation activity. Cancer research. 2003;63:7136–7146. [PubMed] [Google Scholar]
- Li CJ, Averboukh L, Pardee AB. beta-Lapachone, a novel DNA topoisomerase I inhibitor with a mode of action different from camptothecin. The Journal of biological chemistry. 1993;268:22463–22468. [PubMed] [Google Scholar]
- Liu J, Kouzine F, Nie Z, Chung HJ, Elisha-Feil Z, Weber A, Zhao K, Levens D. The FUSE/FBP/FIR/TFIIH system is a molecular machine programming a pulse of c-myc expression. The EMBO journal. 2006;25:2119–2130. doi: 10.1038/sj.emboj.7601101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu LF, Wang JC. Supercoiling of the DNA template during transcription. Proceedings of the National Academy of Sciences of the United States of America. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Ma Q, Wong K, Li W, Ohgi K, Zhang J, Aggarwal AK, Rosenfeld MG. Brd4 and JMJD6-associated anti-pause enhancers in regulation of transcriptional pause release. Cell. 2013;155:1581–1595. doi: 10.1016/j.cell.2013.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Bai L, Wang MD. Transcription under torsion. Science. 2013;340:1580–1583. doi: 10.1126/science.1235441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado E, Drapkin R, Reinberg D. Purification of human RNA polymerase II and general transcription factors. Methods in enzymology. 1996;274:72–100. doi: 10.1016/s0076-6879(96)74009-0. [DOI] [PubMed] [Google Scholar]
- Maruyama T, Farina A, Dey A, Cheong J, Bermudez VP, Tamura T, Sciortino S, Shuman J, Hurwitz J, Ozato K. A Mammalian bromodomain protein, brd4, interacts with replication factor C and inhibits progression to S phase. Molecular and cellular biology. 2002;22:6509–6520. doi: 10.1128/MCB.22.18.6509-6520.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino A, Madden KR, Lane WS, Champoux JJ, Reinberg D. DNA topoisomerase I is involved in both repression and activation of transcription. Nature. 1993;365:227–232. doi: 10.1038/365227a0. [DOI] [PubMed] [Google Scholar]
- Miao ZH, Player A, Shankavaram U, Wang YH, Zimonjic DB, Lorenzi PL, Liao ZY, Liu H, Shimura T, Zhang HL, et al. Nonclassic functions of human topoisomerase I: genome-wide and pharmacologic analyses. Cancer research. 2007;67:8752–8761. doi: 10.1158/0008-5472.CAN-06-4554. [DOI] [PubMed] [Google Scholar]
- Naughton C, Avlonitis N, Corless S, Prendergast JG, Mati IK, Eijk PP, Cockroft SL, Bradley M, Ylstra B, Gilbert N. Transcription forms and remodels supercoiling domains unfolding large-scale chromatin structures. Nature structural & molecular biology. 2013;20:387–395. doi: 10.1038/nsmb.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Z, Hu G, Wei G, Cui K, Yamane A, Resch W, Wang R, Green DR, Tessarollo L, Casellas R, et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012;151:68–79. doi: 10.1016/j.cell.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvin JD, Sharp PA. DNA topology and a minimal set of basal factors for transcription by RNA polymerase II. Cell. 1993;73:533–540. doi: 10.1016/0092-8674(93)90140-l. [DOI] [PubMed] [Google Scholar]
- Patel MC, Debrosse M, Smith M, Dey A, Huynh W, Sarai N, Heightman TD, Tamura T, Ozato K. BRD4 coordinates recruitment of pause release factor P-TEFb and the pausing complex NELF/DSIF to regulate transcription elongation of interferon-stimulated genes. Molecular and cellular biology. 2013;33:2497–2507. doi: 10.1128/MCB.01180-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen JM, Fredsoe J, Roedgaard M, Andreasen L, Mundbjerg K, Kruhoffer M, Brinch M, Schierup MH, Bjergbaek L, Andersen AH. DNA Topoisomerases Maintain Promoters in a State Competent for Transcriptional Activation in Saccharomyces cerevisiae. PLoS genetics. 2012;8:e1003128. doi: 10.1371/journal.pgen.1003128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Molecular cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Pommier Y. Topoisomerase I inhibitors: camptothecins and beyond. Nature reviews Cancer. 2006;6:789–802. doi: 10.1038/nrc1977. [DOI] [PubMed] [Google Scholar]
- Pommier Y. DNA topoisomerase I inhibitors: chemistry, biology, and interfacial inhibition. Chemical reviews. 2009;109:2894–2902. doi: 10.1021/cr900097c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puc J, Kozbial P, Li W, Tan Y, Liu Z, Suter T, Ohgi KA, Zhang J, Aggarwal AK, Rosenfeld MG. Ligand-dependent enhancer activation regulated by topoisomerase-I activity. Cell. 2015;160:367–380. doi: 10.1016/j.cell.2014.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan Y, Rajpara SM, Reza SM, Lees E, Shuman S, Mathews MB, Pe'ery T. Three RNA polymerase II carboxyl-terminal domain kinases display distinct substrate preferences. The Journal of biological chemistry. 2001;276:10913–10920. doi: 10.1074/jbc.M010975200. [DOI] [PubMed] [Google Scholar]
- Redinbo MR, Stewart L, Kuhn P, Champoux JJ, Hol WG. Crystal structures of human topoisomerase I in covalent and noncovalent complexes with DNA. Science. 1998;279:1504–1513. doi: 10.1126/science.279.5356.1504. [DOI] [PubMed] [Google Scholar]
- Roca J. Transcriptional inhibition by DNA torsional stress. Transcription. 2011;2:82–85. doi: 10.4161/trns.2.2.14807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainsbury S, Niesser J, Cramer P. Structure and function of the initially transcribing RNA polymerase II-TFIIB complex. Nature. 2013;493:437–440. doi: 10.1038/nature11715. [DOI] [PubMed] [Google Scholar]
- Shykind BM, Kim J, Stewart L, Champoux JJ, Sharp PA. Topoisomerase I enhances TFIID-TFIIA complex assembly during activation of transcription. Genes & development. 1997;11:397–407. doi: 10.1101/gad.11.3.397. [DOI] [PubMed] [Google Scholar]
- Simmons DT, Roy R, Chen L, Gai D, Trowbridge PW. The activity of topoisomerase I is modulated by large T antigen during unwinding of the SV40 origin. The Journal of biological chemistry. 1998;273:20390–20396. doi: 10.1074/jbc.273.32.20390. [DOI] [PubMed] [Google Scholar]
- Sordet O, Larochelle S, Nicolas E, Stevens EV, Zhang C, Shokat KM, Fisher RP, Pommier Y. Hyperphosphorylation of RNA polymerase II in response to topoisomerase I cleavage complexes and its association with transcription- and BRCA1-dependent degradation of topoisomerase I. Journal of molecular biology. 2008;381:540–549. doi: 10.1016/j.jmb.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangler L, Wang X, Conaway JW, Conaway RC, Dvir A. TFIIH action in transcription initiation and promoter escape requires distinct regions of downstream promoter DNA. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:5544–5549. doi: 10.1073/pnas.101004498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart L, Redinbo MR, Qiu X, Hol WG, Champoux JJ. A model for the mechanism of human topoisomerase I. Science. 1998;279:1534–1541. doi: 10.1126/science.279.5356.1534. [DOI] [PubMed] [Google Scholar]
- Strumberg D, Pilon AA, Smith M, Hickey R, Malkas L, Pommier Y. Conversion of topoisomerase I cleavage complexes on the leading strand of ribosomal DNA into 5'-phosphorylated DNA double-strand breaks by replication runoff. Molecular and cellular biology. 2000;20:3977–3987. doi: 10.1128/mcb.20.11.3977-3987.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teves SS, Henikoff S. Transcription-generated torsional stress destabilizes nucleosomes. Nature structural & molecular biology. 2014;21:88–94. doi: 10.1038/nsmb.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirode F, Busso D, Coin F, Egly JM. Reconstitution of the transcription factor TFIIH: assignment of functions for the three enzymatic subunits, XPB, XPD, and cdk7. Molecular cell. 1999;3:87–95. doi: 10.1016/s1097-2765(00)80177-x. [DOI] [PubMed] [Google Scholar]
- Uemura T, Yanagida M. Isolation of type I and II DNA topoisomerase mutants from fission yeast: single and double mutants show different phenotypes in cell growth and chromatin organization. The EMBO journal. 1984;3:1737–1744. doi: 10.1002/j.1460-2075.1984.tb02040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang MD, Schnitzer MJ, Yin H, Landick R, Gelles J, Block SM. Force and velocity measured for single molecules of RNA polymerase. Science. 1998;282:902–907. doi: 10.1126/science.282.5390.902. [DOI] [PubMed] [Google Scholar]
- Wu J, Phatnani HP, Hsieh TS, Greenleaf AL. The phosphoCTD-interacting domain of Topoisomerase I. Biochem Biophys Res Commun. 2010;397:117–119. doi: 10.1016/j.bbrc.2010.05.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SY, Chiang CM. The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. The Journal of biological chemistry. 2007;282:13141–13145. doi: 10.1074/jbc.R700001200. [DOI] [PubMed] [Google Scholar]
- Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, Zhou Q. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Molecular cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- Zhelkovsky AM, Moore CL. Overexpression of human DNA topoisomerase I in insect cells using a baculovirus vector. Protein expression and purification. 1994;5:364–370. doi: 10.1006/prep.1994.1053. [DOI] [PubMed] [Google Scholar]
- Zobeck KL, Buckley MS, Zipfel WR, Lis JT. Recruitment timing and dynamics of transcription factors at the Hsp70 loci in living cells. Molecular cell. 2010;40:965–975. doi: 10.1016/j.molcel.2010.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.