Abstract
Poliovirus (PV) is the prototypical picornavirus. It is a non-enveloped RNA virus with a small (~7.5 kb) genome of positive polarity. It has long served as a model to study RNA virus biology, pathogenesis, and evolution. cDNA clones of several strains are available, and infectious virus can be produced by the transfection of in vitro transcribed viral genomes into an appropriate host cell. PV infects many human and non-human primate cell lines including HeLa and HeLa S3 cells, and can grow to high titer in culture. Protocols for the production, propagation, quantification, and purification of PV are presented. A separate chapter concerning the generation and characterization of PV mutants will also be presented.
Keywords: poliovirus, picornavirus, plaque assay, titration, virus purification
INTRODUCTION
Poliovirus (PV) is the causative agent of poliomyelitis and the founding member of the Picornaviridae family of small RNA viruses (one of five families of viruses that comprise the order Picornavirales). The picornaviruses are non-enveloped viruses with single stranded RNA genomes of positive polarity. The family consists of twelve genera, including the Enterovirus genus, to which PV belongs. This genus can be further subdivided into ten species. One species, Human enterovirus C, includes the three serotypes of poliovirus (1, 2, and 3) as well as a number of closely related enterovirus and group A coxsackievirus strains (King et al., 2011; Knowles et al., 2010).
A number of infectious cDNA clones of PV are available, and it has become well established as a model to study many aspects of RNA virus biology. The following protocols describe the production of PV from a cDNA clone and its propagation in HeLa or HeLa S3 cell culture, as well as standard methods for quantification of the virus and the production of clonal viral stocks. Support protocols for the growth and maintenance of the cDNA clone and HeLa and HeLa S3 cells are also included. Finally we include a protocol for the purification of PV virions by ultracentrifugation through a sucrose cushion.
CAUTION: Poliovirus (PV) is a Biosafety Level 2 (BSL-2) pathogen. Follow all appropriate guidelines and regulations for the use and handling of pathogenic microorganisms. See UNIT 1A.1 and other pertinent resources (APPENDIX 1B) for more information.
CAUTION: HeLa and HeLa S3 cells contain human papilloma virus (HPV-18), which is a Biosafety Level 2 (BSL-2) pathogen. Follow all appropriate guidelines and regulations for the use and handling of pathogenic microorganisms. See UNIT 1A.1 and other pertinent resources (APPENDIX 1B) for more information.
Basic Protocol 1: GENERATION OF PV FROM cDNA PLASMID
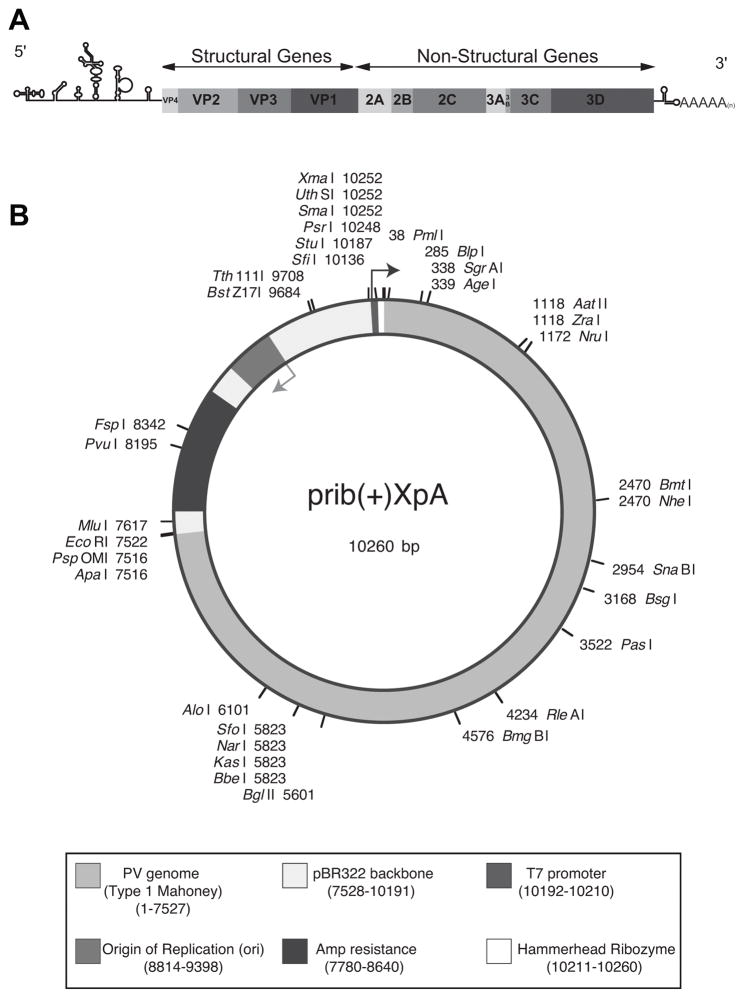
Because PV is a (+) sense RNA virus, its RNA genome (Figure 1A) alone is sufficient to produce infectious virus, provided it can enter a host cell. Entrance (transfection) is normally achieved by electroporation. The genome can then be translated to produce the viral proteins, including the structural (capsid) proteins and the nonstructural proteins involved in replication. Once the replication machinery has been produced, the genome can serve as a template for complementary (−) strand synthesis, which in turn templates the production of further (+) strands, which can be packaged with the capsid proteins into infectious virions. The initial RNA genomes for transfection are produced from a PV cDNA plasmid.
Figure 1.
(A) Schematic of the PV Genome. The PV genome includes a highly structured 5′UTR followed by a single open reading frame and a polyadenylated 3′UTR. The coding region can be separated into structural and non-structural genes. The structural genes are required to form the viral capsid and the non-structural genes are all required for a successful replication cycle. (B) prib(+)XpA plasmid map. PV1 (Mahoney) genome is in the pBR322 backbone. The plasmid contains an ampicillin resistance cassette, a T7 promoter, and a hammerhead ribozyme. Location of single cut restriction sites are shown.
A basic PV cDNA plasmid must contain a PV genome with a T7 promoter upstream and a poly(A) tail downstream. For the poly (A) tail, a minimum of 8 ‘A’s are essential for viral replication, and a longer tail will improve efficiency (Polacek & Andino, unpublished data). This protocol is based on methods used primarily for the plasmid, prib(+)XpA (Figure 1B) (Herold and Andino, 2000). This plasmid contains the Mahoney (PV1) genome in a pBR322 backbone. In addition to the T7 promoter and a long (75 nt) poly(A) tail, it also contains a hammerhead ribozyme sequence between the promoter start site and the genome. The ribozyme ensures an accurate 5′ end, which improves the efficiency of replication (Herold and Andino, 2000), but is not essential. If you are using a plasmid with a short (<20 nt) poly(A) tail and/or no ribozyme, you may want to use more RNA than described here to improve your yield of P0 virus. For linearization, you should choose a unique restriction site as close as possible to the end of the poly (A) tail.
Materials
cDNA plasmid prib(+)XpA (Support Protocol 1)
20 U/μL EcoRI (New England BioLabs)
10× NEBuffer EcoRI (provided with EcoRI)
Nuclease-free water
1% agarose TAE gels and running buffer (Voytas, 2001)
6× DNA Loading Dye
DNA Ladder (e.g. GeneRuler 1 kb Plus, Fermentas)
Phenol:chloroform:IAA (25:24:1)
Chloroform:IAA (24:1)
3 M NaOAc (pH 5.2)
Isopropanol
75% (v/v) EtOH
T7 RNA Polymerase*
5× Transcription Buffer*
40 U/μL RNaseOUT (Invitrogen)
NTP Mix (25 mM each NTP)
2U/μL DNase I (RNase-free; New England BioLabs)
3× LiCl-EDTA
0.5× TE
2× RNA Denaturing Loading Dye
HeLa or HeLa S3 Cells (Support Protocol 2)
D-PBS (PBS w/o Ca2+ or Mg2+)
0.05% Trypsin-EDTA
2.5% NCS DMEM/F-12
37°C heat block or water bath
95°C heat block
Ice
NanoDrop spectrophotometer
37°C and 5% CO2 humidified incubator
Electroporator and 4 mm cuvettes
Tissue culture microscope (inverted phase-contrast)
Plasmid linearization
Poliovirus cDNA plasmids typically contain several unique restriction sites immediately downstream of the encoded poly(A) tail. In the case of our most commonly used plasmid, prib(+)XpA (Herold and Andino, 2000), the best site is EcoRI. A minimum of 10 μg of DNA is required. We typically digest 25 μg in a 500 μL volume. After phenol-chloroform extraction, this yields enough linearized plasmid to template 3–4 in vitro transcription reactions.
-
1
Combine 25 μg plasmid DNA, 50 μL 10× NEBuffer EcoRI, 125 U EcoRI, and nuclease-free water in a 500 μL final volume and incubate 1h at 37°C.
-
2
Remove 5 μL (1%) of digest reaction. Dilute 500 ng undigested plasmid DNA to 5 μL. Add 1 μL 6× DNA Loading Dye to each and analyze by electrophoresis in a 1% agarose TAE gel (Voytas, 2001).
If digestion is complete (all digested DNA should be converted to a single linear fragment of ~10 kbp, distinguishable from the multiple circular DNA forms of the undigested plasmid), proceed to step 4. -
3
If digestion is not complete, redigest with 125 U fresh EcoRI and repeat step 3.
-
4
Perform phenol:chloroform extraction:
Add an equal volume (500 μL) of phenol:chloroform:IAA and mix. Centrifuge 10 min at ≥ 12,000× g. Transfer aqueous phase to a fresh tube.
Add an equal volume (500 μL) of chloroform:IAA and mix. Centrifuge 10 min at ≥ 12,000× g. Transfer aqueous phase to a fresh tube.
To precipitate the DNA, add 0.1 volume (50 μL) 3M NaOAc and 1 volume (500 μL) isopropanol. Incubate at least 20 min at −20°C. To pellet the DNA, centrifuge 20 min at ≥ 12,000× g, 4°C.
Discard the supernatant and wash the pellet in 500 μL 75% EtOH. Centrifuge 5 min at ≥ 7,500× g.
-
Discard the supernatant and remove the residual liquid with a pipette. Resuspend the DNA in 25 μL nuclease-free water. Quantitate the DNA on a NanoDrop Spectrophotometer and adjust the concentration to 500 ng/μL.
Store linearized DNA at −20°C.
In vitro transcription of viral RNA
Most poliovirus cDNA plasmids contain a T7 promoter upstream of the viral genome and the ribozyme if applicable. A minimum of 1 μg linearized DNA is required. We typically use 5 μg template in a 100 μL volume. Yield is highly variable. After lithium chloride precipitation, there is usually enough for 1–4 transfections.
*NOTE: We use an in-house preparation of T7 RNA Polymerase with 5× Transcription Buffer. The enzyme is also available commercially from a number a sources and is typically provided with a 10× Tris-based buffer.
-
5
Combine 5 μg linearized plasmid DNA, 20 μL 5× Transcription Buffer, 30 μL NTP Mix, 40 U RNaseOUT, 1 μL T7 RNA Polymerase, and nuclease-free water in a 100 μL final volume and incubate 4–16 h at 37°C.
Longer incubation times increase yield at the expense of some level of RNA degradation. Overnight incubation produces RNA of sufficient quality to generate virus. If a precise quantification of transcripts is necessary, RNA of higher quality (but lower yield) is obtained from shorter incubations. -
6
To digest the DNA template, add 5U DNase I and incubate 10 min at 37°C.
-
7
To selectively precipitate the RNA transcript (but not the free nucleotides), add 0.5 vol (52.5 μL) 3× LiCl-EDTA and incubate at least 1 h at −20°C. To pellet the RNA, centrifuge 20 min at ≥ 12,000× g, 4°C.
-
8
Discard the supernatant, remove the residual liquid with a pipette, and wash the pellet in 500 μL 75% EtOH. Centrifuge 5 min at ≥ 7,500× g, 4°C.
-
9
Discard the supernatant and remove the residual liquid with a pipette. Resuspend the RNA in 25 μL 0.5× TE. Quantitate a 1:10 dilution of the RNA on a NanoDrop Spectrophotometer and adjust the concentration to 1 μg/μL.
Store in vitro transcribed RNA at −80°C. -
10
Dilute 1 μg RNA to 3 μL and add 3 μL Gel Loading Buffer II. Incubate 5 min at 95°C, then 2 min on ice. Analyze by electrophoresis in a 1% agarose TAE gel (Voytas, 2001).
If RNA quality is good, this “semi-denaturing” gel analysis (denaturing buffer, non-denaturing gel) will show a clean single band. RNA degradation is seen as smearing below the main band. Smearing above the main band is usually due to incomplete linearization of plasmid template. Poor quality RNA will usually still generate virus upon transfection, but should not be used if precise quantification of transcripts is important (e.g. to calculate transfection efficiency or generate a qPCR standard curve).
Transfection of viral RNA
If you are unfamiliar with the appearance of cytopathic effect (CPE) in HeLa or HeLa S3 cells, it is advisable to include a mock-transfected control (using 0.5X TE) to distinguish between the cellular debris/damage caused by electroporation and that due to productive transfection/infection. Many cells are lost during the washes. Typically a confluent T150 flask provides enough cells for 3–5 transfections. We use a BTX Electro Cell Manipulator 600 with the pulse settings described below. Settings may vary depending on the model of electroporator used.
-
11
Wash HeLa or HeLa S3 monolayer with D-PBS and trypsinize cells with 3 mL 0.05% Trypsin-EDTA.
-
12
Resuspend cells in 15 mL D-PBS. Centrifuge 5 min at 200× g. Aspirate or pour off supernatant.
-
13
Repeat step 2 twice more. After the third spin, resuspend the cells in 2.5 mL D-PBS and count on a hemocytometer. Adjust the concentration to 5×106 cells/mL.
-
14
Combine 800 μL of cells and 20 μg of RNA in a chilled 4 mm electroporation cuvette and incubate 20 min on ice.
-
15
Electroporate (voltage = 300 V, capacitance = 1000 μF, resistance = 24 Ω) cells and recover in 7.5 mL 37°C 2.5% NCS DMEM/F12. Transfer to T75 tissue culture flask and incubate at 37°C in 5% CO2.
Recovery of P0 virus
You must allow at least one infectious cycle (~8 h) to occur before checking for CPE. Transfection efficiencies vary, so allowing 2–3 cycles (16–24 h) is more typical. The infectious cycle may be slower in mutant viruses. Also, viable virus can be recovered even in the absence of visible CPE. We typically wait for visible CPE or 24 h post-transfection, whichever comes first. The freeze-thaw cycles lyse any remaining cells and release the virus.
-
16
Check transfected cells for CPE on tissue culture microscope. If no CPE is visible, return cells to incubator. If CPE is visible, or you wish to attempt viral recovery anyway, proceed to step 17.
-
17
Freeze at −20°C or −80°C. Thaw at room temperature or 37°C.
-
18
Repeat step 17 twice more.
-
19
To pellet cellular debris, centrifuge 5 min at 2500× g. Transfer supernatant (P0 virus) to a fresh tube.
This is the only purification necessary for most purposes. Store P0 virus at −20°C.
Support Protocol 1: PROPAGATION AND MAINTENANCE OF cDNA PLASMID
Our most commonly used plasmid is prib(+)XpA (Figure 1B) (Herold and Andino, 2000). This plasmid contains the Mahoney (PV1) genome in a pBR322 (Watson, 1988) backbone. It is a relatively large (10.2 kb) plasmid and many competent cell lines will recombine out all or part of the viral genome. We have found SURE Electroporation-Competent cells (Agilent) to work reliably with this and other PV cDNA plasmids that contain an ampicillin resistance marker. We use a BTX Electro Cell Manipulator 600 with the pulse settings described below. Settings may vary depending on the model of electroporator used. There are a wide variety of midi- and maxi-prep kits available, and which one to use is a matter of personal or lab preference. We use primarily the NucleoBond Xtra Midi Kit from Machery-Nagel.
NOTE: SURE cells are sensitive to ampicillin but resistant to several other antibiotics, so be sure to check before using them.
NOTE: pBR322 is a low copy-number vector with ampR and tetR, but in prib(+)XpA the tetR locus is disrupted by the viral genome.
Materials
prib(+)XpA (or other PV cDNA plasmid; check resistance - see NOTE)
SURE Electroporation-competent Cells (Agilent)
sterile water
LB medium
LB-Amp agar plates (50–100 μg/mL)
LB-Amp medium (50–100 μg/mL)
Midi or Maxi Prep Kit (e.g. NucleoBond Xtra, Machery-Nagel)
1× TE (pH 8.0)
Ice
Electroporator and 1 mm cuvettes
Chill a 1 mm electroporation cuvette and a 1.5 mL tube on ice. Thaw SURE cells on ice.
Dilute prib(+)XpA to ~50 pg/μL in sterile water.
-
In chilled 1.5 mL tube, combine 30 μL sterile water, 10 μL SURE cells, and 1 μL plasmid. Mix well. Transfer to 1 mm cuvette.
After first thaw, re-freeze extra SURE cells in single-use aliquots to avoid repeated freeze-thaw cycles. Electroporate (voltage = 1.8 kV, resistance = 129Ω) cells and recover in 960 mL LB-Amp. Transfer to 1.5 mL tube.
Incubate 1 h at 37°C with shaking.
Plate 100 μL on LB-Amp agar plate and incubate overnight at 37°C.
Use a sterile pipet tip to pick a colony and infect 3 mL LB-Amp starter culture. Incubate overnight at 37°C with shaking.
Dilute 1000-fold (e.g. 200 μL into 200 mL) into LB-Amp to launch culture for midi- or maxi-prep. Consult kit manual to determine the appropriate culture volume for a low copy-number plasmid. Incubate overnight at 37°C with shaking.
-
Perform plasmid prep according to kit instructions. Resuspend plasmid DNA in 1× TE. Quantitate the DNA on a NanoDrop Spectrophotometer and adjust the concentration to 1 μg/μL.
Store DNA at −20°C.
Support Protocol 2: STORAGE, PROPAGATION, AND MAINTENANCE OF HeLa AND HeLa S3 MONOLAYER CULTURES
HeLa and HeLa S3 cells are available from the ATCC and most university cell culture facilities. Either line grows well in monolayer culture and is easily infectable by PV. We tend to use HeLa S3 cells because they can be also be grown in suspension when needed. The choice of cell line for monolayer culture and infection is a matter of personal or lab preference. HeLa and HeLa S3 divide roughly once every 24 h. It is good practice to split between 1:3 and 1:12 every 2–4 days.
NOTE: PV can infect many other primate cell lines. Receptor presence is the main factor limiting infectability, and a few transgenic mouse lines that express the PV receptor (PVR or CD155) are also available. However other cell lines may be less amenable to transfection by electroporation. They may also form poor monolayers for plaque assays. If you want to study PV in another cell line, consider using HeLa or HeLa S3 to generate the P0 virus and to titer experimental samples.
Materials
HeLa or HeLa S3 Cells (ATCC # CCL-2 or # CCL-2.2)
10% NCS DMEM/F-12
D-PBS (PBS w/o Ca2+ or Mg2+)
0.05% Trypsin-EDTA
Freezing DMEM/F-12
37°C and 5% CO2 humidified incubator
Tissue culture microscope (inverted phase-contrast)
Cell freezing container with foam insert and isopropanol
Thaw HeLa or HeLa S3 cells
-
1
Warm 10% NCS DMEM/F-12 to 37°C and aliquot ~10 mL into a 15 mL conical tube.
-
2
Remove a vial of frozen cells from the liquid nitrogen and thaw at 37°C until just liquid.
-
3
Resuspend cells in the warmed growth medium in the 15 mL tube.
-
4
To gently pellet cells, centrifuge 3 min at 400× g. Aspirate or pour off supernatant.
This step removes residual DMSO from the freezing medium. -
5
Resuspend in 30 mL fresh warm 10% NCS DMEM/F-12 and transfer cells to a T150 flask. Incubate 24 h at 37°C and 5% CO2.
The next day, cells should be ~ 50% confluent and adherent. Changing the medium removes dead cells and helps the living ones recover from freezing. -
6
Aspirate old medium and add 30 mL fresh warm 10% NCS DMEM/F-12. Incubate 24 h at 37°C and 5% CO2.
Propagate HeLa or HeLa S3 cells
Typically, after you thaw new cells for the first time, you will want to propagate them for ~ 2 passages to grow enough to freeze down a new batch of aliquots (see below). Maintain the original culture until you have tested one of the new aliquots to confirm the batch is viable and uncontaminated. Ideally, cells should be allowed to divide only ~50 times (less than 2 months) after thawing. At that point you should discard the culture and thaw a fresh aliquot.
-
7
Starting with a confluent T150 flask, wash cell monolayer with 15 mL D-PBS.
-
8
Aspirate D-PBS. Add 3 mL 0.05% Trypsin-EDTA. Tilt flask to cover cell monolayer. Incubate 5–10 minutes at room temperature or 37°C until monolayer detaches from flask surface.
-
9
Add 9 mL warm 10% NCS DMEM/F-12. Pipet down side of flask to remove cells, then up and down against the bottom of the flask to break up clumps.
Work quickly because the serum will inactivate the trypsin. This gives a total volume of 12 mL, which divides easily for almost any level of split (1:2, 1:3, 1:4, 1:6, 1:8, or 1:12). It is better practice to actually count the cells on a hemacytometer and then seed exactly the desired number. -
10
Distribute desired volume of cells to new T150 flasks and add warm 10% NCS DMEM/F-12 to 30 mL. Incubate at 37°C and 5% CO2 until confluent again.
Freeze HeLa or HeLa S3 cells
-
11
Prepare nine 85% confluent T150 flasks. Wash each monolayer with 15 mL D-PBS.
-
12
Aspirate D-PBS. Add 3 mL 0.05% Trypsin-EDTA. Tilt flask to cover cell monolayer. Incubate 5–10 minutes at room temperature or 37°C until monolayer detaches from flask surface.
-
13
Add 9 mL warm 10% NCS DMEM/F-12. Pipet down side of flask to remove cells, then up and down against the bottom of the flask to break up clumps.
-
14
To gently pellet cells, centrifuge 3 min at 400× g. Aspirate or pour off supernatant.
-
15
Label 18 sterile 1.5 mL cryovials.
-
16
Aspirate medium and resuspend cells in 18 mL cold Freezing DMEM/F-12.
-
17
Transfer 1 mL to each cryovial and cap.
-
18
Place cryovials in cell freezing container with foam insert and isopropanol and place container at −80°C overnight.
This allows the cells to freeze slowly (~1°C/minute) which is important for their viability. -
19
Transfer cryovials to liquid nitrogen dewar for long-term storage.
Basic Protocol 2: QUANTIFICATION OF PV TITER BY PLAQUE ASSAY
The infectivity of PV stocks or experimental samples can be determined by plaque assay or by TCID50 assay. Titer is important for planning experiments (e.g. determining MOI). It is also one of the most common ways to assess the results of an experiment (e.g. the titer of a mutant or drug-treated virus compared to WT or untreated). Plaque assays are less labor intensive and have shorter incubation times, so are usually the method of choice. TCID50 assays should be used to compare a mutant with a small plaque phenotype to WT, because small plaques can cause a systematic underestimation of true titer.
NOTE: This quantification is approximate. If more precision is needed, either perform 3 or more replicates to get an accurate measurement of variance, or choose 1–2 appropriate dilutions and repeat the assay in 10 cm dishes. One 10 cm dish has approximately 6-fold more area than one well in a 6-well plate, so scale up the volumes of cells, viral inoculum, and overlay accordingly, or see Basic Protocol 4 for detailed instructions.
Materials
Virus stocks/samples to be titered
HeLa or HeLa S3 cells (Support Protocol 2)
10% NCS DMEM/F12
Serum-free DMEM/F12
2× DMEM/F12
2% (w/v) agarose
2% (w/v) formaldehyde
1% (v/v) bleach solution
0.1% crystal violet stain
6-well plates
37°C and 5% CO2 humidified incubator
Tissue culture microscope (inverted phase-contrast)
37°C water bath
57°C water bath
Prepare cells
-
1
Seed HeLa or HeLa S3 cells in 6-well plates at 1×106 cells in 2 mL of 10% NCS DMEM/F12 per well and incubate 24 h at 37°C in 5% CO2, at which time they should be confluent. Prepare 1 plate for each stock/sample to be titered.
-
2
On the day of the plaque assay, aspirate old medium and replace with serum-free medium. Work with only 4–6 plates at a time, to prevent cells from drying out. Incubate at 37°C in 5% CO2 while diluting virus.
Alternatively, cells can be washed with PBS immediately prior to inoculation. Eliminating this wash step by performing an earlier media change allows infections to be launched more synchronously when doing multiple plaque assays.
Dilute virus and infect cells
-
3
Prepare 2% agarose for overlays, with a separate bottle for each group of 8–12 plates. Prepare 12.5 mL 2% agarose per plate. Melt the agarose in a microwave and cool to 57°C. Warm 2× DMEM/F-12 to 37°C.
By keeping the agarose at 57°C and the medium at 37°C, a mixture at approximately 47°C is created when they are combined in step 5. This temperature will keep the agarose liquid long enough to distribute the overlays but will not harm the cells. -
4
Thaw virus stocks/samples to be titered and make 10-fold serial dilutions in serum-free DMEM/F-12 (111 μL into 1.0 mL works well). Always change tips when performing serial dilutions.
Titering over a 6-log range gives a fair amount of flexibility. 1×108 is the largest dilution necessary for standard PV stocks, which are rarely more concentrated than 1×109 PFU/mL (unless further concentrated after harvest, e.g. Basic Protocol 5). Fewer dilutions may be necessary for samples expected to have lower titers, such as P0 stocks, early time points in growth curves, or high drug concentrations in resistance curves. -
5
Again working with 4–6 plates at a time, aspirate the medium and add 250 μL of virus dilution to each well. Start with the most dilute sample.
Starting with the most dilute sample avoids the need to change pipette tips between dilutions of the same sample. -
6
Distribute the virus by rocking by hand. Incubate plates at least 30 min at 37°C in 5% CO2. Redistribute virus once every 10 min.
-
7
Combine equal volumes of 2% agarose and 2× DMEM/F-12 and swirl to mix. Add 4 mL overlay solution to each well and swirl plate gently to mix inoculum into overlay. Let plates sit at room temperature until overlay solidifies.
-
8
Incubate 40–48 h at 37°C in 5% CO2.
Fix and stain cells
-
9
Add 1 mL 2% formaldehyde to each well, on top of the overlay. Incubate 10 min at room temperature.
-
10
Pour off formaldehyde and remove overlay (flip out into 10% bleach). Add 1 mL 0.1% crystal violet to each well. Incubate 10 min at room temperature.
-
11
To remove stain and disinfect plate, plunge plate into a dilute (~1%) bleach solution. Rinse immediately 3–5 times under gently running water to remove bleach and prevent bleaching of stained cells. Invert on paper towels to drain, air dry and count.
-
12
Count plaques from a well with 10 – 100 plaques. Divide by inoculum volume then multiply by dilution factor to get titer.
Titers are expressed as plaque-forming units (PFU) per mL. Titer (PFU/mL) = well plaque count/0.25 mL × dilution factor.
Alternate Protocol 1: QUANTIFICATION OF PV TITER BY TCID50 ASSAY
TCID50 assays are most commonly used to measure infectivity for viruses that do not form plaques. They are more labor intensive and have longer incubation times than plaque assays. However there is less ambiguity in the count, due to the binary (live/dead) nature of the read out. TCID50 assays should be used instead of plaque assays any time a large variation in plaque size is expected, which would make it difficult to count a similar number of plaques for each sample.
Additional Materials (also see Basic Protocol 2)
96-well plates
Multi-channel 20–200 μL pipette and tips
Sterile pipette reservoirs
Prepare cells, dilute virus and infect cells
-
1
Dilute HeLa or HeLa S3 cells to 1×105 cells/mL in 10% NCS DMEM/F12 and use multi-channel pipette to distribute 100 μL/well to 96-well plates. Allow 1 plate per sample to be titered. Incubate at 37°C in 5% CO2 while diluting virus.
To save time on the day of the TCID50 assay, cells may be plated 24 h in advance, in which case dilute to 5×104 cells/mL. -
2
Thaw virus samples and make 10-fold serial dilutions in serum-free DMEM/F12 (167 μL into 1.5 mL works well). Always change tips when performing serial dilutions.
Titering over an 8-log range gives a fair amount of flexibility. 1×1011 is the largest dilution necessary for standard PV stocks, which are rarely more concentrated than 1×109 PFU/mL (unless further concentrated after harvest, e.g. Basic Protocol 5). Fewer dilutions may be necessary for samples expected to have lower titers, such as P0 stocks, early time points in growth curves, or high drug concentrations in resistance curves. -
3
Pour virus dilution into reservoir and use multichannel pipette to add 100 μL to each well in one row of 96-well plate. Start with the most dilute sample. Aspirate residual virus from reservoir between dilutions.
Starting with the most dilute sample avoids the need to change pipette tips, aspirator pipet, and reservoirs between dilutions of the same sample. However it is critical to change all of these between samples to avoid cross-contamination. -
4
Incubate 5–7 days at 37°C in 5% CO2.
Assess CPE and Calculate Titer
-
5
Score wells as positive or negative for CPE. There are several alternative ways to do this:
Score based on well color (pH). DMEM/F12 contains a pH indicator, phenol red. If the cells are alive, after a week the pH will be more acidic, and the wells will be orange-yellow. If the cells are dead, the wells will be red-pink. This is by far the fastest and easiest way to score, but occasionally you miss wells that died very slowly, giving the cells time to metabolize and alter the pH. This underestimation is most problematic when working with a mutant virus that kills more slowly than WT, since it will cause systematic underestimation of the mutant titer.
Score each well for CPE using an inverted phase-contrast microscope. This is the most accurate method, but also the most labor intensive. It can be combined with the first method, scoring the obvious extreme dilutions as live/dead by color, but examining the 2–3 intermediate rows under the microscope.
-
Score by staining with 0.1% crystal violet. Pour off or aspirate medium from cells. Add 50 μL 0.1% crystal violet to each well. Incubate 10 min at room temperature. Gently bleach and rinse plates as for plaque assays.
It is difficult to thoroughly rinse out the bleach without disturbing the stained monolayers in 96-well plates. Therefore, it is advisable to score wells soon after staining, as residual bleach will tend to destroy the stain over time.
-
6
Calculate titer according to the method of (Reed and Muench, 1938).
A useful Microsoft Excel based calculator is available at: http://www.med.yale.edu/micropath/pdf/Infectivity%20calculator.xls (Lindenbach, 2009).
Basic Protocol 3: PROPAGATION OF PV IN HeLa OR HeLa S3 MONOLAYER CULTURES
The amount of virus produced in a P0 stock (see Basic Protocol 1) is often insufficient for the desired experiments, so propagation of the virus is necessary. P1 and subsequent stocks may be propagated directly from the P0 stock, but it is better practice to generate a clonal stock first (see NOTE). Monolayer infections generate sufficient virus for most purposes. If a large amount of highly concentrated virus is needed (see Basic Protocol 5), a suspension infection can be performed (see Alternate Protocol 2).
NOTE: The plasmid preparation, RNA transcription, and transfection processes offer numerous opportunities for either mutation or cross contamination. If you are working with a strongly attenuated mutant that either reverts easily or was prepped at the same time as a WT stock, it is essential to clonally purify and sequence stocks (see Basic Protocol 4 or Alternate Protocol 3) to confirm the presence of the desired mutation and no others. For WT and other mutants it is still better practice, but not absolutely essential.
NOTE: Titering your infecting stock by plaque assay or TCID50 assay is necessary to achieve a precise Multiplicity of Infection (MOI). If knowledge of MOI is not necessary, infecting with a fixed volume (e.g. 5% of inoculum) and waiting for CPE is adequate.
NOTE: Serum does not significantly inhibit poliovirus infections and can sometimes improve virus yields because it improves to host cell viability. However the presence of serum in virus stocks can have consequences for subsequent experiments, especially in animal models. For this reason we usually recover virus in low serum (2.5% NCS) or serum-free DMEM/F-12, depending on the intended use of the virus.
Materials
HeLa or HeLa S3 cells (Support Protocol 2)
10% NCS DMEM/F-12
P0 or clonal infecting stock
Serum-free DMEM/F-12
PBS
2.5% NCS DMEM/F-12
37°C and 5% CO2 humidified incubator
Tissue culture microscope (inverted phase-contrast)
Prepare and infect cells
-
1
Prepare a confluent T150 of HeLa or HeLa S3 cells. This can be achieved by seeding 7.5×106 cells in 30 mL 10% NCS DMEM/F-12 48 h prior to the infection. Cell count and incubation time can be adjusted as convenient.
If less virus is needed, it can be propagated in a T75 or T25 by scaling a volumes down 2-fold or 6-fold, respectively. -
2
Prepare viral inoculum by diluting infected stock to desired concentration in 7.5 mL of serum-free DMEM/F-12.
If the titer of the infecting stock is unknown, a fixed volume (375 μL works well) is adequate. If the titer of the infecting stock is known, multiplicity of infection (MOI) can be calculated. If precise MOI is important, and you are infecting with stocks that were recovered in 2.5% NCS DMEM/F-12 and that differ significantly in titer, consider diluting to a common intermediate concentration in 2.5% NCS DMEM/F-12, then to a final concentration in serum-free DMEM-F-12. This will ensure that the infections are not affected by different concentrations of serum. -
3
Aspirate old medium from cells. Wash monolayer with 15 mL PBS. Aspirate PBS and cover with 7.5 mL inoculum.
-
4
Distribute the virus by rocking by hand. Incubate flask at least 30 min at 37°C in 5% CO2. Redistribute virus once every 10 min.
-
5
Aspirate the inoculum. Cover with 15 mL 2.5% NCS. Incubate at 37°C in 5% CO2.
Recover P1 virus
Allow at least one to three infectious cycles (8–24 h) to occur, depending on MOI and purpose of experiment. Check for CPE. The infectious cycle may be slower in mutant viruses. Also sometimes viable virus can be recovered even in the absence of visible CPE. We typically wait for visible CPE or 24 h post-infection, whichever comes first. The freeze-thaw cycles lyse any remaining cells and release the virus.
-
6
Check infected cells for CPE on tissue culture microscope. If no CPE is visible, return cells to incubator. If CPE is visible, or you wish to attempt viral recovery anyway, proceed to step 7.
-
7
Freeze at −20°C or −70°C. Thaw at room temperature or 37°C.
-
8
Repeat step 7 twice more.
-
9
To pellet cellular debris, centrifuge 5 min at 2500× g. Transfer supernatant (P1 virus) to a fresh tube.
This is the only purification necessary for most purposes. Store P1 virus at −20°C.
NOTE: To make P2 (or subsequent) stocks, repeat the protocol above using a P1 (or subsequent) stock as your infecting stock.
Alternate Protocol 2: PROPAGATION OF PV IN HeLa S3 SUSPENSION CULTURES
If a large amount of highly concentrated virus is needed (see Basic Protocol 5), a high density suspension infection can be performed. Your infecting stock should be P1 or subsequent with a as high a titer as possible (~1×109 PFU/mL is about the maximum; it is often one to several logs lower for attenuated mutants.)
Materials
HeLa S3 cells in suspension (Support Protocol 3)
Serum-free SMEM
P1 or subsequent infecting stock
37°C and 5% CO2 humidified incubator
500 mL spinner flask and magnetic stir plate
Tissue culture microscope (inverted phase-contrast)
Grow 3 L of HeLa S3 suspension culture at ~5×105 cells/mL (see Support Protocol 3).
-
Prepare inoculum, ideally 30 mL at 5×108 PFU/mL in serum-free SMEM.
This gives an MOI of 10, ensuring that > 99.995% of cells are infected in the first infectious cycle. If your infecting viral titer is ≤ 5×108 PFU/mL, simply use undiluted virus, calculate MOI, and allow a second infectious cycle if necessary. To get a high enough volume of infecting virus, it may be necessary to combine P1 or subsequent stocks of the same virus if they are derived from the same P0 or clonal stock. To gently pellet cells, centrifuge 5 min at 200× g. Aspirate or pour off supernatant. Resuspend in inoculum. Incubate 30 min at 37°C, inverting tube every 10 min to mix cells with inoculum.
Expand to a final volume of 300 mL with serum-free SMEM in 500 mL spinner flask. Incubate 8–16 h at 37°C and 5% CO2.
Check for CPE on hemacytometer.
Freeze viral culture in eight 50-mL conical tubes −20°C or −70°C. Thaw at room temperature or 37°C.
Repeat step 6 twice more.
-
To pellet cellular debris, centrifuge 5 min at 2500× g. Transfer supernatant (virus) to a fresh tubes.
Store virus at −20°C.
Support Protocol 3: PREPARATION AND GROWTH OF HeLa S3 SUSPENSION CULTURES
Suspension cultures are useful when it is necessary to grow or maintain a large volume of cells. HeLa S3 cells are HeLa cells specially adapted for growth in suspension.
Materials
HeLa S3 cells in monolayers (Support Protocol 2)
D-PBS (PBS w/o Ca2+ or Mg2+)
0.05% Trypsin-EDTA
10% FBS SMEM
5% FBS/5% NCS SMEM
10% NCS SMEM
37°C and 5% CO2 humidified incubator
250 mL, 1 L, and 3 L spinner flasks and magnetic stir plate
Tissue culture microscope (inverted phase-contrast)
Prepare four 85% confluent T150 flasks. Wash each monolayer with 15 mL D-PBS.
Aspirate D-PBS. Add 3 mL 0.05% Trypsin-EDTA. Tilt flask to cover cell monolayer. Incubate 5–10 minutes at room temperature or 37°C until monolayer detaches from flask surface.
-
Add 9 mL warm 10% FBS SMEM. Pipet down side of flask to remove cells, then up and down against the bottom of the flask to break up clumps.
Work quickly because the serum will inactivate the trypsin. Count cells on a hemacytometer. Adjust to 5×105 cells/mL in 10% FBS SMEM.
Place ~100 mL in a 250 mL spinner flask on a magnetic stir plate and incubate at 37°C in 5% CO2 overnight.
Add 100 mL fresh warm 10% FBS SMEM and incubate at 37°C in 5% CO2 overnight.
-
Transfer to a 1 L spinner flask and add 200 mL 10% FBS SMEM.
You can continue to propagate or maintain the culture by splitting 1:1 with fresh medium every day. Once culture volume gets above 1 L you can start to wean it off FBS and onto NCS.. Start with 1 L of 5%FBS/5%NCS SMEM and when that is used up switch to 10% NCS SMEM. It is a good idea to check your cell count every few days to confirm it is between 5×105 and 1×106 cells/mL.
Basic Protocol 4: GENERATION OF CLONAL PV STOCK BY PLAQUE PURIFICATION AND AMPLIFICATION
It is best practice to generate and sequence a clonal stock for every new virus that is made. If you are working with a strongly attenuated mutant that either reverts easily or was prepped at the same time as a WT stock, it is essential. To sequence the viral genome, we typically perform a basic oligo-dT/random hexamer primed RT followed by three separate PCR reaction. These reactions produce overlapping amplicons of ~2.6 kb, each of which can be sequenced in 8 reactions (4 forward, 4 reverse). Useful PCR and sequencing primers for PV1 (Mahoney) are listed in Table 1. This method provides sequence information for all but the terminal ~75 nt at each end, which is usually sufficient. If knowledge of the terminal sequences is critical, 5′- and/or 3′-RACE PCR can be performed (Strachan and Read, 1999).
Table 1.
Useful PV1 Primers
| Primer Name | Sequence |
|---|---|
| 51For | GTACTCCGGTATTGCGGTACCCTTGTACGCC |
| 695For | CTCTAAGTACAATTTCAACAGTTATTTCAATCAGACAA |
| 1337For | ATGTGTCTGGCCGGGGATAGCAACACCACTACCATGCA |
| 1997For | ACAGACGATCCCATACTCTGCCTGTCACTCTCTCCAGC |
| 2439For | TGCGAGATACCACACATATAGAGC |
| 3077For | GGTATTTCGAACGCCTATTCACACTTTTACGACGGT |
| 3728For | TGTCACCACGGGGTGATAGGGATCATTACTGCTGGT |
| 4379For | CAGTCTAAGAGGTTTGCCCCTCTTTACGCAGTGGAA |
| 4790For | AACTCAAGCAGAATTTCCCCCCCCACTGTG |
| 5434For | ACAAGGACCAGGGTTCGATTACGCAGTGGCTATGGCT |
| 6077For | CACTATGTGTTTGAAGGGGTGAAGGAACCAGCAGTCC |
| 6736For | GATGGTGCTTGAGAAAATCGGATTCGGAGACAGAGTT |
| 698Rev | GTACTTAGAGTAAACACACTCAATGGAGCGG |
| 1356Rev | TAGTGGTGTTGCTATCCCCGGCCAGACACATCTCTG |
| 2002Rev | GGGATCGTCTGTATGTGGTTTGTCACTTAAC |
| 2651Rev | AAGGGACTAGTGGATTTGTGGCCCC |
| 3092Rev | AAAGTGTGAATAGGCGTTCGAAATACCAACATACGG |
| 3743Rev | AATGATCCCTATCACCCCGTGGTGACATCTGAGTAT |
| 4393Rev | AGGGGCAAACCTCTTAGACTGGATGGATAAC |
| 5050Rev | CATTGTAGTGATCTGGTCAATACTG |
| 5443Rev | TAATCGAACCCTGGTCCTTGTACCTTTGCTGTCCGA |
| 6094Rev | GGTTCCTTCACCCCTTCAAACACATAGTGGAAAGCA |
| 6753Rev | AATCCGATTTTCTCAAGCACCATCTTTAGTG |
| 7400Rev | CCAATCCAATTCGACTGAGGTAGGG |
Materials
HeLa or HeLa S3 cells (Support Protocol 2)
10% NCS DMEM/F-12
P0 or other infecting stocks of known titer
Serum-free DMEM/F-12
PBS
2× DMEM/F12
2% (w/v) agarose
TRIzol LS Reagent (Invitrogen)
Chloroform
20 μg/μL Nuclease-free glycogen
Isopropanol
75% (v/v) EtOH
Nuclease-free water
50 μM Oligo (dT)20 (provided with ThermoScript RT)
50 ng/μL Random hexamers (provided with ThermoScript RT)
10 mM dNTP Mix (provided with ThermoScript RT)
15 U/μL ThermoScript RT (Invitrogen)
40 U/μL RNaseOUT (provided with ThermoScript RT)
5× cDNA Synthesis Buffer (provided with ThermoScript RT)
0.1 M DTT (provided with ThermoScript RT)
10 μM PCR primers (51For & 2651Rev, 2439For & 5050Rev, 4790For & 7400Rev; Table 1)
5U/μL Taq DNA Polymerase
10× PCR buffer with Mg2+ (provided with Taq DNA Polymerase)
1% agarose TAE gels and running buffer (Voytas, 2001)
6× DNA Loading Dye
DNA Ladder (e.g. GeneRuler 1 kb Plus, Fermentas)
PCR Clean-up Kit (e.g. Nucleospin Gel and PCR Clean-up, Machery-Nagel)
2.5% NCS DMEM/F-12
37°C and 5% CO2 humidified incubator
Tissue culture microscope (inverted phase-contrast)
37°C water bath
57°C water bath
Ice
PCR Machine
NanoDrop spectrophotometer
Prepare plaque assays
-
1
Seed HeLa or HeLa S3 cells in 10 cm dishes at 6×106 cells in 12 mL of 10% NCS DMEM/F12 per dish and incubate 24 h at 37°C in 5% CO2, at which time they should be confluent. Prepare 1 plate for each infecting stock.
If stock is believed to be strongly attenuated, contaminated with WT, or otherwise likely to be difficult to isolate a good clone from, you may wish to prepare extra plates. -
2
Prepare 2% agarose for overlays, with a separate bottle for each group of 8–12 dishes. Prepare 12.5 mL 2% agarose per dish. Melt the agarose in a microwave and cool to 57°C. Warm 2× DMEM/F-12 to 37°C.
By keeping the agarose at 57°C and the medium at 37°C, a mixture at approximately 47°C is created when they are combined in step 5. This temperature will keep the agarose liquid long enough to distribute the overlays but will not harm the cells. -
3
Thaw P0 or other infecting stock and dilute to ~25 PFU/mL in serum-free DMEM/F-12. Allow 2 mL inoculum per dish.
Since starting titers are often ~1×108 PFU/mL or higher, it is most accurate to dilute to, for example, 2.5 × 107 PFU/mL and then perform 10-fold serial dilutions rather than trying to dilute nearly one million-fold in one step. Always change tips when performing serial dilutions. -
4
Working with 8–12 dishes at a time to keep cells from drying out, aspirate the medium and wash with PBS.
-
5
Aspirate PBS and add 1.5 mL of viral inoculum each dish.
-
6
Distribute the virus by rocking by hand. Incubate dishes at least 30 min at 37°C in 5% CO2. Redistribute virus by hand once every 10 min.
-
7
Combine equal volumes of 2% agarose and 2× DMEM/F-12 and swirl to mix. Add 24 mL overlay solution to each dish and swirl gently to mix inoculum into overlay. Let dishes sit at room temperature until overlay solidifies.
-
8
Incubate 72 h at 37°C in 5% CO2.
If the virus of interest is a small plaque mutant, you may need to incubate for longer (96 h).
Isolate and amplify plaques
-
9
24 h before isolating plaques, seed HeLa or HeLa S3 cells in 24-well plates at 2.5×105 cells in 0.5 mL of 10% NCS DMEM/F12 per well and incubate 24 h at 37°C in 5% CO2, at which time they should be confluent. Prepare 1 plate for each plaque assay.
-
10
On the day of the isolation, aspirate old medium and replace with serum-free medium.
-
11
Identify and mark plaques. To do this, hold the plaque assay up to a light source and look through the bottom of the dish. Plaques appear as “cloudy” patches in the monolayer. Circle with an ethanol-resistant marker.
-
12
Use a 10 μL pipette tip to “pick” a plaque and transfer it to a 24-well plate. Set the volume of the pipette to full (10 μL). Plunge tip through the agarose overlay to touch the bottom of the plate in the center of the circle marking the plaque. Withdraw pipette tip and pipet up and down several times into one well of the 24-well plate to infect it.
-
13
Repeat step 4 and isolate up to 24 plaques per virus.
-
14
Incubate 24-well plates 24–48 h at 37°C in 5% CO2. Check for CPE.
If the virus of interest is attenuated, you may need to incubate for longer (72 h). -
15
Freeze plates at −20°C or −70°C. Thaw at room temperature or 37°C.
-
16
Repeat step 15 twice more and transfer each clone (0.5 mL) to a clean 1.5 mL tube.
-
17
To pellet cellular debris, centrifuge 5 min at 2500× g. Transfer supernatant to a fresh tube.
Store virus at −20°C.
Sequence clones
-
18
Extract viral RNA:
A less toxic and easier (but more expensive) alternative to TRIzol is to use a kit for the isolation of viral RNA, such as the ZR Viral RNA Kit.For each clone, combine 250 μL of supernatant with 750 μL TRIzol LS. Pipet up and down to mix. Incubate 5 min at room temperature.
Add 200 μL chloroform. Shake vigorously by hand for 15 s. Incubate 5 min at room temperature. To separate phases, centrifuge 15 min at 12,000× g, 4°C.
Transfer aqueous (top) phase (~775 μL) to clean 1.5 mL tube. Add 1 μL glycogen and 500 μL isopropanol. Incubate 10 min at room temperature. To pellet RNA, centrifuge 10 min at 12,000× g, 4°C.
Carefully remove supernatant. Wash pellet with 1 mL 75% EtOH. Vortex to mix. Centrifuge 5 min at 7500× g, 4°C.
-
Carefully remove supernatant and air-dry pellet 5–10 min. Resuspend in 12 μL nuclease-free water. Quantitate the RNA on a NanoDrop Spectrophotometer.
Typical yields are 100–600 ng/μL of total RNA. Store RNA at −70°C.
-
19
Perform cDNA synthesis:
Always work with RNA on ice.-
Prepare a master mix for sample denaturation. For 24 clones, prepare enough for 25 reactions. Mix (per reaction):
2.0 μL 10 mM dNTP mix
1.0 μL 50 μM oligo (dT)20
1.0 μL 50 ng/μL random hexamers
-
For each clone add 4 μL sample denaturation mix to 9 μL viral RNA. Incubate 5 min at 65°C. Place on ice.
The Thermoscript kit can use up to 5 μg of total RNA as template for the reaction. The total RNA concentration is usually such that 9 μL is less than 5 μg. However it is best practice to dilute all samples to the same concentration of total RNA and use a fixed amount as template. -
Prepare a master mix for cDNA synthesis. For 24 clones, prepare enough for 25 reactions. Mix (per reaction):
4.0 μL 5× cDNA synthesis buffer
1.0 μL 0.1 M DTT
1.0 μL 15 U/μL ThermoScript RT
1.0 μL 40 U/μL RNaseOUT
-
Add 7 μL cDNA synthesis mix to each tube. Incubate 10 min at 25°C, 50 min at 50°C, and 5 min at 85°C.
Store cDNA at −20°C.
-
-
20
Perform and clean-up PCR:
-
Prepare 3 separate master mixes for PCR, one with each of these primer pairs: 51For & 2651Rev, 2439For & 5050Rev, 4790For & 7400Rev. For 24 clones, prepare enough for 25 reactions. Mix (per reaction):
5.0 μL 10× PCR buffer with Mg2+
1.0 μL 10 mM dNTP mix
1.0 μL 10 μM For primer
1.0 μL 10 μM Rev primer
0.5 μL 5U/μL Taq DNA Polymerase
39.5 μL nuclease-free water
-
For each clone add 2 μL cDNA synthesis reaction to 48 μL each PCR mix. Mix well and perform PCR:
First step: 2:00 94°C (initial denaturation) 30 cycles: 0:30 94°C (denaturation) 0:30 55°C (annealing) 3:00 72°C (extension) Final step 10:00 72°C (final extension) Run 5 μL of the PCR reaction with 1 μL 6× DNA loading dye on a 1% agarose TAE gel (Voytas, 2001) and compare to DNA ladder to confirm size.
Perform PCR clean-up using a spin column kit such as the Nucleospin Gel and PCR Clean-up Kit (Machery-Nagel). Elute in 50 μL elution buffer.
Check DNA concentration on NanoDrop spectrophotometer.
-
-
21
Sequence PCR products. Each PCR product can be sequenced in 8 reactions (4 forward, 4 reverse) using the primers from Table 1. Adjust template and primer concentrations according to the instructions from your sequencing facility.
Amplify and recover clone(s) of interest
-
22
Examine your sequencing results and identify clones that contain the desired sequence and no additional mutations.
-
23
Prepare one confluent T75 of HeLa or HeLa S3 cells for each clone to be amplified. This can be achieved by seeding 3.75×106 cells in 15 mL 10% NCS DMEM/F-12 48 h prior to the infection.
If less virus is needed, it can be amplified in a T25 by scaling all volumes down 3-fold. -
24
Prepare viral inoculum by diluting 150 μL of the remaining viral supernatant from the clone(s) of interest into 3.75 mL of serum-free DMEM/F-12.
-
25
Aspirate old medium from cells. Wash monolayer with 7.5 mL PBS. Aspirate PBS and cover with 3.75 mL inoculum.
-
26
Distribute the virus by rocking by hand. Incubate flask at least 30 min at 37°C in 5% CO2. Redistribute virus once every 10 min.
-
27
Aspirate the inoculum. Cover with 7.5 mL 2.5% NCS. Incubate at 37°C in 5% CO2.
-
28
Check infected cells for CPE on tissue culture microscope. If no CPE is visible, return cells to incubator. If CPE is visible, or you wish to attempt viral recovery anyway, proceed to step 29.
-
29
Freeze at −20°C or −70°C. Thaw at room temperature or 37°C.
-
30
Repeat step 29 twice more.
-
31
To pellet cellular debris, centrifuge 5 min at 2500× g. Transfer supernatant (clonal PV stock) to a fresh tube.
This is the only purification necessary for most purposes. Store virus at −20°C.
Alternate Protocol 3: GENERATION OF CLONAL PV STOCK BY LIMITING DILUTION AND AMPLIFICATION
Whether to generate clonal stocks by plaque purification or limiting dilution is a matter of personal preference. Plaque purification is slightly faster, but some people find it difficult to accurately pick individual plaques through the agarose. Limiting dilution is especially useful if working with a small plaque mutant.
Additional Materials (see also Basic Protocol 4)
Viral RNA Isolation Kit (e.g. ZR Viral RNA Kit, ZymoResearch)
96-well plates
Multi-channel 20–200 μL pipette and tips
Sterile pipette reservoirs
Prepare limiting dilutions
-
1
Dilute HeLa or HeLa S3 cells to 1×105 cells/mL in 10% NCS DMEM/F12 and use multi-channel pipette to distribute 100 μL/well to 96-well plates. Prepare 1 plate per infecting stock.
To save time on the day of the assay, cells may be plated 24 h in advance, in which case dilute to 5×104 cells/mL. If stock is believed to be strongly attenuated, contaminated with WT, or otherwise likely to be difficult to isolate a good clone from, you may wish to prepare extra plates. -
2
Thaw P0 or other infecting stock and dilute to ~2.5 TCID50/mL in serum-free DMEM/F-12. Always change tips when performing serial dilutions. Allow 10 mL inoculum per plate.
Since starting titers are often ~1×108 PFU/mL or higher, it is most accurate to dilute to, for example, 2.5 × 107 PFU/mL and then perform 10-fold serial dilutions rather than trying to dilute nearly one million-fold in one step. Always change tips when performing serial dilutions. -
3
Pour virus dilution into reservoir and use multichannel pipette to add 100 μL to each well of 96-well plate.
-
4
Incubate 7 days at 37°C in 5% CO2.
Isolate clones
-
5
Check each well for CPE on the microscope and mark positive wells.
-
6
Freeze plates at −20°C or −70°C. Thaw at room temperature or 37°C.
-
7
Repeat step 10 twice more and transfer each clone (0.2 mL) to a clean 0.5 mL tube.
-
8
To pellet cellular debris, centrifuge 5 min at 2500× g. Transfer supernatant to a fresh tube.
Store virus at −20°C. Save supernatant from all positive wells until clone containing the desired sequence and no additional mutations has been identified and successfully amplified. You may sequence a few clones at a time to save reagents.
Sequence clones
-
9
Extract viral RNA from 100 μL viral supernatant. Use a spin-column based viral RNA isolation kit such as the ZR Viral RNA Kit (Zymo Research). Elute in 12 μL nuclease-free water. Quantitate the RNA on a NanoDrop Spectrophotometer.
When working with such a small volume of viral supernatant, it is difficult to ensure an adequate yield of RNA using TRIzol, so use of a kit is strongly advised. Store RNA at −70°C. Typical yields are 40–200 ng/μL of total RNA. Store RNA at −70°C. -
10
Perform cDNA synthesis, PCR, and sequencing: see steps 19–21 of Basic Protocol 4.
Amplify and recover clone(s) of interest
-
11
Examine your sequencing results and identify clones that contain the desired sequence and no additional mutations.
-
12
Prepare one confluent T25 of HeLa or HeLa S3 cells for each clone to be amplified. This can be achieved by seeding 1.25×106 cells in 5 mL 10% NCS DMEM/F-12 48 h prior to the infection.
-
13
Prepare viral inoculum by diluting 50 μL of the remaining viral supernatant from the clone(s) of interest into 1.25 mL of serum-free DMEM/F-12.
-
14
Aspirate old medium from cells. Wash monolayer with 2.5 mL PBS. Aspirate PBS and cover with 1.25 mL inoculum.
-
15
Distribute the virus by rocking by hand. Incubate flask at least 30 min at 37°C in 5% CO2. Redistribute virus by hand once every 10 min.
-
16
Aspirate the inoculum. Cover with 3.75 mL 2.5% NCS. Incubate at 37°C in 5% CO2.
-
17
Check infected cells for CPE on tissue culture microscope. If no CPE is visible, return cells to incubator. If CPE is visible, or you wish to attempt viral recovery, proceed to step 18.
-
18
Freeze at −20°C or −70°C. Thaw at room temperature or 37°C.
-
19
Repeat step 18 twice more.
-
20
To pellet cellular debris, centrifuge 5 min at 2500× g. Transfer supernatant (clonal PV stock) to a fresh tube.
This is the only purification necessary for most purposes. Store virus at −20°C.
Basic Protocol 5: PURIFICATION OF PV BY ULTRACENTRIFUGATION THROUGH SUCROSE CUSHION
This protocol is adapted from one that was kindly shared with us by members of Eckard Wimmer’s lab at SUNY Stonybrook. The level of purification is only necessary when you need to separate virions from unpackaged viral RNA or cellular contaminants present in the viral supernatant, for example to calculate particle:PFU ratio or to isolate only properly packaged viral genomes.
Materials
~200 mL of high titer viral stock (see Alternate Protocol 2)
10 mg/mL RNase A
1× DMEM/F12/5% SDS/20 mM EDTA
30% (w/v) sucrose in HBSS
HBSS
Virion Storage Buffer
1 × 3½ in. Ultra-Clear Centrifuge Tubes (Beckman #344058)
Fill five 1 × 3½ in. Ultra-Clear Centrifuge Tubes to the top (~38.5 mL) with high titer viral stock. Place in five swinging buckets of SW28 rotor (Beckman). Fill sixth tube with HBSS as blank and place in sixth bucket. Balance tubes.
Centrifuge 10 min at 14,000 rpm, 20°C in SW28.
Transfer supernatant to clean 50 mL tubes and add 0.001 volume (38.5 μL per tube) 10 mg/mL RNase A. Incubate 1 h at room temperature.
Prepare six fresh 1 × 3½ in. Ultra-Clear Centrifuge Tubes with 6 mL each of 30% sucrose in HBSS.
It is helpful to have a color contrast between your viral stock and your sucrose cushion. If your viral stock is in medium containing a pH indicator, prepare your sucrose cushion in HBSS without an indicator.
To stop RNase A treatment, add 0.1 volume (4.28 mL per tube) 1× DMEM/F12/5% SDS/20 mM EDTA. Mix well but gently (try to avoid bubbles).
Gently pipette the virus supernatant onto the sucrose cushions. Fill tubes to the top.
Centrifuge 4 h at 28,000 rpm, 20°C in SW28.
Carefully aspirate supernatant but leave sucrose cushion in place.
To remove any debris at the interface, gently wash the top of the sucrose cushion twice with 1 mL HBSS.
Tilt the tube and remove the sucrose with a pipette.
-
Resuspend the virus in 1.2 mL (200 μL/tube) of virion storage buffer.
Store virus at −20°C. Depending on the intended use of the purified virus, you may wish to resuspend in a different buffer or a larger volume. If SDS will not interfere with downstream applications, a low concentration (0.2%) can make resuspension easier.
REAGENTS AND SOLUTIONS
Crystal violet stain
Prepare a stock solution 1% (w/v) in EtOH.
To make 500 mL of 0.1% working solution, combine:
50 mL 1% stock
25 mL EtOH
425 mL water
Protect from light. Store at room temperature for up to a year.
Cell culture medium and supplements
DMEM/F-12 is used for monolayer cultures. It is a 1:1 mixture of Dulbecco’s Modified Eagle Medium and Ham’s F-12. It is available commercially from several companies. We purchase 450 mL bottles for most purposes; to make the 2× concentration for plaque assays we purchase powder in packages designed to make 1 L from Invitrogen (cat. no. 12400-024). SMEM (Jolik-Modified) is used for suspension cultures. We purchase 1 L bottles from Lonza (cat no. 04-719Q). Like most tissue culture medium, both of these contain L-glutamine, which breaks down in solution over time. One approach is to supplement each bottle with fresh L-glutamine, as we do. There are also commercially available media that use a more stable form, such as a glutamine dipeptide, which obviates the need for such supplementation. It is also possible to supplement with antibiotics and anti-fungals, to reduce contamination. We generally supplement with a 100× mixture of penicillin, streptomycin, and L-glutamine, which is commercially available. Newborn Calf Serum (NCS) is cheaper than Fetal Bovine Serum (FBS) and more than adequate for HeLa and HeLa S3 monolayer growth. FBS is preferable for starting suspension cultures, but successful suspensions may be slowly weaned onto NCS over time.
Serum-free DMEM/F-12
450 mL DMEM/F-12 medium
4.5 mL 100× penicillin-streptomycin-glutamine
Store at 4°C for 1 month.
2.5% NCS DMEM/F-12
450 mL DMEM/F12 medium
11.5 mL Newborn Calf Serum
4.6 mL 100× penicillin-streptomycin-glutamine
Store at 4°C for 1 month.
10% NCS DMEM/F-12
450 mL DMEM/F-12 medium
50 mL Newborn Calf Serum
5.0 mL 100× penicillin-streptomycin-glutamine
Store at 4°C for 1 month.
Freezing DMEM/F-12 (20 mL)
Combine:
16 mL 10% NCS DMEM/F-12
2 mL Newborn Calf Serum
2 mL DMSO
Filter-sterilize through a 0.2 μm filter. Store at 4°C for 1 month.
2× DMEM/F-12 (500 mL)
Combine:
1 packet powdered DMEM/F-12 (normally makes 1 L)
1.2 g NaHCO3
10 mL 100× penicillin-streptomycin-glutamine
Add water to 500 mL. Filter-sterilize through a 0.2 μm filter. Store at 4°C for 1 month.
1× DMEM/F-12/5% SDS/20 mM EDTA (1 L)
Combine:
1 packet powdered DMEM/F-12 (makes 1 L)
1.2 g NaHCO3
10 mL 100× penicillin-streptomycin-glutamine
40 mL 500 mM EDTA (pH 8.0)
50 g SDS
Add water to 1 L and stir to dissolve SDS. Filter-sterilize through a 0.2 μm filter. Store at 4°C for 1 month. Warm room temperature before use to redissolve SDS.
Serum-free SMEM
1 L SMEM (Jolik-Modified)
10 mL 100× penicillin-streptomycin-glutamine
Store at 4°C for 1 month.
10% FBS SMEM
1 L SMEM (Jolik-Modified)
110 mL Fetal Bovine Serum
11 mL 100× penicillin-streptomycin-glutamine
Store at 4°C for 1 month.
5% FBS/5% NCS SMEM
1 L SMEM (Jolik-Modified)
55 mL Fetal Bovine Serum
55 mL Newborn Calf Serum
11 mL 100× penicillin-streptomycin-glutamine
Store at 4°C for 1 month.
10% NCS SMEM
1 L SMEM (Jolik-Modified)
110 mL Newborn Calf Serum
11 mL 100× penicillin-streptomycin-glutamine
Store at 4°C for 1 month.
3× LiCl-EDTA
7.5 M LiCl
50 mM EDTA (pH 8.0)
0.5× TE
5 mM Tris-HCl (pH 8.0)
0.5 mM EDTA (pH 8.0)
1× TE
10 mM Tris-HCl (pH 8.0)
1 mM EDTA (pH 8.0)
5× Transcription Buffer
400 mM HEPES-KOH (pH 7.5)
120 mM MgCl
10 mM spermidine
200 mM DTT
Virion Storage Buffer
50 mM HEPES (pH 8.0)
200 mM NaCl
3 mM MgCl2
COMMENTARY
Background Information
Poliovirus (PV) was identified as the causative agent of paralytic poliomyelitis in 1909. The disease reached epidemic proportions in the United States in the early 20th century and remained so until the development of first the inactivated Salk vaccine in 1952 and then the live attenuated Sabin vaccine in 1963. Disease incidence in the developed world plummeted, and since 1988 it has been the target of a global WHO eradication campaign. As of 2012, PV is endemic to only three countries: Afghanistan, Nigeria, and Pakistan. [For a review of the history of poliomyelitis, see (De Jesus, 2007).]
The first full length genome sequences and infectious cDNA clones of PV were produced in 1981 (Kitamura et al., 1981; Racaniello and Baltimore, 1981). Humans are the only natural host for PV infections, but other primates can also be infected. In tissue culture, PV infects a wide range of human and non-human primate cell lines. The factor limiting host range is expression of the poliovirus receptor (PVR or CD155), and transgenic mice and mouse cell lines that express the receptor are available (Mendelsohn et al., 1989; Ren et al., 1990).
In general, unless a cell line with other characteristics is required, HeLa or HeLa S3 cells are used for the propagation of PV, due to the ease of growing and maintaining them in culture, as well as their transfectability and ability to form good monolayers for plaque assays. In HeLa and HeLa S3 cells, PV undergoes a classic lytic infection cycle of ~ 8 h. Cytopathic effect (CPE) is visible as the rounding and detachment of cells in monolayers prior to cell lysis. During an infection, high yields of both viral proteins and genomes are produced. These yields ensure synthesis of up to 10,000 virions per cell (Kew et al., 2005).
The poliovirus genome is 7440 nucleotides long, excluding a poly(A) tail of a variable length at the 3′ end. It can be divided into a 742-nucleotide 5′ untranslated region (5′UTR), a single long open reading frame encoding the viral polyprotein, a 68-nucleotide 3′ untranslated region (3′UTR). A small viral protein of 22 amino acids, VPg (3B), is covalently attached to the 5′ end of the genome. The mature virion consists of an icosahedral protein shell, composed of four capsid proteins (VP1, VP2, VP3, and VP4), which encapsidates the genome (Hogle et al., 1985). The viral genome serves as an mRNA (which is why transfection of in vitro transcribed genomes produces virus), and the entire viral life cycle takes place in the cytoplasm, with replication occurring on specialized membraneous structures derived from the host cell organelles.
Critical Parameters and Troubleshooting
Tissue culture problems
The most common problem is contamination with bacteria or fungi. This can be reduced by the use of careful aseptic technique. Work should always be performed in a biosafety cabinet, and surfaces and supplies wiped down with 75% ethanol. Routine use of penicillin and streptomycin in culture medium also reduces bacterial contamination. Also, if tissue culture work and bacterial work (e.g. Support Protocol 1) are to be performed on the same day, always do the tissue culture work first.
Contamination of attenuated mutants with WT virus
This contamination typically occurs in the molecular biology stages of virus production. If plasmid preps or in vitro transcriptions contain even trace amounts of WT plasmid or RNA, small amounts of WT virus will be produced upon transfection. If your mutant is attenuated compared to WT, the WT virus will rapidly overtake the population. The best way to prevent this is to routinely isolate clonal stocks of new viruses. Good aseptic technique generally prevents re-contamination with WT virus at the cell culture stage. It should be noted that PV is a non-enveloped virus, so decontamination of equipment with ethanol is ineffective. A solution of dilute bleach (1:10) works well.
Quantification
When choosing a method for quantification (Basic Protocol 2 or Alternate Protocol 1), consider that the theoretical relationship between TCID50 and PFU is based on the Poisson distribution: TCID50/mL = (PFU/mL)*(ln2). This is based on the assumption that the conditions that allow a productive infection are the same for the plaque assay and the TCID50 assay, i.e. any “unit” capable of forming a plaque will also be capable of causing CPE in a well. In practice, we do not find this to be the case with PV. Titer by plaque assay is typically ~5-fold higher than would be predicted based on TCID50 and the theoretical relationship.
Anticipated Results
Basic Protocol 3 produces viral titers as high a 1×109 PFU/mL for WT PV. For attenuated mutants, titers are often reduced by a log or more. Alternate Protocol 2 can produce titers up to 5×109 PFU/mL. For sucrose cushion purification (Basic Protocol 5), it is desirable to start with as concentrated a stock as possible. Yield is typically ~50%, i.e. starting with 200 mL of stock at 5×109 PFU/mL should produced 1.2 mL of purified, concentrated virions at ~4×1011 PFU/mL.
Time Considerations
Starting (on “day 1”) from a small amount of PV cDNA plasmid, transformation and propagation of the plasmid (Support Protocol 1) typically requires three overnight incubations (initial transformation onto agar plate; starter culture growth; midi- or maxi-culture growth). The midi- or maxi-prep can be performed on the morning of day 4 and followed by plasmid linearization in the afternoon and in vitro transcription overnight (Basic Protocol 1). If HeLa or HeLa S3 cells are available on day 5, the RNA can be transfected and P0 virus available by day 6. Because the freeze-thaw cycles for virus harvest are time-consuming, quantification of the P0 stock usually takes place on day 7. Titers will be known on day 9 if a plaque assay (Basic Protocol 2) is performed or on day 12–14 if a TCID50 is performed (Alternate Protocol 1). Thus 10 days to 2 weeks to generate a P0 stock of known titer.
If MOI is not a critical concern, P1 and subsequent stocks can be generated (Basic Protocol 3 or Alternate Protocol 2) without waiting to quantify the P0 stock. However isolation and amplification of a clonal stock (Basic Protocol 4 or Alternate Protocol 3) is strongly recommended. Starting with a P0 stock of known titer, to isolate, sequence, and amplify clones takes a further week to 10 days.
Starting with either a P0 or clonal stock, each subsequent infection takes 1–2 days, depending on MOI. If MOI is critical, you must allow time between viral passages for quantification. Purification of virions over a sucrose cushion (Basic Protocol 5) takes a full day, including a 4 h ultracentrifugation step.
Acknowledgments
We wish to thank members of the Andino laboratory for comments on the manuscripts. This work was supported by NIH (R01, AI36178, AI40085, P01 AI091575, 5T32AI060537) and the University of California (CCADD).
Literature Cited
- De Jesus NH. Epidemics to eradication: the modern history of poliomyelitis. Virol J. 2007;4:70. doi: 10.1186/1743-422X-4-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold J, Andino R. Poliovirus requires a precise 5′ end for efficient positive-strand RNA synthesis. Journal of virology. 2000;74:6394–6400. doi: 10.1128/jvi.74.14.6394-6400.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogle JM, Chow M, Filman DJ. Three-dimensional structure of poliovirus at 2.9 A resolution. Science. 1985;229:1358–1365. doi: 10.1126/science.2994218. [DOI] [PubMed] [Google Scholar]
- Kew OM, Sutter RW, de Gourville EM, Dowdle WR, Pallansch MA. Vaccine-derived polioviruses and the endgame strategy for global polio eradication. Annu Rev Microbiol. 2005;59:587–635. doi: 10.1146/annurev.micro.58.030603.123625. [DOI] [PubMed] [Google Scholar]
- King AMQ, Lefkowitz E, Adams MJ, Carstens EB, editors. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. Academic Press, Elsevier, Inc; San Diego, CA: 2011. [Google Scholar]
- Kitamura N, Semler BL, Rothberg PG, Larsen GR, Adler CJ, Dorner AJ, Emini EA, Hanecak R, Lee JJ, van der Werf S, Anderson CW, Wimmer E. Primary structure, gene organization and polypeptide expression of poliovirus RNA. Nature. 1981;291:547–553. doi: 10.1038/291547a0. [DOI] [PubMed] [Google Scholar]
- Knowles NJ, Hovi T, King AMQ, Stanway G. Overview of Taxonomy. In: Ehrenfeld E, Domingo E, Roos RP, editors. The Picornaviruses. ASM Press; Washington, DC: 2010. pp. 19–32. [Google Scholar]
- Lindenbach BD. Measuring HCV infectivity produced in cell culture and in vivo. Methods Mol Biol. 2009;510:329–336. doi: 10.1007/978-1-59745-394-3_24. [DOI] [PubMed] [Google Scholar]
- Mendelsohn CL, Wimmer E, Racaniello VR. Cellular receptor for poliovirus: molecular cloning, nucleotide sequence, and expression of a new member of the immunoglobulin superfamily. Cell. 1989;56:855–865. doi: 10.1016/0092-8674(89)90690-9. [DOI] [PubMed] [Google Scholar]
- Racaniello VR, Baltimore D. Cloned poliovirus complementary DNA is infectious in mammalian cells. Science. 1981;214:916–919. doi: 10.1126/science.6272391. [DOI] [PubMed] [Google Scholar]
- Reed LJ, Muench H. A simple method of estimating fifty per cent endpoints. Am J Hygiene. 1938;27:493–497. [Google Scholar]
- Ren RB, Costantini F, Gorgacz EJ, Lee JJ, Racaniello VR. Transgenic mice expressing a human poliovirus receptor: a new model for poliomyelitis. Cell. 1990;63:353–362. doi: 10.1016/0092-8674(90)90168-e. [DOI] [PubMed] [Google Scholar]
- Strachan T, Read AP. Human Molecular Genetics. 2. Wiley-Liss; New York: 1999. Figure 20.1. RACE-PCR facilitates the isolation of 5′ and 3′ end sequences form cDNA. [Google Scholar]
- Voytas D. Agarose gel electrophoresis. Curr Protoc Mol Biol. 2001;Chapter 2(Unit2):5A. doi: 10.1002/0471142727.mb0205as51. [DOI] [PubMed] [Google Scholar]
- Watson N. A new revision of the sequence of plasmid pBR322. Gene. 1988;70:399–403. doi: 10.1016/0378-1119(88)90212-0. [DOI] [PubMed] [Google Scholar]