Abstract
Poly(ADP-ribose) polymerase I (PARP1) is a primary DNA damage sensor whose (ADP-ribose) polymerase activity is acutely regulated by interaction with DNA breaks. Upon activation at sites of DNA damage, PARP1 modifies itself and other proteins by covalent addition of long branched polymers of ADP-ribose, which in turn recruit downstream DNA repair and chromatin remodelling factors. PARP1 recognizes DNA damage through its N-terminal DNA-binding domain (DBD), which consists of a tandem repeat of an unusual zinc-finger (ZnF) domain. We have now determined the crystal structure of the human PARP1-DBD bound to a DNA break. Along with functional analysis of PARP1 recruitment to sites of DNA damage in vivo, the structure reveals a dimeric assembly whereby ZnF1 and ZnF2 domains from separate PARP1 molecules form a strand-break recognition module that helps activate PARP1 by facilitating its dimerization and consequent trans-automodification.
Short-patch repair of DNA single-strand breaks is initiated by poly(ADP-ribose) polymerase-1 (PARP1) – a multi-domain enzyme activated by binding of its N-terminal DNA-binding domain (DBD) to DNA breaks 1-5. Activated PARP1 utilises NAD+ to generate long branched chains of poly(ADP-ribose) (PAR)6, covalently attached to lysines on PARP1 itself 7 as well as chromatin-components such as histones 8-10.
PAR chains may act as DNA competitors, displacing histones and other proteins in the vicinity of a lesion to facilitate access 11,12. Specific PAR binding has been demonstrated for the macrodomain 13, occurring in the histone variant macroH2A 14 and the DNA helicase Alc1 15,16, and the PBZ domain, found in aprataxin and PNK-like factor (APLF) and CHFR 17. The existence of specific PAR-binding domains suggests that PAR acts as a bona fide post-translational ‘signalling’ modification analogous to poly-ubiquitination.
Due to its function as a primary DNA damage sensor, PARP1 is a target for therapeutic intervention in cancer, with NAD+ competitive inhibitors giving substantial tumour-selective killing in genetic backgrounds with defective homologous recombination 18,19. However, despite recent work 20-22 the biochemical and structural mechanism by which PARP1 recognises and is activated by DNA gaps and breaks remains obscure. A substantial literature suggests that PARP1 activation involves dimerisation mediated by N-terminal and central domains of the protein, with consequent automodification occurring in trans as an intermolecular reaction 1,7,23-26. How binding to DNA breaks facilitates PARP1 trans-automodification is unknown.
We have now determined the crystal structure of the DNA-binding domain of PARP1 (PARP1-DBD) encompassing the first two zinc-finger (ZnF) domains, bound to a DNA break. In contrast with structural analysis of the separated domains 22, we show that DNA binding by both zinc-finger domains is essential to damage recruitment in vivo, and that ZnF1 and ZnF2 domains from separate PARP1 molecules act as a functional unit to generate a dimeric binding module that specifically recognizes the single-strand / double-strand transition at a recessed DNA break. Mutational analysis in vitro and in cells demonstrates the functional requirement for zinc-finger dimerisation and reveals a mechanism for bringing two PARP1 molecules into close proximity at a DNA break as a prerequisite for trans-modification.
RESULTS
Structure of the PARP1-DBD – DNA Complex
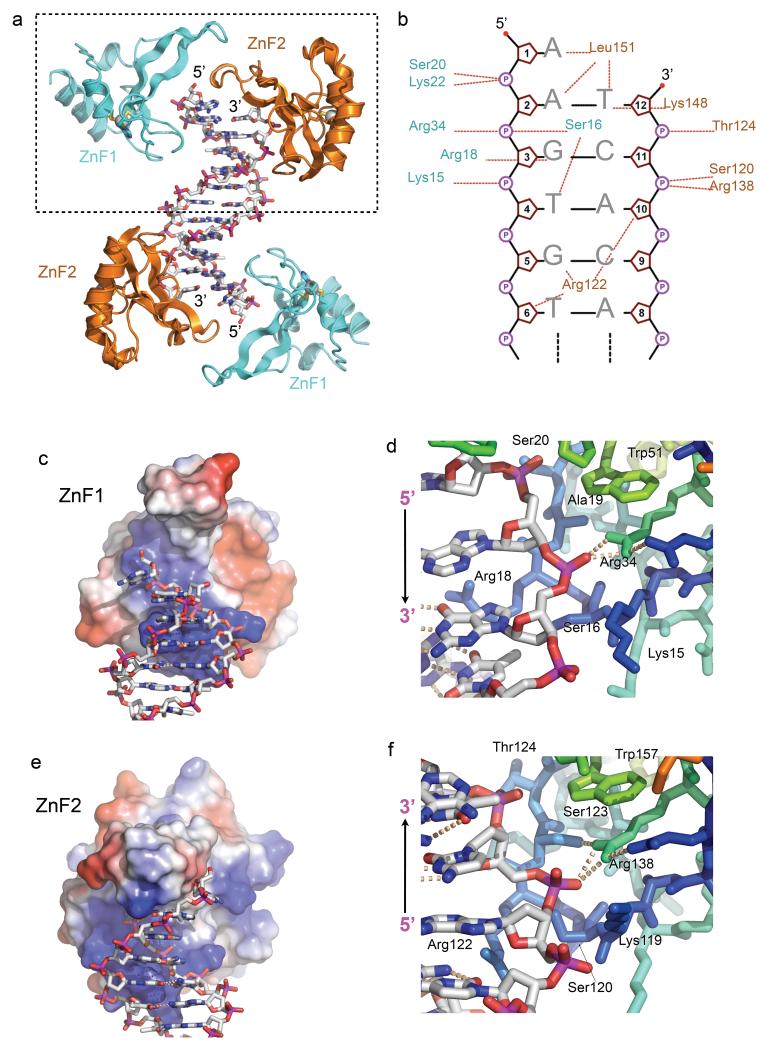
An N-terminal segment of human PARP1 (residues 5-202) was expressed in insect cells and purified by column chromatography. Screening with a range of DNA molecules yielded diffracting co-crystals with a duplex DNA molecule of 11 base-pairs and a single base 5′ overhang on each end. The structure was solved by molecular replacement using a B-form DNA model and refined at 3.1Å resolution (TABLE 1). The asymmetric unit ls contains a single DNA duplex with a ZnF1-ZnF2 pair, bound at each end (FIGURE 1a).
Table 1. Data collection and refinement statistics.
| PARP1 DBD / DNA complex | |
|---|---|
| Data collection ‡ | |
| Space group | C2 |
| Cell dimensions | |
| a, b, c (Å) | 163.98, 59.50, 61.58 |
| α, β, γ (°) | 90.00, 101.18, 90.00 |
| Resolution (Å) | 44.35 – 3.10 (3.27-3.10)* |
| R merge | 0.128 (0.468) |
| Rpim | 0.082 (0.287) |
| I / σI | 7.4 (2.2) |
| Completeness (%) | 92.2 (95.3) |
| Redundancy | 3.5 (3.6) |
| Refinement | |
| Resolution (Å) | 44.35– 3.10 |
| No. reflections | 9863 |
| Rwork / Rfree | 0.23 / 0.25 |
| No. atoms | |
| Protein | 2810 |
| DNA | 486 |
| Ligand/ion | 4 (Zn) |
| Water | 44 |
| B-factors (average) | |
| Protein | 69.53 |
| DNA | 45.21 |
| Ligand/ion | 81.54 (Zn) |
| Water | 46.05 |
| Rm.s. deviations | |
| Bond lengths (Å) | 0.01 |
| Bond angles (°) | 0.66 |
Data were collected from a single crystal..
Values in parentheses are for highest-resolution shell.
FIGURE 1. PARP1-DNA interactions.
a) Structure of the PARP1 DNA-binding domain (DBD) bound to a DNA duplex with a 5’-overhang. Each end of the non-self-complementary duplex is bound by a complex of the PARP1 ZnF1 and ZnF2 domains.
b) Schematic of the interactions of PARP1 DBD with the DNA duplex within the dashed box in a). Dotted lines indicate interactions with phosphate (circled P), sugar (pentagon) or base (letters).
c) ZnF1 DNA-interacting surface coloured by electrostatic potential (+ve – blue to −ve – red) interacts with the sugar-phosphate backbone of the overhanging strand and the major groove.
d) Details of ZnF1-DNA interactions, centred on the polar interaction of Arg34 and a DNA phosphate group.
e) As c) but for ZnF2, which interacts with the sugar-phosphate backbone of the recessed strand and the minor groove.
f) Details of ZnF2-DNA interactions, centred on the polar interaction of Arg138 and a DNA phosphate group. The conserved DNA interacting groups in the ZnF1 and ZnF2 domains, are able to interact with a DNA strand in either polarity.
PARP1 ZnF1 and ZnF2 bind together at the end of the DNA, with both domains interacting with the sugar-phosphate backbone and with the edges of base pairs via the major or minor grooves, depending on which ZnF domain is involved (FIGURE 1b). ZnF1 (visible from residue 6-91) accommodates the sugar-phosphate backbone of the DNA in a basic helical groove, formed by the extended zinc-binding loop connecting β-strands 1 and 2, and residues from the concave face of the main β-sheet. Residues 16 - Ser Gly Arg Ala - 19 project into the major groove, interacting with the inner surfaces of the backbone, and with the side chain of Arg18 contacting the edges of the base pairs (FIGURE 1c,d). ZnF2 (visible from residue 107-202) utilises the topologically equivalent residues to those in ZnF1 to interact with the backbone of the complementary DNA strand, which traverses the basic groove on ZnF2 with the opposite polarity. In ZnF2, residues 120 - Ser Asn Arg Ser -123 interact with the minor groove, with the side chain of Arg122, topologically equivalent to ZnF1-Arg 18, snaking in to make hydrogen bonding interactions with base-pair edges (FIGURE 1e,f).
ZnF1 and ZnF2 form a functional unit
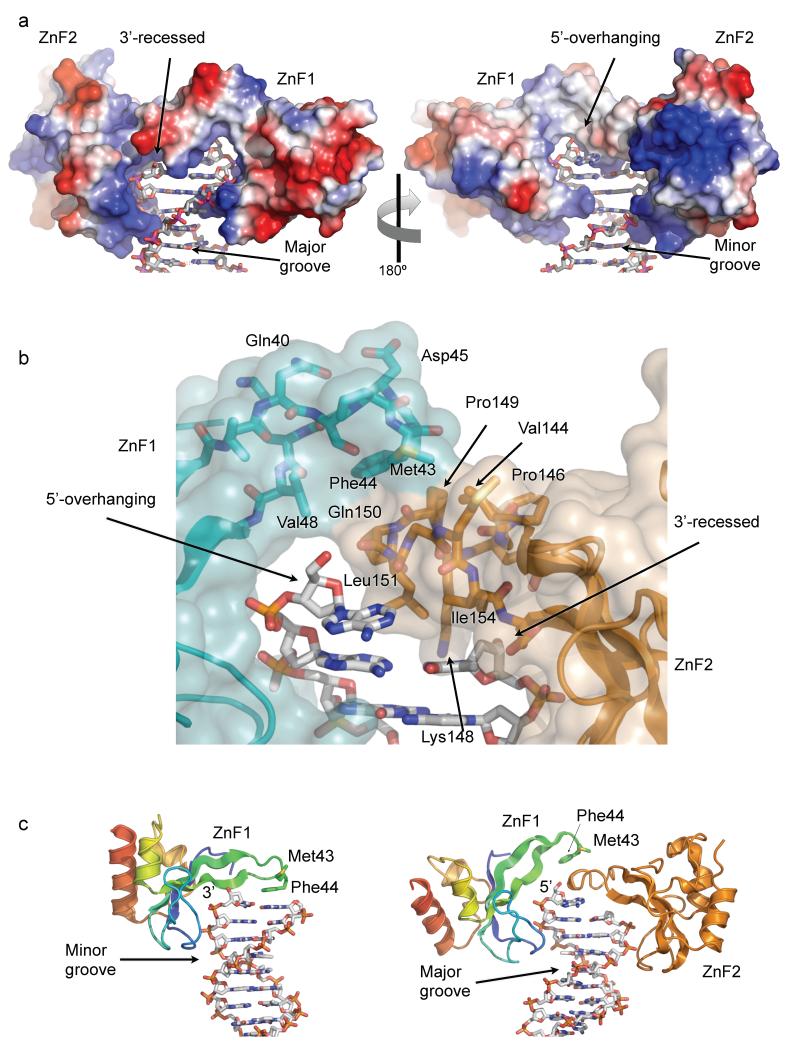
The ZnF domains, bound to opposite grooves of the DNA, together form a continuous binding surface that interacts with multiple features of the DNA (FIGURE 2a). Formation of the ZnF1-ZnF2 functional unit involves the loop between β-strands 2 and 3, which differs in structure between the two ZnF domains. This loop in ZnF2, which is structured by a pair of proline residues and a turn of 310 helix, projects into the body of the DNA over the recessed 3′ nucleotide. The side chains of Leu151 and Ile154 form a hydrophobic platform that penetrates the DNA from the minor groove and packs onto the face of the final base-pair.
FIGURE 2. DNA end-binding by the ZnF1-ZnF2 complex.
a) Electrostatic-colored (blue – basic; red –acidic) molecular surface of the DNA-end binding structure formed by PARP1 ZnF1 and ZnF2 domains, extending across the surface of the DNA end and interacting with both grooves of the duplex. The 3’-recessed end of one strand is on the left – the 5’-overhanging end of the other is on the right.
b) Detail of the interface between the tips of the β2-3 connecting loops of ZnF1 and ZnF2, which form the bridge overlying the terminal base-pair of the duplex. Transparent molecular surface and carbon atoms are colored by domain; ZnF1 – cyan, ZnF2 – gold.
c) Comparison of the interaction of isolated PARP1-ZnF1 with a blunt-end oligonucleotide 22 (left) and PARP1-ZnF1 in the context of the PARP1-DBD construct used in this work (right). In the two different structures PARP1-ZnF1 uses essentially the same residues to interact with the sugar-phosphate backbone in a 3’-5’ orientation via the minor groove (ZnF1 only) or in a 5’-3’ orientation via the major groove (ZnF1-ZnF2 complex).
The equivalent loop in ZnF1 makes no direct contact with DNA, but overlies the projecting loop of ZnF2, forming a hydrophobic interface involving ZnF1 residues Met43, Phe44 and Val48, packing against ZnF2 residues Val144, Pro149, Gln150, Gly152 and Met 153 (FIGURE 2b). This inter-domain interface buries ~300Å2 of molecular surface, which is at the lower end of the range commonly observed for reversible protein-protein interactions observed in many regulatory systems. However the effective affinity of this interaction would be substantially increased by the scaffold effect of the two domains interacting simultaneous with the DNA as a functional unit. Thus, while the loop between the β-strands 2 and 3 of ZnF2 stacks onto the final base pair of the DNA oligonucleotide, the equivalent region in ZnF1 mediates protein-protein interactions with the ZnF2 domain.
While this work was in progress, structures of the separate PARP1 ZnF domains bound to blunt-ended DNA oligonucleotides were reported 22. The DNA interactions made by ZnF2 in our study closely resembles those of the isolated domain, however the behavior of ZnF1 is radically different (see DISCUSSION). As an isolated domain it binds the minor groove of the DNA with Phe 44 interacting with the exposed surface of the bases at the blunt end of the oligonucleotide, whereas in the context of the intact DBD it binds on the major groove side of the helix, with Phe 44 involved in a protein-protein interaction with ZnF2 (FIGURE 2c)
The projecting segment of ZnF2, coming from the minor groove side of the DNA duplex, stacks against the terminal base-pair, blocking the trajectory of any continuation of the recessed strand as duplex DNA (FIGURE 3a,b). This interaction of ZnF2 with DNA is therefore absolutely contingent on a gap in the canonical B-form base-paired DNA duplex structure on at least one of the strands, giving ZnF2 the major role in DNA damage recognition, as previously shown 23. The projecting segment of ZnF1, coming from the major groove side, stacks on top of the ZnF2 projecting segment, rather than the DNA itself, and thereby opens a channel that accommodates the overhanging nucleotide (FIGURE 2b). The base of the overhanging nucleotide is pushed out from the duplex by the main chain of Gln 150, Leu151 and Gly 152 at the tip of the ZnF2 projection, but remains partly stacked with the preceding base. Unlike the recessed strand, the path of the DNA backbone is unimpeded and there is no obstacle to continuation of that strand beyond the site of interaction with the PARP1 ZnF domains (FIGURE 3c). Thus, the combination of the two ZnF domains, with their distinctive modes of binding the DNA from opposite grooves, provides an elegant mechanism for recognition of the single-strand / double-strand transition at a DNA break.
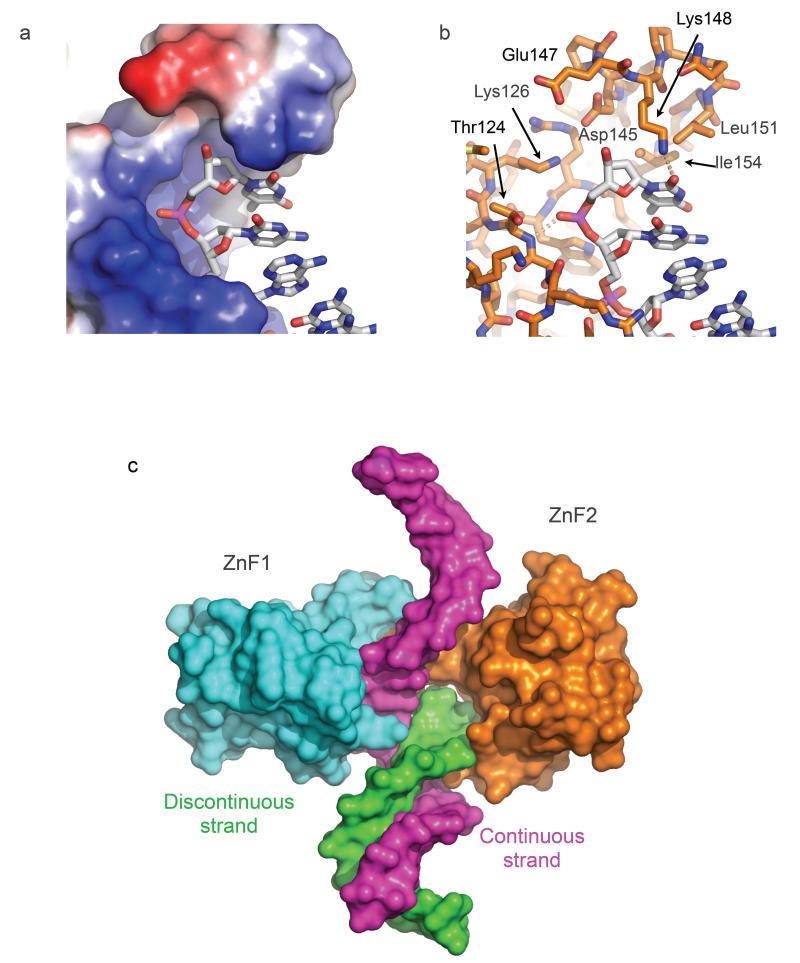
FIGURE 3. DNA break detection by PARP1 ZnF2.
a) The β2-3 loop of PARP1 ZnF2 projects over the basic DNA-binding groove and blocks continuation of the DNA strand, thereby conferring binding specificity to DNA ends.
b) Detail of the end-binding structure of ZnF2. Leu151 and Ile154 provide a hydrophobic wedge that overlies the terminal base on the recessed strand. The polar pocket formed by the side chains of Lys126, Asp145, Glu147 and Arg156 would be capable of accommodating a range of end chemistries, including 3’-PO4, 5’-OH and 5’- PO4, as well as the 3’-OH in the present crystal structure.
c) Model of gap recognition by the combined PARP1 ZnF1 and ZnF2 structure. Specific recognition of the discontinuity in one strand (green) is provided by the β2-3 loop of ZnF2 which completely blocks the end of the DNA-binding groove, while the other strand (magenta) can continue comfortably through the open DNA-binding groove of ZnF1 with only a small kink introduced.
Damage-dependent recruitment of PARP1-DBD in vivo
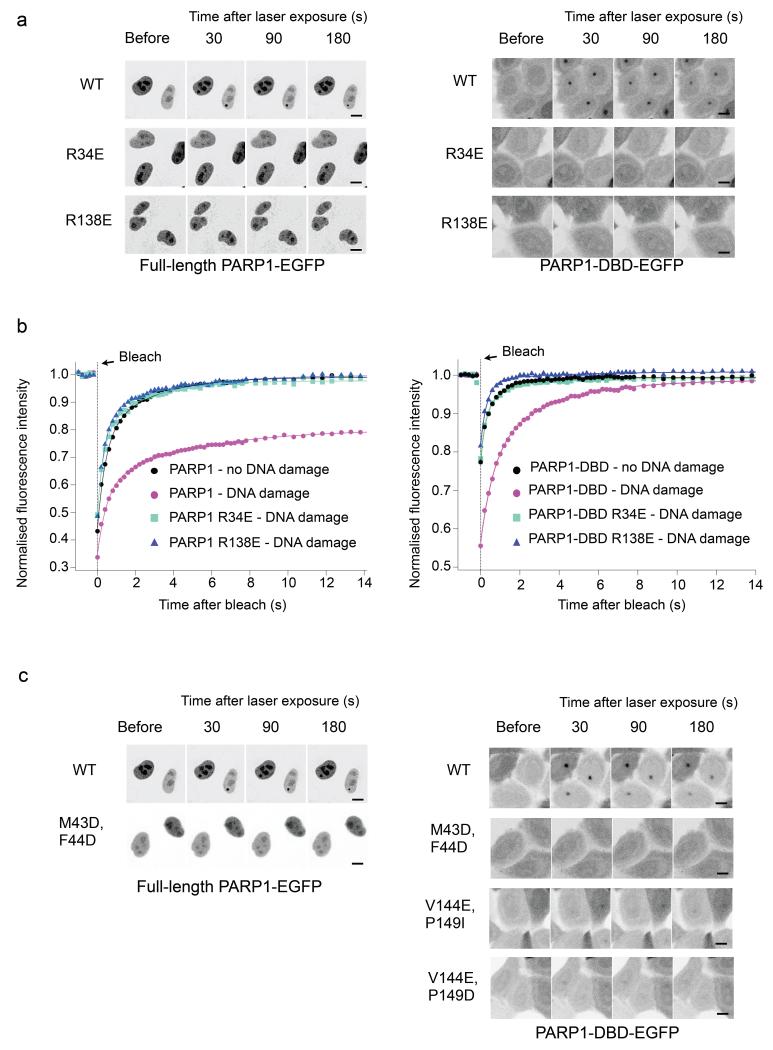
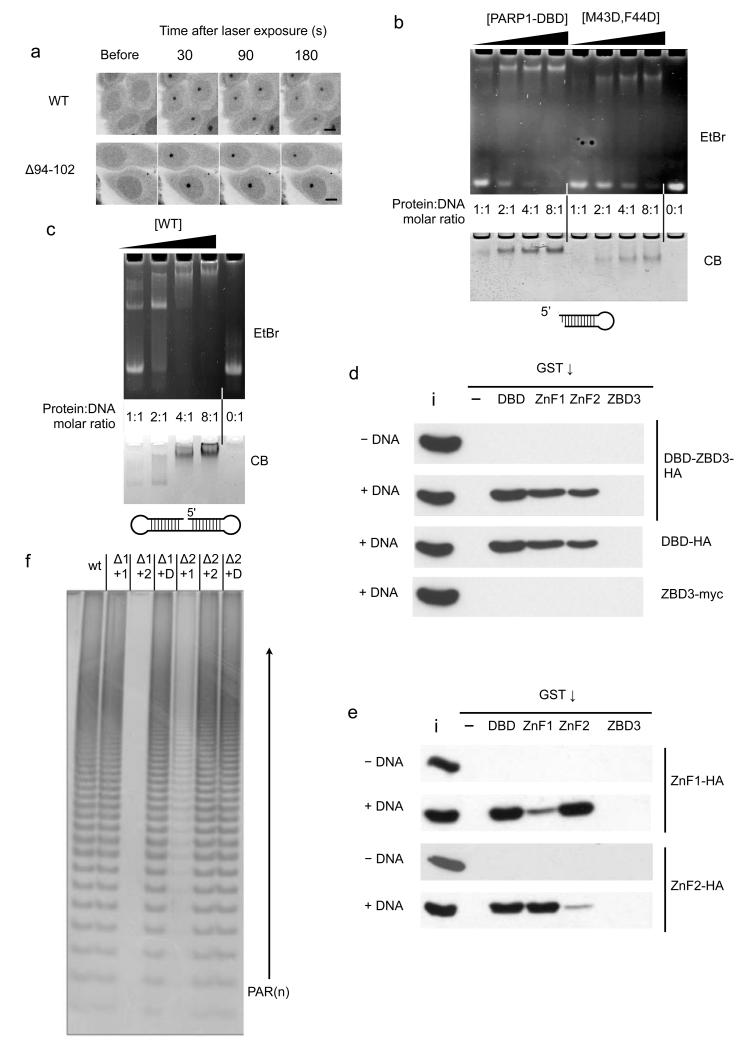
To test the biological significance of the DNA-binding mode revealed by the structure, we established a laser-damage microscopy assay for PARP1 recruitment. Briefly, we measured formation of fluorescent foci by EGFP-fused PARP1 constructs in response to DNA damage in live HeLa cells with an siRNA mediated knock-down of endogenous PARP1. We found that EGFP-fusions of both the full-length PARP1 and the DNA-binding domain construct used for structural studies (PARP1-DBD) readily formed fluorescent foci at sites of DNA damage upon laser irradiation (FIGURE 4a). Focus formation by both constructs was substantially reduced by mutations designed to disrupt DNA binding by ZnF1 (R34E) or by ZnF2 (R138E), confirming the biological relevance of the observed interactions.
FIGURE 4. DNA damage focus formation by PARP1 and its mobility.
a) Recruitment of C-terminal EGFP fusions with full-length PARP1 (left) or PARP1-DBD (right) to damage foci following laser damage. Fluorescent foci are evident 30 seconds post-irradiation in the nuclei of cells expressing the wild-type full-length PARP1 (only the bottom, right nucleus was laser-damaged) or PARP1-DBD (all nuclei damaged) constructs and diminish in intensity over subsequent minutes. No significant foci are formed by either full-length or DBD constructs harbouring mutations designed to disrupt DNA binding by ZnF1 (R34E) or by ZnF2 (R138E), showing a clear requirement for both zinc-finger domains in DNA damage recognition. Scale bar indicates 10 μm.
b) FRAP analysis shows that an intact PARP1-DBD is required for PARP1 retention at damaged DNA. In the absence of DNA damage, wild-type PARP1 full-length and PARP1-DBD are not immobilized on DNA. In contrast, under DNA damage conditions, wild-type PARP1 constructs are immobilized, while disruption of DNA binding by either ZnF1 or ZnF2 abolishes retention.
c) Recruitment of full-length PARP1 (left) or PARP1-DBD (right) to damage foci is abolished by polar mutations of Met 43 and Phe 44 in the ZnF1 domain, or of Val 144 and Pro 149 in the ZnF2 domain (not determined for full-length PARP1), which together form the hydrophobic protein-protein interface between ZnF1 and ZnF2. Scale bar indicates 10 μm.
We also looked at the retention of PARP1 on chromatin, using fluorescence recovery after photobleaching (FRAP). In the absence of DNA damage, fluorescence recovered rapidly for both EGFP-fused PARP1 and PARP1-DBD. When the whole nucleus was subjected to laser-damage, fluorescence recovery was substantially delayed, consistent with retention at sites of DNA damage. However, with the R34E and R138E mutations, fluorescence recovery in the presence of DNA damage, showed virtually identical kinetics to the wild-type in the absence of DNA damage, suggesting a substantial loss of damage-induced retention.
These data show that DNA interaction by each of PARP1 ZnF1 and ZnF2 domains is both necessary and sufficient for recruitment to and retention at sites of DNA damage in vivo, regardless of any ability of the isolated domains to interact with DNA in vitro or the putative involvement of any other domains in catalytic activation and other downstream events subsequent to DNA damage recruitment.
Within the structure, ZnF1 and ZnF2 interact to form a functional unit tailored to recognition of a discontinuity on at least one DNA strand. To test the biological significance of this interaction we constructed EGFP fusions of full-length PARP1 and PARP1-DBD with mutations of residues essential to the hydrophobic interface between ZnF1 and ZnF2, and tested their recruitment to damage foci in vivo (FIGURE 4c).
Mutation of Met 43 and Phe 44 in ZnF1 to polar residues that would disrupt the hydrophobic interface (M43D F44D), severely decreased PARP1-DBD recruitment to foci compared to wild-type. A similar effect was seen using the PARP1-DBD construct with mutations of Pro 149 and Val 144 in ZnF2 (V144E P149D, V144E P149I), against which Met 43 and Phe 44 pack. While the ZnF1 mutations did affect DNA binding by the isolated domain in vitro, the ZnF2 mutant protein retained the ability to bind DNA (SUPPLEMENTARY FIGURE 1). The strong effect of these disruptive mutations on either side of the interface highlights a critical functional role for this protein-protein interaction in PARP1 recruitment in vivo.
DNA damage-dependent dimerization of PARP1-DBD
Our data identify a functional requirement for interaction between ZnF1 and ZnF2 domains in DNA strand-break recognition. The two ZnF domains themselves are well resolved and ordered in our crystals, however no electron density is visible for the linker segment that connects them (PARP1 residues 92-106), which contains a high proportion of glycine and polar residues consistent with inherent flexibility. In the absence of clear electron density for the linker, the structure does not define whether the interaction between the zinc finger domains bound to the same DNA break is an intramolecular interaction between ZnF1 and ZnF2 domains from a single PARP1 molecule, or an intermolecular interaction between ZnF1 and ZnF2 domains from separate PARP1 molecules. The visible C-terminus of ZnF1 and the visible N-terminus of ZnF2 within the functional unit bound to a DNA break, are separated by ~54Å – a distance that could not be spanned by the 15 residues of linker separating ZnF1 and ZnF2 in the primary structure, without serious steric clashes. Similarly, the non-interacting ZnF1 and ZnF2 domains bound at opposite ends of the oligonucleotide are >60Å apart. However the C-terminus of ZnF1 and the N-terminus of ZnF2 domains bound to different DNA molecules in the lattice, are separated by as little as 19Å with no steric problems. Thus the functional interaction between ZnF1 and ZnF2 observed in the crystals appears to be an intermolecular interaction involving two PARP1 molecules rather than an intramolecular arrangement within the same polypeptide.
To test this hypothesis, we constructed an EGFP-fusion of PARP1-DBD with residues 94-102 in the linker segment deleted. This leaves only six residues between the two ZnF domains, making it impossible to form the DNA break-binding functional unit via an intramolecular interaction. Despite the much shorter linker, this construct was essentially wild-type in its recruitment to DNA damage foci (FIGURE 5a), supporting that notion that an intermolecular functional unit is required for binding to DNA breaks in vivo. The functionality of the Δ94-102 PARP1-DBD construct is consistent with the poor conservation of the inter-domain linker amongst PARP1 homologues (see SUPPLEMENTARY FIGURE 2) and suggests that its function is to tether ZnF1 and ZnF2 domains in proximity at sites of damage, but leaving enough flexibility to permit interaction with other PARP1 molecules.
FIGURE 5. Intermolecular dimerisation of PARP1 Znf1 and ZnF2 domain.
a) A nine residue deletion in the linker connecting ZnF1 and ZnF2 has no effect on the ability of the PARP1 - DBD to localise to damage foci. Scale bar indicates 10 μm.
b) Wild-type PARP1-DBD (WT) forms a strong interaction with the DNA in an electrophoretic mobility assay (EMSA) that is only saturated at a 2:1 protein:DNA ratio. DNA binding of the M43D, F44D PARP1-DBD mutant is substantially impaired.
c) EMSA of a ‘dumbbell’ oligonucleotide shows some complex formation at 1:1 protein-DNA ratio, but a 2:1 ratio is required to achieve saturation.
d) Pull-down assay showing that the DBD and isolated DNA-binding zinc-finger domains, but not the third zinc-binding domain (ZBD3), are able to co-precipitate constructs incorporating the DBD in a DNA dependent manner. ‘i’ = 20% input, ‘-‘ = empty beads.
e) Pull-down assay showing that either of the isolated tagged DNA-binding zinc-finger domains can be efficiently co-precipitated in a DNA-dependent manner, by the GST-DBD fusion or the GST-fusion with the other zinc-finger domain, but not by the same zinc-finger domain or by GST-ZBD3. ‘i’ = 5% input, ‘-‘ = empty beads.
f) Catalytic activity of PARP1 constructs deleted for either ZnF1 (Δ1) or ZnF2 (Δ2) domain can be complemented by presentation of the deleted domain in trans (+1, +2, or D = +1 and 2), consistent with the requirement for both DNA-binding zinc-finger domains to form an intermolecular interaction for PARP1 activation at sites of DNA damage.
The functional unit observed in the structure and the behaviour of the interface and linker mutants in vivo, suggest a mechanism of PARP1 dimerisation at ‘damaged’ DNA structures such as the 3’-recessed DSB used in the crystals. PARP1 dimerisation at sites of DNA damage has been implicated in multiple previous studies of PARP1 activation 1,7,26-28, although this has not been universally observed 20,21.
We investigated the stoichiometry of PARP1-DBD binding to DNA, using a stable DNA hairpin with a single 3’-recessed DSB in an electrophoretic mobility shift assay (EMSA). Using the wild-type PARP1-DBD construct, we observed only slight DNA binding at a stoichiometry of 1:1, but a robust shift of the majority of the DNA at a protein:DNA molar ratio of 2:1, consistent with preferential binding of a PARP1-DBD dimer. No significant bands of intermediate mobility were observed, suggesting strong cooperativity to form a single bound species. Despite the presence of a ZnF2 domain fully competent for DNA interaction, the ZnF1 M43D/F44D mutant showed much weaker binding, consistent with a role for these residues in forming a functional unit with ZnF2 (FIGURE 5b). We also conducted an EMSA assay with a double-hairpin DNA molecule that provides a single ‘nick’ as a potential binding site for PARP1. In a previous study 21 it was suggested that this DNA bound only a single PARP1 molecule under the conditions used. While we did observe some binding of PARP1-DBD at a protein:DNA ratio of 1:1, in our hands a protein:DNA ratio of 2:1 was required to fully shift the DNA (FIGURE 5c).
We also performed a series of ‘pull-down’ assays where we determined the ability of different GST-tagged PARP1 constructs to co-precipitate PARP1-DBD or a longer PARP1-DBD-ZBD3 construct (residues 1-372) that also includes the structurally unrelated third zinc-binding domain (FIGURE 5d). In the absence of DNA, we observed no interaction between GST fusions of the isolated ZnF1, ZnF2 or ZnBD3 domains, or the DBD construct, and PARP11-372. However, in the presence of a hairpin DNA molecule with a ‘damaged’ 3’-recessed end we observed robust co-precipitation of PARP11-372 and PARP1-DBD by GST-PARP1-DBD and by GST-ZnF1 and GST-ZnF2. We did not observe any dimeric interactions, in the presence or absence of DNA, involving ZnBD3, which had been previously suggested to mediate PARP1 dimerisation 29.
To verify whether these DNA dependent co-precipitations involved intermolecular interactions of ZnF1 and ZnF2, we tested the ability of the various GST constructs to co-precipitate isolated ZnF1 and ZnF2 domains (FIGURE 5e). We found that ZnF1 was efficiently co-precipitated by GST-PARP1-DBD and GST-ZnF2, but only weakly by GST-ZnF1. Conversely, ZnF2 was efficiently co-precipitated by GST-PARP1-DBD and GST-ZnF1, but not by ZnF2. No pull-down was observed in the absence of DNA or with GST-ZnBD3.
Finally we looked at the ability of the ZnF domains to complement catalytic activation of PARP1 mutants in trans. Wild-type PARP1 was robustly activated by a DNA break, generating poly-ADP-ribose chains. A PARP1 construct from which ZnF1 was deleted (ΔZnF1) was only catalytically active when the isolated ZnF1 domain or the PARP1-DBD construct containing the ZnF1 domain were added to the reaction, but not isolated ZnF2. Similarly, the activity of a ZnF2-deleted construct (ΔZnF2) was complemented by ZnF2 or PARP1-DBD, but not by ZnF1.
DISCUSSION
Our data show unambiguously that the DNA binding domain of PARP1, comprised of both N-terminal zinc-finger domains ZnF1 and ZnF2, is both necessary and sufficient for recruitment to and retention of PARP1 at sites of DNA damage in vivo. Furthermore, mutational disruption of the ability of either zinc-finger to interact with DNA or to form an intermolecular protein-protein interaction with the other, impairs DNA damage recruitment. Taken together, our data are fully consistent with the idea that PARP1 is able to dimerise and be activated at DNA breaks via formation of an intermolecular complex involving a protein-protein interaction between ZnF1 and ZnF2 domains from separate molecules.
DNA Break Recognition
The structure of the complex formed by ZnF1 and ZnF2 with each other and with the opposing grooves of the DNA, provides a satisfying explanation for the ability of PARP1 to recognise a wide range of damaged DNA structures, including DSBs, SSBs, gaps, and a range of non-B form structures in which the base-stacking continuity of at least one strand is disrupted 30-34. The DNA backbone interactions made by the two ZnF domains use structurally equivalent residues, but are effectively blind to the polarity of the strand, so that the same ZnF1-ZnF2 functional unit could accommodate a 5’-recessed end as well as it does the 3’-recessed end in the present crystals, and could potentially accommodate a terminal phosphate group of either polarity.
Structures of the isolated PARP1 ZnF1 and ZnF2 domains have been determined in complex with blunt-ended DNA oligonucleotides 22. The structure of the isolated ZnF2-DNA complex is consistent with our observation that ZnF2 in the intact PARP1-DBD complex can only bind to duplex DNA at strand discontinuities, and with previous work suggesting that ZnF2 plays the major role in gap-recognition 23,35. By contrast, ZnF1 shows a quite different DNA binding mode in isolation, where it binds via the minor groove, to the intact PARP1-DBD, where it binds via the major groove. Remarkably, these different binding modes involve the same residues in the basic groove on the concave face of the ZnF1 domain, but interacting with the DNA backbone in the opposite polarity. (FIGURE 2c).
Different chemical shift perturbations were seen in an NMR study, between isolated ZnF1 binding to DNA, and its binding in the context of the intact PARP1-DBD, consistent with DNA-dependent ‘interfinger’ contacts 21. Significantly, the major differences observed map to the loop containing Met 43 and Phe 44, which interacts with the faces of the terminal base-pair in the context of the isolated ZnF1, but forms the protein-protein interface with ZnF2 in the context of the intact PARP1-DBD. If ZnF1 has a role of mediating nonspecific PARP1 interaction with undamaged DNA, as has been suggested 21,22, then this may be mediated by Phe 44 and the 2-3 loop of ZnF1, independently of ZnF2. However our in vivo data show unambiguously that recruitment of PARP1 to damaged DNA is critically dependent on the ability of both ZnF domains to interact with DNA, and with each other.
A Mechanism for PARP1 activation at DNA breaks
PARP1 is an abundant nuclear protein that is constitutively associated with chromatin in a predominantly inactive state 36-38 but catalytically activated by DNA damage. Overactivation of PARP1 can have devastating consequences for a cell by triggering PAR-mediated cell death – parthanatos 39. Activation must therefore be tightly regulated and restricted to sites of DNA damage, with minimal activation by its association with undamaged DNA, which nonetheless ensures a high concentration of PARP1 on chromatin and thereby enables its very rapid recruitment to sites of damage as a primary sensor. Data from Drosophila 40 suggest that ZnF1 and ZnF2 have differential roles in mediating chromatin association, but our data show that both zinc-finger domains are required for human PARP1 to be localized to and retained at sites of DNA damage in human cells.
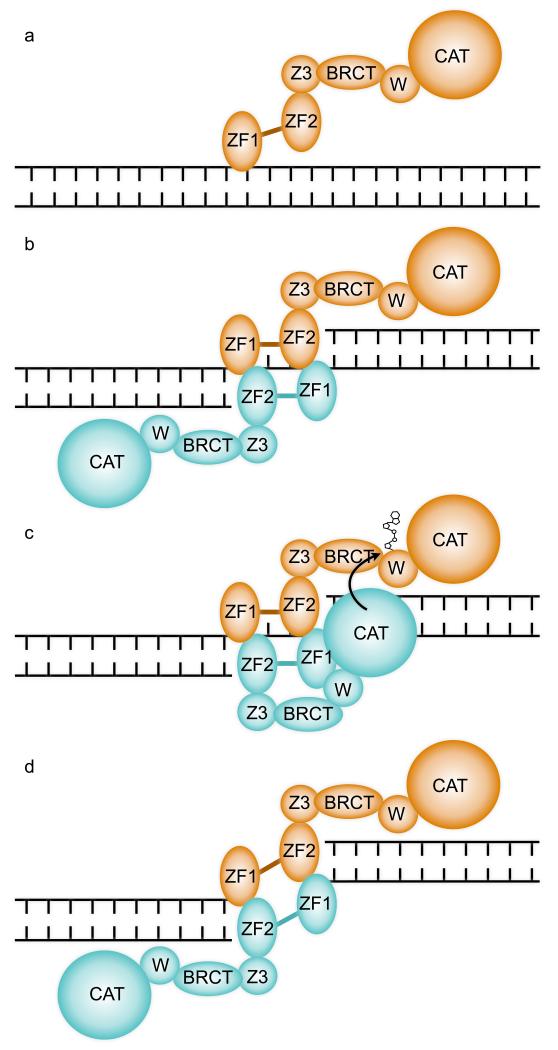
A substantial body of data suggest that PARP1 becomes activated at sites of DNA damage by trans-modification of one PARP1 molecule by another, with formation at least transiently, of a dimeric interaction between DNA-bound PARP1 molecules 1,7,26-28. Two recent studies 20,21, while observing DNA binding modes consistent with dimerization or oligomerization in some circumstances, were able to define buffer conditions and DNA molecules in which 1:1 protein:DNA complexes predominated. These apparently contradictory observations may be rationalized in terms of PARP1 having (at least) two modes of interaction with DNA – a monomeric low activity interaction providing chromatin localization (FIGURE 6a), and a dimeric (or oligomeric) interaction coupled to strand break-recognition, that brings PARP1 molecules into close proximity and thereby facilitates activation and trans-modification (FIGURE 6b,c). The functionally essential DNA-binding-coupled interaction between DBD domains from separate PARP1 molecules we describe here, is fully consistent with the latter, while the monomeric interactions observed in solution studies 20,21 may represent the former. The different DNA interactions displayed by ZnF1 in isolation 22 and in the intact PARP1-DBD, suggest that this domain plays a flexible role in both binding modes (FIGURE 2c).
FIGURE 6. A Mechanism for DNA Damage Dependent trans-automodification by PARP1.
a) PARP1 is constitutively associated with chromatin in the absence of DNA damage 36-38 and may associate with undamaged DNA, via the generally weak interaction of ZnF1 with the sugar-phosphate backbone. ZF1 and ZF2 – first and second zinc-finger domains; Z3 – zinc-binding domain; BRCT – BRCA1 C-terminal homology domain; W – WGR domain; CAT – catalytic domain.
b) A discontinuity in the DNA backbone permits additional binding of ZnF2 and formation of a functional break-recognition module via dimerization with a second PARP1 molecule. An inter-molecular ZnF1-ZnF2 complex could form at both margins if the single-strand gap is sufficiently large.
c) The close proximity of two PARP1 molecules bound cooperatively at a site of DNA damage, facilitates access of the catalytic domain of one to the poly-(ADP-ribose) acceptor sites on the other to achieve the well-described trans-automodification that initiates PARP1 signalling. Formation of a productive trans-autoribosylation complex is likely to involve conformational changes and multiple inter- and intra-molecular interactions of the other domains of PARP1, which are yet to be elucidated.
d) Formation of two break recognition modules formed by inter-molecular dimerization of ZnF1 and ZnF2 domains from two PARP1 molecules, explains the ability of PARP1 to maintain synapsis of DNA double-strand breaks and thereby facilitate a Ku-independent end-joining reaction 41-43.
Formation of a dimeric break recognition module by the ZnF1 domain of one molecule and the ZnF2 domain of a second, would permit a cooperating pair of PARP1 molecules to interact simultaneous at both margins of a single-strand gap of sufficient size, or with two separate DNA ends (FIGURE 6c,d). Consistent with this, PARP1 has been found to facilitate synapsis of double-strand breaks, and can mediate a Ku-independent mechanism of micro-homology end joining 41-43.
Ligand-dependent dimerization (and sometimes oligomerization) is an effective regulatory mechanism employed by evolution in signaling systems such as protein kinases for example, where ligand-dependent dimerization directly facilitates trans-phosphorylation by bringing otherwise monomeric enzyme molecules into proximity 44-46. The DNA damage-dependent dimerization of the PARP1-DBD we describe here is precisely analogous and fulfills the essential requirement of bringing two PARP1 molecules into close proximity, to allow trans-ADP-ribosylation. This is likely to be accompanied by substantial conformational rearrangements of the whole molecule, involving intra- and inter-molecular cooperation between other PARP1 domains (see for example 29) that ultimately enable the catalytic domain of one molecule to access and ADP-ribosylate the PAR-acceptor sites on the other. Elucidation of the detailed interactions and conformational states that facilitate specific trans-automodification of PARP1 and subsequent hetero-modification of down-stream targets remains a significant challenge.
Supplementary Material
ACKNOWLEDGMENTS
We thank Mark Roe for assistance with X-ray data collection and Keith Caldecott for useful discussion. We are grateful to the Diamond Light Source Ltd., Didcot, UK, for access to synchrotron radiation. We thank the Advanced Light Microscopy Facility (ALMF) at the European Molecular Biology Laboratory (EMBL) and Olympus Europe for supporting EMBL’s ALMF. This work was supported by EMBL, the Human Frontiers Science Program and the EU FP6 Marie Curie Research and Training Network “Chromatin Plasticity” to A.G.L. and a Cancer Research UK Programme Grant (C302/A8265) to L.H.P. L.H.P. is a Wellcome Trust Senior Investigator.
Footnotes
ACCESSION CODE
4AV1
REFERENCES
- 1.Pion E, et al. DNA-induced dimerization of poly(ADP-ribose) polymerase-1 triggers its activation. Biochemistry. 2005;44:14670–81. doi: 10.1021/bi050755o. [DOI] [PubMed] [Google Scholar]
- 2.Kirsten E, Kun E, Mendeleyev J, Ordahl CP. Activity assays for poly-ADP ribose polymerase. Methods Mol Biol. 2004;287:137–49. doi: 10.1385/1-59259-828-5:137. [DOI] [PubMed] [Google Scholar]
- 3.Huang K, et al. Analysis of nucleotide sequence-dependent DNA binding of poly(ADP-ribose) polymerase in a purified system. Biochemistry. 2004;43:217–23. doi: 10.1021/bi0301800. [DOI] [PubMed] [Google Scholar]
- 4.Shall S, de Murcia G. Poly(ADP-ribose) polymerase-1: what have we learned from the deficient mouse model? Mutation Research-Dna Repair. 2000;460:1–15. doi: 10.1016/s0921-8777(00)00016-1. [DOI] [PubMed] [Google Scholar]
- 5.Ame JC, Spenlehauer C, de Murcia G. The PARP superfamily. Bioessays. 2004;26:882–93. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- 6.D'Amours D, Desnoyers S, D'Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochemical Journal. 1999;342:249–268. [PMC free article] [PubMed] [Google Scholar]
- 7.Altmeyer M, Messner S, Hassa PO, Fey M, Hottiger MO. Molecular mechanism of poly(ADP-ribosyl)ation by PARP1 and identification of lysine residues as ADP-ribose acceptor sites. Nucleic Acids Res. 2009;37:3723–38. doi: 10.1093/nar/gkp229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fontan-Lozano A, et al. Histone H1 poly[ADP]-ribosylation regulates the chromatin alterations required for learning consolidation. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2010;30:13305–13. doi: 10.1523/JNEUROSCI.3010-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Messner S, et al. PARP1 ADP-ribosylates lysine residues of the core histone tails. Nucleic Acids Research. 2010;38:6350–62. doi: 10.1093/nar/gkq463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Messner S, Hottiger MO. Histone ADP-ribosylation in DNA repair, replication and transcription. Trends in Cell Biology. 2011 doi: 10.1016/j.tcb.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 11.Mathis G, Althaus FR. Release of Core Dna From Nucleosomal Core Particles Following (Adp-Ribose)N-Modification Invitro. Biochemical and Biophysical Research Communications. 1987;143:1049–1054. doi: 10.1016/0006-291x(87)90358-5. [DOI] [PubMed] [Google Scholar]
- 12.Althaus FR. Poly (Adp-Ribose) and Chromatin Organization in Dna Excision Repair. British Journal of Cancer. 1987;56:176–176. [Google Scholar]
- 13.Karras GI, et al. The macro domain is an ADP-ribose binding module. Embo J. 2005;24:1911–20. doi: 10.1038/sj.emboj.7600664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Timinszky G, et al. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat Struct Mol Biol. 2009;16:923–9. doi: 10.1038/nsmb.1664. [DOI] [PubMed] [Google Scholar]
- 15.Ahel D, et al. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science. 2009;325:1240–3. doi: 10.1126/science.1177321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gottschalk AJ, et al. Poly(ADP-ribosyl)ation directs recruitment and activation of an ATP-dependent chromatin remodeler. Proc Natl Acad Sci USA. 2009;106:13770–4. doi: 10.1073/pnas.0906920106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahel I, et al. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature. 2008;451:81–5. doi: 10.1038/nature06420. [DOI] [PubMed] [Google Scholar]
- 18.Bryant HE, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–7. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 19.Farmer H, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–21. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 20.Lilyestrom W, van der Woerd MJ, Clark N, Luger K. Structural and biophysical studies of human PARP-1 in complex with damaged DNA. J Mol Biol. 2010;395:983–94. doi: 10.1016/j.jmb.2009.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eustermann S, et al. The DNA-binding domain of human PARP-1 interacts with DNA single-strand breaks as a monomer through its second zinc finger. Journal of Molecular Biology. 2011;407:149–70. doi: 10.1016/j.jmb.2011.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Langelier MF, Planck JL, Roy S, Pascal JM. Crystal structures of poly(ADP-ribose) polymerase-1 (PARP-1) zinc fingers bound to DNA: structural and functional insights into DNA-dependent PARP-1 activity. The Journal of biological chemistry. 2011;286:10690–701. doi: 10.1074/jbc.M110.202507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gradwohl G, et al. The second zinc-finger domain of poly(ADP-ribose) polymerase determines specificity for single-stranded breaks in DNA. Proc Natl Acad Sci USA. 1990;87:2990–4. doi: 10.1073/pnas.87.8.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Panzeter PL, Althaus FR. Dna Strand Break-Mediated Partitioning of Poly(Adp-Ribose) Polymerase Function. Biochemistry. 1994;33:9600–9605. doi: 10.1021/bi00198a028. [DOI] [PubMed] [Google Scholar]
- 25.Kim JW, Kim K, Kang K, Joe CO. Inhibition of homodimerization of poly(ADP-ribose) polymerase by its C-terminal cleavage products produced during apoptosis. Journal of Biological Chemistry. 2000;275:8121–8125. doi: 10.1074/jbc.275.11.8121. [DOI] [PubMed] [Google Scholar]
- 26.Mendoza-Alvarez H, Alvarez-Gonzalez R. Poly(ADP-ribose) polymerase is a catalytic dimer and the automodification reaction is intermolecular. J Biol Chem. 1993;268:22575–80. [PubMed] [Google Scholar]
- 27.Bauer PI, Buki KG, Hakam A, Kun E. Macromolecular association of ADP-ribosyltransferase and its correlation with enzymic activity. Biochem J. 1990;270:17–26. doi: 10.1042/bj2700017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pion E, et al. Poly(ADP-ribose) polymerase-1 dimerizes at a 5' recessed DNA end in vitro: a fluorescence study. Biochemistry. 2003;42:12409–17. doi: 10.1021/bi0344432. [DOI] [PubMed] [Google Scholar]
- 29.Langelier MF, Ruhl DD, Planck JL, Kraus WL, Pascal JM. The Zn3 domain of human poly(ADP-ribose) polymerase-1 (PARP-1) functions in both DNA-dependent poly(ADP-ribose) synthesis activity and chromatin compaction. The Journal of biological chemistry. 2010;285:18877–87. doi: 10.1074/jbc.M110.105668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Murcia G, Menissier -de Murcia J. Poly(ADP-ribose) polymerase: a molecular nick sensor. Trends in Biochemical Sciences. 1994;19:172–176. doi: 10.1016/0968-0004(94)90280-1. [DOI] [PubMed] [Google Scholar]
- 31.D'Silva I, et al. Relative affinities of poly(ADP-ribose) polymerase and DNA-dependent protein kinase for DNA strand interruptions. Biochimica Et Biophysica Acta-Protein Structure and Molecular Enzymology. 1999;1430:119–126. doi: 10.1016/s0167-4838(98)00278-7. [DOI] [PubMed] [Google Scholar]
- 32.Potaman VN, Shlyakhtenko LS, Oussatcheva EA, Lyubchenko YL, Soldatenkov VA. Specific binding of poly(ADP-ribose) polymerase-1 to cruciform hairpins. J Mol Biol. 2005;348:609–15. doi: 10.1016/j.jmb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 33.Lonskaya I, et al. Regulation of poly(ADP-ribose) polymerase-1 by DNA structure-specific binding. J Biol Chem. 2005;280:17076–83. doi: 10.1074/jbc.M413483200. [DOI] [PubMed] [Google Scholar]
- 34.Bryant HE, et al. PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination. Embo J. 2009;28:2601–15. doi: 10.1038/emboj.2009.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ikejima M, et al. The zinc fingers of human poly(ADP-ribose) polymerase are differentially required for the recognition of DNA breaks and nicks and the consequent enzyme activation. Other structures recognize intact DNA. The Journal of biological chemistry. 1990;265:21907–13. [PubMed] [Google Scholar]
- 36.Kim MY, Mauro S, Gevry N, Lis JT, Kraus WL. NAD+-dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1. Cell. 2004;119:803–14. doi: 10.1016/j.cell.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 37.Tulin A, Spradling A. Chromatin loosening by poly(ADP)-ribose polymerase (PARP) at Drosophila puff loci. Science. 2003;299:560–2. doi: 10.1126/science.1078764. [DOI] [PubMed] [Google Scholar]
- 38.Ju BG, et al. Activating the PARP-1 sensor component of the groucho/ TLE1 corepressor complex mediates a CaMKinase IIdelta-dependent neurogenic gene activation pathway. Cell. 2004;119:815–29. doi: 10.1016/j.cell.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 39.David KK, Andrabi SA, Dawson TM, Dawson VL. Parthanatos, a messenger of death. Front Biosci. 2009;14:1116–28. doi: 10.2741/3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kotova E, Jarnik M, Tulin AV. Uncoupling of the transactivation and transrepression functions of PARP1 protein. Proc Natl Acad Sci U S A. 2010;107:6406–11. doi: 10.1073/pnas.0914152107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Audebert M, Salles B, Calsou P. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. The Journal of biological chemistry. 2004;279:55117–26. doi: 10.1074/jbc.M404524200. [DOI] [PubMed] [Google Scholar]
- 42.Audebert M, Salles B, Weinfeld M, Calsou P. Involvement of polynucleotide kinase in a poly(ADP-ribose) polymerase-1-dependent DNA double-strand breaks rejoining pathway. J Mol Biol. 2006;356:257–65. doi: 10.1016/j.jmb.2005.11.028. [DOI] [PubMed] [Google Scholar]
- 43.Audebert M, Salles B, Calsou P. Effect of double-strand break DNA sequence on the PARP-1 NHEJ pathway. Biochem Biophys Res Commun. 2008;369:982–8. doi: 10.1016/j.bbrc.2007.11.132. [DOI] [PubMed] [Google Scholar]
- 44.Cock JM, Vanoosthuyse V, Gaude T. Receptor kinase signalling in plants and animals: distinct molecular systems with mechanistic similarities. Current Opinion in Cell Biology. 2002;14:230–6. doi: 10.1016/s0955-0674(02)00305-8. [DOI] [PubMed] [Google Scholar]
- 45.Pellegrini L. Role of heparan sulfate in fibroblast growth factor signalling: a structural view. Curr Opin Struct Biol. 2001;11:629–34. doi: 10.1016/s0959-440x(00)00258-x. [DOI] [PubMed] [Google Scholar]
- 46.Ali MM, et al. Structure of the Ire1 autophosphorylation complex and implications for the unfolded protein response. Embo J. 2011;30:894–905. doi: 10.1038/emboj.2011.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.