SUMMARY
Gene duplication is a major evolutionary force driving adaptation and speciation, as it allows for the acquisition of new functions and can augment or diversify existing functions. Here, we report a gene duplication event that yielded another outcome – the generation of antagonistic functions. One product of this duplication event – UPF3B – is critical for the nonsense-mediated RNA decay (NMD) pathway, while its autosomal counterpart – UPF3A – encodes an enigmatic protein previously shown to have trace NMD activity. Using loss-of-function approaches in vitro and in vivo, we discovered that UPF3A acts primarily as a potent NMD inhibitor that stabilizes hundreds of transcripts. Evidence suggests that UPF3A acquired repressor activity through simple impairment of a critical domain, a rapid mechanism that may have been widely used in evolution. Mice conditionally lacking UPF3A exhibit “hyper” NMD and display defects in embryogenesis and gametogenesis, consistent with UPF3A serving as a molecular rheostat that directs developmental events.
Graphical Abstract

INTRODUCTION
Gene duplication is an innovative platform for the diversification and adaptation of species. While most duplicated genes are of no immediate selective value and thus undergo degeneration, in rare instances, a duplicated gene provides a useful function and thus it persists over evolutionary time (Innan et al., 2010). The specific selective forces that lead to the retention and subsequent alterations in duplicated genes have been widely studied. A clear-cut example of a duplicated gene that is of immediate selective value is when it increases the level of a rate-limiting gene product (Kondrashov et al., 2002). Gene duplication can also lead to a scenario in which the gene paralogs acquire mutations that ultimately subdivide their functions or expression patterns between the paralogs, a process called subfunctionalization (Hittinger et al., 2007; Innan et al., 2010). For example, if the original protein has two or more functions that cannot be independently improved upon, paralogs can provide a division of labor such that each paralog specializes and optimizes the functions independently. Another example of subfunctionalization occurs when there is a divergence in the expression pattern of redundant gene paralogs; this leads to strong selection to maintain all such gene paralogs because each paralog becomes essential for the particular cell types and developmental stages that it is expressed in. In other cases, some gene paralogs acquire new functions, a process called neo-functionalization (Innan et al., 2010; Teshima et al., 2008). For example, a protein domain may be created in a newly formed gene paralog that increases the fitness of an organism. While intuitively attractive, the neofunctionalization model suffers from the fact that selection processes act on current functions, not promises of future functions. Thus, it is unclear whether new functions can be sculpted rapidly enough (through mutation) for selective forces to act upon them. In the absence of selection, gene paralogs become non-functional; e.g., deteriorate into pseudogenes.
Here, we report an evolutionary outcome of gene duplication – functional antagonism – that can rapidly integrate into existing regulatory circuitry to provide biological benefit. In particular, we identify a gene paralog pair that has evolved to encode proteins with opposing functions. The pathway regulated by this paralog pair is nonsense-mediated RNA decay (NMD), a highly conserved RNA degradation pathway that has dual roles. One role is to serve as a RNA surveillance pathway that degrades aberrant mRNAs harboring premature termination (nonsense) codons (PTCs). This quality control role is important, as PTC-bearing mRNAs are commonly generated by mutation and biosynthetic errors, including alternative splicing. By reducing the levels of such PTC-containing mRNAs, NMD reduces the expression of truncated proteins, some of which have dominant-negative effects (Chang et al., 2007; Rebbapragada and Lykke-Andersen, 2009). The second role of NMD is to degrade a subset of normal mRNAs. Studies in which factors crucial for NMD have been depleted or completely ablated in species spanning the phylogenetic scale have revealed dysregulation of a large subset of the “normal transcriptome” (~3 to 15% of mRNAs) (Lykke-Andersen and Jensen, 2015). This suggests that NMD serves as an important regulator of normal gene expression.
Several contexts have been defined that allow a stop codon in a normal mRNA to trigger mRNA decay. In mammals, the best-established NMD-inducing context is when there is at least one exon-exon junction downstream of the stop codon – a dEJ. This allows a set of NMD-enhancing proteins recruited near exon-exon junctions—collectively called the exon-junction complex (EJC)—to avoid being displaced by translating ribosomes (Lykke-Andersen and Jensen, 2015). The EJC directly binds with the UPF3B protein (also known as “UPF3X”), which serves as an adaptor protein that interacts with other NMD factors to trigger rapid RNA decay (Buchwald et al., 2010; Kadlec et al., 2004).
Interest in the NMD adaptor protein, UPF3B, intensified when it was discovered that UPF3B mutations cause intellectual disability in humans (Tarpey et al., 2007). Intellectual disability patients with UPF3B mutations also commonly suffer from autism or schizophrenia, suggesting that UPF3B, and by implication, NMD, is also important for normal psychiatric behavior (Nguyen et al., 2014). The underlying basis for UPF3B function in the nervous system is poorly understood, but one function is to coordinate differentiation decisions in neural stem cells and progenitors. Depletion of UPF3B in neural progenitor cells (NPCs) increases their proliferation and impairs neurite formation, suggesting that NMD promotes the differentiation of already committed neural progenitor cells (Jolly et al., 2013). However, substantial evidence indicates that NMD has the opposite effect on neural stem cells – it promotes the stem-like state and proliferation (Lou et al., 2014). Because NMD is a branched pathway, it is possible that these different functions emanate from different branches, each of which are known to regulate different subsets of NMD substrate mRNAs (Lykke-Andersen and Jensen, 2015; Huang et al., 2012).
UPF3B is unique among known NMD factors in having a related sister protein – UPF3A. UPF3A and UPF3B are encoded by an evolutionarily ancient paralog pair that exists in most, if not all, vertebrates, including all sequenced mammals, frogs, fish, and birds. It is not known why this gene paralog pair has persisted since the origin of vertebrates. It has been hypothesized that UPF3A and UPF3B have redundant functions (Chan et al., 2007; Kunz et al., 2006; Lykke-Andersen et al., 2000; Nguyen et al., 2012), a notion supported by the fact that UPF3A is dramatically upregulated when UPF3B is downregulated or eliminated (Chan et al., 2009) and the association between the magnitude of this UPF3A upregulatory response and the severity of neurological symptoms in intellectual disability patients with UFP3B mutations (Nguyen et al., 2012).
If indeed UPF3A and UPF3B act redundantly in NMD, it is critical that they also each have unique properties that have allowed them to both persist over evolutionary time. One possibility is that UPF3A and UPF3B have unique expression patterns that allow them to be independently selected for, in accordance with the subfunctionalization model. In support of this possibility, Upf3a is much more highly expressed in the testis than other adult organs (Serin et al., 2001; Zetoune et al., 2008). In contrast, Upf3b, while ubiquitously expressed in most tissues, is a candidate to be transcriptionally silenced in testicular germ cells given that it is an X-linked gene and thus would be predicted to be subject to meiotic sex chromosome inactivation (MSCI), which transcriptionally silences genes on the X and Y chromosomes during male meiosis (Turner et al., 2007). If indeed Upf3b is transcriptionally silenced in meiotic germ cells, this raises the possibility that Upf3a, which is present on an autosome, might serve to replace Upf3b function in meiotic germ cells, thereby explaining the high expression of Upf3a in the testis and providing a justification for the persistence of these two paralogs over evolutionary time.
While potentially attractive, the subfunctionalization model for explaining the long-term persistence of the UPF3A/UPF3B paralog pair suffers from the uncertainty as to whether UPF3A is actually an NMD factor. The only evidence that UPF3A is a NMD factor comes from gain-of-function studies in which UPF3A was tethered downstream of a stop codon in reporter RNAs using the high-affinity RNA-binding proteins, MS2 and λ N. Such UPF3A-fusion proteins only elicited trace NMD activity (~20% downregulation), as judged by reporter RNA analysis (Kunz et al., 2006; Lykke-Andersen et al., 2000). In contrast, other human NMD proteins, including UPF3B, exhibited strong NMD activity in this tethering assay. This weak ability of UPF3A to promote NMD is surprising given that it is encoded by an ancient gene (~500 million years old) that presumably has had ample time to be selected to encode a protein with strong NMD activity. Indeed, UPF3A is poised for such a role, as a single amino-acid substitution is sufficient to convert UPF3A into a strong NMD factor, comparable in activity with UPF3B (Kunz et al., 2006).
In this communication, we addressed this paradox by re-evaluating the function of UPF3A using loss-of-function approaches. Our analysis revealed that UPF3A is actually a broadly acting NMD inhibitor. This discovery implies that UPF3A and UPF3B do not primarily work in a complementary or redundant manner as previously supposed; instead, they oppose each other, allowing this paralog pair to serve as a molecular rheostat to modulate the level of gene expression during development.
RESULTS
UPF3A is a NMD repressor
We addressed the function of UPF3A by performing loss-of-function experiments. Surprisingly, we found UPF3A depletion led to downregulation, not upregulation, of 6 of the 8 NMD substrates we examined in mouse P19 cells (Figure 1A), raising the possibility that UPF3A is a NMD repressor. Since the hallmark of NMD is that it destabilizes its target mRNAs (Maderazo et al., 2003), we next examined whether UPF3A antagonizes this destabilization. Indeed, we found that depletion of UPF3A destabilized multiple NMD target mRNAs, implying that UPF3A stabilizes NMD target RNAs (Figures 1B and S1A). UPF3A depletion also increased NMD magnitude as judged using a NMD reporter system (Figures 1C, S1B and S1D) (Boelz et al., 2006). As a converse experiment, we overexpressed UPF3A and found that this impaired the magnitude of NMD (Figure S1E). To determine whether the ability of UPF3A to repress NMD is a peculiarity of P19 cells, we examined mouse embryonic fibroblasts (mEFs) and found that UPF3A also suppressed the downregulation of most NMD substrates in these cells (Figure 1D).
Figure 1. UPF3A is a NMD inhibitor.
(A) qPCR analysis of NMD substrates in mouse P19 cells transfected with a Upf3a shRNA or a negative control (Ctrl) shRNA construct, the latter of which was given a value of “1.”
(B) NMD substrate half-lives determined by qPCR analysis. P19 cells were transfected with the constructs described in panel A, followed by ActD treatment for the indicated times.
(C) Depletion of Upf3a leads to destabilization of a NMD reporter. P19 cells were transfected with the constructs described in panel A and analyzed by the NMD reporter described in the Materials and Methods.
(D) qPCR analysis of NMD substrates in mEFs electroporated with either Upf3a shRNA (shUpf3a) or a control shRNA (Ctrl).
(E) qPCR analysis of NMD substrate mRNAs in Upf3b-null mNSCs transfected as in panel A.
Graphs are represented as mean and standard error (SEM) of replicates.
*P<0.05; **P<0.01; ***P<0.001; ****P<0.0001
Our evidence that UPF3A is a NMD repressor was at apparent odds with the previous evidence that UPF3A is a weak NMD factor (Kunz et al., 2006; Lykke-Andersen et al., 2000). To reconcile this, we considered the possibility that UPF3A is not a NMD repressor per se, but instead it reduces NMD activity conferred by its paralog, UPF3B. By replacing its paralog in molecular interactions with other NMD factors, UPF3A might “repress” NMD merely by virtue of being a weaker NMD factor than UPF3B, which acts in a specific branch of the NMD pathway (Chan et al., 2007; Huang et al., 2011; Tarpey et al., 2007). This hypothesis predicts that depletion of UPF3A in cells already lacking UPF3B would weaken NMD. Alternatively, if UPF3A is a bona fide NMD repressor, depletion of UPF3A would strengthen NMD, even in UPF3B-deficient cells. To test this hypothesis, we generated Upf3b-KO mouse neural stem cells (mNSCs) and matching control cells. We found that depletion of UPF3A in these Upf3b-KO mNSCs decreased the level of all NMD substrates tested (Figure 1E). As a control, we depleted the core NMD factor, UPF1, which triggered the opposite effect, as expected (Figure 1E). This evidence strongly supported the notion that UPF3A is a bona fide NMD repressor in vitro.
We found that a subset of NMD substrates escaped UPF3A-mediated NMD repression in some contexts. For example, Gas5 and Snord22 RNA were not downregulated in UPF3A-depleted P19 cells, indicating that these “non-coding” NMD substrate transcripts can escape UPF3A-mediated repression of NMD (Figure 1A). However, both of these transcripts were downregulated in response to UPF3A depletion in mEFs and mNSCs (Figures 1D and 1E). This suggested that the ability of Gas5 and Snord22 RNA to escape UPF3A-mediated repression is not an intrinsic feature of these transcripts, but rather a cell type-specific response. We conclude that while UPF3A is a broadly acting NMD suppressor, some NMD substrate transcripts can escape its activity in specific cellular contexts, a possibility we explore further below.
Genome-wide Impact of UPF3A
We performed RNA-seq half-life analysis to globally define mRNAs regulated by UPF3A. If UPF3A is primarily a NMD repressor, this predicts that more RNAs would be destabilized than stabilized in response to UPF3A depletion. Consistent with this prediction, we found that 83% of the mRNAs displaying significantly altered stability were destabilized in response to UPF3A depletion (Supplemental Table 1 and Figures 2B–C). Because destabilization after UPF3A depletion implies these mRNAs are stabilized by UPF3A, we will refer to these mRNAs as “UPF3A-stabilized.” Conversely, mRNAs stabilized after UPF3A depletion will be referred to as “UPF3A-destabilized.” Figures S2C and S2D show examples of UPF3A-stabilized and –destabilized mRNAs, respectively.
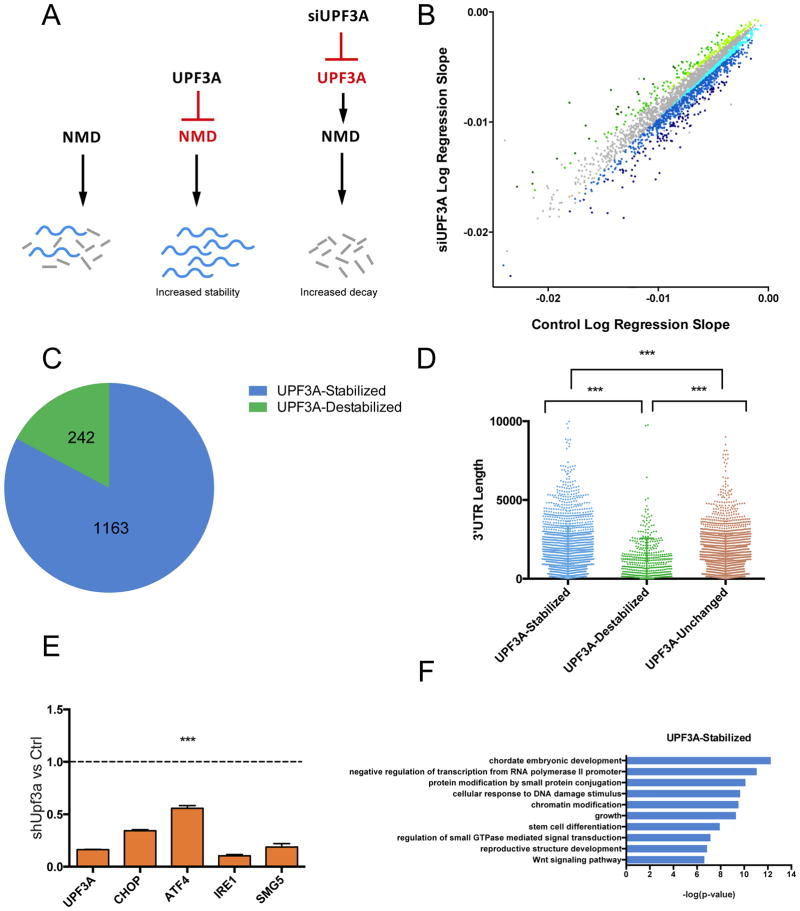
Figure 2. Genome-wide Half-Life Analysis of UPF3A-Regulated mRNAs.
(A) Model: UPF3A stabilizes NMD target transcripts by inhibiting NMD. Depletion of UPF3A stimulates NMD.
(B) Scatter plot of RNA half-life slope of UPF3A-depleted (UPF3A siRNA transfected) vs. Control (control siRNA transfected) P19 cells (R>0.7). UPF3A-stabilized and -destabilized transcripts are blue and green, respectively. Transcripts not exhibiting altered stability are gray. Darker shades of blue and green convey progressively increasing regulation.
(C) Proportion of significantly UPF3A-stabilized and –destabilized transcripts (destabilized and stabilized, respectively, in response to UPF3A depletion (R>0.7) in the transfected cells described in panel B.
(D) The distribution of 3′ UTR length in UPF3A-destabilized and -stabilized transcripts in the transfected cells described in panel B. ***P<0.001 (unpaired Student’s t-test).
(E) qPCR analysis of NMD substrates in stably UPF3A-depleted (shUPF3A) HeLa cells vs Control (shLuc) HeLa cells.
(F) GO analysis of functional categories overrepresented (p<0.05) in the UPF3A- stabilized mRNAs defined in panel C.
To assess whether a significant proportion of the UPF3A-stabilized mRNAs are normally degraded by NMD, we compiled high-confidence NMD substrates identified from previously published studies and determined whether any of these overlapped with the UPF3A-stablized RNAs we identified. This analysis revealed that 350 of the mouse UPF3A-stabilized transcripts in P19 cells are high-confidence NMD substrates (Supplemental Table 2). We regarded this as a remarkably large number of mRNAs given that few high confidence NMD substrates have been defined so far. As evidence that UPF3A can also act as a NMD factor, we found that 64 of the UPF3A-destabilized transcripts are also high-confidence NMD substrates (Supplemental Table 2). Given that our results were obtained with mouse P19 cells, whereas the vast majority of high-confidence NMD substrates were defined in human cells (Supplementary Table 2), our results suggest that UPF3A acts as both a NMD repressor and NMD factor on conserved NMD target mRNAs. As further evidence for this, human orthologs of mouse mRNAs exhibiting altered stability in UPF3A-depleted mouse P19 cells also exhibited altered mRNA level in UPF3A-depleted human HeLa cells (Figure 2E).
NMD is elicited when the stop codon defining the end of the main ORF is present in specific contexts, including when a stop codon is followed by a dEJ (see Introduction). Consistent with UPF3A acting on mRNAs degraded NMD, the frequency of dEJs was higher for both UPF3A-stabilized and -destabilized mRNAs than mRNAs not exhibiting altered stability in response to UPF3A depletion (Figure S2B). However, this trend was not statistically significant, suggesting that another NMD-inducing feature might also contribute. Indeed, we found that UPF3A-stabilized mRNAs had significantly longer 3′ UTR length than did control mRNAs or UPF3A-destabilized mRNAs (P<0.0001; Figure 2D), which is consistent with previous findings that long 3′ UTRs can elicit NMD (Lykke-Andersen and Jensen, 2015). This finding raised the possibility that 3′ UTR length is one determinant that dictates whether UPF3A stabilizes or destabilizes its targets.
Gene ontology (GO) analysis (Sherman et al., 2007) revealed that the proteins encoded by UPF3A-stabilized and -destabilized RNAs were statistically enriched for several processes, including functions related to transcription, cell cycle, and embryonic development (Figures 2F and S2A).
Conservation and Molecular Mechanism of UPF3A Repressor Activity
Analysis of available databases revealed that invertebrates harbor only one copy of the Upf3 gene, while vertebrates have two copies – Upf3a and Upf3b (Figure 3A). This suggests that the Upf3 duplication event occurred approximately when the vertebrate lineage first emerged. In support of this, phylogenetic analysis showed that Upf3a and Upf3b cluster separately in all vertebrates we examined (Figure S3A).
Figure 3. Evolution and Functional Analysis of UPF3A.
(A) Two copies of upf3 appear to have emerged at the dawn of vertebrates. See Figure S3A for a phylogenetic tree showing the evolution of these paralogs.
(B, F) Functional analysis of human UPF3A (hUPF3A). P19 cells were transiently transfected with shRNAs (mouse Upf3a [shUpf3a] or negative control [Ctrl] shRNAs) and the hUPF3A expression constructs (WT or the mutants depicted in panel E) or empty vector (EV). NMD magnitude was assessed using the NMD reporter described in Figure 1C.
(C) qPCR analysis of NMD substrates in HeLa cells, comparing expression in cells transiently transfected with UPF3A expression vector or empty vector, the latter of which was set to “1.”
(D) Model: UPF3B promotes NMD by bridging the EJC with UPF1 and UPF2.
(E) Schematic of hUPF3A constructs used in panel F. Both of the mutant UPF3A proteins were previously shown to be expressed at levels similar to that of the wild-type UPF3A protein (Kunz et al., 2006).
(G) Amino-acid similarity between mouse and human UPF3A/UPF3B. Values below 70% are shown in red.
*P<0.05; ***P<0.001
The experiments described above were performed on mouse UPF3A. To determine whether the ability of UPF3A to repress NMD is conserved, we tested the function of human UPF3A. In support of human UPF3A acting as a NMD repressor, we found that exogenous expression of human UPF3A restored NMD inhibition in mouse UPF3A-depleted P19 cells (Figure 3B) and overexpression of UPF3A in HeLa cells significantly upregulated the majority of NMD substrates we tested (Figure 3C). This data, along with our finding that both mouse and human orthologs of NMD substrates are targeted by UPF3A (Figures 1A and 2E), suggests that the ability of UPF3A to repress NMD is conserved.
To address how UPF3A represses NMD, we first considered the action of UPF3A’s paralog, UPF3B, which serves as an adapter that links the NMD protein, UPF2, with the EJC (Figure 3D). The UPF2- and EJC-interacting regions of UPF3B are in its N- and C-terminal halves, respectively (Buchwald et al., 2010; Kadlec et al., 2004; Kunz et al., 2006). UPF3A has been shown to have the same basic organization as UPF3B (Lykke-Andersen et al., 2000) and thus we examined its UPF2- and EJC-interacting regions to determine whether they had a role in the ability of UPF3A to repress NMD. To test the UPF2-interaction region, we examined the NMD activity of UPF3A- Δ ex4, a natural splice variant of UPF3A mRNA lacking exon 4 (Figure 3E) (Kadlec et al., 2004). This form of UPF3A (also called UPF3A-S), while stable, fails to interact with UPF2, based on co-immunoprecipitation experiments (Kunz et al., 2006). We found that it also failed to inhibit NMD, even when expressed at high levels (Figure S3B), and that it did not significantly rescue NMD repression in P19 cells depleted of Upf3a (Figure 3F). The notion that UPF3A must interact with UPF2 to silence NMD was further supported by the fact that 7 of the 8 amino acid residues in UPF3B known to be critical for UPF2 interaction are present in both mouse and human UPF3A (Kadlec et al., 2004). Given the functional difference we uncovered for UPF3A and UPF3A-S, we examined their expression in different tissues. We found that the UPF3A/UPFA-S ratio shifted in different tissues (Figures S3C–E), suggesting tissue-specific regulation.
To elucidate the role of the EJC-interaction domain of UPF3A, we tested the UPF3A mutant, UPF3A Δ 434–447, which lacks the ability to interact with the EJC (Kunz et al., 2006). Dose-response analysis showed that this UPF3A mutant was indistinguishable from wild-type UPF3A in its ability to silence NMD (Figure S3B). Moreover, this UPF3A Δ 434–447 mutant was as capable as wild-type UPF3A in restoring NMD silencing in mouse P19 cells depleted of mouse UPF3A (Figure 3F). The dispensability of the EJC interaction domain for UPF3A repressor activity is consistent with the fact that the EJC-interacting half of UPF3A is extremely poorly conserved (only 60% similar between mice and humans; Figure 3G). In striking contrast, the EJC-interacting half of UPF3B is relatively highly conserved (92%), as are the UPF2-interacting halves of both UPF3A and UPF3B (94% and 96% similarity, respectively) (Figure 3G).
These findings supported a model in which UPF3A acts as a molecular decoy that discourages UPF2 from forming an NMD-promoting complex with the EJC (Figure 4A). Consistent with this model, several previous studies have shown that UPF3A interacts more poorly than UPF3B with the EJC, as shown by several methods, including co-immunoprecipitation (Kim et al., 2001; Kunz et al., 2006), fluorescence anisotropy (Buchwald et al., 2010), and surface plasmon resonance analyses (Buchwald et al., 2010). To further evaluate this model, we tested the effect of other UPF3 mutations. Our model predicts that swapping UPF3A’s weak EJC-interaction domain with the strong EJC-interaction domain of UPF3B would eliminate UPF3A’s NMD repressor activity. Indeed, we found that a hybrid UPF3A/UPF3B protein with this swap (Figure 4B) no longer had NMD-inhibiting ability and instead exhibited modest, but statistically significant, NMD-promoting activity (Figure 4C). Indeed, a single amino-acid substitution in the EJC-interaction domain of UPF3A (UPF3A-A432R) previously shown to increase UPF3A’s interaction with the EJC (Kunz et al., 2006) was sufficient to convert UPF3A into a NMD activator (Figure 4D). The converse prediction of our model is that UPF3B can be converted into a NMD inhibitor by simply ablating its EJC-interaction domain. Indeed, we found this to be the case (hUPF3B Δ 421–434 in Figure 4C).
Figure 4. Mechanism of UPF3A Action.
(A) Model: UPF3A sequesters UPF2 away from the NMD machinery in order to silence NMD.
(B) Schematic of different hUPF3A and hUPF3B constructs used in panels C and D. These UPF3 mutants were previously shown to be expressed at levels similar to that of the corresponding wild-type UPF3 proteins (Kunz et al., 2006).
(C, D) A single amino acid in the EJC-interaction domain is critical for NMD repressor activity. Experiments were performed as in Figure 3B with the constructs shown in Figure 4B.
(E) Schematic of an mRNA reporter with 6 MS-binding sites or a control reporter lacking the MS-binding sites (Ctrl).
(F) Left: Northern blot analysis of HeLa cells co-transfected with the indicated expression vectors and siRNAs, along with both the reporters described in panel E. Right: Quantification of the mRNA reporter level were normalized to the levels of the Ctrl reporter mRNA in 3 independent experiments.
(G) Evidence that UPF3A inhibits UPF1-EJC interactions. Left: Co-IP analysis of UPF1 with the EJC components, MLN51 and MAGOH, in UPF3A-depleted P19 cells (shUpf3a) or P19 cells transfected with a negative-control shRNA (Ctrl). Right: Quantification of MLN51- and MAGOH-UPF1 interactions in UPF3A-depleted cells relative to control cells (n=3).
*P<0.05
UPF3A could inhibit NMD by sequestering UPF2 from an NMD substrate RNA or it could prevent the interaction of UPF2 with other NMD factors after UPF2 is already assembled on a NMD substrate RNA. To distinguish between these possibilities, we asked whether UPF3A influenced NMD triggered by UPF2 tethered to a reporter mRNA (Figure 4E–F). We found that strong UPF3A depletion (Figure S4A) did not significantly affect the ability of tethered UPF2 to elicit NMD (Figure 4F). As a control, we tested the effect of UPF3B depletion (Figure S4A) and found that this ablated the ability of tethered UPF2 to elicit NMD (Figure 4F). This suggested that UPF3A does not inhibit the action of UPF2 once it is already recruited to an mRNA and instead supported the notion that UPF3A sequesters UPF2 from NMD substrates.
As a final test of our model, we examined whether UPF3A suppresses the generation of a productive NMD mRNP complex. Our model predicts that depletion of UPF3A should reduce the sequestration of UPF2 from a productive NMD-promoting complex and thus increase the interaction of the EJC with the central NMD factor, UPF1 (Figure 4A). In agreement with this prediction, UPF3A depletion (Figure S4B) significantly increased the interaction of UPF1 with the EJC components, MLN51 and MAGOH, as assessed by co-immunoprecipitation analysis (Figure 4G). We conclude that UPF3A is a conserved NMD silencing factor that requires the N-terminal domain but not the C-terminal domain and likely acts by sequestering the essential NMD factor, UPF2, from the NMD machinery.
UPF3A is Essential for Early Embryogenesis
To investigate the physiological role of UPF3A in vivo, we generated Upf3a- floxed (fl) mice (Figure 5A, 5B, S5A, and S5B). To generate complete knockout Upf3a mice, we bred Upf3afl/+ mice with EIIa-Cre mice, which express CRE beginning in early embryogenesis. Only heterozygous (Upf3a+/−) and wild-type (Upf3a+/+) mice progeny were generated, implying an embryonic lethal phenotype. We traced the lethality as occurring between embryonic day 4.5 (E4.5) and E8.5 (Figure 5C). Defects were observable as early as E3.5, as Upf3a−/− embryos with poor morphology were enriched at this time point, relative to Upf3a+/− and Upf3a+/+ embryos (Figure 5D).
Figure 5. Upf3a is Required for Early Embryogenesis.
(A) Upf3a conditional knockout scheme and location of primers used for the detection of the floxed and knockout alleles at the Upf3a locus.
(B) Genomic PCR analysis of tails from mice with the indicated genotypes. The data show successful insertion of the targeted allele harboring loxP sites.
(C) Genotypes of the progeny from Upf3a+/− breeding pairs at the indicated embryonic and postnatal time points.
(D) E3.5 embryos isolated from superovulated Upf3a+/− female mice bred with Upf3a+/− male mice. The embryos were manually flushed out of fallopian tubes of superovulated mice and the distribution of genotypes in different morphological groups is shown.
(E–F) Immunofluorescence analysis of UPF3A (red) and nuclear LAMIN (green) expression in Panel E shows mouse blastocysts and Panel F shows a mouse 1-cell embryo with polar body. DAPI staining (blue) shows the position of nuclei.
*P<0.05; ****P<0.0001
Consistent with a role in early embryogenesis, we found that UPF3A protein is highly expressed in the inner cell mass of E3.5 blastocysts (Figure 5E). UPF3A is also highly expressed in earlier stages, including the 1-cell embryo stage, where we found it concentrated in the cytoplasm (Figure 5F). In contrast to the high expression of UPF3A in early embryogenesis, UPF3A protein is undetectable or lowly expressed in most adult tissues (Figure S6A). This low level of UPF3A protein occurs despite ubiquitous expression of the Upf3a gene in all adult tissues (Figure 6A). The low expression of UPF3A probably results from a cross-regulatory pathway we previously uncovered that destabilizes UPF3A protein when it cannot interact with UPF2, an NMD factor for which both UPF3A and UPF3B compete (Chan et al., 2009). Because of this cross-regulatory pathway, UPF3A protein levels would be predicted to be greatly elevated in the absence of UPF3B. Indeed, we found that mice lacking UPF3B (Upf3b-null mice) (Huang et al., 2011) had greatly elevated levels of UPF3A in all tissues we examined (Figures S6B and S6C).
Figure 6. UPF3A Expression Pattern.
(A–B) Expression pattern of Upf3a and Upf3b mRNA, as determined by qPCR analysis. Panel A shows adult mouse tissues and Panel B shows germ cell subsets. Values are relative to that in epididymis, which was given a value of “1.”
(C) Immunohistochemical analysis of UPF3A and UPF3B protein expression in adult mouse testes. Pre-immune IgG serves as a negative control. L, leptotene spermatocyte; P, pachytene spermatocyte; S, spermatid.
(D) H&E staining of Upf3a-cKO and littermate control testis at different postnatal time points. Most seminiferous tubules in the mutant had delayed spermatogenesis at P28, as evidenced by the presence of pachytene spermatocytes (P) and round spermatids (RS) near the lumen. Wild-type tubules typically have elongated spermatids (ES) at the lumen. Green arrows points to vacuoles, which were commonly present in mutant testes.
UPF3A Functions in Gametogenesis
We elected to study the function of UPF3A in detail in the testis, as we found it is the only adult tissue in which UPF3A protein and Upf3a mRNA is highly expressed (Figures 6A and S6A), the latter of which was previously reported (Serin et al., 2001; Zetoune et al., 2008). We hypothesized that one reason why UPF3A is abundant in the testis is because its paralog, the X-linked gene, Upf3b, is transcriptionally inactivated in meiotic germ cells (spermatocytes) as a result of the epigenetic silencing mechanism, MSCI (see Introduction). If indeed Upf3b were transcriptionally silenced by MSCI, this would relieve the destabilizing effect of UPF3B on UPF3A (Chan et al., 2009), which would increase UPF3A protein levels. In support of MSCI operating on the Upf3b gene, we found that Upf3b mRNA levels were exceedingly low during leptotene-to-pachytene (Figure 6B; note the log scale), the stages of meiosis when MSCI is known to be most active (Turner et al., 2007). As further evidence, we found that UPF3B protein was not visibly expressed in pachytene spermatocytes (Figure 6C). In striking contrast, UPF3A protein is highly expressed in spermatocytes, including at the leptotene and pachytene stages (Figure 6C), consistent with its relief from UPF3B-mediated destabilization. Also likely contributing to the high expression of UPF3A protein in spermatocytes is the high level of Upf3a mRNA in these cells. Analysis of Upf3a mRNA expression across different germ cell subsets revealed that Upf3a mRNA is strongly enriched in pre-leptotene spermatocytes (>10-fold higher than whole testis) and this high expression persists through all subsequent meiotic germ cell stages (Figure 6B).
Given the high expression of UPF3A in male germ cells, we examined whether it functions in this cell lineage by crossing Upf3a-floxed mice with Stra8-Cre mice, which first express CRE in male germ cells on postnatal day ~5, just prior to the initiation of meiosis (Sadate-Ngatchou et al., 2008). These Stra8-Cre;Upf3afl/fl mice (hereafter called Upf3a-conditional [c] KO mice) had no gross level abnormalities, and were of normal body weight (Figure S6D), but had a significant (~10-fold) reduction in sperm count relative to wild-type littermates (Figure S6E). To determine whether specific germ cell subsets were affected by loss of UFP3A, we evaluated the expression of germ cell markers in dissociated cells obtained from purified seminiferous tubules from Upf3a-cKO and control littermate mice. This analysis revealed that the general germ cell marker genes, Ldhc and Rec8, were significantly downregulated in Upf3a-cKO mice, as were the spermatocyte-specific marker genes, Spo11 and Sycp3 (Figure 7A). The former confirmed our hypothesis that spermatogenesis is impacted by loss of UPF3A and the latter suggested that the generation or survival of spermatocytes was specifically impacted, a possibility consistent with high UPF3A expression in these cells (Figure 6B and C). If this were the case, post-meiotic cells would also be predicted to decline. In support of this notion, the spermatid-specific markers, Tp2 and Prm2, were strongly downregulated in Upf3a-cKO mice (Figure 7A). In contrast, Stra8, which is expressed at earlier stages (from the pro-spermatogonium to early leptotene spermatocyte stages), was upregulated in Upf3a-cKO mice, consistent with accumulation of these early germ cells as a result of a partial blockade or delay in progression of spermatocyes through mid-to-late meiosis (Figure 7A). To directly assess this possibility, we used the marker, phospho- γ H2AX, which permits quantification of leptotene/zygotene spermatocytes (which concentrate phospho- γ H2AX in condensed chromosomes) and pachytene spermatocytes (which concentrate phospho-γ H2AX in XY bodies) (Turner et al., 2004). This revealed that Upf3a-cKO mice had a significant reduction in pachytene spermatocytes, but not leptotene/zygotene spermatocytes, as measured by two independent means (Figures 7B and S7A), confirming that these mutant mice have a defect in spermatocyte progression.
Figure 7. Loss of UPF3A in Male Germ Cells Elevates NMD and Perturbs Spermatogenesis.
(A) qPCR analysis of markers representing different germ cell stages in 4-week-old Upf3a-cKO vs. control (Ctrl) littermate mice (n=2–3 per genotype). “1” represents expression level normalized to Ctrl level.
(B) Spermatocytes per tubule, as quantified by γ-H2AX expression pattern in testis cross-sections (see text) from 4-week-old Upf3a-cKO vs. control littermate mice (n=2–3 per genotype).
(C) FACS analysis of Hoescht 33342-stained cells from 4-week-old Upf3a-cKO vs. littermate control seminiferous tubules. The data are plotted as Hoescht-Blue (DNA content) versus Hoescht-Red (DNA condensation) and the numbers indicate percentage of gated live cells. Population A, leptotene spermatocytes; population B, zygotene-pachytene-diplotene spermatocytes; and population C, round and elongated spermatids.
(D) FACS analysis of population C from panel C, showing strong deficit in both round spermatids (RS) and elongated spermatids (ES), which are distinguished by size (forward scatter [FSC]).
(E–G) qPCR analysis of 4-week-old Upf3a-cKO vs. littermate control mice. Panel E shows dramatically reduced levels of spermatocyte markers (Stra8, Spo11, and Sypc3) in purified 4N spermatocytes from cKO mice. The 4N spermatocytes were purified as in panel C. Panel F shows expression of known NMD substrates in spermatocytes isolated as in panel E. Panel G shows expression of known NMD substrates in the olfactory epithelium of Krt5-Cre;Upf3afl/fl mice. n=2–3 per genotype for all 3 panels. “1” represents expression level normalized to Ctrl level.
(H) Model: Cell A has high levels of UPF3B and consequently high NMD. Cell B has high levels of UPF3A and thus has low NMD, allowing accumulation of NMD substrates.
*P<0.05; **P<0.01; ***P<0.001
As another test of whether Upf3a-cKO mice have a spermatocyte progression defect, we used FACS analysis coupled with Hoescht 33342 staining. Hoescht blue emission quantifies DNA content, allowing one to distinguish haploid, diploid, and tetraploid cells, while Hoescht red emission quantifies the degree of chromatin condensation, allowing one to segregate different primary spermatocyte subsets (Bastos et al., 2005). In agreement with the results we obtained using phopho-H2AX staining, Hoescht 33342/FACS analysis showed ~6-fold fewer pachytene spermatocytes in Upf3a-cKO mice than control mice (Figure 7C, population B). Round and elongated spermatids were reduced by a similar amount (Figure 7C, population C; and Figure 7D), suggesting their decline was a secondary result of fewer spermatocytes, the cells that give rise to these post-meiotic cells.
As a final assay of the defects resulting from ablation of Upf3a in germ cells, we molecularly analyzed Hoescht/FACS-purified spermatocytes from Upf3a-cKO and control testes. Upf3a-cKO spermatocytes had significantly reduced expression levels of all 3 spermatocyte markers compared to control spermatocytes (Figure 7E), confirming that loss of UPF3A perturbs spermatocyte progression (Figures 7A–D). Of note, analysis of these isolated spermatocytes revealed that the excision of Upf3a did not occur in all Upf3a-cKO cells, as there was still 30% residual Upf3a expression in Upf3a-cKO spermatocytes (Figure S6F). This putative mosaicism suggests that the severity of the defects we observed is an underestimate of the defects resulting from loss of Upf3a in male germ cells.
To assess whether UPF3A might function prior to meiosis, we examined the effect of UPF3A loss on pre-meiotic germ cells – spermatogonia. The number of spermatogonia was not significantly different in Upf3a-cKO and control mice, as assayed at 1 and 2 weeks of age, when spermatogonia are particularly abundant (Figures 6D and S7B). The number of apoptotic testicular cells at 2 weeks of age was also not significantly different between Upf3a-cKO and control mice (Figure S7C). Defects became apparent at 4 weeks of age, as shown by the reduced fraction of seminiferous tubules containing elongating spermatids in Upf3a-cKO mice as compared to control mice (Figures 6D and S7D). Furthermore, there was a ~3-fold increase in apoptotic (TUNEL+) germ cells in 4-week-old Upf3a-cKO mice (Figure S7E).
We next examined whether UPF3A dose is critical for spermatogenesis. To this end, we generated and examined mice conditionally lacking one functional copy of Upf3a specifically in male germ cells (Stra8-Cre;Upf3afl/+ mice). As expected, these Upf3a-cHet mice had ~1/2 the normal level of Upf3a expression in the testis (Figure S7F). At 4 weeks of age, their testes exhibited decreased expression of several spermatogenic markers (Figure S7G). A similar pattern of spermatogenic marker expression profile defects was also observed in global Upf3a-heterozygous (Upf3a+/−) mice testis (data not shown). Hoescht 33342 FACS sorting revealed that Upf3a-cHet mice also had fewer pachytene spermatocytes and postmeiotic germ cells (Figure S7H). Spermatocytes purified from Upf3a-cHet mice also had decreased levels of several spermatocyte markers (Figure S7I). This reduction in meiotic markers persisted, as it was also observed in older Upf3a-cHet mice (Figure S7J). Together, these results suggest that even a modest reduction in the level of UPF3A in germ cells is sufficient to impair spermatocyte progression. We conclude that UPF3A level must be delicately controlled to allow normal spermatogenesis.
UPF3A is a NMD Repressor In Vivo
The finding that UPF3A is critical for spermatogenesis raised the possibility that it serves as a NMD repressor not only in vitro (Figures 1–4), but also in vivo. To test this, we examined the levels of NMD substrates in spermatocytes purified from Upf3a-cKO and control testes. We found that most NMD substrate RNAs were downregulated in Upf3a-cKO spermatocytes (Figure 7F), analogous to what occurred when UPF3A was depleted in cultured cells (Figures 1A, 1D, and 1E). The two NMD substrates that escaped UPF3A regulation were the non-coding RNAs, Gas5 and Snord22 (Figure 7F), which also escaped UPF3A-mediated suppression in P19 cells (Figure 1A). In addition, we observed that NMD substrates were downregulated in purified spermatocytes from Upf3a-cHet mice (Figure S7K), thereby providing an explanation for why these heterozygous mice have spermatogenesis defects. Given that Upf3a-cHet mice have only a ~50% reduction of UPF3A levels (Figure S7F), these data support the notion that UPF3A acts as a molecular ratchet to control the magnitude of NMD in vivo.
As described above, Upf3b is transcriptionally silenced in spermatocytes through MSCI. This unique regulatory phenomenon provided an opportunity to ask whether UPF3A functions independently of UPF3B in vivo. In particular, it allowed us to distinguish between UPF3A acting as a bona fide NMD repressor versus acting as a weak NMD factor that merely replaces its stronger paralog, UPF3B. If UPF3A serves as a weak NMD factor that competes with UPF3B, this predicts that loss of Upf3a in spermatocytes would have a reduced magnitude of NMD, as these cells also lack Upf3b. Instead we observed that NMD was stronger when Upf3a was ablated from spermatocytes (Figures 7F and S7K), providing strong support for the notion that UPF3A is a bona fide NMD repressor.
To determine whether UPF3A’s repressor activity is restricted to germ cells or acts more broadly, we generated mice that conditionally ablate Upf3a in the olfactory epithelium, which has the highest Upf3a expression of all the non-testicular tissues we tested (Figure 6A). Olfactory epithelium from Krt5-Cre;Upf3afl/fl mice had reduced levels of most NMD substrates (Figure 7G). This result coupled with our finding that the same outcome occurred in response to loss or depletion of UPF3A in other cells—spermatocytes (Figure 7F) and three cell lines representing different cell lineages and developmental stages (Figures 1A, 1D, and 1E)—leads us to conclude that UPF3A is a broadly acting NMD repressor.
To decipher the in vivo targets of UPF3A during male meiosis, we in silico extracted mRNAs that are enriched in pachytene spermatocytes from microarray analysis that was performed previously (Namekawa et al., 2006). We focused on identifying pachytene-enriched transcripts harboring dEJs, as a dEJ is the only well-established and reliable NMD-inducing feature (see Introduction). From this database, we identified 119 pachytene spermatocyte transcripts with dEJs (Supplementary Table 3), at least 10 of which are stabilized by UPF3A in P19 cells (Supplemental Tables 1 and 3). This data, coupled with our finding that UPF3A is essential for the normal progression of spermatocytes, supports a model in which UPF3A is highly expressed in spermatocytes to suppress NMD and thereby stabilize NMD substrate RNAs critical for spermatocyte developmental progression, including meiosis (Figure 7H).
DISCUSSION
How gene duplications provide biological innovation has long intrigued biologists. Here, we provide several lines of evidence for a gene duplication event that yielded functionally antagonistic gene products. This duplication event, which occurred approximately at the dawn of the vertebrate lineage, acted upon the UPF3 gene, which is essential for the NMD RNA degradation pathway in modern-day invertebrates (Serin et al., 2001). The vertebrate paralog, UPF3B, has maintained this function (Chan et al., 2007), while we demonstrated here that that its sister paralog, UPF3A, encodes a potent NMD repressor. Thus, unlike all other known NMD factors, UPF3A is a positive regulator of gene expression. We suggest that this role provides an explanation why UPF3A was selected for over evolutionary time. By acquiring the ability to repress NMD, UPF3A became a volume control to dictate the level of transcripts normally degraded by NMD in order to influence biological events.
While previous examples of antagonistic gene paralogs have been reported, the evolutionary origins and underlying basis for their antagonism has been poorly understood (Stoick-Cooper et al., 2007; Zhang et al., 2013). Our study identified a simple mechanism by which antagonistic paralog pairs arise – conversion of an activator into a repressor by simple perturbation (through mutation) of a functional domain. This loss-of-function “strategy” allows for rapid selection of the repressor paralog, thereby permitting escape from gene deterioration. In particular, we obtained several lines of evidence that the particular route taken by UPF3A to become a repressor was to lose the ability to efficiently interact with the EJC. This permitted UPF3A to serve as a molecular decoy that sequesters UPF2 from the NMD machinery and thus impairs NMD function (Figure 4A). Of note, however, UPF3A has retained the ability to weakly interact with the EJC (Buchwald et al., 2010; Kim et al., 2001; Kunz et al., 2006), which provides an explanation for why some NMD substrates escape UPF3A-mediated repression of NMD. Indeed, this may explain how UPF3A can sometimes act as a NMD factor. Thus, we suggest that whether UPF3A serves as a NMD activator or repressor is likely to depend on context.
While Upf3a is ubiquitously expressed, we found it is developmentally regulated in germ cells and exhibits altered expression in different adult tissues. The contribution of transcriptional and post-transcriptional mechanisms to this regulation is not known. It will be interesting to examine the role of its 3′ UTR in this regulation given that its paralog, Upf3b, has a relatively long 3′ UTR that confers regulation in response to neurally expressed miRNAs (Lou et al., 2014; Bruno et al., 2011). Another contributing factor to Upf3a regulation is alternative splicing. We found that Upf3a alternative splicing is regulated in a tissue-specific manner, which may have functional consequences given that only one UPF3A isoform is capable of repressing NMD. Another level of regulation is protein stability, as we previously showed that UPF3A protein is dramatically destabilized in the presence of its paralog, UPF3B (Chan et al., 2009). Escape from this destabilization mechanism is probably largely responsible for dictating the high levels of UPF3A protein in meiotic germ cells, as we found these cells largely lack UPF3B. Given that the Upf3a and Upf3b paralogs separated ~500 million years ago, only modest selective forces are likely to have been sufficient to lead to their unique expression patterns. We suggest that the ubiquitous but differential expression patterns of these antagonistic paralogs allow for regulated NMD in a wide variety of biological contexts.
Several lines of evidence suggest that UPF3A is a molecular rheostat. First, UPF3A is a potent NMD repressor that acts on a large number of NMD substrates. Using genome-wide analysis, we found that knockdown of UPF3A altered the stability of over 1000 mRNAs, many of which were previously defined as high-confidence NMD substrates. Second, UPF3A exhibits dose-dependent effects. Even a ~50% reduction (i.e., in Upf3a+/− mice) was sufficient to cause shifts in NMD substrate levels and defects in spermatocytes. We suggest that this is because UPF3A normally tightly regulates the levels of key target mRNAs and thus only a modest deviation in their level is detrimental. Finally, we found that UPF3A represses NMD even when UPF3B is essentially absent, both in vitro and in vivo. Thus, UPF3A acts as a general NMD repressor, not specifically for the UPF3B dependent-branch of NMD.
One biological scenario in which Upf3a likely acts as a molecular rheostat is developing male germ cells. The least mature male germ cell subset in the testis—spermatogonia—exhibit a high Upf3b/Upf3a ratio and thus would be predicted to have strong NMD (Figure 7H, cell A). After several rounds of cell proliferation, spermatogonia become spermatocytes, which we found had upregulated Upf3a expression and silenced Upf3b transcription (because of MSCI), resulting in a very low Upf3b/Upf3a ratio that is ~40-fold less than in spermatogonia (Figure 7H, cell B). We suggest that this dramatic ratio change serves as a switch to suppress NMD and thus strongly upregulate NMD target transcripts important for spermatocytes. Consistent with this, we found that loss of UPF3A in male germ cells causes a large reduction of mid-to-late spermatocytes and all subsequent stages of male germ cells.
What transcripts must be stabilized by UPF3A in spermatocytes to permit progression through this stage of germ cell development? GO analysis of UPF3A-stabilized transcripts identified “reproduction” as statistically overrepresented (Figure 2F), and many of these transcripts encode proteins that are essential for male fertility. Several function in meiosis, including ING2, EXO1, SGOL2, TERF1, and UBE2B (Supplemental Table 6). Thus, these transcripts are candidates to require UPF3A-mediated stabilization to drive meiotic progression. Another class of potentially important UPF3A targets is mRNAs encoding proteins involved in signaling pathways required for spermatogenesis (e.g., TGFβ, WNT, and NOTCH). Another class is mRNAs encoding apoptotic regulators, as we identified 35 UPF3A-stabilized mRNAs encoding proteins involved in apoptosis (Supplemental Tables 1 and 6). This finding coupled with the fact that apoptosis is an obligate requirement for normal spermatogenesis (Print and Loveland, 2000) and Upf3a-cKO mice exhibit increased germ cell apoptosis, raises the possibility that UPF3A controls the normal balance of mRNAs encoding pro- and anti-apoptotic regulators in germ cells to influence their survival. Other candidate UPF3A target mRNAs are the dEJ-containing mRNAs we identified by in silico analysis as being enriched in pachytene spermatocytes (Supplemental Table 3). Many of these high-confidence NMD substrate mRNAs encode proteins involved in RNA metabolism, raising the possibility that UPF3A influences post-transcriptional networks in germ cells. Among the UPF3A-regulated dEJ-containing spermatocyte-expressed mRNAs are those encoding factors acting in the AU-rich element-mediated RNA decay pathway (Supplemental Table 6). Given that this pathway is critical for the rapid induction and repression of such mRNAs, UPF3A may have a role in such events. Another potential target for this type of regulation is Arc, which is transiently induced by synaptic activity in neurons and is critical for learning and memory. It will be interesting to determine the functional significance of Arc’s expression and regulation by UPF3A in germ cells (Supplemental Table 6).
Another site of UPF3A action is the early embryo. Thus, it will be of interest to determine the identity of transcripts that require UPF3A regulation to permit pre-implantation embryonic development. Intriguingly, GO analysis of UPF3A-stabilized transcripts identified “chordate embryonic development” as the most statistically enriched category (Figure 2F). Among the sub-categories enriched were “embryonic organ development” and “neural tube development,” which include genes, such as Cdx2 and Sox2 (Supplemental Table 6), that are candidates to function downstream of UPF3A in early embryonic development. Other significantly enriched GO categories included “stem cell differentiation” and “growth” (Figure 2F), reinforcing the notion that UPF3A is critical for early development. Given that several UPF3A-stabilized mRNAs encode signaling pathway components, we suggest that these are also good candidates to act downstream of UPF3A. A potentially important site of UPF3A action, in general, is in transcriptional networks, as “negative transcriptional regulation” and “chromatin modifications” were also statistically enriched categories of UPF3A-stabilized mRNAs (Figure 2F). This raises the possibility that UPF3A influences the fate of specific cell types at both the transcriptional and post-transcriptional level. By regulating gene expression at both levels, UPF3A could more dramatically induce critical RNAs during development, and then more rapidly repress their expression once their functions are completed. The ability of UPF3A to dictate the stability of mRNAs encoding transcriptional regulators may also confer robustness to transcriptional circuits, thereby locking in specific developmental states.
The discovery that UPF3A serves as a NMD rheostat has potential clinical implications. For example, genetic diseases caused by dominant-negative proteins produced from genes harboring nonsense and frameshift mutations are candidates to be treated with UPF3A inhibitors. Such agents would enhance NMD activity, leading to reduced expression of deleterious dominant-negative proteins. In conclusion, we suggest that the discovery that UPF3A is a NMD repressor has widespread implications for evolutionary biology, RNA biology, and medicine.
MAIN EXPERIMENTAL PROCEDURES (Please see Supplemental Experimental Procedures for further information on all sections)
Mammalian Cell Culture and Transfection
P19 and HeLa cells were transfected using Lipofectamine 2000 (Invitrogen) and TransiT-2020 (Mirus), respectively. mEFs and mNSCs were electroporated using Nucleofector Kit for Mouse Neural Stem Cells (Lonza) according to manufacturer’s protocol. shUpf3a and shControls were from Thermofisher. siUpf3a and siControls were from GE Dharmacon (ON-TARGETplus SMART-pool siRNA). Stable HeLa shUPF3A knockdown cells described in Chan et al., 2007.
Generation of Upf3a-Mutant Mice
Upf3a exon 3 was selected to be flanked by loxP sites. The targeting vector included a Neomycin resistance cassette flanked by a FRT site, which upon recombination by CRE generated a truncated from of UPF3A protein lacking both the UPF2 and EJC-interacting domains (Kadlec et al., 2004). Primers used to generate Southern blot probes and to genotype Upf3a-mutants are listed in Supplemental Table 4.
RNA, Luciferase, and Protein Analysis
Total cellular RNA was isolated from cells and tissues using Trizol (Invitrogen), as described (Lou et al., 2014). NMD activity was measured using the NMD reporter plasmids pCI-Neo-WT PTC (−) and pCI-Neo-NS39 PTC (Boelz et al., 2006). They were cotransfected with pCI-Neo-FLY, a Firefly luciferase control plasmid. Western blot analysis was performed as previously described (Chan et al., 2007). Histology and immunofluorescence of testis sections were performed as previously described (Song et al., 2012). Expression vectors were obtained from the Andreas Kulozik Laboratory (Kunz et al., 2006).
RNA-Seq Library Construction and Data Analysis
RNA-seq analysis was performed on RNA half-life samples isolated from P19 cells that were transfected with siControl or siUpf3a and then subsequently actinomycin D. RNA quality was assessed using an Agilent Bioanalyzer. Libraries were generated using Illumina’s TruSeq RNA Sample Preparation Kit. Raw data analysis was performed using the Tuxedo suite and RStudio for subsequent procedures (Trapnell et al., 2010).
Phylogenetic Tree Construction
Phylogenetic analysis of UPF3 protein-coding sequences was done using 17 representative animal taxa. Values above 90% are indicated at the respective nodes. The tree is rooted on the five protostome taxa.
Coimmunoprecipitation (CoIP) Assay
CoIP was performed as described in Frank et al., 2010.
Germ Cell Profiling
Isolation and dissociation of cells from seminiferous tubules were performed as previously described (Bastos et al., 2005; McCarrey et al., 1992). FACS analysis was performed at the UCSD Human Embryonic Stem Cell Core Facility with a BD Influx Cell Sorter (BD Biosciences).
Supplementary Material
HIGHLIGHTS.
Functional antagonism can be a consequence of gene duplication
UPF3A inhibits NMD while its paralog, UPF3B, activates NMD
UPF3A suppresses NMD by sequestering UPF2 from the NMD machinery
Upf3a-null mice have hyper NMD and defects in embryogenesis and gametogenesis
Acknowledgments
This work was supported by the National Institute of Health (GM111838 and HD001259), the Howard Hughes Medical Institute, the Interfaces Scholar program, and the Women’s Reproductive Health Research grant (K12 HD001259). We thank Jens Lykke-Andersen (UCSD), Sebastien Durand (UCSD), and Mary Ann Handel (Jackson Laboratories) for their valuable scientific input, antibodies and samples.
Footnotes
AUTHOR CONTRIBUTIONS
E.S. and M.W. conceived of most of the experiments. The majority of the experiments were performed and analyzed by E.S. and S.J. Additional experiments were performed by A.S., M.R., M.K., W.C., C.L., H.S., M.P., L.H., J.E., J.N.C., and H.C. Testicular germ cell subset RNA was provided by J.M. Help with phylogenetic and testicular analyses was provided by K.P. and D.D., respectively. The manuscript was written by E.S., S.J., J.N.C., and M.W.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bastos H, Lassalle B, Chicheportiche A, Riou L, Testart J, Allemand I, Fouchet P. Flow cytometric characterization of viable meiotic and postmeiotic cells by Hoechst 33342 in mouse spermatogenesis. 2005 doi: 10.1002/cyto.a.20129. [DOI] [PubMed] [Google Scholar]
- Boelz S, Neu-Yilik G, Gehring NH, Hentze MW, Kulozik AE. A chemiluminescence-based reporter system to monitor nonsense-mediated mRNA decay. 2006 doi: 10.1016/j.bbrc.2006.08.017. [DOI] [PubMed] [Google Scholar]
- Bruno IG, Karam R, Huang L, Bhardwaj A, Lou CH, Shum EY, … Wilkinson MF. Identification of a microRNA that activates gene expression by repressing nonsense-mediated RNA decay. 2011 doi: 10.1016/j.molcel.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchwald G, Ebert J, Basquin C, Sauliere J, Jayachandran U, Bono F, Le Hir H, Conti E. Insights into the recruitment of the NMD machinery from the crystal structure of a core EJC-UPF3b complex. 2010 doi: 10.1073/pnas.1000993107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan W-K, Huang L, Gudikote JP, Chang YF, Imam JS, MacLean JA, 2nd, Wilkinson MF. An alternative branch of the nonsense-mediated decay pathway. 2007 doi: 10.1038/sj.emboj.7601628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan W-K, Bhalla AD, Le Hir H, Nguyen LS, Huang L, Gécz J, Wilkinson MF. A UPF3-mediated regulatory switch that maintains RNA surveillance. 2009 doi: 10.1038/nsmb.1612. [DOI] [PubMed] [Google Scholar]
- Chang Y-F, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. 2007 doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- Hittinger CT, Carroll SB. Gene duplication and the adaptive evolution of a classic genetic switch. 2007 doi: 10.1038/nature06151. [DOI] [PubMed] [Google Scholar]
- Gehring NH, Kunz JB, Neu-Yilik G, Breit S, Viegas MH, Hentze MW, Kulozik AE. Exon-Junction Complex Components Specify Distinct Routes of Nonsense-Mediated mRNA Decay with Differential Cofactor Requirements. 2005 doi: 10.1016/j.molcel.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Huang L, Wilkinson MF. Regulation of nonsense-mediated mRNA decay. 2012 doi: 10.1002/wrna.1137. [DOI] [PubMed] [Google Scholar]
- Huang L, Lou C-HH, Chan W, Shum EY, Shao A, Stone E, Karam R, Song H-WW, Wilkinson MF. RNA homeostasis governed by cell type-specific and branched feedback loops acting on NMD. 2011 doi: 10.1016/j.molcel.2011.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan H, Kondrashov F. The evolution of gene duplications: classifying and distinguishing between models. 2010 doi: 10.1038/nrg2689. [DOI] [PubMed] [Google Scholar]
- Jolly LA, Homan CC, Jacob R, Barry S, Gecz J. The UPF3B gene, implicated in intellectual disability, autism, ADHD and childhood onset schizophrenia regulates neural progenitor cell behaviour and neuronal outgrowth. 2013 doi: 10.1093/hmg/ddt315. [DOI] [PubMed] [Google Scholar]
- Kadlec J, Izaurralde E, Cusack S. The structural basis for the interaction between nonsense-mediated mRNA decay factors UPF2 and UPF3. 2004 doi: 10.1038/nsmb741. [DOI] [PubMed] [Google Scholar]
- Karam R, Wengrod J, Gardner LB, Wilkinson MF. Regulation of nonsense-mediated mRNA decay: implications for physiology and disease. 2013 doi: 10.1016/j.bbagrm.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karam R, Lou C-H, Kroeger H, Huang L, Lin JH, Wilkinson MF. The unfolded protein response is shaped by the NMD pathway. 2015 doi: 10.15252/embr.201439696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Kataoka N, Dreyfuss G. Role of the nonsense-mediated decay factor hUpf3 in the splicing-dependent exon-exon junction complex. 2001 doi: 10.1126/science.1062829. [DOI] [PubMed] [Google Scholar]
- Kondrashov FA, Rogozin IB, Wolf YI, Koonin EV. Selection in the evolution of gene duplications. 2002 doi: 10.1186/gb-2002-3-2-research0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz JB, Neu-yilik G, Hentze MW, Kulozik AE, Gehring NH. and translation Functions of hUpf3a and hUpf3b in nonsense-mediated mRNA decay and translation. 2006 doi: 10.1261/rna.12506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou CH, Shao A, Shum EY, Espinoza JL, Huang L, Karam R, Wilkinson MF. Posttranscriptional control of the stem cell and neurogenic programs by the nonsense-mediated RNA decay pathway. 2014 doi: 10.1016/j.celrep.2014.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J, Shu MD, Steitz JA. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. 2000 doi: 10.1016/s0092-8674(00)00214-2. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen S, Jensen TH. Nonsense-mediated mRNA decay: an intricate machinery that shapes transcriptomes. 2015 doi: 10.1038/nrm4063. [DOI] [PubMed] [Google Scholar]
- Maderazo AB, Belk JP, He F, Jacobson A. Nonsense- Containing mRNAs That Accumulate in the Absence of a Functional Nonsense-Mediated mRNA Decay Pathway Are Destabilized Rapidly upon Its Restitution. 2003 doi: 10.1128/MCB.23.3.842-851.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namekawa SH, Park PJ, Zhang L-F, Shima JE, McCarrey JR, Griswold MD, Lee JT. Postmeiotic sex chromatin in the male germline of mice. 2006 doi: 10.1016/j.cub.2006.01.066. [DOI] [PubMed] [Google Scholar]
- Nguyen LS, Jolly L, Shoubridge C, Chan WK, Huang L, Laumonnier F, Raynaud M, Hackett A, Field M, Rodriguez J, et al. Transcriptome profiling of UPF3B/NMD-deficient lymphoblastoid cells from patients with various forms of intellectual disability. 2012 doi: 10.1038/mp.2011.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LS, Wilkinson MF, Gecz J. Nonsense-mediated mRNA decay: inter-individual variability and human disease. 2014 doi: 10.1016/j.neubiorev.2013.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Print CG, Loveland KL. Germ cell suicide: new insights into apoptosis during spermatogenesis. 2000 doi: 10.1002/(SICI)1521-1878(200005)22:5<423::AID-BIES4>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Rebbapragada I, Lykke-Andersen J. Execution of nonsense-mediated mRNA decay: what defines a substrate? 2009 doi: 10.1016/j.ceb.2009.02.007. [DOI] [PubMed] [Google Scholar]
- Sadate-Ngatchou PI, Payne CJ, Dearth AT, Braun RE. Cre recombinase activity specific to postnatal, premeiotic male germ cells in transgenic mice. 2008 doi: 10.1002/dvg.20437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serin G, Gersappe A, Black JD, Aronoff R, Maquat LE. Identification and characterization of human orthologues to Saccharomyces cerevisiae Upf2 protein and Upf3 protein (Caenorhabditis elegans SMG-4) 2001 doi: 10.1128/MCB.21.1.209-223.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman BT, Huang DW, Tan Q, Guo Y, Bour S, Liu D, Stephens R, Baseler MW, Lane HC, Lempicki RA. DAVID Knowledgebase: a gene-centered database integrating heterogeneous gene annotation resources to facilitate high-throughput gene functional analysis. 2007 doi: 10.1186/1471-2105-8-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H-W, Dann CT, McCarrey JR, Meistrich ML, Cornwall GA, Wilkinson MF. Dynamic expression pattern and subcellular localization of the Rhox10 homeobox transcription factor during early germ cell development. 2012 doi: 10.1530/REP-11-0479. [DOI] [PubMed] [Google Scholar]
- Stoick-Cooper CL, Weidinger G, Riehle KJ, Hubbert C, Major MB, Fausto N, Moon RT. Distinct Wnt signaling pathways have opposing roles in appendage regeneration. 2007 doi: 10.1242/dev.001123. [DOI] [PubMed] [Google Scholar]
- Tarpey PS, Raymond FL, Nguyen LS, Rodriguez J, Hackett A, Vandeleur L, Smith R, Shoubridge C, Edkins S, Stevens C, et al. Mutations in UPF3B, a member of the nonsense-mediated mRNA decay complex, cause syndromic and nonsyndromic mental retardation. 2007 doi: 10.1038/ng2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teshima KM, Innan H. Neofunctionalization of duplicated genes under the pressure of gene conversion. 2008 doi: 10.1534/genetics.107.082933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JMA. Meiotic sex chromosome inactivation. 2007 doi: 10.1242/dev.000018. [DOI] [PubMed] [Google Scholar]
- Turner JMA, Aprelikova O, Xu X, Wang R, Kim S, Chandramouli GVR, Barrett JC, Burgoyne PS, Deng C-X. BRCA1, histone H2AX phosphorylation, and male meiotic sex chromosome inactivation. 2004 doi: 10.1016/j.cub.2004.11.032. [DOI] [PubMed] [Google Scholar]
- Zetoune AB, Fontanière S, Magnin D, Anczuków O, Buisson M, Zhang CX, Mazoyer S. Comparison of nonsense-mediated mRNA decay efficiency in various murine tissues. 2008 doi: 10.1186/1471-2156-9-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Duc A-CE, Rao S, Sun X-L, Bilbee AN, Rhodes M, Li Q, Kappes DJ, Rhodes J, Wiest DL. Control of hematopoietic stem cell emergence by antagonistic functions of ribosomal protein paralogs. 2013 doi: 10.1016/j.devcel.2013.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.