Summary
The existence of adult pancreatic progenitor cells has been debated. While some favor the concept of facultative progenitors involved in homeostasis and repair, neither a location nor markers for such cells have been defined. Using genetic lineage tracing, we show that Doublecortin like kinase-1 (Dclk1) labels a rare population of long-lived, quiescent pancreatic cells. In vitro, Dclk1+ cells proliferate readily and sustain pancreatic organoid growth. In vivo, Dclk1+ cells are necessary for pancreatic regeneration following injury and chronic inflammation. Accordingly, their loss has detrimental effects after cerulein-induced pancreatitis. Expression of mutant Kras in Dclk1+ cells does not affect their quiescence or longevity. However, experimental pancreatitis converts Kras mutant Dclk1+ cells into potent cancer initiating cells. As a potential effector of Kras, Dclk1 contributes functionally to the pathogenesis of pancreatic cancer. Taken together, Dclk1 marks quiescent pancreatic progenitors that are candidates for the origin of pancreatic cancer.
Introduction
Studies to date have excluded the existence of stem cells in the pancreas. Nevertheless, the pancreas displays a slow but steady cellular turnover and significant capacity for regeneration following injury. Acinar cells show a high degree of plasticity, while less plasticity has been shown for centroacinar and ductal cells, although acinar to ductal metaplasia (ADM) likely requires reprogramming not only within the acinar compartment but perhaps in ductal cells as well. It has not been determined whether all acinar cells have the same ability to dedifferentiate, or if there is a specific subset with greater plasticity (Bailey et al., 2015; Kong et al., 2011; Puri et al., 2015; Reichert et al., 2013; Yanger and Stanger, 2011; Yanger et al., 2013; Ziv et al., 2013). While some investigators support the notion of committed progenitors, others propose that during injury, acinar cells dedifferentiate to act as facultative progenitor cells (Kong et al., 2011; Mills and Sansom, 2015). In theory, such facultative progenitors should demonstrate considerable plasticity in response to stress (Valdez et al., 2015). Another explanation for the regenerative capacities of the pancreas could be the presence of quiescent or reserve progenitors (Li and Clevers, 2010; Tian et al., 2011).
Numerous studies in mice have demonstrated a predominant role for acinar cells in the development of pancreatic intraepithelial neoplasia (PanINs) (De La et al., 2008; Habbe et al., 2008; Houbracken et al., 2011; Kopp et al., 2011; Maitra and Leach, 2012; Strobel et al., 2007; Zhu et al., 2007), but it is uncertain if all acinar cells are equally competent in initiating pancreatic tumorigenesis. The possibility that the pancreas harbors acinar cells with a higher capacity to participate in transformation, for example facultative progenitor cells, has not been thoroughly examined, but may have important implications for diagnosis and prevention of pancreatic ductal adenocarcinoma (PDAC) (Kong et al., 2011). Such a subpopulation of cells may not be distinguishable based upon conventional morphology but would require genetic lineage tracing.
Doublecortin like kinase-1 (Dclk1) has been proposed as a marker of pancreatic progenitors (May et al., 2010). Recent studies suggest that Dclk1+ cells are involved in the development of a variety of gastrointestinal tumors. Using Dclk1 CreERT knockin mice, it has been shown that Dclk1 identifies stem-like cells within ApcMin/+ adenomas (Nakanishi et al., 2012). Colonic Dclk1+ cells are long-lived and quiescent even in the setting of oncogenic mutations, but can be activated by injury to initiate colorectal cancer (Westphalen et al., 2014). Rare Dclk1+ cells can be found in the healthy pancreas, primarily in the ductal epithelium, and are markedly increased in ADM and murine PanINs (mPanIN) (Delgiorno et al., 2014). Moreover, Dclk1+ cells in preinvasive lesions and pancreatic cancer cell lines display a considerable amount of stemness (Bailey et al., 2014).
Thus, although earlier studies have suggested a role for Dclk1+ cells in PDAC development, their role in homeostasis, repair and initiation of PDAC has not been clearly established (Kopp and Sander, 2014). Using genetic lineage tracing, we show that Dclk1 labels a population of quiescent cells activated by pancreatic injury. Dclk1+ cells are largely resistant to an oncogenic Kras mutation, but act as potent cancer initiating cells following injury. Finally, Dclk1 gene function is involved in pancreatic tumorigenesis, as Dclk1 is a potential Kras effector protein.
Results
Dclk1 labels quiescent cells in the murine pancreas
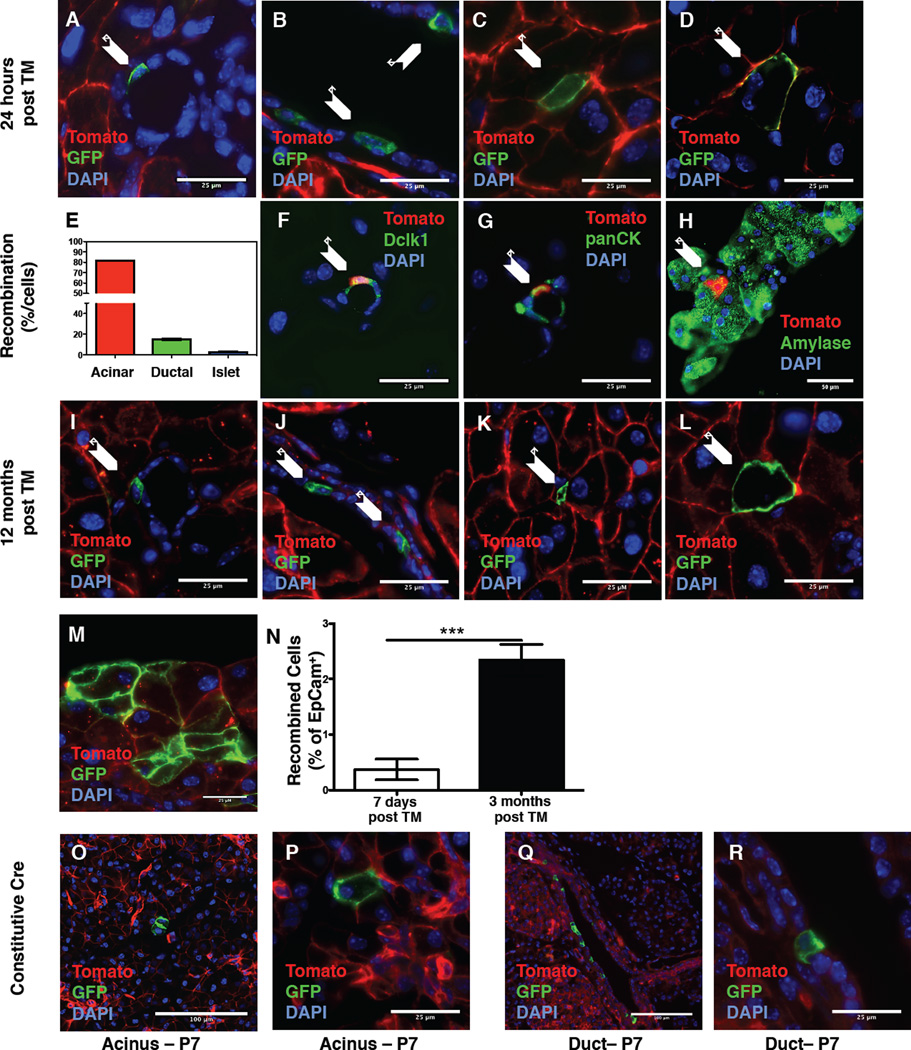
Dclk1 has been proposed to mark pancreatic progenitor cells (May et al., 2010). To test this hypothesis, we performed lineage tracing studies using Dclk1 BAC CreERT (Dclk1 CreERT) mice (Westphalen et al., 2014) (Figure S1A & Table S1) crossed to R26 mTmG reporter mice (Figure 1A–D & I–L, Figure S2H). In uninduced mice, uniform expression of membranous red fluorescent protein indicated absence of Cre recombinase activity at baseline (Figure S1C&D). Twenty-four hours post induction with Tamoxifen, recombination was not confined to one compartment but occurred in acinar, centroacinar and duct cells (Figure 1A–D & Figure S1E). Immunofluorescence (IF) on Dclk1 R26-tdTomato (tdTom) mice showed uniform expression of EpCam, which marks all pancreatic epithelial cells (Khan et al., 2011), in Dclk1-TdTomato+ cells (Figure S1F&G). One week after induction, flow cytometry revealed recombination in 0.1–0.5% of pancreatic epithelial cells isolated from Dclk1 tdTom mice (Figure S1H). The majority of Dclk1- TdTomato+ cells (82%) were located in the acinar compartment, whereas ~15% of recombination occurred in pancreatic ducts and terminal duct/centroacinar cells. Of note, a fraction of 15% recombined duct cells shows a slight enrichment of ductal cells compared to the pancreas as a whole. Recombination in islets was rare (Figure 1E).
Figure 1. Pancreatic Dclk1+ cells are largely quiescent.
A–D) Recombination in small (A) and large ducts (B), terminal duct/centroacinar (C) and acinar (D) cells in Dclk1 mTmG mice 24 hours post induction with Tamoxifen (n>5 mice). Arrows indicate recombined cells. E) Quantification of recombined cells by cellular compartment. F–H) Representative Immunofluorescence (green) for Dclk1 (F), pan-Cytokeratin (G) and Amylase (H) in Dclk1 tdTom mice. Arrows indicate recombined cells staining positive. I-L) Recombination in small (I) and large ducts (J), terminal duct/centroacinar (K) and acinar (L) cells in Dclk1 mTmG mice 12 months post induction with Tamoxifen (n>5 mice). Arrows indicate recombined cells. M) Traced acinar cluster 3 months post Tamoxifen N) Quantification of EpCam+/Dclk1+ cells in Dclk1 tdTom mice at baseline and 3 months after induction with Tamoxifen. Data represented as mean ± SEM, (n=4 mice) O–R) Recombination in the acinar (O&P) and ductal compartment (Q&R) in Dclk1-Cre mTmG mice at P7 (n=3 mice).
To characterize the Dclk1+ lineage, we isolated recombined cells from Dclk1 tdTom mice by flow cytometry and subjected them to RNAseq analysis. Analysis by qPCR confirmed higher levels of Dclk1 in tdTomato+ cells when compared to tdTomato- cells (Figure S1I). Both fractions (tdTomato+ and tdTomato- cells) contained a heterogeneous mix of acinar, ductal and other cell types. We analyzed our RNAseq results, versus published results on pure pancreatic acinar (Krah et al., 2015) and ductal (Ferreira et al., 2015) cells, to identify the dominant cell type in our fractions. We used average counts/million ≥1 as a criterion (Smyth et al., 2015) for gene expression and performed Venn analysis. Both Dclk1+ and Dclk1- cells expressed 8% more genes that are expressed in acinar cells but not ductal cells, compared to genes that are expressed in ductal cells but not acinar cells (p= 0.0003) (Table S1, Figure S1I–M), indicating that both populations were primarily acinar in composition. Thus, while the Dclk1 lineage contains both acinar and ductal cells, the acinar cells predominate as shown by gene expression as well as morphology.
Pathway analysis of Dclk1-TdTomato+ versus Dclk1-TdTomato- cells (Supplementary Information) revealed a quiescent phenotype in Dclk1+ cells caused by significant inactivation of genes involved in cell proliferation in the Ras (Bryant et al., 2014), PI3K-AKT (Isenovic et al., 2009) and HIPPO (Liu et al., 2012) pathways (Table S3). Specifically, Myc (Strom et al., 2007), AFP (Liu et al., 2007) and FasL (Reinehr et al., 2008) were downregulated in Dclk1-TdTomato+ cells (Table S4). Gene ontology analysis (Supplementary Information) revealed that Dab2ip (Min et al., 2015) and Fzd8 (Sugimura et al., 2012; Wang et al., 2015), two genes negatively affecting cell cycle progression were upregulated in Dclk1-TdTomato+ cells (Table S4). Confirmatory qPCR revealed differential expression of all genes tested with the exception of Fzd8 (p=0,07) (Figure S1N–R).
To improve identification of recombined cells and allow for staining using a green fluophore, we conducted IF staining on Dclk1 tdTom mice. Very few recombined ductal and centroacinar cells stained positive for Dclk1 (Figure 1F). Ductal Dclk1-TdTomato+ cells stained positive for Cytokeratin 19 (Figure 1G) while Dclk1-TdTomato+ cells within the acinar compartment stained positive for amylase (Figure 1H). Based on morphology and the absence of Dclk1 and cytoplasmic acetylated-tubulin staining (not shown), the vast majority of recombined cells did not qualify as tuft cells. These findings of a discrepancy between Dclk1 gene expression (based on Cre recombination) and protein expression are in line with previous reports showing a paucity of Dclk1+ tuft cells in the healthy pancreas (Delgiorno et al., 2014).
Recombined cells rarely stained positive for proliferation markers such as Ki67 and Phospho-Histone H3, and long-term lineage tracing revealed that most recombined cells persisted as single cells over 12 months (Figure 1I–L & Figure S2H). Over longer periods of observation, we detected occasional clusters of recombined acinar cells (Figure 1M), indicative of a small degree of lineage tracing. Such clusters were rarely seen in ducts (not shown). Flow cytometry of pancreata from Dclk1 tdTom mice 1-week and 3-months post induction revealed a small but significant expansion (~5-fold) of recombined epithelial (Dclk1+/EpCam+) cells over 3 months (Figure 1N). Analysis of the constitutive Dclk1 BAC Cre line (Figure S1B) on postnatal day 7 (P7) revealed single acinar and ductal cells, arguing against a major role of the Dclk1+ lineage in pancreatic development (Figure 1O&P).
Dclk1+ cells display increased proliferation potential in vitro
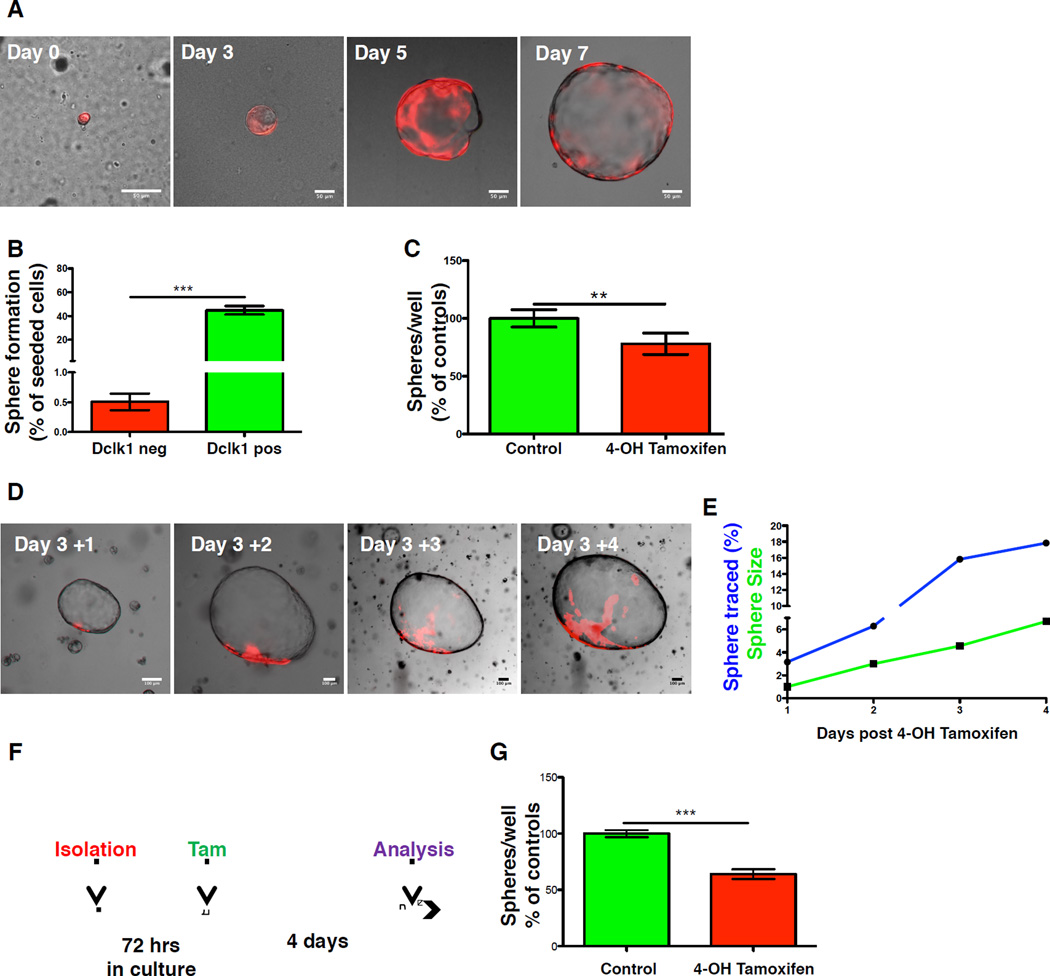
Three-dimensional (3D) pancreatic spheroid cultures have been used to study the characteristics of adult pancreatic stem cells (Huch et al., 2013; Wang et al., 2013), as well as ADM (Avila et al., 2012). Dclk1 tdTom mice were treated with Tamoxifen and pancreatic primary cultures were established as described (Wescott et al., 2009). In these experiments, 10,000 cells were seeded in each well and recombined (tdTom positive) cells were counted immediately after seeding. Under these conditions, Dclk1-TdTomato+ cells readily formed spheres (Figure 2A), and were roughly 70 times more efficient in forming spheres than Dclk1-TdTomato- cells, indicating a considerable degree of proliferative potential (Figure 2B).
Figure 2. Dclk1+ cells display increased proliferation potential in vitro.
A) Representative photographs of single cells (Day 0) and resulting spheres (Day 3–7) isolated from Dclk1 tdTom mice B) Sphere forming ability of tdTomato+ (red) and tdTomato- (green) cells shown as spheres/cell plated (n=4). C) Number of resulting spheres from Dclk1 DTA mice cultured in the absence (green) and presence of Tamoxifen (red). Data normalized to untreated controls (n=3). D) Consecutive photographs of the same sphere isolated after in vitro induction at day 3. E) Increase in sphere size (green) and traced pixels (blue) of the sphere depicted in D. F) Experimental setup for the data depicted in G. G) Number of resulting spheres from Dclk1 DTA mice after delayed treatment with Tamoxifen (red) and control mice (green) (n=3). Data normalized to untreated controls. All data represented as mean ± SEM.
To address whether Dclk1+ cells were necessary for spheroid formation in vitro, we generated pancreatic spheres from Dclk1 CreERT;R26-DTA (Dclk1 DTA) mice, where Cre-recombination causes diphtheria toxin A dependent elimination of cells expressing Dclk1 (Buch et al., 2005). Although Dclk1+ cell were present in only a minority of spheres, 4-OH Tamoxifen (Tam) treatment 24 hours post isolation led to a significant decrease (23% reduction) in sphere formation (Figure 2C). Sphere size did not differ between treated spheres and controls, arguing against a toxic effect of Tam (Figure S3A).
To demonstrate that Dclk1-TdTomato+ cells not only initiate but also sustain organoid growth, we conducted lineage tracing of established organoids. Cells were isolated from Dclk1 tdTom mice and spheres were cultured for 3 days prior to Tamoxifen induction. Twelve hours post induction; single TdTomato+ cells were found in a subset of organoids (Figure 2D & S3B–D), but over time fluorescent microscopy and morphometric analysis revealed clonal expansion of these single cells, paralleling organoid growth (Figure 2D&E). After in vitro induction, some organoids showed uniform expression of the tdTom reporter, arguing for an upregulation of Dclk1 mRNA expression during organoid formation (Figure S3E&F). This is in line with a significant upregulation of Dclk1 expression in the process of acinar to ductal metaplasia (Bailey et al., 2014) and could thus explain the reduction in organoid numbers observed after DTA-mediated ablation of Dclk1+ cells. Accordingly, when spheres generated from Dclk1 DTA mice were allowed to grow for 72h before treatment with Tamoxifen (Figure 2F), we observed a significant reduction in sphere numbers after 4 additional days in culture (Figure 2G). Taken together, these data indicate that Dclk1+ cells efficiently form pancreatic organoids and sustain organoid growth.
Dclk1+ cells are critically involved in pancreatic regeneration
Expansion of Dclk1+ tuft cells has been observed in response to tissue damage (Saqui-Salces et al., 2011; Tu et al., 2008). In sections from human chronic pancreatitis patients and a mouse model of chronic pancreatitis (Marrache et al., 2008), Dclk1+ tuft cells were significantly expanded (Figure S4A–D).
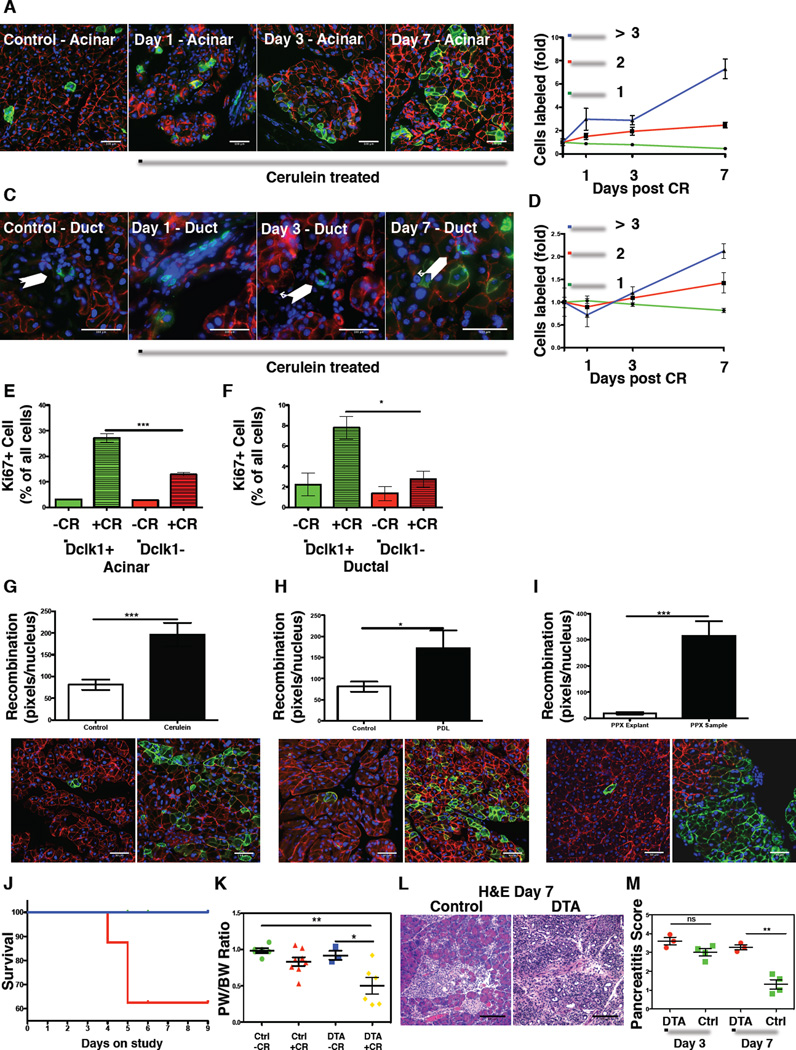
To test if the Dclk1+ lineage responds to injurious stress, Dclk1 mTmG mice were subjected to various forms of pancreatic injury. Cerulein-induced pancreatitis led to a significant expansion (2-fold) of recombined cells over the course of 2 weeks (Figure S4E & Figure 3A–F). As Dclk1+ cells can be found in ductal and acinar compartments, we analyzed the expansion in each compartment by scoring GFP-labeled cells and classifying them as single cells (Figure 3B&D green line), doublets (Figure 3B&D red line) or clones of three or more cells (Figure 3B&D blue line). While there was a modest expansion of ductal clones (2 fold - Figure 3C&D), we observed a marked increase in clonal labeling within the acinar compartment (~2% at day 0 versus ~36% at day 7 - Figure 3A&B). Control mice (Dclk1 mTmG) were induced with Tamoxifen but not treated with cerulein and analyzed in parallel. Furthermore, to exclude the possibilities that the Dclk1 transcript was upregulated through tissue injury or that we mistook a relative survival advantage of Dclk1+ cells as an expansion of the lineage, we treated Dclk1 mTmG mice with cerulein, waited for 24 hours, induced recombination with Tamoxifen and analyzed animals after an additional 12 hours. In these experiments, we observed similar numbers of Dclk1+ cells when compared to mice under resting conditions (Figure S4F–H).
Figure 3. Dclk1+ cells are critically involved in pancreatic regeneration.
A) Fluorescent images of recombined acinar cells from Dclk1 mTmG control mice (left panel) and mice 1, 3 and 7 days after treatment with cerulein. B) Increase in acinar recombination depicted as singlets (green), doublets (red) and clones of three or more cells (blue) over the course of the study. Data normalized to untreated controls. (n≥3 mice/condition) C) Fluorescent images of recombined ductal cells from Dclk1 mTmG control mice and mice 1, 3 and 7 days after treatment with cerulein. D) Increase in ductal recombination depicted as singlets (green), doublets (red) and clones of three or more cells (blue) over the course of the study. Data normalized to untreated controls (n≥3 mice/condition). E) Quantification of Ki67 positive acinar cells in the Dclk1+ (green) & Dclk1- (red) lineage in the presence (+CR) and absence (−CR) of cerulein (n≥3 mice/condition). F) Quantification of Ki67 positive ductal cells in the Dclk1+ (green) & Dclk1- (red) lineage in the presence (+CR) and absence (−CR) of cerulein (n≥3 mice/condition). G–I) Morphometric quantification of recombination and representative fluorescent images from Dclk1 mTmG mice after (G) cerulein treatment (n=6 mice), (H) pancreatic duct ligation (n=4 mice) and I) partial pancreatectomy (n=4 mice) J) Survival curve of Dclk1 DTA (red) mice induced with Tamoxifen and treated with cerulein (n=8) and controls mice (Dclk1 mTmG - blue) treated with cerulein (n=8) K) Pancreatic weight/body weight ratio at time of euthanasia. Green = WT mice without cerulein (n=6), Red: DTA mice without cerulein & Tamoxifen (n=7), Blue: DTA mice without cerulein, treated with Tamoxifen (n=3), Yellow: DTA mice treated with cerulein and Tamoxifen (n=6) Data represented as mean ± SEM L) Representative H&E sections of pancreata from control and Dclk1 DTA mice at time of euthanasia M) Pancreatitis score in Dclk1 DTA mice (red) and controls (green) 3 and 7 days after cerulein treatment. (n≥ 3 mice/condition). All data represented as mean ± SEM.
To investigate proliferation in Dclk1+ and Dclk1- cells, we performed Ki67 staining on sections from Dclk1 mTmG mice after cerulein treatment and scored Ki67+ cells in recombined and non-recombined cells. Seven days after cerulein treatment, Ki67+ acinar (Figure 3E) and ductal (Figure 3F) cells were significantly more frequent in the Dclk1+ compartment, arguing for a proliferative advantage of the Dclk1+ lineage. Finally, specific analysis of areas with ADM 72 hours after cerulein treatment revealed that ~20–40% of ADMs were at least partly traced by previously labeled Dclk1+ cells (Figure S4I&J).
Pancreatic duct ligation led to a 50% increase in lineage tracing when compared to untreated controls (Figure 3H). Lineage tracing was analyzed in the pancreas proximal to the duodenum to assess compensatory growth in the pancreas not affected by pressure necrosis due to the suture. Next, we tested the effect of partial pancreatectomy. Two weeks after surgery, we observed large areas of traced exocrine tissue (Figure 3I). Morphometric analysis of the regenerating pancreas revealed a 15-fold increase in tracing compared to the pancreatic explant that served as an internal control.
To test the importance of Dclk1+ cells in response to injury, Dclk1 DTA and control animals were induced with Tamoxifen, fasted and then subjected to cerulein treatment. Control animals recovered within 1-week from pancreatic injury while Dclk1 DTA mice failed to regain body weight and appeared sickly, leading to a significant mortality (40% in DTA mice vs. 0% in controls) (Figure 3J and S4I). This inability to recover was underscored by significantly increased hunching scores (Figure S4J) (Sevcik et al., 2006) and smaller pancreata in Dclk1 DTA mice (30–36% reduction in pancreatic weight/body weight ratio) (Figure 3K&S4K). Seven days after cerulein treatment, histology confirmed the absence of pancreatic regeneration in Dclk1 DTA mice (Figure 3L), with significantly higher pancreatitis scores (Figure 3M), greater areas of metaplastic ducts (Figure S4L&M), ongoing apoptotic cell death, and comparable numbers of proliferating cells (Figure S4N&O). Thus, these studies indicate that Dclk1+ cells are critically involved in pancreatic regeneration.
Dclk1 gene expression contributes to pancreatic regeneration
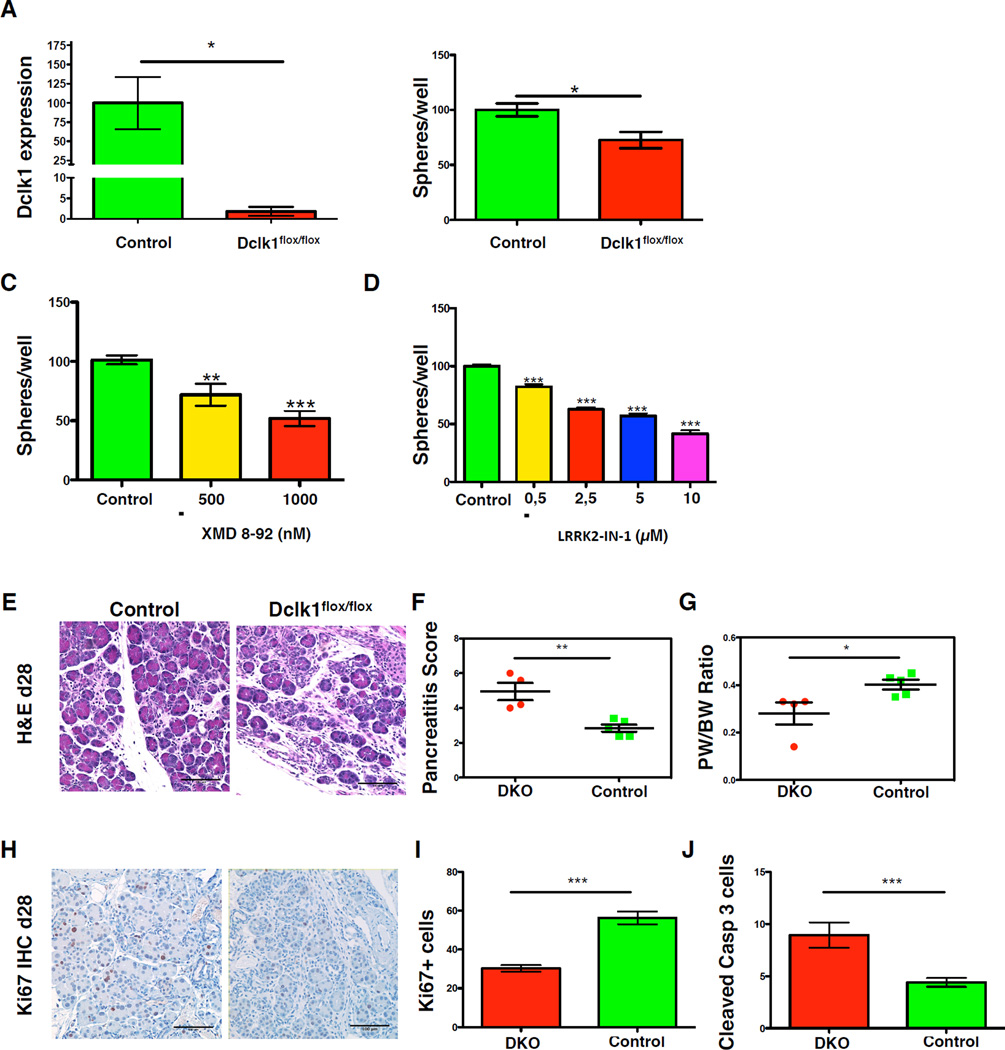
To test whether the Dclk1 gene, which encodes for a serine/threonine kinase, has a functional role in response to pancreatic injury, we crossed Pdx1-Cre mice (Hingorani et al., 2003) with Dclk1flox/flox mice (Koizumi et al., 2006) to generate mice lacking both pancreatic Dclk1 alleles (DKO mice). DKO mice were born at Mendelian ratio and showed no pathology under resting conditions. We confirmed a significant reduction of Dclk1 expression in pancreata of DKO mice compared to controls (Figure 4A). Functionally, cells isolated from DKO pancreata displayed a significant reduction in sphere forming-capacity compared to controls (Figure 4B). We confirm this finding pharmacologically by treating pancreatic organoid cultures with two molecules (XMD 8-92 & LRRK2-IN-1) targeting Dclk1 (Miduturu et al., 2011; Weygant et al., 2014), which led to a reduction in resulting organoids (Figure 4C&D).
Figure 4. Dclk1 gene expression contributes to pancreatic regeneration.
A) Quantification of Dclk1 mRNA expression in Pdx1-Cre (Control – green) and Pdx1-Cre Dclk1flox/flox (Dclk1flox/flox – red) mice (n=3/condition). Data represented as mean ± SEM & normalized to controls. B) Number of resulting spheres from Pdx1-Cre Dclk1flox/flox (DKO) mice (red) and control mice (green) (n=4). Data normalized to controls (Pdx1-Cre mice). C) Number of resulting spheres cultured in the absence (green) and the presence of 500 nm (yellow) or 1000 nm (red) XMD 8–92 (n=3 mice). Data normalized to untreated controls. D) Number of resulting spheres cultured in the absence (green) and the presence of 0,5 µm (yellow), 2,5 µm (red), 5 µm (blue) and 10 µm (violet) LRRK2-IN-1 (n=3 mice). Data normalized to untreated controls. E) Representative photographs of H&E staining in control mice (left) and DKO mice three days post chronic cerulein treatment (n=≥4 mice). F) Pancreatitis score in DKO and controls after chronic cerulein treatment. G) Pancreatic weight/body weight ratio in DKO and controls H) Representative photographs of Ki67 IHC in DKO and control mice. I) Quantification of Ki67 staining depicted in P. J) Quantification of cleaved Caspase 3 staining. All data represented as mean ± SEM.
To investigate the requirement for Dclk1 gene function in response to pancreatic injury, DKO mice were treated with 7 hourly injections of cerulein twice a week for 4 weeks. When compared to controls (Pdx1-Cre mice), DKO mice showed more severe histological signs of pancreatitis (Figure 4E&F) with lower pancreatic/body weight ratio (Figure 4G), a blunted proliferative response (Figure 4H&I), and a higher number of apoptotic cells (Figure 4J). Taken together, our findings indicate that expression of the Dclk1 gene is needed for optimal pancreatic regeneration after cerulein treatment.
Dclk1+ cells efficiently initiate pancreatic tumorigenesis
Dclk1+ cells are expanded in a variety of preneoplastic conditions, and have been suggested to function as cancer stem or initiating cells in the colon (Nakanishi et al., 2013; Westphalen et al., 2014) and the pancreas (Bailey et al., 2014). To test if pancreatic Dclk1+ cells indeed serve as cancer initiating cells, we crossed Dclk1 CreERT mice to LSL-KrasG12D (Jackson et al., 2001) mice (Dclk1 Kras). Following induction, early mPanINs were only infrequently observed after several months (Figure 5A). When Dclk1 Kras mice were crossed to mTmG reporter mice (Dclk1 Kras mTmG), there was neither a significant expansion nor a significant loss of the lineage (Figure 5B), suggesting that oncogenic Kras does not cause a progressive loss of mutant cells as reported (Morton et al., 2010).
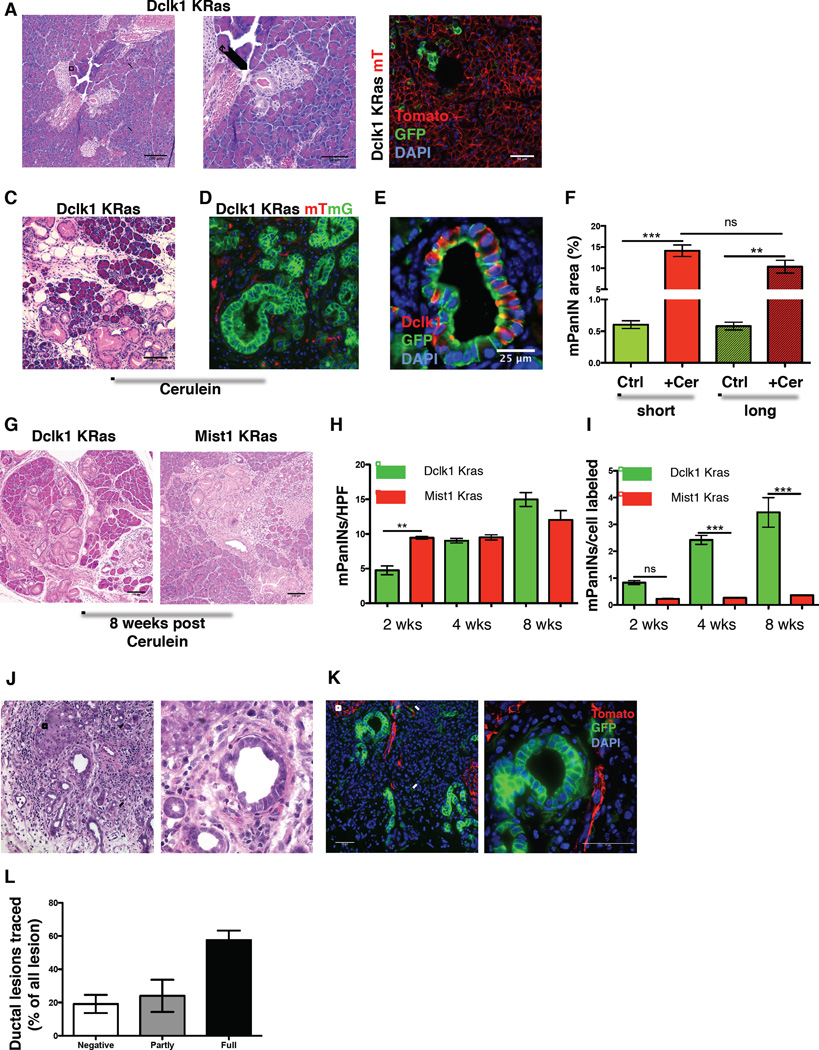
Figure 5. Dclk1+ cells efficiently initiate pancreatic tumorigenesis.
A) Low-grade mPanIN in a Dclk1 CreERT×LSL KrasG12D mouse 3 months post induction with Tamoxifen. Arrow indicates the lesion. B) Representative fluorescent image of a Dclk1 Kras mTmG mouse 6 months after induction with Tamoxifen. C) Histopathology of a Dclk1 Kras mouse 8 weeks post treatment with cerulein. D) Tracing of mPanINs in Dclk1 Kras mTmG mice. E) IF for GFP (green) & Dclk1 (red) in a Dclk1 KRas mTmG mouse. F) Analysis of the area affected by mPanINs in Dclk1 KRas and control mice after short delay (short - 2 weeks) and prolonged delay (long – 4–6 months) of treatment with cerulein (Cer) G) Histopathology of a Dclk1 Kras (left) and a Mist1 Kras (right) mouse 8 weeks post treatment with cerulein H) Number of mPanINs in Dclk1 Kras (green) and Mist1 Kras (red) mice at indicated time points (n ≥ 3 mice/time point) I) Number of mPanINs (Figure 5H) per cell labeled. J) Representative H&E section from a ductal lesion in Dclk1 mTmG mice induced with Tamoxifen followed by implantation of DMBA K) Representative fluorescent image of a ductal lesion in Dclk1 mTmG mice induced with Tamoxifen followed by implantation of DMBA. L) Quantification of ductal lesions labeled in Dclk1 mTmG mice implanted with DMBA (n=4). All data represented as mean ± SEM.
Cerulein treatment of Dclk1 Kras mice led to the rapid development of multiple mPanINs (Figure 5C). Simultaneous lineage tracing revealed uniform expression of GFP within the resulting lesions, proving their descent from previously labeled Dclk1+ cells (Figure 5D). Immunofluorescence for Dclk1 (red) and GFP (green) on paraffin sections from Dclk1 Kras mTmG mice demonstrated the ability of the Dclk1+ lineage to give rise to Dclk1+ tuft cells present in mPanINs (Bailey et al., 2014) (Figure 5E). This result suggests that the expansion of Dclk1+ tuft cells observed in human and murine pancreatitis is at least partly driven by Dclk1+ acinar cells participating in ADM. Since colonic Dclk1+ cells can harbor oncogenic mutations and remain dormant (Westphalen et al., 2014), we induced Cre-recombination with Tamoxifen and extended the duration between induction and injury for up to 6 months. Again, we observed the development of numerous mPanINs. The area affected by mPanINs did not differ between mice stressed 2 weeks after induction and mice in which injury was delayed for up to 6 months (Figure 5F). Thus, pancreatic Dclk1+ cells remain quiescent in the presence of an oncogenic mutation but retain their ability to initiate pancreatic tumorigenesis at any point in time.
To estimate their degree of transformability, we used FACS analysis to quantify the number of recombined cells in Dclk1+ tdTom and Mist1 CreERT tdTom mice. Here, Mist1 CreERT mice were chosen as controls as they label the majority of pancreatic acinar cells (Habbe et al., 2008). In these experiments, we found Mist1 cells to be roughly 12 times more frequent than Dclk1 cells (Figure S5A). After induction with Tamoxifen, Dclk1 Kras and Mist1 Kras mice were treated with cerulein. Two weeks after cerulein treatment, Mist1 Kras mice showed more mPanINs and a greater area covered by mPanINs, probably reflective of the quiescent nature of Dclk1 cells and the abundance of Mist1+ acinar cells. However, as early as 4 weeks after injury, there was no difference in mPanIN numbers and area covered between Dclk1 Kras and Mist1 Kras mice (Figure 5G&H & Figure S5B&C). Based on these results, we calculated the number of resulting mPanINs on the basis of the baseline labeling in both lines. These calculations revealed, that Dclk1+ cells were between three to nine (3 – 9) times more efficient in mPanIN initiation when compared to Mist1+ cells (Figure 5I).
To test if sporadic, carcinogen-induced neoplastic lesions can also arise from previously labeled Dclk1+ cells, we induced Dclk1 mTmG mice with Tamoxifen and then implanted the chemical carcinogen 7,12-Dimethylbenz(a)anthracene (DMBA) into the pancreatic tail (Osvaldt et al., 2006). While DMBA-mediated carcinogenesis induces poorly differentiated sarcomatoid tumors, we concentrated on the metaplastic ductal lesions also observed in this model. After 3 to 5 months, the majority of resulting ductal lesions were partly or fully lineage traced, indicating that they were derived from previously labeled Dclk1+ cells (Figure 5J–L).
Dclk1 expression is important for the progression of early neoplastic lesions
In order to investigate the functional importance of Dclk1 expression in pancreatic tumorigenesis, we decided to target Dclk1 gene function in vitro and in vivo. First, we treated pancreatic sphere cultures from Pdx1-Cre LSL Kras (KC) mice (Hingorani et al., 2003) with increasing doses of the Dclk1 kinase inhibitors, XMD 8-92 or LRRK2-IN-1, and observed a significant decrease in sphere numbers (Figure 6A&B). We then generated KC Dclk1flox/flox (KC DKO) mice. While KrasG12D mutant spheres typically grow in the absence of exogenous EGF, loss of Dclk1 in the absence of EGF led to a significant decrease in spheres when compared to spheres isolated from KC mice (Figure 6C). Addition of EGF to the culture medium, in an attempt to rescue the proliferative defect, did not restore the growth of KC DKO spheres, suggesting a mechanism downstream of receptor tyrosine kinases (Figure 6D).
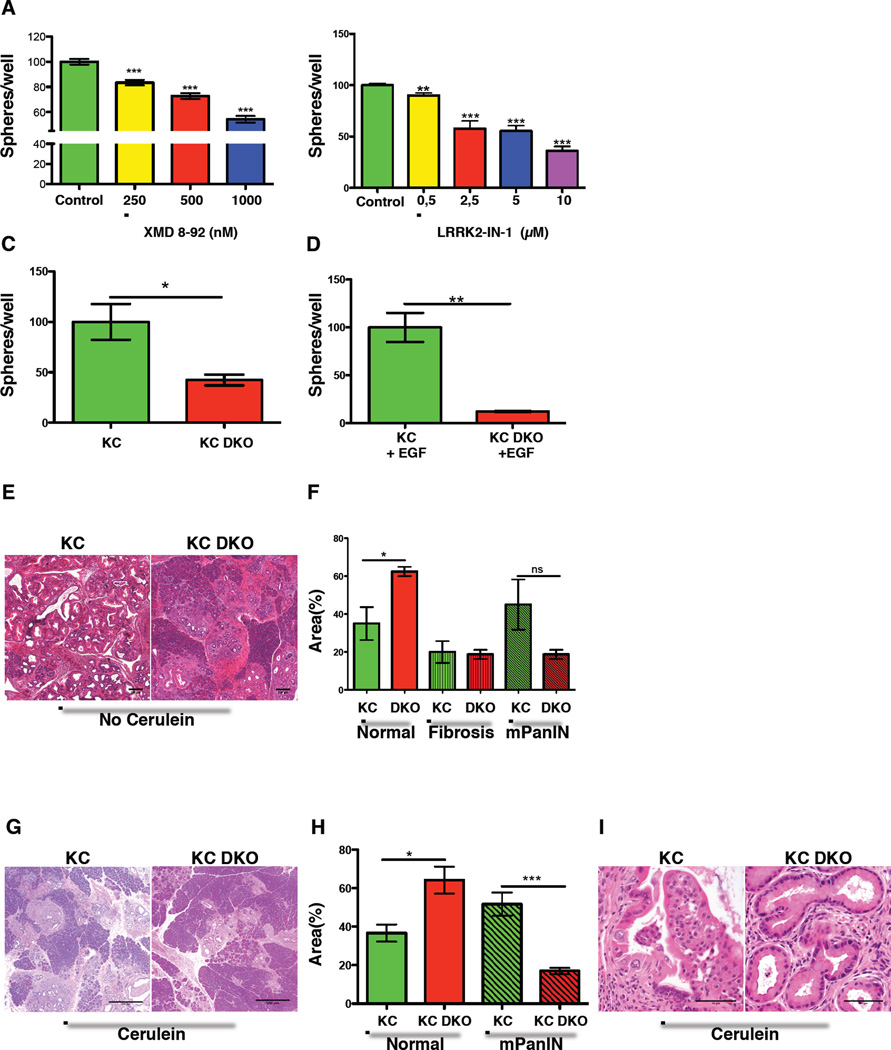
Figure 6. Dclk1 is important for the progression of early neoplastic lesions.
A) Resulting number of spheres isolated from KC mice cultured in the absence (green) and the presence of 250 nm (yellow), 500nm (red) or 1000nm (blue) XMD 8–92. Normalized to untreated controls (n=3 mice). B) Resulting number of spheres isolated from KC mice in the absence (green) and the presence of 0,5 µm (yellow), 2,5 µm (red), 5 µm (blue) and 10 µm (violet) LRRK2-IN-1. Normalized to untreated controls (n=3 mice). C) Resulting spheres isolated from KC mice (green) and KC DKO mice (red). Data normalized to controls (n=3). D) Resulting number of spheres isolated from KC mice (green) and KC DKO mice (red) treated with EGF. Data normalized to controls (n=≥3 mice). E) H&E sections from KC and KC DKO at 32 weeks of age. F) Area covered (percentage of the area analyzed) by unaffected pancreas, fibrosis and mPanINs in KC (green) and KC DKO mice at 32 weeks of age (n=4 mice). G) H&E sections from KC and KC DKO mice 16 weeks after treatment with cerulein. H) Analysis of normal and preneoplastic pancreatic tissue in KC and KC DKO mice (n=5). I) Typical lesions in KC (left) and KC DKO mice (right) 16 weeks after treatment with cerulein. All data represented as mean ± SEM.
To address the importance of Dclk1 function in early pancreatic tumorigenesis, KC DKO mice were followed for 8 months. Histopathology revealed that Dclk1 deficiency caused a reduction in early preneoplastic lesions in KC DKO mice when compared to KC mice. Furthermore, pancreata from KC DKO mice had significantly more unaffected tissue when compared to KC mice (Figure 6E&F & Table S4). IHC analysis of the resulting mPanINs revealed absent Dclk1 and significantly reduced acetylated-tubulin immunoreactivity (Figure S5D). To accelerate tumorigenesis, we subjected KC and KC DKO mice to cerulein treatment. Pancreata from KC mice showed more severe histological changes compared to pancreata from KC DKO mice, with greater inflammatory changes, larger mPanIN area and less remaining normal pancreatic tissue. Morphometrics confirmed that KC DKO mice had more normal pancreatic tissue and less mPanINs compared to KC mice (Figure 6G&H). Furthermore, careful analysis of the resulting lesions revealed that KC mice displayed multiple mPanIN2 lesions, while the most of the lesions in KC DKO mice were low grade (Figure 6I & Table S5). These data indicate that Dclk1 deficiency inhibits early neoplastic changes in the setting of mutant Kras. As Dclk1+ cells appear in ADM and early mPanINs and decline during mPanIN progression, these effects on early pancreatic tumorigenesis are in line with previous studies (Bailey et al., 2014).
Dclk1 is a potential Kras effector protein
Previous studies suggested an effect of Dclk1 on Kras signaling (Sureban et al., 2013). We applied two computational methods to detect whether Dclk1 might be a potential Kras effector. The first, PrePPI, is a database of predicted protein-protein interactions derived from the integration of multiple evidence sources, such as co-expression, phylogenetic profiles and sequence orthology (Zhang et al., 2012). PrePPI integrates three-dimensional structure information with non-structural clues, and provides a structural model for each interaction where it deems a direct physical interaction likely. PrePPI contains predictions for over 1.3 million interactions in the human proteome and provides estimated probabilities for each. We searched the PrePPI database (https://bhapp.c2b2.columbia.edu/PrePPI/) and found that the N-terminal domain of Dclk1 has a probability of 0.57 to interact with Kras. Moreover, there is strong evidence for a direct physical interaction based on the PrePPI-inferred structural homology between this domain and that of RaIGDS, whose interaction with Kras has been determined crystallographically. A model for this interaction appears in Supplemental Figure 5A. Compared with a background probability of about 1 in 200 million for two randomly chosen human proteins to physically interact, a probability of 0.5 is considered as a highly reliable prediction. Indeed, of 18 tested interactions at this threshold, 15 were experimentally validated in previous studies (Zhang et al., 2012).
The second, the DeMAND algorithm (Woo et al., 2015), uses network-based analysis of gene expression profiles to predict whether specific perturbations, either small-molecule or genetically induced, may modulate the interaction of a protein of interest (Dclk1 in this case) with its cognate binding partners and transcriptional targets. In this case, we used the gene expression of cells harboring an activating Kras mutation vs. those with wild-type Kras to model the perturbation of interest, using protein-protein and protein-DNA interaction networks that had been previously published (Lefebvre et al., 2010). When comparing the expression of cells with activated vs. wild-type Kras, the interactions of Dclk1 with its signaling partners were significantly dysregulated (p=7.61e-20), making this protein the 97th most dysregulated out of 17,248 tested ones. Taken together, these computational results support the hypothesis that Kras may engage in a functionally relevant protein-protein interaction with Dclk1 and that aberrant Kras activity may contribute to Dclk1 dysregulation.
Accordingly, we tested the hypothesis that Dclk1 affects Kras driven tumorigenesis by direct interaction with Kras using co-immunoprecipitation. In these experiments, MiaPaCa2 cells were co-transfected with vectors expressing Dclk1 and either wild type or mutant Kras. Both Kras constructs carried a flag-tag and the pull-down was conducted using agarose beads conjugated with anti-flag antibodies. These experiments revealed a potential physical interaction between Dclk1 and mutant, and to a lesser extent wild type Kras (Figure S5B), thus providing a possible explanation for the phenotype observed in KC DKO mice.
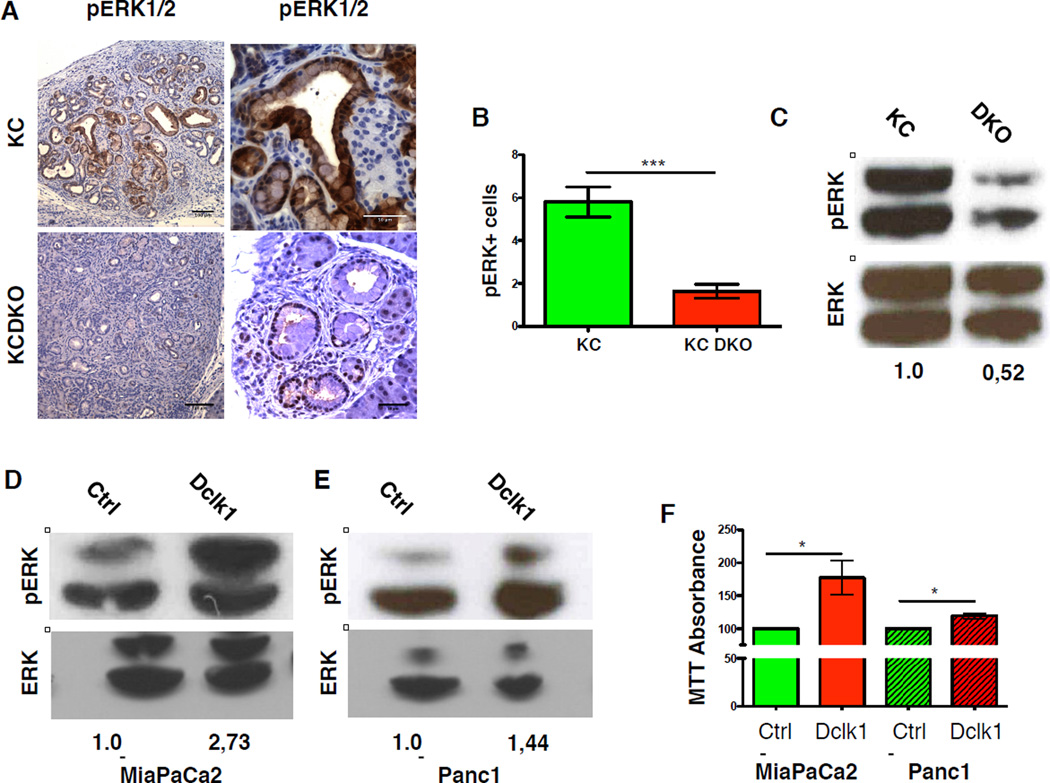
To test if loss of Dclk1 affects Kras signaling, we stained KC and KC DKO samples for phosphorylated ERK1/2 and counted pERK+ cells in mPanINs. Compared to KC mice, loss of Dclk1 led to a significant reduction of pERK1/2+ cells within mPanINs of KC DKO mice (Figure 7C&D). pERK levels were lower in pancreata taken from KC DKO mice when compared to controls (Figure 7E). Thus, loss of Dclk1 inhibits the ability of mutant Kras to activate ERK signaling in the pancreas. Consistent with this observation, overexpression of Dclk1 in two human pancreatic cancer cell lines led to increased pERK levels (Figure 7F–G) and promoted cellular proliferation (Figure 7H)., Transfection efficiency in cells viable at thirty-six hours post-transfection was assessed based on an EGFP reporter in the Dclk1 expression vector and ranged between 40–60% (not shown).
Figure 7. Dclk1 is a potential Kras effector protein.
A) IHC for pERK in mPanINs of KC and KC DKO mice B) Quantification of pERK staining in mPanINs of KC and KC DKO mice (n=5). C) Western Blot for ERK and pERK in KC and KC DKO mice (data of two independent experiments – images cropped). D&E) Western blot for ERK and pERK in MiaPaCa2 (E) and Panc1 (F) cells upon overexpression of Dclk1 (data of two independent experiments – images cropped) F) MTT assays of human pancreatic cancer cell lines upon overexpression of Dclk1 (data of three independent experiments). All data represented as mean ± SEM.
Discussion
The possibility that acinar cells are heterogeneous in their gene expression or plasticity has not been well addressed. Further, the existence of dedicated or facultative pancreatic progenitor cells remains unresolved, although a variety of markers and cell types have been proposed (Kong et al., 2011; Stanger and Hebrok, 2013; Yanger and Stanger, 2011). Using genetic lineage tracing, we demonstrate the existence of a predominantly acinar Dclk1+ cell type that is quiescent under resting conditions but highly efficient in forming spheres in vitro. In previous studies, Dclk1+ cells isolated from mPanINs or cancer cell lines showed a high degree of stemness (Bailey et al., 2014), supporting our observations in untransformed cells. The finding that untransformed Dclk1+ cells are largely quiescent in vivo but act as bona fide stem cells in organoids and premalignant lesions is consistent with properties of quiescent progenitor cells (Li and Clevers, 2010). As Dclk1+ cells are activated by pancreatic injury to induce a regenerative and dedifferentiation program, it is not surprising that 3D culture conditions lead to a similar activation of quiescent Dclk1+ cells (Buczacki et al., 2013).
In vivo, Dclk1+ cells are increased in pancreatitis, and the Dclk1+ lineage was critical for recovery. While comprising only a small proportion of pancreatic cells, ablation of Dclk1+ cells abrogated regeneration following cerulein-induced pancreatic injury. The concept of special subsets of acinar cells has been proposed before (Kong et al., 2011). Bmi1+ cells were proposed as one such subset, but these studies did not determine if regeneration occurred from a pool of existing or newly generated Bmi1+ cells (Sangiorgi and Capecchi, 2009). Here, we labeled Dclk1+ cells prior to pancreatic injury and can thus conclude that pancreatic regeneration occurred from preexisting Dclk1+ cells. Dclk1+ cells can be found in both the acinar and the ductal compartment, but careful analysis of the expansion of Dclk1+ cells during pancreatic injury revealed that the regenerative response occurred predominantly in the acinar compartment. Ki67 staining revealed a proliferative advantage of the Dclk1+ lineage over Dclk1- cells, a phenotype that was more pronounced in acinar compartment. Taken together, the data indicate the presence of a distinct subpopulation of Dclk1+ acinar cells that are specifically activated by pancreatic injury and thus qualify as quiescent progenitor cells. Nevertheless, we cannot exclude that the Dclk1+ population within the ductal compartment has important functional properties that have been underestimated.
Due to its histological appearance, pancreatic cancer was for years presumed to arise from the ductal epithelium (Murtaugh and Leach, 2007). While many pancreatic lineages harbor the ability to initiate PDAC under the right conditions (Gidekel Friedlander et al., 2009), recent studies suggest the origin of PDAC within the acinar compartment (Maitra and Leach, 2012). When targeted with a mutant Kras oncogene, acinar cells readily give rise to PanINs and are 100-fold more susceptible to Kras transformation than the ductal compartment (Kopp JL et al, 2012). However, given the large number of acinar cells, it is not clear if cancer indeed arises to the same degree from all acinar cells, or if there is a subset of acinar cells with a higher degree of transformability (Ziv et al., 2013).
Dclk1 has been proposed as a marker for cancer stem cells in PDAC (Bailey et al., 2014). While Kras mutant Dclk1+ cells remained quiescent under resting conditions, the combination of injury and Kras mutation was sufficient to induce pancreatic tumorigenesis. Dclk1+ cells were significantly more efficient in forming mPanINs than the majority of adult Mist1+ acinar cells. This remarkable susceptibility of quiescent Dclk1+ cells to the combination of an oncogenic hit and cellular stress can be explained by their remarkable proliferative potential, demonstrated in sphere cultures and during regeneration, a possible prerequisite for PDAC development (Mills and Sansom, 2015; Puri et al., 2015). While adult acinar cells are relatively resistant to malignant transformation in the absence of a secondary hit (Guerra et al., 2007), the longevity of such a mutant cell population has not been addressed. In fact, Kras mutant cells are progressively lost in some mouse models of pancreatic cancer (Morton et al., 2010). Accordingly, our observations that quiescent cells in the adult pancreas may harbor Kras mutant cells for extended periods could inform our understanding regarding the timing of initiation and rate of progression of pancreatic cancer.
Previous studies suggested a functional role for Dclk1 expression in gastrointestinal cancer (Bailey et al., 2014; Weygant et al., 2014). The PrePPI algorithm (Zhang et al., 2013) predicted a physical interaction between Dclk1 and Kras, and our in vitro data indicates a physical interaction between Dclk1 and Kras. Furthermore, loss of Dclk1 affected Kras signaling and early pancreatic tumorigenesis and overexpression of Dclk1 led to an increase in pERK levels and proliferation. Given the appearance of Dclk1+ tuft cells during the earliest steps of pancreatic tumorigenesis (Delgiorno et al., 2014) and the exceptional role of Kras in pancreatic cancer (Eser et al., 2014), our findings shed new light on the role of Dclk1 expression and Dclk1+ cells in PDAC. While it is difficult to inhibit Kras therapeutically (Zimmermann et al., 2013), Dclk1 is a potentially targetable kinase, and preclinical studies exploiting Dclk1 as a therapeutic target in cancer have yielded early promising results (Sureban et al., 2015; Sureban et al., 2013; Weygant et al., 2014).
In summary, Dclk1 labels a quiescent pancreatic progenitor population activated by cellular stress. Lineage tracing results demonstrate the ability of Dclk1+ cells to participate in pancreatic repair. Moreover, their absence has detrimental effects after injury. Quiescent in the setting of oncogenic mutation, Dclk1+ cells can be activated by injury to act as potent source of mPanINs. Moreover, Dclk1+ cells displayed a significantly higher degree of transformability than most acinar cells, possible due to the fact that Dclk1 is a potential interaction partner of Kras. In view of their ability to harbor mutations over prolonged periods of time, these long-lived progenitor cells could potentially serve as an important origin of pancreatic cancer.
Experimental Procedures
Additional information can be found in the supplementary material.
Animal studies
Studies were carried out in accord with institutional guidelines. Dclk1 CreERT and Dclk1-CreGFP mice were crossed to Rosa26;LacZ (LacZ), Rosa26;mTomato/mGFP (mTmG) Rosa26;tdTomato (tdTom), Rosa26;LSL-DTA (DTA) and LSL-KrasG12D mice. For lineage tracing experiments, 6-week-old mice were given 6 mg of Tamoxifen (Sigma) via oral gavage. To quantify recombination in Dclk1-CreERT tdTomato and Mist1-CreERT tdTomato mice through flow cytometry, animals received 3 doses of Tamoxifen (6mg) to ensure maximal recombination efficiency. Animals expressing LSL-KrasG12D driven by Dclk1-CreERT or Mist1-CreERT received 3 doses of Tamoxifen (6mg) to ensure maximal recombination efficiency. Acute murine pancreatitis was induced as described (Reichert et al., 2013). Control animals were treated with Tamoxifen, did not receive Cerulein and were analyzed in parallel. Treated animals were sacrificed 3 and 7 days after the last injection of cerulein or at endpoint criteria. Chronic pancreatitis was induced with 7 hourly injections of cerulein twice weekly over the course of 4 consecutive weeks. The hunching score was recorded at the end of the study.
Quantification of recombination
In lineage tracing studies 5 random high power fields were analyzed from Dclk1 mTmG mice. For each field, one fluorescent picture was taken in both the green and the blue channel to visualize recombination and 4,6-Diamidin-2-phenylindol (DAPI) positive nuclei, respectively. Morphometric analysis was done using ImageJ. After adjusting the image threshold, DAPI+ nuclei were counted automatically. After adjusting the color threshold, the percentage of green (recombined) pixels per field was measured. To control for cellular density, the number of recombined pixels was divided by the number of nuclei. To analyze the compartmental expansion of the Dclk1+ lineage in Dclk1 mTmG mice after cerulein induced pancreatitis, 80–100 recombined cells were randomly selected and grouped as single cells (1), doublets (2) and clones of three or more cells (>3). Data were normalized group-wise to untreated controls.
Supplementary Material
Acknowledgments
Thanks to the entire Wang & Olive laboratories. T. C. Wang received grants from the NIH (RO1 DK097016), the Pancreatic Cancer Action Network - ACCR (Innovative Grant) and the Lustgarten Foundation. C.B. Westphalen and B.W. Renz were supported by a grant from the German Research Foundation (Deutsche Forschungsgemeinschaft). Y. Hayakawa was supported by a grant from JSPS Postdoctoral Fellowships for Research Abroad. Y. Hayakawa and T. Tanaka were supported by the Uehara memorial foundation. S. Asfaha was supported by a “Canadian Institute for Health Research Clinician Scientist Phase I Award”. M. Quante was supported by a grant from the Max Eder Program of the Deutsche Krebshilfe. B. Honig’s research was supported by NIH grant GM30518. Joshua Broyde was supported by GM30518 and by training grant T32 GM082797. A. Califano’s research was supported by U01CA168426.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
C.B.W. and T.C.W designed the studies and wrote the manuscript. T.C.W. supervised all studies and received funding. C.B.W., Y.T., T.T., M.M., Z.J., B.W.R., X.C., K.N., Y.T., Y.C., S.A., D.L.W., Y.H., A.M.U., M.R., J.B., P.S.S. conducted experiments, were involved in data analysis and manuscript drafting. M.Q. generated Dclk1 mice and was involved in manuscript drafting. H.R. and S.O. were responsible for pathological analysis. G.H.S. supplied reagents. R.A.F. was involved in data analysis and statistical testing. C.W.H. and R.M. supplied reagents and conducted experiments. A.K.R. was involved in data analysis and manuscript drafting. K.P.O. supplied reagents and was involved in study design and manuscript drafting.
References
- Avila JL, Troutman S, Durham A, Kissil JL. Notch1 is not required for acinar-to-ductal metaplasia in a model of Kras-induced pancreatic ductal adenocarcinoma. PLoS One. 2012;7:e52133. doi: 10.1371/journal.pone.0052133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JM, Alsina J, Rasheed ZA, McAllister FM, Fu YY, Plentz R, Zhang H, Pasricha PJ, Bardeesy N, Matsui W, et al. DCLK1 marks a morphologically distinct subpopulation of cells with stem cell properties in preinvasive pancreatic cancer. Gastroenterology. 2014;146:245–256. doi: 10.1053/j.gastro.2013.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JM, Hendley AM, Lafaro KJ, Pruski MA, Jones NC, Alsina J, Younes M, Maitra A, McAllister F, Iacobuzio-Donahue CA, et al. p53 mutations cooperate with oncogenic Kras to promote adenocarcinoma from pancreatic ductal cells. Oncogene. 2015 doi: 10.1038/onc.2015.441. [DOI] [PubMed] [Google Scholar]
- Bryant KL, Mancias JD, Kimmelman AC, Der CJ. KRAS: feeding pancreatic cancer proliferation. Trends Biochem Sci. 2014;39:91–100. doi: 10.1016/j.tibs.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buch T, Heppner FL, Tertilt C, Heinen TJ, Kremer M, Wunderlich FT, Jung S, Waisman A. A Cre-inducible diphtheria toxin receptor mediates cell lineage ablation after toxin administration. Nat Methods. 2005;2:419–426. doi: 10.1038/nmeth762. [DOI] [PubMed] [Google Scholar]
- Buczacki SJA, Zecchini HI, Nicholson AM, Russell R, Vermeulen L, Kemp R, Winton DJ. Intestinal label-retaining cells are secretory precursors expressing Lgr5. Nature. 2013;495:65–69. doi: 10.1038/nature11965. [DOI] [PubMed] [Google Scholar]
- De La OJ, Emerson LL, Goodman JL, Froebe SC, Illum BE, Curtis AB, Murtaugh LC. Notch and Kras reprogram pancreatic acinar cells to ductal intraepithelial neoplasia. Proc Natl Acad Sci U S A. 2008;105:18907–18912. doi: 10.1073/pnas.0810111105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgiorno KE, Hall JC, Takeuchi KK, Pan FC, Halbrook CJ, Washington MK, Olive KP, Spence JR, Sipos B, Wright CV, et al. Identification and manipulation of biliary metaplasia in pancreatic tumors. Gastroenterology. 2014;146:233–244. e235. doi: 10.1053/j.gastro.2013.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eser S, Schnieke A, Schneider G, Saur D. Oncogenic KRAS signalling in pancreatic cancer. British Journal of Cancer. 2014;111:817–822. doi: 10.1038/bjc.2014.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira MJ, McKenna LB, Zhang J, Reichert M, Bakir B, Buza EL, Furth EE, Bogue CW, Rustgi AK, Kaestner KH. Spontaneous Pancreatitis Caused by Tissue-Specific Gene Ablation of in Mice. Cellular and molecular gastroenterology and hepatology. 2015;1:550–569. doi: 10.1016/j.jcmgh.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidekel Friedlander SY, Chu GC, Snyder EL, Girnius N, Dibelius G, Crowley D, Vasile E, Depinho RA, Jacks T. Context-dependent transformation of adult pancreatic cells by oncogenic K-Ras. Cancer Cell. 2009;16:379–389. doi: 10.1016/j.ccr.2009.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra C, Schuhmacher AJ, Cañamero M, Grippo PJ, Verdaguer L, Pérez-Gallego L, Dubus P, Sandgren EP, Barbacid M. Chronic pancreatitis is essential for induction of pancreatic ductal adenocarcinoma by K-Ras oncogenes in adult mice. Cancer Cell. 2007;11:291–302. doi: 10.1016/j.ccr.2007.01.012. [DOI] [PubMed] [Google Scholar]
- Habbe N, Shi G, Meguid RA, Fendrich V, Esni F, Chen H, Feldmann G, Stoffers DA, Konieczny SF, Leach SD, et al. Spontaneous induction of murine pancreatic intraepithelial neoplasia (mPanIN) by acinar cell targeting of oncogenic Kras in adult mice. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:18913–18918. doi: 10.1073/pnas.0810097105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, Ross S, Conrads TP, Veenstra TD, Hitt BA, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4:437–450. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- Houbracken I, Waele ED, Lardon J, Ling Z, Heimberg H, Rooman I, Bouwens L. Lineage Tracing Evidence for Transdifferentiation of Acinar to Duct Cells and Plasticity of Human Pancreas. Gastroenterology. 2011 doi: 10.1053/j.gastro.2011.04.050. [DOI] [PubMed] [Google Scholar]
- Huch M, Bonfanti P, Boj SF, Sato T, Loomans CJM, van de Wetering M, Sojoodi M, Li VSW, Schuijers J, Gracanin A, et al. Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. The EMBO Journal. 2013;32:2708–2721. doi: 10.1038/emboj.2013.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isenovic ER, Kedees MH, Tepavcevic S, Milosavljevic T, Koricanac G, Trpkovic A, Marche P. Role of PI3K/AKT, cPLA2 and ERK1/2 signaling pathways in insulin regulation of vascular smooth muscle cells proliferation. Cardiovascular & hematological disorders drug targets. 2009;9:172–180. doi: 10.2174/187152909789007034. [DOI] [PubMed] [Google Scholar]
- Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, Jacks T, Tuveson DA. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes & development. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MS, Tsigani T, Rashid M, Rabouhans JS, Yu D, Luong TV, Caplin M, Meyer T. Circulating tumor cells and EpCAM expression in neuroendocrine tumors. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17:337–345. doi: 10.1158/1078-0432.CCR-10-1776. [DOI] [PubMed] [Google Scholar]
- Koizumi H, Tanaka T, Gleeson JG. Doublecortin-like kinase functions with doublecortin to mediate fiber tract decussation and neuronal migration. Neuron. 2006;49:55–66. doi: 10.1016/j.neuron.2005.10.040. [DOI] [PubMed] [Google Scholar]
- Kong B, Michalski CW, Erkan M, Friess H, Kleeff J. From tissue turnover to the cell of origin for pancreatic cancer. Nature reviews Gastroenterology & hepatology. 2011 doi: 10.1038/nrgastro.2011.114. [DOI] [PubMed] [Google Scholar]
- Kopp JL, Dubois CL, Schaffer AE, Hao E, Shih HP, Seymour PA, Ma J, Sander M. Sox9+ ductal cells are multipotent progenitors throughout development but do not produce new endocrine cells in the normal or injured adult pancreas. Development (Cambridge, England) 2011;138:653–665. doi: 10.1242/dev.056499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp JL, Sander M. New Insights Into the Cell Lineage of Pancreatic Ductal Adenocarcinoma: Evidence for Tumor Stem Cells in Premalignant   Lesions? Gastroenterology. 2014;146:24–26. doi: 10.1053/j.gastro.2013.11.023. [DOI] [PubMed] [Google Scholar]
- Krah NM, De La OJ, Swift GH, Hoang CQ, Willet SG, Chen Pan F, Cash GM, Bronner MP, Wright CV, MacDonald RJ, et al. The acinar differentiation determinant PTF1A inhibits initiation of pancreatic ductal adenocarcinoma. eLife. 2015;4 doi: 10.7554/eLife.07125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre C, Rajbhandari P, Alvarez MJ, Bandaru P, Lim WK, Sato M, Wang K, Sumazin P, Kustagi M, Bisikirska BC, et al. A human B-cell interactome identifies MYB and FOXM1 as master regulators of proliferation in germinal centers. Molecular systems biology. 2010;6:377. doi: 10.1038/msb.2010.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Clevers H. Coexistence of quiescent and active adult stem cells in mammals. Science. 2010;327:542–545. doi: 10.1126/science.1180794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Jiang D, Chi F, Zhao B. The Hippo pathway regulates stem cell proliferation, self-renewal, and differentiation. Protein & cell. 2012;3:291–304. doi: 10.1007/s13238-012-2919-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Guo J, Yuan L, Cheng M, Cao L, Shi H, Tong H, Wang N, De W. Alpha-fetoprotein is dynamically expressed in rat pancreas during development. Development, growth & differentiation. 2007;49:669–681. doi: 10.1111/j.1440-169X.2007.00961.x. [DOI] [PubMed] [Google Scholar]
- Maitra A, Leach SD. Disputed paternity: the uncertain ancestry of pancreatic ductal neoplasia. Cancer Cell. 2012;22:701–703. doi: 10.1016/j.ccr.2012.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrache F, Tu SP, Bhagat G, Pendyala S, Osterreicher CH, Gordon S, Ramanathan V, Penz-Osterreicher M, Betz KS, Song Z, et al. Overexpression of Interleukin-1β in the Murine Pancreas Results in Chronic Pancreatitis. Gastroenterology. 2008;135:1277–1287. doi: 10.1053/j.gastro.2008.06.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May R, Sureban SM, Lightfoot SA, Hoskins AB, Brackett DJ, Postier RG, Ramanujam R, Rao CV, Wyche JH, Anant S, et al. Identification of a novel putative pancreatic stem/progenitor cell marker DCAMKL-1 in normal mouse pancreas. AJP: Gastrointestinal and Liver Physiology. 2010;299:G303–G310. doi: 10.1152/ajpgi.00146.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miduturu CV, Deng X, Kwiatkowski N, Yang W, Brault L, Filippakopoulos P, Chung E, Yang Q, Schwaller J, Knapp S, et al. High-Throughput Kinase Profiling: A More Efficient Approach toward the Discovery of New Kinase Inhibitors. Chemistry & Biology. 2011;18:868–879. doi: 10.1016/j.chembiol.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills JC, Sansom OJ. Reserve stem cells: Differentiated cells reprogram to fuel repair, metaplasia, and neoplasia in the adult gastrointestinal tract. Science Signaling. 2015;8:re8. doi: 10.1126/scisignal.aaa7540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J, Liu L, Li X, Jiang J, Wang J, Zhang B, Cao D, Yu D, Tao D, Hu J, et al. Absence of DAB2IP promotes cancer stem cell like signatures and indicates poor survival outcome in colorectal cancer. Sci Rep. 2015;5:16578. doi: 10.1038/srep16578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton JP, Timpson P, Karim SA, Ridgway RA, Athineos D, Doyle B, Jamieson NB, Oien KA, Lowy AM, Brunton VG, et al. Mutant p53 drives metastasis and overcomes growth arrest/senescence in pancreatic cancer. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:246–251. doi: 10.1073/pnas.0908428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtaugh LC, Leach SD. A case of mistaken identity? Nonductal origins of pancreatic "ductal" cancers. Cancer Cell. 2007;11:211–213. doi: 10.1016/j.ccr.2007.02.020. [DOI] [PubMed] [Google Scholar]
- Nakanishi Y, Seno H, Fukuoka A, Ueo T, Yamaga Y, Maruno T, Nakanishi N, Kanda K, Komekado H, Kawada M, et al. Dclk1 distinguishes between tumor and normal stem cells in the intestine. Nature Genetics. 2012 doi: 10.1038/ng.2481. [DOI] [PubMed] [Google Scholar]
- Osvaldt AB, Wendt LR, Bersch VP, Backes AN, de Cássia A, Schumacher R, Edelweiss MIA, Rohde L. Pancreatic intraepithelial neoplasia and ductal adenocarcinoma induced by DMBA in mice. Surgery. 2006;140:803–809. doi: 10.1016/j.surg.2006.02.012. [DOI] [PubMed] [Google Scholar]
- Puri S, Folias AE, Hebrok M. Plasticity and Dedifferentiation within the Pancreas: Development, Homeostasis, and Disease. Cell stem cell. 2015;16:18–31. doi: 10.1016/j.stem.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichert M, Takano S, von Burstin J, Kim S-B, Lee J-S, Ihida-Stansbury K, Hahn C, Heeg S, Schneider G, Rhim AD, et al. The Prrx1 homeodomain transcription factor plays a central role in pancreatic regeneration and carcinogenesis. Genes & Development. 2013;27:288–300. doi: 10.1101/gad.204453.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinehr R, Sommerfeld A, Haussinger D. CD95 ligand is a proliferative and antiapoptotic signal in quiescent hepatic stellate cells. Gastroenterology. 2008;134:1494–1506. doi: 10.1053/j.gastro.2008.02.021. [DOI] [PubMed] [Google Scholar]
- Sangiorgi E, Capecchi MR. Bmi1 lineage tracing identifies a self-renewing pancreatic acinar cell subpopulation capable of maintaining pancreatic organ homeostasis. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:7101–7106. doi: 10.1073/pnas.0902508106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saqui-Salces M, Keeley TM, Grosse AS, Qiao XT, El-Zaatari M, Gumucio DL, Samuelson LC, Merchant JL. Gastric tuft cells express DCLK1 and are expanded in hyperplasia. Histochemistry and Cell Biology. 2011;136:191–204. doi: 10.1007/s00418-011-0831-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevcik MA, Jonas BM, Lindsay TH, Halvorson KG, Ghilardi JR, Kuskowski MA, Mukherjee P, Maggio JE, Mantyh PW. Endogenous opioids inhibit early-stage pancreatic pain in a mouse model of pancreatic cancer. Gastroenterology. 2006;131:900–910. doi: 10.1053/j.gastro.2006.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK, Ritchie M, Thorne N, Wettenhall J, Shi W, Hu Y. Limma: Linear Models for Microarray and RNA-Seq Data User's Guide. Melbourne, Australia: Bioinformatics Division, The Walter and Eliza Hall Institute of Medical Research; 2015. [Google Scholar]
- Stanger BZ, Hebrok M. Control of cell identity in pancreas development and regeneration. Gastroenterology. 2013;144:1170–1179. doi: 10.1053/j.gastro.2013.01.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel O, Dor Y, Alsina J, Stirman A, Lauwers G, Trainor A, Fernández-Del Castillo C, Warshaw AL, Thayer SP. In vivo lineage tracing defines the role of acinar-to-ductal transdifferentiation in inflammatory ductal metaplasia. Gastroenterology. 2007;133:1999–2009. doi: 10.1053/j.gastro.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom A, Bonal C, Ashery-Padan R, Hashimoto N, Campos ML, Trumpp A, Noda T, Kido Y, Real FX, Thorel F, et al. Unique mechanisms of growth regulation and tumor suppression upon Apc inactivation in the pancreas. Development. 2007;134:2719–2725. doi: 10.1242/dev.02875. [DOI] [PubMed] [Google Scholar]
- Sugimura R, He XC, Venkatraman A, Arai F, Box A, Semerad C, Haug JS, Peng L, Zhong XB, Suda T, et al. Noncanonical Wnt signaling maintains hematopoietic stem cells in the niche. Cell. 2012;150:351–365. doi: 10.1016/j.cell.2012.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sureban SM, Madhoun MF, May R, Qu D, Ali N, Fazili J, Weygant N, Chandrakesan P, Ding K, Lightfoot SA, et al. Plasma DCLK1 is a marker of hepatocellular carcinoma (HCC): Targeting DCLK1 prevents HCC tumor xenograft growth via a microRNA-dependent mechanism. Oncotarget. 2015 doi: 10.18632/oncotarget.5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sureban SM, May R, Qu D, Weygant N, Chandrakesan P, Ali N, Lightfoot SA, Pantazis P, Rao CV, Postier RG, et al. DCLK1 Regulates Pluripotency and Angiogenic Factors via microRNA-Dependent Mechanisms in Pancreatic Cancer. PLoS ONE. 2013;8:e73940. doi: 10.1371/journal.pone.0073940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H, Biehs B, Warming S, Leong KG, Rangell L, Klein OD, De Sauvage FJ. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature. 2011 doi: 10.1038/nature10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu S, Bhagat G, Cui G, Takaishi S, Kurt-Jones EA, Rickman B, Betz KS, Penz-Oesterreicher M, Bjorkdahl O, Fox JG, et al. Overexpression of interleukin-1beta induces gastric inflammation and cancer and mobilizes myeloid-derived suppressor cells in mice. Cancer Cell. 2008;14:408–419. doi: 10.1016/j.ccr.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdez IA, Teo AK, Kulkarni RN. Cellular stress drives pancreatic plasticity. Sci Transl Med. 2015;7:273ps272. doi: 10.1126/scitranslmed.3010577. [DOI] [PubMed] [Google Scholar]
- Wang MT, Holderfield M, Galeas J, Delrosario R, To MD, Balmain A, McCormick F. K-Ras Promotes Tumorigenicity through Suppression of Non-canonical Wnt Signaling. Cell. 2015;163:1237–1251. doi: 10.1016/j.cell.2015.10.041. [DOI] [PubMed] [Google Scholar]
- Wang YJ, Bailey JM, Rovira M, Leach SD. Sphere-forming assays for assessment of benign and malignant pancreatic stem cells. Methods in molecular biology (Clifton, NJ) 2013;980:281–290. doi: 10.1007/978-1-62703-287-2_15. [DOI] [PubMed] [Google Scholar]
- Wescott MP, Rovira M, Reichert M, von Burstin J, Means A, Leach SD, Rustgi AK. Pancreatic ductal morphogenesis and the Pdx1 homeodomain transcription factor. Molecular biology of the cell. 2009;20:4838–4844. doi: 10.1091/mbc.E09-03-0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphalen CB, Asfaha S, Hayakawa Y, Takemoto Y, Lukin DJ, Nuber AH, Brandtner A, Setlik W, Remotti H, Muley A, et al. Long-lived intestinal tuft cells serve as colon cancer-initiating cells. The Journal of clinical investigation. 2014 doi: 10.1172/JCI73434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weygant N, Qu D, Berry WL, May R, Chandrakesan P, Owen DB, Sureban SM, Ali N, Janknecht R, Houchen CW. Small molecule kinase inhibitor LRRK2-IN-1 demonstrates potent activity against colorectal and pancreatic cancer through inhibition of doublecortin-like kinase 1. Molecular cancer. 2014;13:103. doi: 10.1186/1476-4598-13-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo JH, Shimoni Y, Yang WS, Subramaniam P, Iyer A, Nicoletti P, Rodriguez Martinez M, Lopez G, Mattioli M, Realubit R, et al. Elucidating Compound Mechanism of Action by Network Perturbation Analysis. Cell. 2015;162:441–451. doi: 10.1016/j.cell.2015.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanger K, Stanger BZ. Facultative stem cells in liver and pancreas: Fact and fancy. Developmental dynamics : an official publication of the American Association of Anatomists. 2011 doi: 10.1002/dvdy.22561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanger K, Zong Y, Maggs LR, Shapira SN, Maddipati R, Aiello NM, Thung SN, Wells RG, Greenbaum LE, Stanger BZ. Robust cellular reprogramming occurs spontaneously during liver regeneration. Genes & Development. 2013 doi: 10.1101/gad.207803.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang QC, Petrey D, Deng L, Qiang L, Shi Y, Thu CA, Bisikirska B, Lefebvre C, Accili D, Hunter T, et al. Structure-based prediction of protein-protein interactions on a genome-wide scale. Nature. 2012;490:556–560. doi: 10.1038/nature11503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang QC, Petrey D, Garzon JI, Deng L, Honig B. PrePPI: a structure-informed database of protein-protein interactions. Nucleic acids research. 2013;41:D828–D833. doi: 10.1093/nar/gks1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Shi G, Schmidt CM, Hruban RH, Konieczny SF. Acinar cells contribute to the molecular heterogeneity of pancreatic intraepithelial neoplasia. The American Journal of Pathology. 2007;171:263–273. doi: 10.2353/ajpath.2007.061176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann G, Papke B, Ismail S, Vartak N, Chandra A, Hoffmann M, Hahn SA, Triola G, Wittinghofer A, Bastiaens PIH, et al. Small molecule inhibition of the KRAS-PDEδ interaction impairs oncogenic KRAS signalling. Nature. 2013 doi: 10.1038/nature12205. [DOI] [PubMed] [Google Scholar]
- Ziv O, Glaser B, Dor Y. The plastic pancreas. Developmental Cell. 2013;26:3–7. doi: 10.1016/j.devcel.2013.06.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.