SUMMARY
Nuclear DNA wraps around core histones to form nucleosomes, which restricts the binding of transcription factors to gene regulatory sequences. Pioneer transcription factors can bind DNA sites on nucleosomes and initiate gene regulatory events, often leading to the local opening of chromatin. However, the nucleosomal configuration of open chromatin and the basis for its regulation is unclear. We combined low and high levels of MNase digestion along with core histone mapping to assess the nucleosomal configuration at enhancers and promoters in mouse liver. We find that MNase-accessible nucleosomes, bound by transcription factors, are retained more at liver-specific enhancers than at promoters and ubiquitous enhancers. The pioneer factor FoxA displaces linker histone H1, thereby keeping enhancer nucleosomes accessible in chromatin and allowing other liver-specific transcription factors to bind and stimulate transcription. Thus, nucleosomes are not exclusively repressive to gene regulation when they are retained with, and exposed by, pioneer factors.
INTRODUCTION
Chromatin consists of a fundamental repeating unit, the nucleosome, which contains genomic DNA wrapped around an octamer of the core histones H2A, H2B, H3, and H4. Nucleosome organization provides steric constraints on how transcription factors (TFs) can bind gene regulatory sequences and thereby impacts diverse processes, from cell differentiation to disease progression (Jiang and Pugh, 2009; Voss and Hager, 2014). Although most TFs cannot access nucleosomal DNA on their own, a subset of TFs possess a special ability to engage their target sites on nucleosomal DNA and are thus referred to as “pioneer” factors (Iwafuchi-Doi and Zaret, 2014; Soufi et al., 2015). In higher eukaryotes, gene expression is regulated by the coordinated action of enhancers and promoters (Levine et al., 2014). Active enhancers and promoters share diverse features, including TF occupancy, open chromatin, as seen by DNase I-hypersensitivity (Thurman et al., 2012), and active histone modifications (Creyghton et al., 2010; Heintzman et al., 2009; Rada-Iglesias A, 2011; Shen et al., 2012). In mammalian genomes, enhancers tend to be active in a tissue-specific manner, while promoters are more likely to be used in a more ubiquitous fashion (Heintzman et al., 2009; Thurman et al., 2012). Diverse lines of evidence indicate that the open chromatin structure at promoters primarily consists of a nucleosome- and histone-free region established by DNA sequence, general TFs, chromatin remodelers, and the basal transcription machinery (Floer et al., 2010; Hughes et al., 2012; Jiang and Pugh, 2009; Rhee et al., 2014; Struhl and Segal, 2013).
In contrast, there are conflicting reports on the presence of nucleosomes at active enhancers. Single locus studies with titrations of enzyme probes, sequential ChIP, and in vivo footprinting indicate an open chromatin structure at a liver-specific enhancer that retains phased nucleosomes, bound by the pioneer factor FoxA and other TFs (Chaya et al., 2001; McPherson et al., 1993). A genome-wide micrococcal nuclease (MNase) mapping study showed that at least half of FoxA2 binding sites were nucleosome-free, while the remainder of the of sites were with MNase-resistant fragments (Li et al., 2011). Yet the prevailing genome-wide view, based on MNase-seq mapping studies, is that DNase-hypersensitive, open chromatin structures at enhancers are essentially nucleosome-free (Chai et al., 2013; Gaffney et al., 2012; Kim and Shiekhattar, 2015). Pioneer factors are candidates to modulate nucleosome configuration, since in vitro studies showed that FoxA can open a local domain of chromatin without the help of an ATP-dependent chromatin remodeler (Cirillo et al., 2002; McPherson et al., 1993; Shim et al., 1998). Thus, the typical nucleosome configuration at enhancers, how it is regulated in vivo, and how it associates with tissue-specific or ubiquitous gene regulation remains unclear.
Nucleosome positions have been mapped by digesting chromatin with MNase. MNase initially cuts chromatin between nucleosomes to generate 180- to 200-bp fragments from mammalian chromatin; further digestion trims the DNA to 147 bp and then degrades the nucleosome core particle itself (Axel, 1975). Thus, in a high level digestion reaction, nucleosomes in open chromatin are destroyed, whereas nucleosomes in closed chromatin are not. Most genome-wide nucleosome mapping studies apply high level MNase digestion to maximize mononucleosome yield in bulk chromatin (e.g. (Chai et al., 2013; Gaffney et al., 2012)), and thus could destroy nucleosomes in open chromatin. A prior nucleosome mapping study, using relatively high levels of MNase digestion, reported no differences in nucleosome occupancy at selected sites in the absence of FoxA1 and FoxA2, possibly because nucleosomes at open chromatin regions had been missed (Li et al., 2011). A recent study, using substantially lower levels of chromatin digestion with methidiumpropyl-EDTA coupled with core histone ChIP, revealed more apparent nucleosome retention than had been revealed by prior high-level MNase studies (Ishii et al., 2015). In yeast, low levels of MNase digestion recovered DNA fragments within previously determined “nucleosome-free” regions (200 bp upstream of the TSS) in a subset of promoters in addition to within the −1 nucleosome position (upstream of the “nucleosome-free” region), and renamed them as “fragile nucleosomes” (Rhee et al., 2014; Weiner et al., 2010; Xi et al., 2011). However, a recent high-resolution core histone mapping study indicates “nucleosome-free” regions at promoters are indeed histone-free (Rhee et al., 2014).
Here, we sought to assess the occupancy and accessibility of nucleosomes at tissue-specific and ubiquitous regulatory sequences in a native animal tissue, and how nucleosome accessibility and its consequences are controlled. We mapped nucleosomes in adult mouse liver by low-level and high-level MNase digestion in conjunction with mapping core histones. We found that, contrary to the predominant assumption in the field, active, tissue-specific enhancers retain MNase-accessible nucleosomes significantly more than promoters and ubiquitous enhancers. Furthermore, FoxA2 is enriched near the dyad axis of accessible nucleosomes and displaces linker histones, thereby keeping nucleosomes accessible at liver-specific enhancers and helping other TFs to bind. We suggest that nucleosomes are not exclusively repressive to gene regulation when they are retained with, and exposed by, pioneer factors to stimulate tissue-specific gene activation.
RESULTS
Low-MNase digestion preferentially recovers nucleosome-size fragments in open chromatin regions
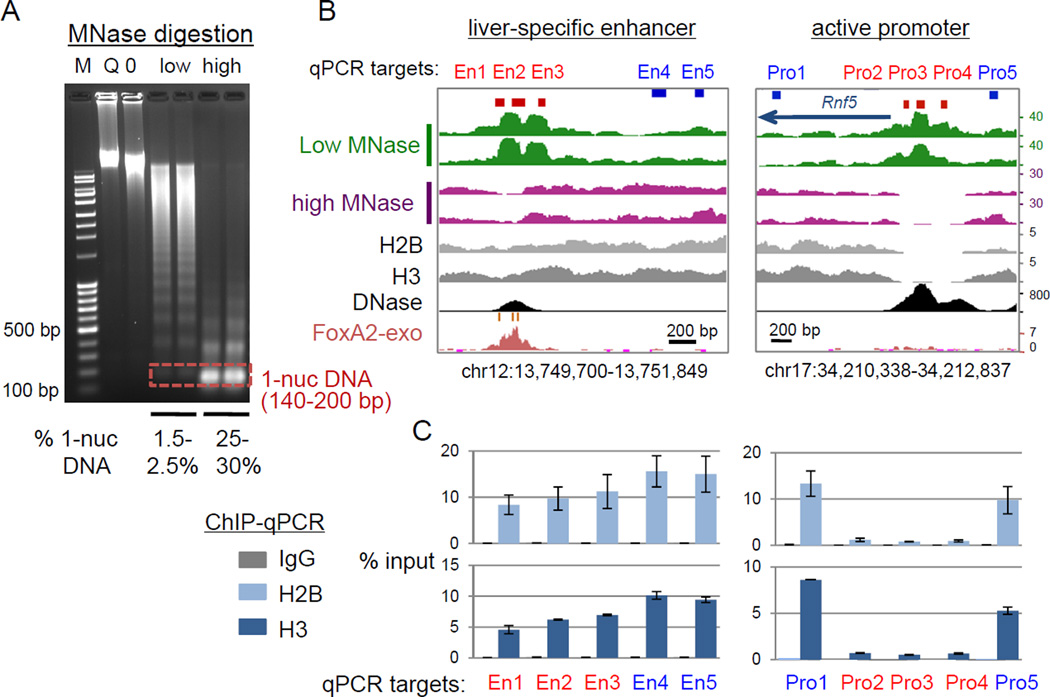
We first tested how the level of MNase digestion affects the recovery of nucleosomal-size DNA fragments at the Alb1 enhancer, which is DNase-hypersensitive in liver but not hypersensitive in spleen (Chaya et al., 2001; McPherson et al., 1993). We prepared native nuclei from adult mouse liver and spleen in a buffer containing 10 mM NaCl, to prevent salt-dependent dissociation of nucleosome cores (Jin and Felsenfeld, 2007), and digested the nuclei with a low concentration of MNase (0.5–1 U/ml) as well as a high concentration (20 U/ml). The low-MNase digestion generated large polynucleosomal fragments with a small fraction of mononucleosomal-size DNA (about 1.5∼2.5% of input chromatin, average size 175 bp), while the high-MNase digestion generated predominantly mononucleosomal-size DNA (25∼30% of input chromatin, average size 160 bp) (Fig. 1A). The latter is similar to that seen in most conventional MNase digestion for nucleosome mapping. We isolated four sizes of DNA fragments from both digests: sub-nucleosomal (25–140 bp) DNA, mononucleosomal (140–200 bp) DNA, 1.5 nuc DNA (200–260 bp), and dinucleosomal (260–400 bp) DNA (Fig. S1A). We then assessed the recovery of the DNase hypersensitive N1 nucleosome, compared to the adjacent non-hypersensitive N2 nucleosome at the liver-specific Alb1 enhancer (Chaya et al., 2001; McPherson et al., 1993) by qPCR (Fig. S1B). The low-MNase digestion enriched for the hypersensitive N1 nucleosome in the mononucleosomal-size fraction from liver, but not from spleen, and modestly enriched for the N2 nucleosome (Fig. S1C). In contrast, the conventional high-MNase digestion markedly degraded the hypersensitive N1 nucleosome (Fig. S1C). Thus, the low-MNase digestion preferentially recovers MNase-accessible and DNase-hypersensitive nucleosomal-size DNA, whereas conventional, high-MNase digestion degrades it.
Figure 1. Liver-specific enhancers retain MNase-accessible nucleosomes.
(A) marker (M), MNase-untreated controls, Q (not warmed) and 0 (warmed), and low- or high-MNase digested DNAs on an agarose gel. (B) Signal tracks of MNase/ChIP/DNase-seq reads; two biological replicates for MNase-seq. Red boxes, sites of low MNase tag enrichment tested by qPCR in C; blue boxes, control sites. (C) Core histone ChIP-qPCR enrichment (means +/− SD of two biological replicates). (See also Figure S1)
Liver-specific enhancers retain MNase accessible nucleosomes more than promoters and ubiquitous enhancers
We mapped the genome-wide distribution of MNase-resistant fragments (140–200 bp) from two biological replicates of the low-MNase digestion (total >195 million low-MNase-seq paired-end reads) and the high-MNase digestion (total >270 million high-MNase-seq paired-end reads) in mouse liver (Table S1). Shen and colleagues predicted tissue-specific and ubiquitous (active in many cell types) enhancers based on p300 binding distal to TSSs, along with H3K4me1 but not H3K4me3, in the mouse genome (Shen et al., 2012). We further filtered these enhancers with H3K27ac peaks (ENCODE, GSM1000140), because H3K27ac predominantly marks active regulatory sequences, while H3K4me1 marks both poised and active enhancers (Creyghton et al., 2010; Rada-Iglesias A, 2011), and centered the enhancers by DNase hypersensitive sites (ENCODE, GSM1014195), where TFs are bound.
Two examples of active promoters and enhancers, respectively (red blocks), showed greater low-MNase-seq signals (green tracks) than high-MNase-seq signals (purple tracks) (Fig. 1B, S1D), indicating the presence of MNase-accessible chromatin. To test whether the low-MNase-seq signals represent nucleosomes, we performed ChIP-qPCR for core histones with crosslinked and highly sonicated chromatin, in order to avoid an MNase digestion bias. Notably, the two examples of liver-specific enhancers were markedly less depleted for core histones H2B and H3 than the active promoters (Fig. 1C and S1E), suggesting that low-MNase signals at liver-specific enhancers represent nucleosomes, but those at promoters do not. Genome-wide analysis also confirmed greater low-MNase enrichment than high-MNase-seq signals over active regulatory sequences (Fig. 2A for averaged signal; p-value < 2.2e-16, and 2B for individual signals).
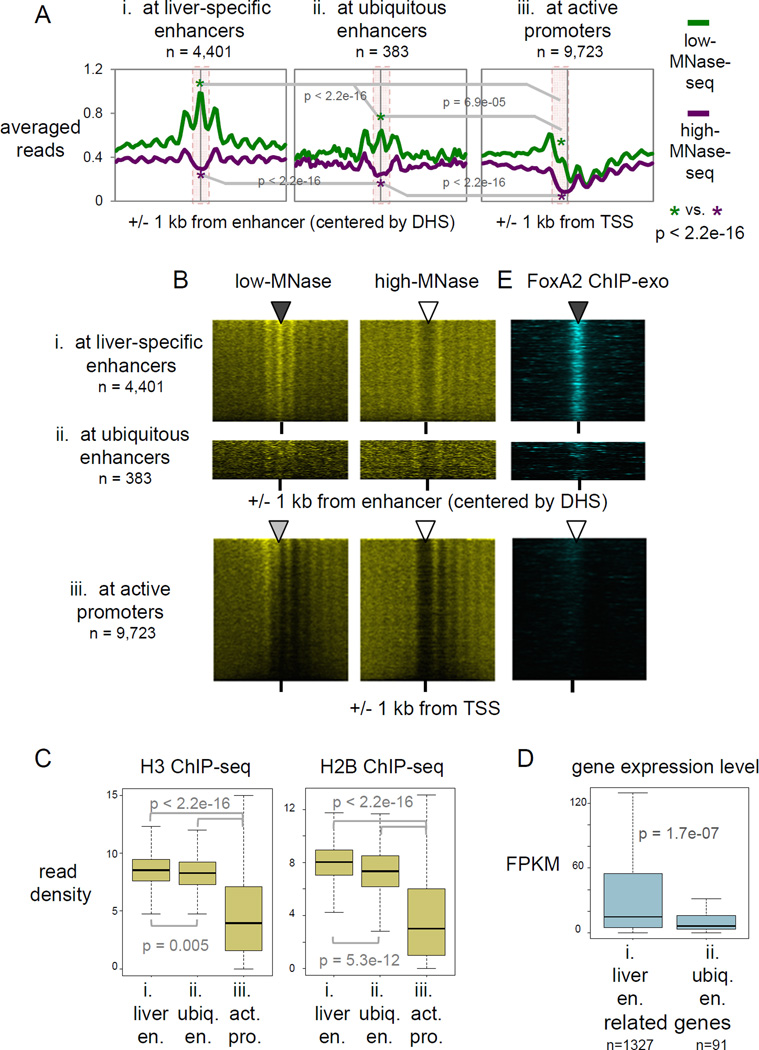
Figure 2. Liver-specific enhancers retain MNase-accessible nucleosomes genome-wide.
(A) Averaged profiles of low- and high-MNase-seq signal enrichments at, liver-specific enhancers ubiquitous enhancers, and active promoters. p-values by Wilcoxon rank-sum test at central 200 bp of enhancers and at upstream 200 bp from active TSS (pink box area). (B, E) Heatmaps of MNase-seqs and FoxA2 ChIP-exo at liver-specific enhancers, ubiquitous enhancers, and active promoters, rank ordered by low-MNase-seq read density. (C) Box and whisker plots show H3 and H2B ChIP-seq signal enrichment at central 200 bp of liver-specific and ubiquitous enhancers and at upstream 200 bp from active TSS. p-value by Wilcoxon rank sum test. (D) Gene expression levels of target genes of liver-specific and ubiquitous enhancers, based on predicted enhancer-promoter pairs (Shen et al., 2012). (See also Figure S2)
Furthermore, we found that low-MNase-seq signals were most enriched at liver-specific enhancers (central 200 bp), compared to ubiquitous enhancers (central 200 bp) and active promoters (upstream 200 bp of TSS) (Fig. 2A; p-value < 2.2e-16). Since the number of ubiquitous enhancers is much smaller (n=383) than the other groups, we randomly sampled 383 liver-specific enhancers in order to compensate for the difference. We confirmed that the sampled enhancers have similar MNase profiles as all liver-specific enhancers, as well as a higher enrichment of the low-MNase-seq signals than at ubiquitous enhancers (p-value < 2.2e-16) (data not shown; see Extended Experimental Procedures, Meta-locus plot section), suggesting that our findings are not due to the difference of the number of sites tested. Notably, genome-wide ChIP-seq showed that the liver-specific enhancers (central 200 bp) were slightly depleted for core histones H2B and H3, but significantly less so than the ubiquitous enhancers (central 200 bp) (p-value 0.005 for H3 and 5.3e-12 for H2B) and markedly less depleted for the histones than were the active promoters (upstream 200 bp of TSS) (p-value < 2.2e-16 for H2B and H3) (Fig. 2C, S2C).
We also curated active promoters into liver-specific and ubiquitous promoters based on RNA polymerase II binding patterns (Shen et al., 2012), and the majority of promoters appeared to be active in other tissues, designated as ubiquitous (n=5,010), rather than liver-specific (n=169). Although the liver-specific promoters had more low-MNase signal and less histone depletion, compared to ubiquitous promoters, they were significantly more depleted of core histones than liver-specific enhancers (p-value < 2.2e-16 for H3 and H2B) (Fig. S2A, S2B). Since the liver-specific enhancers are more stably or frequently occupied by nucleosomes in accessible chromatin, in contrast to ubiquitous enhancers and active promoters, we refer to the former nucleosomes as “exposed” or “accessible”. Previous studies had reported low-MNase signal enrichment at subset of active promoters (upstream 200 bp of TSS) in yeast, which they designated as “fragile nucleosomes” (Weiner et al., 2010; Xi et al., 2011). However, we found that such low-MNase enrichment at active promoters in mammalian tissue can represent primarily a histone depleted state, likely by the stochastic production of mono-nucleosomal size fragments even on nucleosome-free DNA. Regardless, there is a marked difference in histone retention between active liver-specific enhancers and ubiquitous enhancers and promoters.
Histone modifications and variants that do not correlate with accessible enhancer nucleosomes
To address which factors are associated with accessible nucleosomes in native liver, we compared several active chromatin features. Surprisingly, the active histone modification H3K27ac (ENCODE; GSM1000140), and the enhancer histone modification H3K4me1 (ENCODE; GSM769015) were not markedly enriched with accessible nucleosomes at liver specific enhancers (compare Fig. 2A and S2D). Similarly, histone variants H2A.Z (Menet et al., 2014) (GSE47143) and H3.3 (Bargaje et al., 2012); ERR234277) did not show proportional enrichment directly over accessible nucleosomes at liver-specific enhancers, though there was modest enrichment nearby (compare Fig. 2A and S2D).
Since the accessible nucleosomes in liver-specific enhancers were in a low modification state, we tested whether the expression levels of the corresponding target genes was also low. Shen and colleagues developed an algorithm to detect co-regulated promoters-enhancer clusters, and effectively predicted enhancer-promoter pairs in various tissues, including adult liver (Shen et al., 2012). Based on this prediction and liver RNA-seq data (ENCODE; ENCSR000AJU), we assessed the gene expression levels of target genes of liver-specific and ubiquitous enhancers. Remarkably, the liver-specific enhancers, enriched for accessible nucleosomes, drove higher gene expression levels than the ubiquitous enhancers (Fig. 2D), even though the former exhibited lower “active” modification states. Furthermore, when we compared the same number of enhancers from the liver-specific and ubiquitous group exhibiting similar ranges of active H3K27ac enrichment, we still observed higher enrichment of low-MNase and core histone signals at the liver-specific enhancers than at the ubiquitous enhancers (Fig. S2E). These results indicate that active histone modifications known to be enriched at enhancers (largely from cell line studies) do not drive nucleosome accessibility or target gene expression at active enhancers in adult liver tissue. Our results are consistent with recent findings that developmentally regulated genes in embryos and tissue-specific genes show much lower enrichment for active histone modifications, compared to ubiquitously expressed genes, even though the former genes are expressed at comparable or higher levels than ubiquitous genes (Perez-Lluch et al., 2015).
The pioneer factor FoxA2 is enriched at the dyad axis of accessible nucleosomes, along with other liver-enriched TFs
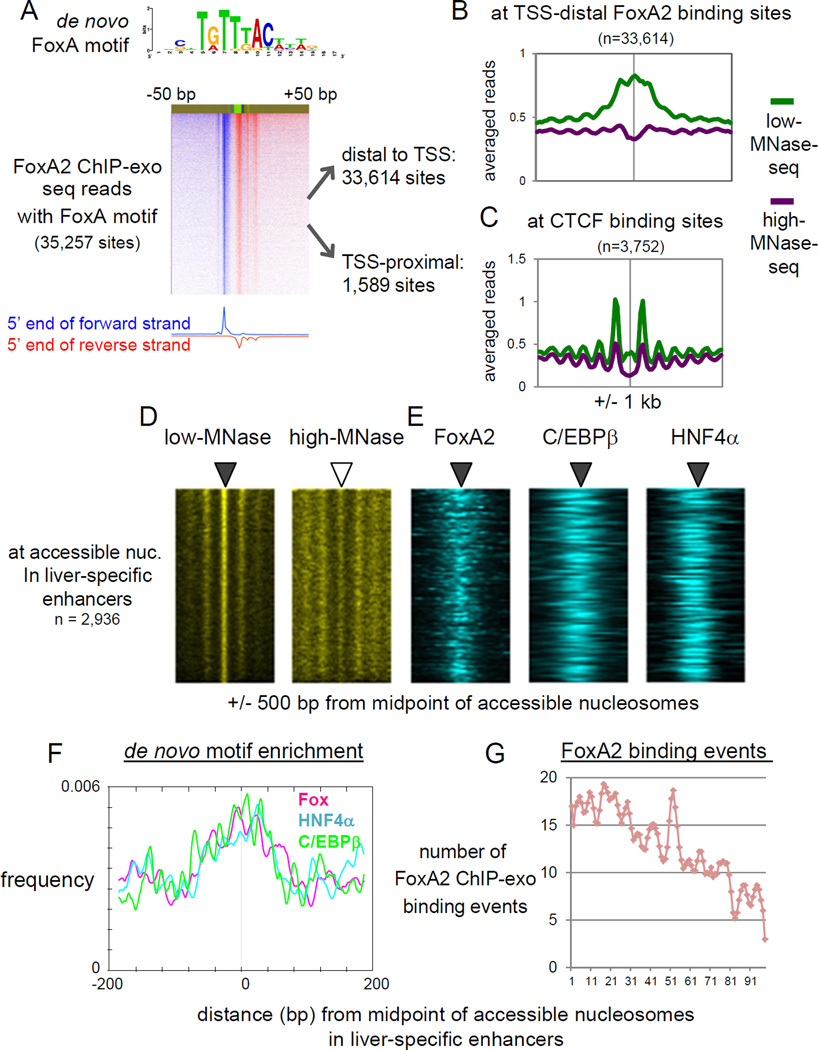
We next assessed the relationship between binding of the pioneer factor FoxA, which regulates many liver-specific genes (Li et al., 2012), and nucleosome configuration in vivo. Since FoxA1 and FoxA2 exhibit quantitatively similar overlap of binding to many target sites and both factors are, for the most part, functionally redundant in adult liver (Bochkis et al., 2012; Watts et al., 2011), here we focused on FoxA2. We identified 35,257 FoxA2 binding sites harboring the FoxA binding motif in adult mouse liver, at base-pair resolution, by ChIP with lambda exonuclease digestion followed by sequencing (ChIP-exo-seq) (Rhee and Pugh, 2011) (Fig. 3A). The vast majority of FoxA2 binding sites (95%; 33,614 sites) were located distal to transcription start sites (TSS) of RefSeq genes (> +/− 500 bp distal to TSS) and only a minority of FoxA2 binding sites (4.5%; 1,589 sites) were located proximal to TSSs (< +/− 500 bp proximal to TSS) (Fig. 3A), suggesting that FoxA2 is primarily involved in gene regulation through distal regulatory regions, i.e. enhancers. Both distal and TSS-proximal FoxA2 peaks mostly overlapped with DHS (ENCODE, GSM1014195) (86% and 96%, respectively). We found that binding of FoxA2 was highly enriched for accessible nucleosomes at liver-specific enhancers, but not at ubiquitous enhancers and active promoters (Fig. 2E). We compared our MNase and core histone enrichments at the FoxA2 binding sites with such enrichments at CTCF binding sites, which are known to be nucleosome-free and flanked by well-positioned nucleosomes (Carone et al., 2014), in active chromatin (as assessed by association with H3K27ac peaks). We found that both low MNase and core histone signals were much more enriched at FoxA2 binding sites compared with CTCF binding sites (Fig. 3B, 3C, S3A; p-value < 2.2e-16). This supports our finding that the accessible nucleosome configuration at FoxA2 binding sites represents nucleosomes rather than nucleosome-free DNA, with the latter seen at CTCF sites.
Figure 3. Pioneer factor FoxA2 and liver TFs are co-localized at accessible nucleosomes in liver-specific enhancers.
(A) FoxA2 ChIP-exo tag 5’ end distribution centered by de novo FoxA motif midpoint and sorted by FoxA2 occupancy level. (B) Averaged profiles of low- and high-MNase-seq at TSS-distal FoxA2 binding sites, and (C) at CTCF binding sites overlapping with H3K27ac peaks. (D. E) Heatmaps of MNase-seqs, and FoxA2 ChIP-exo, C/EBβ and HNF4α ChIP-seq at MNase-accessible nucleosomes in liver-specific enhancers, rank ordered by low-MNase-seq read density. (F) Profiles of de novo motifs enrichment relative to midpoint of accessible nucleosome in enhancers. (G) Histogram of FoxA2 ChIP-exo binding locations relative to midpoint of accessible nucleosomes. The midpoint approximating the position of the respective nucleosome dyad axis. (See also Figure S3)
To obtain insight into which factors associate with nucleosomes in an unbiased manner, we conducted de novo motif analysis of these regulatory sequences. We found that the ubiquitous enhancers were enriched for the motifs of ubiquitous TFs (Sp1 and Ets factors) and Fox (Fig. S3B). The liver-specific enhancers were enriched for the Fox motif, which is very similar to the de novo motif from our FoxA2 ChIP-exo analysis, and also enriched for the motifs of other liver TFs (HNF4a, C/EBP, and Onecut) and certain ubiquitous TF motifs (NF-I and STAT) (Fig. S3B). The ubiquitous factor Sp1 recruits the SWI/SNF chromatin remodeling complex to facilitate chromatin remodeling and transcriptional activation (Kadam and Emerson, 2003), consistent with Sp elements occurring at more nucleosome depleted ubiquitous enhancers (Fig. S3B).
Next, we analyzed how the de novo motifs were localized relative to the apparent dyad axis of the accessible nucleosomes at enhancers. We defined a class of accessible nucleosomes at positions where the 145-bp window counts (centered around MNase-seq fragment midpoints) in the low MNase-seq data were significantly higher than in high MNase-seq data (Fig. 3D). Ubiquitous enhancers had scattered TF motifs throughout the MNase-accessible sites (Fig. S3C). By contrast, the liver-specific enhancers showed FoxA2, C/EPBβ (Jakobsen et al., 2013) and HNF4α (Hoffman et al., 2010) binding events (Fig. 3E) and these motifs focused around the nucleosome dyad axis (Fig. 3F). Our single nucleotide-resolution mapping revealed FoxA2 binding events near the nucleosome dyad axis (Fig. 3G), as had originally been reported for FoxA at the Alb1 enhancer by in vivo footprinting studies (McPherson et al., 1993). Altogether, our results suggest that FoxA and other liver enriched TFs are cooperatively bound to the accessible nucleosomes at liver-specific enhancers.
FoxA2 binds more with core histones than other liver TFs at enhancers
To assess whether FoxA or other liver TFs are bound to accessible nucleosomes in vivo or if there are separate populations of TFs bound to free DNA, we performed sequential ChIP-qPCR (re-ChIP-qPCR) for FoxA2, C/EBβ, and HNF4α, followed by ChIP for different core histones. To minimize false positive ChIP signals from large DNA fragments, we fragmented the liver chromatin to about 200 bp by sonication (Fig. 4A). We selected qPCR target sites in promoters and enhancers where MNase-accessible regions overlap with TF binding (e.g. Fig. 4B). As expected, the first ChIP signals for TFs were enriched at their target sites similarly at promoters and enhancers (Fig. S4). However, the secondary ChIP revealed that FoxA2 was significantly more enriched with core histones at enhancers, compared to promoters (Fig. 4C, pink bars, compare P vs E target sites). C/EBββ and HNF4α were overall less enriched with core histones, but still showed a statistically significant enrichment at active enhancers compared to active promoters (Fig. 4C, green and blue bars). FoxA2 co-occupancy with core histones was also seen in other studies (Chaya et al., 2001; Li et al., 2011), and here we show that FoxA2 binds more stably or frequently with core histones on accessible nucleosomes at enhancers than the other liver TFs. These results directly show that FoxA and other TFs can be bound to accessible nucleosomes.
Figure 4. FoxA2 binds more with core histones than other liver TFs at enhancers.
(A) Biological replicates of sonicated liver chromatin on agarose gel. (B) Examples of re-ChIP-qPCR target sites with MNase/ChIP-seq signals. (C) Box and whisker plots show re-ChIP-qPCR enrichment of core histone H2B and H3 over IgG at the promoter and enhancer target sites. Numbers in parentheses indicate number of sites tested. p-value by Wilcoxon rank sum test. (See also Figure S4)
FoxA2 binding is required to keep nucleosomes accessible in chromatin
We addressed whether FoxA can regulate the nucleosome state in vivo using the genome-wide low- and high-MNase method. We deleted FoxA1 and FoxA2 in a hepatocyte-specific manner (see Experimental Procedure), purified hepatocytes from the mutant livers, along with from livers of wild type controls, and confirmed depletion of FoxA1 and FoxA2, with residual FoxA3 expression (-Fig. S5A). Previous MNase-seq data in this study was derived from whole liver, which, considering the polyploidy nature of mature hepatocytes, is 90% comprised of chromatin signals from hepatocytes.
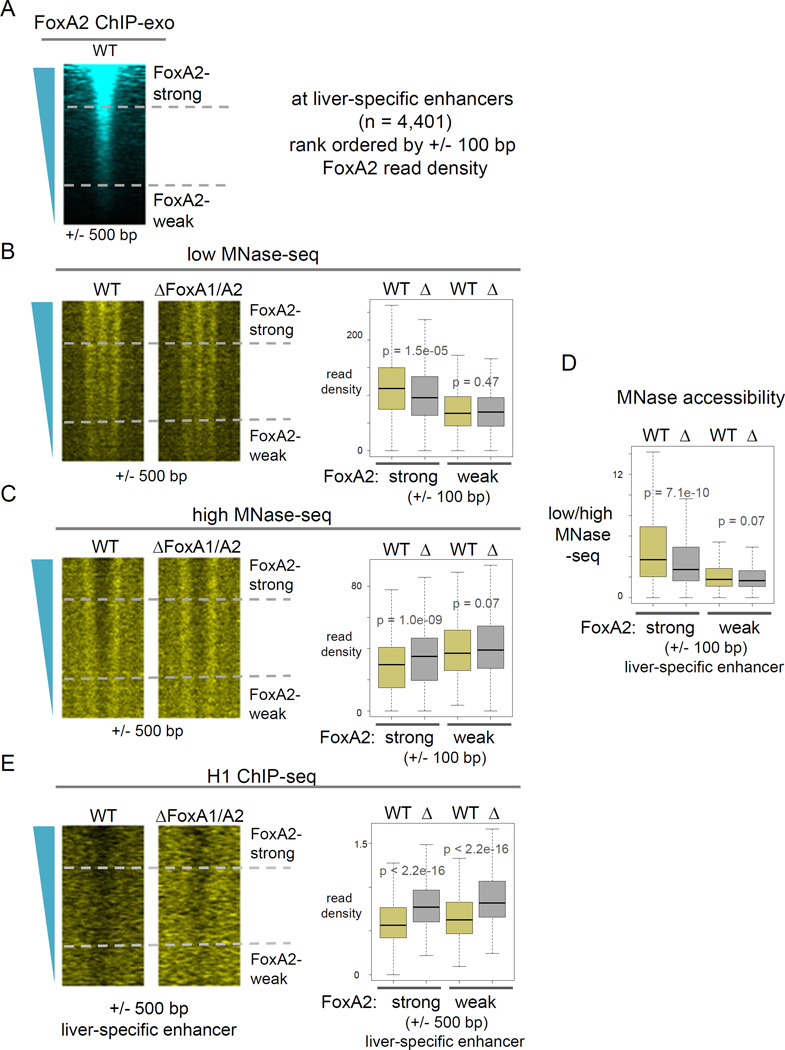
We rank ordered the liver-specific enhancers based on FoxA2 ChIP-exo read density and curated them into top 25% (FoxA2-strong) and bottom 25% (FoxA2-weak) FoxA-bound enhancers (Fig. 5A). In the wild-type controls, FoxA2-strong enhancers showed a more accessible nucleosome configuration than FoxA2-weak enhancers (Fig. 5D, yellow boxes at FoxA2-strong vs. FoxA2-weak), which is due to more low-MNase-seq signals (Fig. 5B, yellow boxes) and fewer high-MNase-seq signals at FoxA2-strong enhancers (Fig. 5C, yellow boxes). In FoxA1/A2 deletion (Δ) mutants, FoxA2-strong enhancers showed a significant reduction of nucleosome accessibility (Fig. 5D, left yellow vs. gray boxes), as shown by reduced low-MNase signals (Fig. 5B left yellow vs. gray boxes) and increased high-MNase signals (Fig. 5C, left yellow vs. gray boxes). However, FoxA2-weak enhancers and silent enhancers in liver (e.g., heart-specific enhancers (Shen et al., 2012)), where FoxA did not bind, showed marginal or no effects on nucleosome accessibility in the mutants (Fig. 5B–D, right yellow vs. gray boxes; Fig. S7B, C, yellow vs. gray boxes), as expected.
Figure 5. FoxA2 binding is required to keep nucleosomes accessible in chromatin.
Heatmap of (A) FoxA2 ChIP-exo in wild type, (B) low-MNase-seq, (C) high-MNase-seq, (D) low- over high-MNase-seq, and (E) H1 ChIP-seq in wild type (WT) and the FoxA1/A2 deletion mutant (Δ) at liver-specific enhancers, rank ordered by FoxA2 ChIP-exo read density. Box and whisker plots show at central 200 bp or 1000 bp of the top 25% FoxA2 occupied (FoxA2-strong) enhancers and at the bottom 25% FoxA2 occupied (FoxA2-weak) enhancers. p-values by Wilcoxon rank-sum test. (See also Figure S5–S7).
Next, we used qPCR to more quantitatively assess 12 control sites that lacked FoxA2 binding: 6 sites at non-accessible nucleosomes, but within 2 kb proximal to FoxA2-bound accessible nucleosomes (e.g. Fig. S6A), and 6 sites at accessible nucleosomes not bound by FoxA2 (e.g. Fig. S6B). Most control sites did not change their MNase accessibility in the FoxA1/A2 mutants (Fig. S6D, E). Thus, the FoxA factors do not affect nucleosome accessibility nonspecifically at sites where they do not bind. By contrast, all 12 accessible nucleosomes with FoxA2 binding events at enhancers (e.g. Fig. S6C) had significantly decreased MNase accessibility in the mutants (Fig. S6F). Furthermore, 7 sites have their nearest gene exhibiting diminished expression of more than 2 fold in FoxA mutants (Li et al., 2011) (Fig. S6G), associating FoxA binding with gene activation. Taken together, low- and high-MNase digestions reveal that the pioneer factors FoxA1 and FoxA2 help maintain an accessible nucleosome configuration at liver-specific enhancers in vivo.
FoxA binding displaces linker histone H1 from nucleosomes
Interestingly, the “winged helix” DNA-binding domain structure of FoxA proteins highly resembles that of linker histone H1 (Clark et al., 1993; Ramakrishnan et al., 1993), the latter of which binds near the dyad axis of the nucleosome (Goytisolo et al., 1996), as does FoxA2 (Fig. 3E). A single locus study showed that induction of FoxA caused reduction of H1 occupancy at Alb1 enhancer (Taube et al., 2010), similar to what had been reported from in vitro studies with purified proteins (Cirillo et al., 2002; Cirillo et al., 1998). Here we assessed the genome-wide occupancy of linker histone H1 in mouse hepatocytes of wild type controls and FoxA1/A2 deletion mutants (Fig. S5B). First, in the wild type, we found that higher FoxA2 occupancy correlates with more H1 depletion (Fig. 5E for +/− 500 bp, p-value = 2.1e-06; S7E for +/− 100 bp, p-value = 5.5e-11; compare yellow boxes at FoxA2-strong vs. FoxA2-weak), suggesting that FoxA binding is involved in displacement of linker histone. In the deletion mutants, enhancers showed a striking increase in H1 deposition (p-value < 2.2e-16) (Fig. 5E for +/− 500 bp, S7E for +/− 100 bp; yellow vs. gray boxes). In contrast, silent heart enhancers, where FoxA2 was not bound, did not show an effect of FoxA deletion on H1 deposition level (Fig. S7D). These in vivo genetic results indicate that FoxA binding displaces linker histones from the local chromatin, providing a simple explanation for the underlying nucleosomes becoming more accessible.
FoxA binding helps other transcription factors bind to their target sites
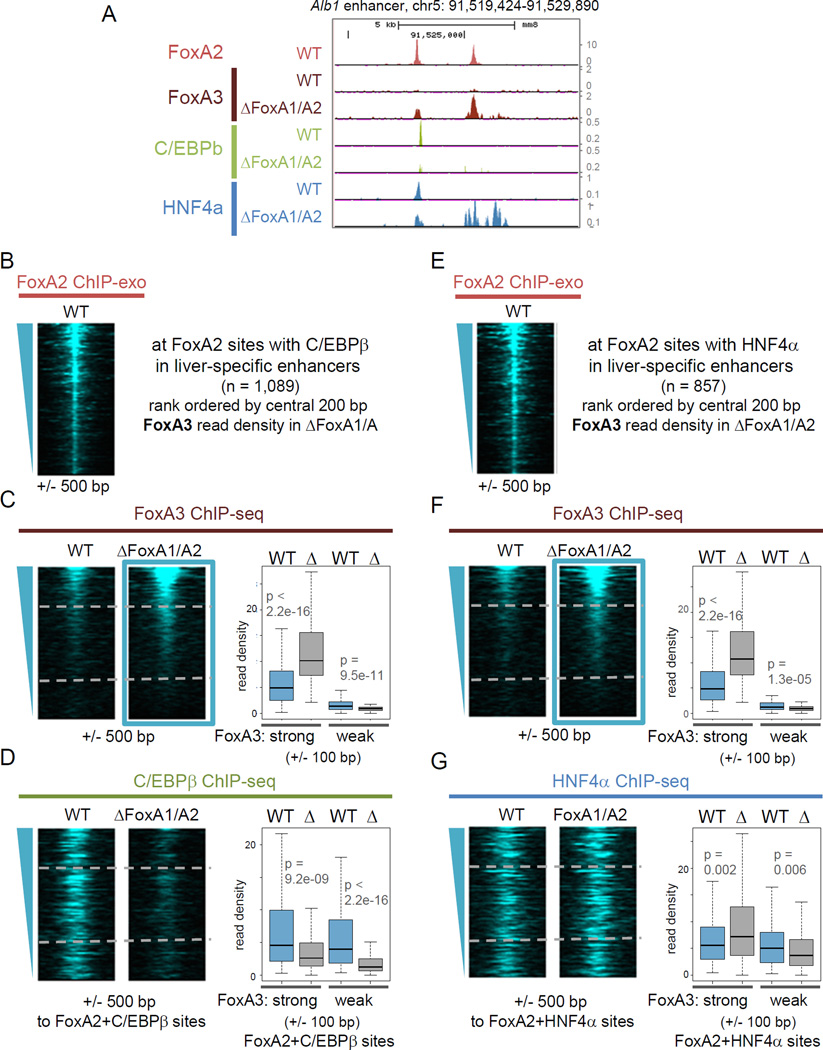
Earlier studies indicated that binding of TFs, such as CREB, GR, and ER, are dependent on pioneer factor FoxA binding (Carroll et al., 2005; Li et al., 2012; Zhang et al., 2005). Here, we examined the genome-wide consequences of FoxA1/A2 deletion on C/EBββ and HNF4α binding, using the FoxA1/A2 deletion mutants (Fig. S5B). We also performed ChIP-seq for FoxA3, the third of the three FoxAs expressed in liver, in order to define compensatory effects in the FoxA1/A2 deletion background. Microarray results showed that C/EBββ, HNF4α, and FoxA3 expression were not changed beyond 2-fold in the FoxA1/A2 deletion mutants (Li et al., 2011), and Western blot analysis showed a slight reduction of HNF4β level and no difference of C/EBββ and FoxA3 level in the mutants (Fig. S5B). Around the Alb1 enhancer, there were two FoxA2 binding peaks but no FoxA3 signal in the wild type (Fig. 6A). However, in the FoxA1/A2 mutants, we found modest emergence of FoxA3 signal at an 5’ site, where C/EBββ binding was diminished and HNF4α binding was slightly decreased, and a strong emergence of FoxA3 signal at the 3’ site, where de novo HNF4α binding occurred (Fig. 6A). These findings reveal partial compensation by FoxA3.
Figure 6. FoxA binding allows other liver-enriched TFs to bind.
(A) FoxA2 ChIP-exo in wild type, and FoxA3, C/EBβ and HNF4α ChIP-seq signal tracks in wild type and the FoxA1/FoxA2 deletion mutant. (B–D) Heatmaps of FoxA2, FoxA3 and C/EBβ ChIP-seq in wild type (WT) and the FoxA1/A2 mutant (Δ) at FoxA2 binding sites overlapped with C/EBβ binding site in liver-specific enhancers, rank ordered by FoxA3 ChIP-seq signal in the FoxA1/A2 mutant. (E–G) Heatmaps of FoxA2, FoxA3 and HNF4α ChIP-seq in wild type and the FoxA1/A2 mutant at FoxA2 binding sites overlapped with HNF4α binding site in liver-specific enhancers, rank ordered by FoxA3 ChIP-seq read density in the FoxA1/A2 mutant. Box and whisker plots show at central 200 bp of the top 25% FoxA3 occupied (FoxA3-strong) enhancers and at the bottom 25% FoxA3 occupied (FoxA3-weak) enhancers. p-values by Wilcoxon rank-sum test. (See also Figure S6).
Next, we analyzed the genome-wide effects of FoxA1/A2 deletion at those FoxA2 binding sites that overlapped with C/EBββ binding (Fig. 6B–D) or with HNF4α binding (Fig. 6E–G) at liver-specific enhancers. We rank ordered the sites by FoxA3 ChIP-seq read density in the FoxA1/A2 mutants and curated the top 25% of sites (FoxA3-strong) where FoxA3 binding was significantly increased and could compensate FoxA1/A2 loss in the mutants, and the bottom 25% sites (FoxA3-weak), where FoxA3 compensation was minimal (Fig. 6C, F). We found a striking reduction of C/EBββ binding at FoxA3-weak sites (Fig. 6D, right blue vs. gray boxes) and FoxA3-strong sites (Fig. 6D, left blue vs. gray boxes) in the FoxA1/A2 mutants, indicating that FoxA1 and FoxA2 are required for C/EBββ recruitment to their target sites.
As was seen at the Alb1 enhancer sites, genome-wide HNF4α binding showed an increase upon FoxA1/A2 deletion at FoxA3-strong sites (Fig. 6G, left blue vs. gray boxes), but not at FoxA3-weak sites (Fig. 6G, right blue vs. gray boxes), suggesting that FoxA3 recruits HNF4α more efficiently than FoxA1/A2. We conclude that the FoxA family of pioneer factors enhances the binding of different liver-specific transcription factors to liver enhancers.
Maintenance of accessible nucleosomes by FoxA for liver-specific gene activation
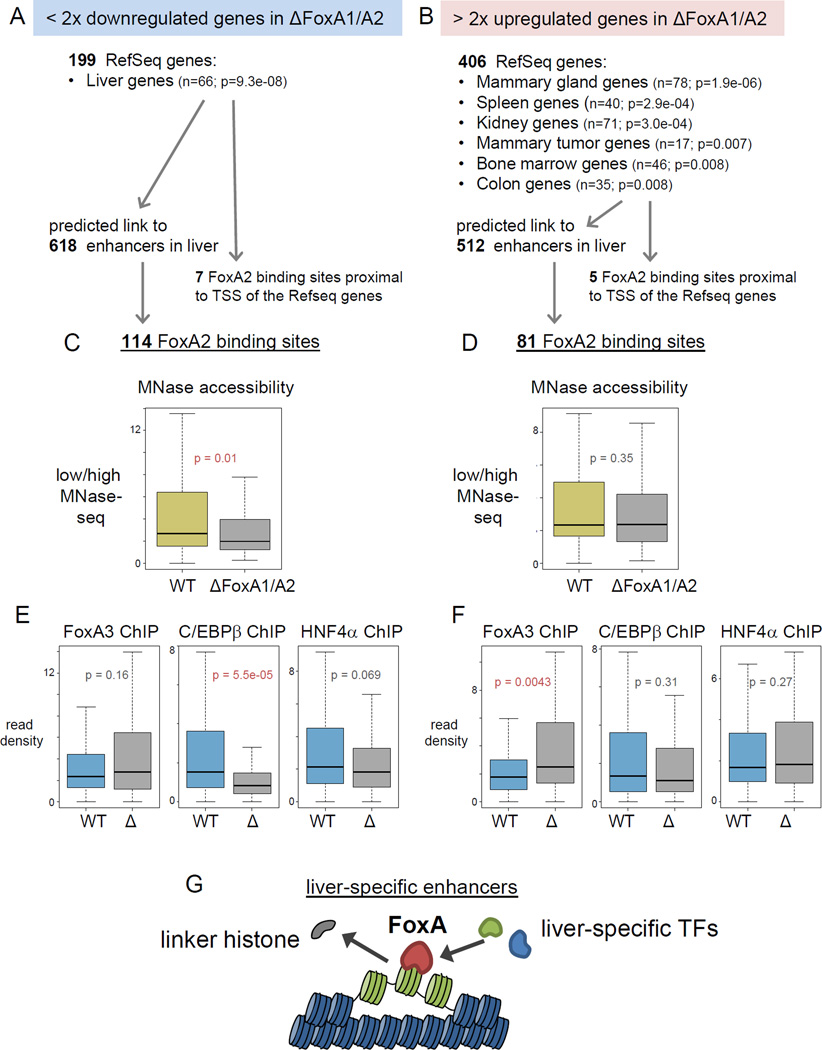
We assessed how the maintainance of nucleosome accessibility by FoxA relates to the regulation of target genes. We curated published gene expression profiles of the liver (Li et al., 2011) into upregulated genes and downregulated genes in the FoxA1/A2 mutants. We found 199 genes that were at least two-fold downregulated (Fig. 7A) and 406 genes that were at least a two-fold upregulated (Fig. 7B, Supplemental Table 3) in the FoxA1/A2 deletion mutants. Tissue expression analysis and KEGG pathway analysis (DAVID Bioinformatics Resources, UniProt tissue expression database, (Huang da et al., 2009)) indicated that the downregulated genes were related to liver function, including various metabolic pathways (Fig. 7A, Supplemental Table 3), as expected for FoxA targets, whereas the upregulated genes were related to non-liver lineages (e.g. mammary gland, spleen and kidney) (Fig. 7B, Supplemental Table 3), consistent with previous studies showing that FoxA occupies certain sites that can be repressed in the adult liver (Watts et al., 2011). We found only a few FoxA2 binding sites proximal to TSSs (< +/− 300 bp) at these target genes (Fig. 7A, B), indicating that FoxA2 binding at the promoter has a minimal effect on the expression of these genes. We assessed the predicted enhancer-promoter pairs in liver (Shen et al., 2012) to find enhancers that linked to differentially expresssed genes in the FoxA1/A2 deletion mutants. We found 618 enhancers associated with downregulated genes and 512 enhancers associated with upregulated genes in the deletion mutants (Fig. 7A, B). We found 114 FoxA2 binding sites in the enhancers linked to downregulated genes, and 81 FoxA2 binding sites in the enhancers linked to upregulated genes (Fig. 7A, B).
Figure 7. Maintenance of accessible nucleosomes by FoxA is required for liver-specific gene activation.
(A, B) Tissue Expression analysis for > two-fold upregulated or downregulated genes in the FoxA1/A2 deletion mutant (Li et al., 2011) (See also Table 3). Number of enhancers that potentially link to the target genes (Shen et al., 2012) and number of FoxA2 ChIP-exo sites in the enhancers. (C, D) Box and whiskers plot show MNase accessibility (low/high MNase-seq) in wild type and the FoxA1/A2 mutant at central 200 bp of downregulated genes related FoxA2 sites and (D) at upregulated genes related FoxA2 sites (See also Figure S7). (E) Box and whiskers plot show FoxA3, C/EBβ and HNF4α ChIP-seq in wild type (WT) and the FoxA1/A2 mutant (Δ) at central 200 bp of downregulated genes related FoxA2 sites and (F) at upregulated genes related FoxA2 sites. p-values by Wilcoxon rank-sum test. (G) Models of nucleosome configuration at liver-specific enhancers. FoxA2 binding is required to displace linker histones, keep nucleosome accessible, and help recruitment of other liver enriched TFs, which stimulate liver gene transcription.
Next, we focused on these FoxA2 target sites and analyzed how the nucleosomal state was changed in the FoxA1/A2 mutants by low- and high-MNase-seq. At FoxA2 binding sites linked to downregulated target genes, nucleosome accessibility was decreased significantly in the FoxA1/A2 deletion mutants (p-value = 0.01) (Fig. 7C), as shown by reduced low-MNase signals and increased high-MNase signals (Fig. S7F). By contrast, at FoxA2 binding sites linked to upregulated target genes, nucleosome accessibility did not change significantly in the FoxA1/A2 deletion mutants (p-value = 0.353) (Fig. 7D, S7G).
We also analyzed how TF binding was affected at the FoxA2 binding sites linked to down- or up-regulated target genes in the FoxA1/A2 deletion mutants. At the FoxA2 sites linked to down-regulated genes, where FoxA3 binding did not increase significantly and was therefore unable to compensate for FoxA1/A2 loss, nucleosome accessibility decreased in the mutants and C/EBβ binding was significantly decreased, while HNF4α binding was modestly decreased (Fig. 7C, 7E). Thus, C/EBβ was more sensitive to the change in nucleosome accessibility than was HNF4α. In contrast, at the FoxA2 sites linked to up-regulated genes, where FoxA3 binding was significantly increased and therefore could compensate for FoxA1/A2 loss, nucleosome accessibility did not change in the mutants, and C/EPBb and HNF4α binding did not change (Fig. 7D, 7F). Thus FoxA3 can, to some extent, compensate for FoxA1/A2 loss to maintain nucleosome accessibility. Furthermore, we performed de novo motif analysis around the FoxA2 binding sites (+/− 100 bp) linked to upregulated genes, and revealed ER/Rfx motifs (E-value = 3.8e-06), Zfp/E2F6/Sp motifs (E-value = 4.2e-03), and an RAR motif (E-value = 1.2e-02); each of which can function as a repressors, e.g. ER in the absence of ligand (Li et al., 2012). Altogether, FoxA binding is required to maintain an accessible nucleosome configuration and recruits other liver-enriched TFs at functional liver enhancers.
DISCUSSION
The distinct mechanistic features of tissue-specific versus ubiquitous regulatory sequences are critical to understand gene regulation (Zabidi et al., 2015), yet structural features that distinguish these classes have been elusive. Here, we find that liver-specific enhancers retain MNase-accessible nucleosomes more than active promoters and ubiquitous enhancers. Furthermore, the basis by which FoxA proteins, and possibly other pioneer factors, initially open chromatin at target sites, genome-wide, had been unclear (Iwafuchi-Doi and Zaret, 2014). Here, we find that FoxA1 and FoxA2 binding displaces linker histones, thereby helping to keep target nucleosomes accessible, which in turn helps recruit other liver enriched TFs to stimulate liver gene transcription (Fig. 7G).
Linker histones normally promote a closed nucleosome configuration. The DNA-binding domain of the linker histone highly resembles that of FoxA (Clark et al., 1993; Ramakrishnan et al., 1993) and binds near the dyad axis of the nucleosome (Goytisolo et al., 1996), as we found for FoxA (Fig. 3E). Therefore, the displacement of H1 can be accomplished by competition between FoxA and linker histone binding near the nucleosome dyad. Interestingly, we found that H1 is displaced not only at FoxA high-occupied liver enhancers, but also at FoxA low-occupied liver enhancers. Previous studies had reported that a FoxA mutant that disrupts sequence specific binding, yet still retains nonspecific DNA and nucleosome binding, can scan chromatin in cells (Sekiya et al., 2009) and remain bound to chromosomes in mitosis (Caravaca et al., 2013). Such scanning could model a first step to make nucleosomes accessible at both weak and strong binding sites, with a second, more specific step to create a stably accessible nucleosome at strong sites. Regardless, our findings suggest that it would be useful for those studying other pioneer factors in development, cell reprogramming and cancer (Zaret and Carroll, 2011) to investigate the role of H1 and its displacement in activating target genes.
Although the cooperative binding of TFs can be based on their protein-protein interactions, nucleosomes can facilitate cooperative binding without the direct factors’ interactions; referred to as nucleosome-mediated cooperativity between TFs (Chavez and Beato, 1997). For example, nucleosome depletion at the MMTV promoter leads to better accessibility for individual TFs, such as NF-I and GR, whereas the functional synergism between these TFs, which is required for strong hormonal induction, is only observed on nucleosomal DNA (Chavez and Beato, 1997). Our finding that the target genes of liver-specific enhancers exhibited higher expression levels than those of ubiquitous enhancers, even though the liver-specific enhancers have a low histone modification state, agrees with what has been observed at developmentally regulated genes in vivo (Perez-Lluch et al., 2015). Taken together, it indicates that the accessible nucleosomes can participate actively in regulatory processes by facilitating cooperativity between TFs and consequent strong liver gene induction. In contrast, ubiquitous enhancers retained fewer nucleosomes and were mainly enriched for ubiquitous TF motifs, some of which (e.g. Sp1) are known to recruit chromatin remodeling factors. The major structural differences between liver-specific and ubiquitous enhancers therefore associate with the different kinds of gene regulation.
Computational studies on mammalian genomes show that clusters of TF binding sites at enhancers are enriched for DNA sequences that encode higher intrinsic nucleosome occupancy compared to flanking sequences (Gaffney et al., 2012; Lidor Nili et al., 2010; Tillo et al., 2010). Furthermore, other TFs such as Sox2, Oct3/4, Klf4, p53, PU.1 and PR, can target binding sites with high nucleosome occupancy (Ballare et al., 2013; Barozzi et al., 2014; Lidor Nili et al., 2010; Soufi et al., 2015). Most of them are also known as pioneer factors, which can allow other TFs to access their target sites (Iwafuchi-Doi and Zaret, 2014). Given that pioneer factors on their own can bind nucleosomal DNA and create accessible nucleosomes, we suggest that pioneer factors can initiate nucleosome-mediated cooperativity between TFs for strong tissue-specific gene activation.
MNase-qPCR has been applied with 16 levels of MNase digestion and the titration curve predicted the fraction of nucleosome occupancy in a cell population (Floer et al., 2010). While this is a powerful quantitative method to analyze a single target locus, it has not yet been scaled to the genome-wide level. Our genome-wide method, comparing low and high MNase-seq coupled with histone signals, was sufficient to reveal specific features of ubiquitous vs. tissue-specific gene regulatory sequences in open chromatin that were previously thought to be nucleosome-free; the latter due to an experimental bias in high MNase-seq nucleosome mapping. Our method provides a more accurate view of how transcription factors bind and function in chromatin and led to the unexpected insight that tissue-specific and ubiquitous enhancers exhibit structural differences. Given that transcriptional enhancers are increasingly being appreciated as contributing markedly to human health and disease (Kasowski et al., 2013), it is critical to obtain a better understanding of their structure and mechanism of action.
EXPERIMENTAL PROCEDURES
(see Supplemental Information for details)
MNase treatment of nuclei and mono-nucleosomal DNA extraction
Animal use was approved by an IACUC committee. Mouse liver nuclei were suspended in RSB (10 mM Tris [pH7.4], 10 mM NaCl, 3 mM MgCl2, 10 mM sodium butyrate, 0.5 mg/ml aprotinin, 0.5 mg/ml leupeptin, 1 mg/ml pepstatin, 3mM CaCl2) was added and the nuclear suspension was pre-warmed for 1.5 min at 37°C and treated with MNase (Worthington Biochemicals) with 0.5 or 1 U/ml for low digestion and 20 U/ml for high digestion for 2 min at 37°C. After purifying DNA, t he mono-nucleosomal DNA (140 bp∼200 bp) was extracted.
MNase-seq
Two biological replicates of MNase-seq libraries were sequenced as 100-bp paired-end fragments, and MNase-seq libraries from FoxA1/FoxA2-flox;AAV-Cre mutant and WT;AAV-Cre were sequenced as 37-bp paired-end fragments. The sequence tags were aligned using Bowtie v0.12.5 (parameters -m 1, --best, paired-end alignment) to assembly NCBI v36 of the mouse genome (mm8).
Supplementary Material
Acknowledgments
We thank M. Teta-Bissett and T. Bernard-Banks for FoxA mice, B. Ren and F. Yue for enhancer data, B. Hoffman and P. Hoodless for HNF4α ChIP-seq data, K.Y. Chan and S.K.H. Han for ChIP-exo preparation, N. Bramswig, S. Shin, P. Sen for experimental advice, M. Lazar, A. Soufi, and J. Kim for comments, and E. Hulme for manuscript preparation. M. I.-D. was supported by postdoctoral fellowships from JSPS and Naito, Astellas, and Uehara Foundations. B.F.P. has a financial interest in Peconic, LLC, which utilizes the ChIP-exo technology implemented in this study and could potentially benefit from the outcomes of this research. The research was supported by NIH grants R37GM36477 to K.S.Z., P01DK049210 to K.H.K, and HG004160 to B.F.P.
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBER: Data have been uploaded to GEO (GSE57559).
AUTHOR CONTRIBUTIONS:
Conceptualization - M.I.-D. and K.S.Z.; Investigation - M.I-D; Bioinformatics - G.D, M.I-D, A.K., S.M., and B.F.P.; Resources - J.W., D.L. and K.H.K; Writing - M.I.-D. and K.S.Z.; Funding Acquisition - K.S.Z, K.H.K, B.F.P.; Supervision - K.S.Z
REFERENCES
- Axel R. Cleavage of DNA in nuclei and chromatin with staphylococcal nuclease. Biochemistry. 1975;14:2921–2925. doi: 10.1021/bi00684a020. [DOI] [PubMed] [Google Scholar]
- Ballare C, Castellano G, Gaveglia L, Althammer S, Gonzalez-Vallinas J, Eyras E, Le Dily F, Zaurin R, Soronellas D, Vicent GP, et al. Nucleosome-driven transcription factor binding and gene regulation. Mol Cell. 2013;49:67–79. doi: 10.1016/j.molcel.2012.10.019. [DOI] [PubMed] [Google Scholar]
- Bargaje R, Alam MP, Patowary A, Sarkar M, Ali T, Gupta S, Garg M, Singh M, Purkanti R, Scaria V, et al. Proximity of H2A.Z containing nucleosome to the transcription start site influences gene expression levels in the mammalian liver and brain. Nucleic Acids Res. 2012;40:8965–8978. doi: 10.1093/nar/gks665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barozzi I, Simonatto M, Bonifacio S, Yang L, Rohs R, Ghisletti S, Natoli G. Coregulation of transcription factor binding and nucleosome occupancy through DNA features of mammalian enhancers. Mol Cell. 2014;54:844–857. doi: 10.1016/j.molcel.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochkis IM, Schug J, Ye DZ, Kurinna S, Stratton SA, Barton MC, Kaestner KH. Genome-wide location analysis reveals distinct transcriptional circuitry by paralogous regulators Foxa1 and Foxa2. PLoS Genet. 2012;8:e1002770. doi: 10.1371/journal.pgen.1002770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caravaca JM, Donahue G, Becker JS, He X, Vinson C, Zaret KS. Bookmarking by specific and nonspecific binding of FoxA1 pioneer factor to mitotic chromosomes. Genes Dev. 2013;27:251–260. doi: 10.1101/gad.206458.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carone BR, Hung JH, Hainer SJ, Chou MT, Carone DM, Weng Z, Fazzio TG, Rando OJ. High-resolution mapping of chromatin packaging in mouse embryonic stem cells and sperm. Dev Cell. 2014;30:11–22. doi: 10.1016/j.devcel.2014.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Chai X, Nagarajan S, Kim K, Lee K, Choi JK. Regulation of the boundaries of accessible chromatin. PLoS Genet. 2013;9:e1003778. doi: 10.1371/journal.pgen.1003778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez S, Beato M. Nucleosome-mediated synergism between transcription factors on the mouse mammary tumor virus promoter. Proc Natl Acad Sci U S A. 1997;94:2885–2890. doi: 10.1073/pnas.94.7.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaya D, Hayamizu T, Bustin M, Zaret KS. Transcription factor FoxA (HNF3) on a nucleosome at an enhancer complex in liver chromatin. J. Biol. Chem. 2001;276:44385–44389. doi: 10.1074/jbc.M108214200. [DOI] [PubMed] [Google Scholar]
- Cirillo L, Lin FR, Cuesta I, Jarnik M, Friedman D, Zaret K. Opening of compacted chromatin by early developmental transcription factors HNF3 (FOXA) and GATA-4. Mol. Cell. 2002;9:279–289. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- Cirillo LA, McPherson CE, Bossard P, Stevens K, Cherian S, Shim E-Y, Clark EA, Burley SK, Zaret KS. Binding of the winged-helix transcription factor HNF3 to a linker histone site on the nucleosome. EMBO J. 1998;17:244–254. doi: 10.1093/emboj/17.1.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark KL, Halay ED, Lai E, Burley SK. Co-crystal structure of the HNF3/fork head DNA recognition motif resembles histone H5. Nature. 1993;364:412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floer M, Wang X, Prabhu V, Berrozpe G, Narayan S, Spagna D, Alvarez D, Kendall J, Krasnitz A, Stepansky A, et al. A RSC/nucleosome complex determines chromatin architecture and facilitates activator binding. Cell. 2010;141:407–418. doi: 10.1016/j.cell.2010.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney DJ, McVicker G, Pai AA, Fondufe-Mittendorf YN, Lewellen N, Michelini K, Widom J, Gilad Y, Pritchard JK. Controls of nucleosome positioning in the human genome. PLoS Genet. 2012;8:e1003036. doi: 10.1371/journal.pgen.1003036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goytisolo FA, Gerchman SE, Yu X, Rees C, Graziano V, Ramakrishnan V, Thomas JO. Identification of two DNA-binding sites on the globular domain of histone H5. EMBO J. 1996;15:3421–3429. [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman BG, Robertson G, Zavaglia B, Beach M, Cullum R, Lee S, Soukhatcheva G, Li L, Wederell ED, Thiessen N, et al. Locus co-occupancy, nucleosome positioning, and H3K4me1 regulate the functionality of FOXA2-, HNF4α-, and PDX1-bound loci in islets and liver. Genome Res. 2010;20:1037–1051. doi: 10.1101/gr.104356.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Hughes AL, Jin Y, Rando OJ, Struhl K. A functional evolutionary approach to identify determinants of nucleosome positioning: a unifying model for establishing the genome-wide pattern. Mol Cell. 2012;48:5–15. doi: 10.1016/j.molcel.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii H, Kadonaga JT, Ren B. MPE-seq, a new method for the genome-wide analysis of chromatin structure. Proc Natl Acad Sci U S A. 2015;112:E3457–3465. doi: 10.1073/pnas.1424804112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwafuchi-Doi M, Zaret KS. Pioneer transcription factors in cell reprogramming. Genes Dev. 2014;28:2679–2692. doi: 10.1101/gad.253443.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsen JS, Waage J, Rapin N, Bisgaard HC, Larsen FS, Porse BT. Temporal mapping of CEBPA and CEBPB binding during liver regeneration reveals dynamic occupancy and specific regulatory codes for homeostatic and cell cycle gene batteries. Genome Res. 2013;23:592–603. doi: 10.1101/gr.146399.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Pugh BF. Nucleosome positioning and gene regulation: advances through genomics. Nat Rev Genet. 2009;10:161–172. doi: 10.1038/nrg2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Felsenfeld G. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev. 2007;21:1519–1529. doi: 10.1101/gad.1547707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadam S, Emerson BM. Transcriptional specificity of human SWI/SNF BRG1 and BRM chromatin remodeling complexes. Mol Cell. 2003;11:377–389. doi: 10.1016/s1097-2765(03)00034-0. [DOI] [PubMed] [Google Scholar]
- Kasowski M, Kyriazopoulou-Panagiotopoulou S, Grubert F, Zaugg JB, Kundaje A, Liu Y, Boyle AP, Zhang QC, Zakharia F, Spacek DV, et al. Extensive variation in chromatin states across humans. Science. 2013;342:750–752. doi: 10.1126/science.1242510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Shiekhattar R. Architectural and Functional Commonalities between Enhancers and Promoters. Cell. 2015;162:948–959. doi: 10.1016/j.cell.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M, Cattoglio C, Tjian R. Looping Back to Leap Forward: Transcription Enters a New Era. Cell. 2014;157:13–25. doi: 10.1016/j.cell.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Schug J, Tuteja G, White P, Kaestner KH. The nucleosome map of the mammalian liver. Nat Struct Mol Biol. 2011;18:742–746. doi: 10.1038/nsmb.2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Tuteja G, Schug J, Kaestner KH. Foxa1 and Foxa2 are essential for sexual dimorphism in liver cancer. Cell. 2012;148:72–83. doi: 10.1016/j.cell.2011.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidor Nili E, Field Y, Lubling Y, Widom J, Oren M, Segal E. p53 binds preferentially to genomic regions with high DNA-encoded nucleosome occupancy. Genome Res. 2010;20:1361–1368. doi: 10.1101/gr.103945.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson CE, Shim E-Y, Friedman DS, Zaret KS. An active tissue-specific enhancer and bound transcription factors existing in a precisely postioned nucleosomal array. Cell. 1993;75:387–398. doi: 10.1016/0092-8674(93)80079-t. [DOI] [PubMed] [Google Scholar]
- Menet JS, Pescatore S, Rosbash M. CLOCK:BMAL1 is a pioneer-like transcription factor. Genes Dev. 2014;28:8–13. doi: 10.1101/gad.228536.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Lluch S, Blanco E, Tilgner H, Curado J, Ruiz-Romero M, Corominas M, Guigo R. Absence of canonical marks of active chromatin in developmentally regulated genes. Nat Genet. 2015;47:1158–1167. doi: 10.1038/ng.3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias ABR, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan V, Finch JT, Graziano V, Lee PL, Sweet RM. Crystal structure of globular domain of histone H5 and its implications for nucleosome binding. Nature. 1993;362:219–224. doi: 10.1038/362219a0. [DOI] [PubMed] [Google Scholar]
- Rhee HS, Bataille AR, Zhang L, Pugh BF. Subnucleosomal structures and nucleosome asymmetry across a genome. Cell. 2014;159:1377–1388. doi: 10.1016/j.cell.2014.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee HS, Pugh BF. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell. 2011;147:1408–1419. doi: 10.1016/j.cell.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya T, Muthurajan UM, Luger K, Tulin AV, Zaret KS. Nucleosome-binding affinity as a primary determinant of the nuclear mobility of the pioneer transcription factor FoxA. Genes Dev. 2009;23:804–809. doi: 10.1101/gad.1775509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim EY, Woodcock C, Zaret KS. Nucleosome positioning by the winged-helix transcription factor HNF3. Genes Dev. 1998;12:5–10. doi: 10.1101/gad.12.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A, Garcia MF, Jaroszewicz A, Osman N, Pellegrini M, Zaret KS. Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell. 2015;161:555–568. doi: 10.1016/j.cell.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K, Segal E. Determinants of nucleosome positioning. Nat Struct Mol Biol. 2013;20:267–273. doi: 10.1038/nsmb.2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taube JH, Allton K, Duncan SA, Shen L, Barton MC. Foxa1 functions as a pioneer transcription factor at transposable elements to activate Afp during differentiation of embryonic stem cells. J Biol Chem. 2010;285:16135–16144. doi: 10.1074/jbc.M109.088096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillo D, Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Field Y, Lieb JD, Widom J, Segal E, Hughes TR. High nucleosome occupancy is encoded at human regulatory sequences. PLoS One. 2010;5:e9129. doi: 10.1371/journal.pone.0009129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss TC, Hager GL. Dynamic regulation of transcriptional states by chromatin and transcription factors. Nat Rev Genet. 2014;15:69–81. doi: 10.1038/nrg3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts JA, Zhang C, Klein-Szanto AJ, Kormish JD, Fu J, Zhang MQ, Zaret KS. Study of FoxA pioneer factor at silent genes reveals Rfx-repressed enhancer at Cdx2 and a potential indicator of esophageal adenocarcinoma development. PLoS Genet. 2011;7:e1002277. doi: 10.1371/journal.pgen.1002277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner A, Hughes A, Yassour M, Rando OJ, Friedman N. High-resolution nucleosome mapping reveals transcription-dependent promoter packaging. Genome Res. 2010;20:90–100. doi: 10.1101/gr.098509.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Y, Yao J, Chen R, Li W, He X. Nucleosome fragility reveals novel functional states of chromatin and poises genes for activation. Genome Res. 2011;21:718–724. doi: 10.1101/gr.117101.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabidi MA, Arnold CD, Schernhuber K, Pagani M, Rath M, Frank O, Stark A. Enhancer-core-promoter specificity separates developmental and housekeeping gene regulation. Nature. 2015;518:556–559. doi: 10.1038/nature13994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Rubins NE, Ahima RS, Greenbaum LE, Kaestner KH. Foxa2 integrates the transcriptional response of the hepatocyte to fasting. Cell Metab. 2005;2:141–148. doi: 10.1016/j.cmet.2005.07.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.