Abstract
N6-methylation of adenosine (m6A) is the most abundant post-transcriptional modification within the coding region of mRNA, but its role during translation remains unknown. Here, we used bulk kinetic and single-molecule methods to probe the effect of m6A in mRNA decoding. Although m6A base pairs with uridine during decoding as shown by x-ray crystallographic analyses of Thermus thermophilus ribosomal complexes, our measurements employing an Escherichia coli translation system revealed that m6A modification of mRNA can act as a barrier to tRNA accommodation and translation elongation. The interaction between an m6A-modified codon and cognate tRNA echoes the interaction between a near-cognate codon and tRNA, as delay in tRNA accommodation depends on the position and context of m6A within codons and on the accuracy level of translation. Overall, our results demonstrate that chemical modification of mRNA can change translational dynamics.
Expression of genetic information in biology is regulated at various levels, among which a control of gene expression at the post-transcriptional level offers distinct advantages. By this mechanism, prompt responses to stimuli are attained without perturbation of overall cellular translational dynamics by bypassing of time consuming mRNA transcription and executing localized control prior to or during protein synthesis1. Recently discovered evidences on N6-methyladenosine (m6A), the most abundant post-transcriptional modification occurring in bacterial2 and eukaryotic mRNA3,4, suggest a potential role of m6A modification in modulating translation elongation dynamics at the post-transcriptional level.
While m6A modification occurs throughout mRNA, most modifications occur within the coding regions and are present during translation3–5. The modification sites for m6A within mammalian cell are conserved and dynamically regulated by m6A methyltransferases and demethylases3,4. Further, perturbing m6A modification disrupts wide variety of cellular functions such as translational efficiency5, cell viability6–8, and development4–14, although precise roles of m6A for these processes remain unknown. Compared to unmodified adenosine (A), m6A has less stable base pairing with uridine (U) and destabilizes local RNA structures18, features which could play important roles in m6A-induced cellular functions19,20.
Central to translation is selection of cognate and discrimination against non-cognate codon:anticodon RNA duplex structures, which define the information transfer from the gene sequence encoded in the mRNA to the protein sequence. The translation machinery has universally evolved to include a sophisticated two-step tRNA selection process involving irreversible chemical reactions: first, initial selection of tRNA prior to GTP-hydrolysis and second, kinetic proofreading of selected tRNA prior to peptide bond formation. This two-step process serves to amplify the accuracy impact of the free energy differences among cognate, near-cognate and non-cognate codon:anticodon interactions to ensure precise information transfer during translation. However, the potential perturbation of these processes by m6A modifications within the mRNA coding sequence and the resulting effects on the accuracy and rate of translation remain unknown. Here, we set out to monitor the effects of m6A modifications on translation elongation dynamics using a combined structural, kinetic and single-molecule approaches.
Results
Codons with m6A has slower translation elongation dynamics
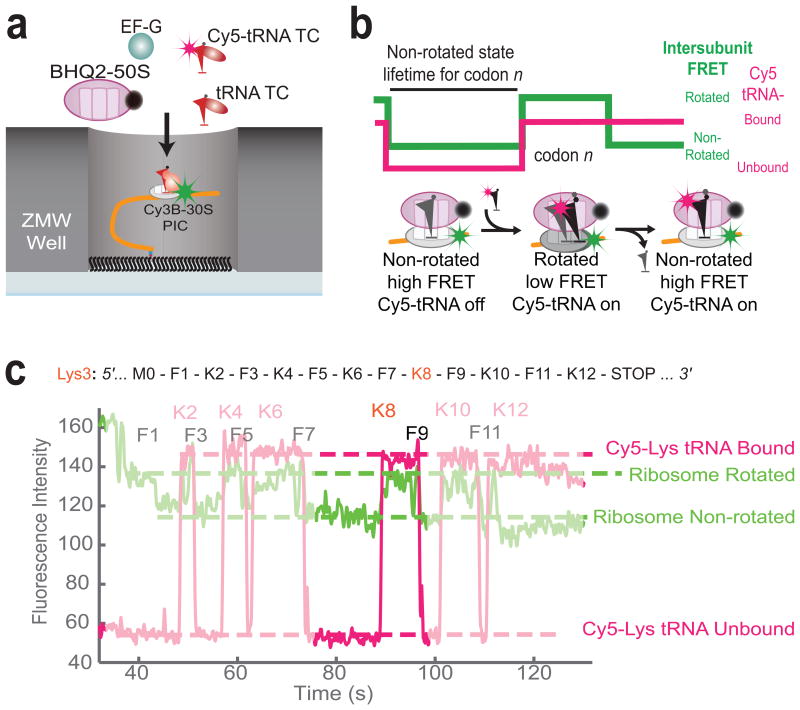
To probe the effect of m6A in tRNA selection dynamics over multiple codons, we directly observed an Escherichia coli (E. coli) ribosome translating an mRNA molecule containing a single-base m6A-modification using single-molecule fluorescence and zero-mode waveguide (ZMW) instrumentation21 (Schematics shown in Figure 1a). The E. coli translation system has been used as a model system to study translational decoding22,23, and has several advantages, including established purification methods for site-specifically labeled factors and a reduced number of factors involved during translation compared to a eukaryotic system. We monitored inter-subunit Förster Resonance Energy Transfer (FRET) between Cy3B and BHQ-2 (a non-fluorescent energy transfer quencher), site-specifically attached to E. coli 30S and 50S subunits, respectively, to observe global conformation changes of a ribosome during translation. For this, we tracked lifetimes of: (1) a non-rotated state prior to an intersubunit rotation of the 30S subunit relative to the 50S subunit upon cognate tRNA accommodation to the A site and peptidyl transfer, and (2) a rotated state prior to a reverse rotation upon EF-G-catalyzed translocation during elongation21,24,25, as shown in the sample trace in Figure 1c. Further, by using fluorescently labeled E. coli lysine tRNA, Lys-(Cy5)tRNALys, we simultaneously tracked the time between binding, ribosomal passage and dissociation of cognate Lys-(Cy5)tRNALys to unmodified and m6A-modified lysine codons in the A site of a translating ribosome. We observed near-simultaneous Lys-(Cy5)tRNALys binding and ribosomal intersubunit rotation, which indicated cognate decoding of a lysine codon by Lys-(Cy5)tRNALys accommodation and peptidyl transfer, while rare short Cy5 fluorescent pulses uncorrelated with Cy3B-BHQ-2 FRET efficiency indicated a transient sampling of Lys-(Cy5)tRNALys to the decoding complex. Correlation between fluorescently labeled tRNA pulses and inter-subunit FRET signal allowed us to identify true translational complexes within ZMWs with substantial confidence.
Figure 1.
Single-molecule assay for observing translational dynamics on m6A-modified mRNA. (a) Experimental setup for single-molecule assay21,24,25. Pre-Initiation Complex (PIC) containing Cy3B labeled 30S ribosomal subunit, Initiation Factor 2 (IF2), fMet-tRNAfMet, and 5′-biotinylated mRNA of interest is immobilized to partially neutravidin-biotinylated polyethylene glycol covered zero-mode waveguides (ZMWs). Experiment is started by illuminating ZMWs with green and red lasers and delivering BHQ-2 labeled 50S, Elongation Factor G (EF-G), Cy5-labeled and unlabeled tRNA ternary complexes to immobilized PICs. (b) Expected sequences of fluorescence signals using Cy3B-BHQ-2 FRET and Cy5 pulses. Cy3B signal reports the rotational state of ribosomal subunits for translating each codon, while long Cy5 pulses indicating stable binding of Cy5-tRNA to translational complex. (c) Sample fluorescent signal observed during experiment using m6A-modified mRNA sequence shown. Each Cy5 pulses are correlated with two Cy3B low-high-low cycle, corresponding to two rounds of elongation during tRNA is bound to translational complex (A-to-P and P-to-E movement in the perspective of transiting tRNA).
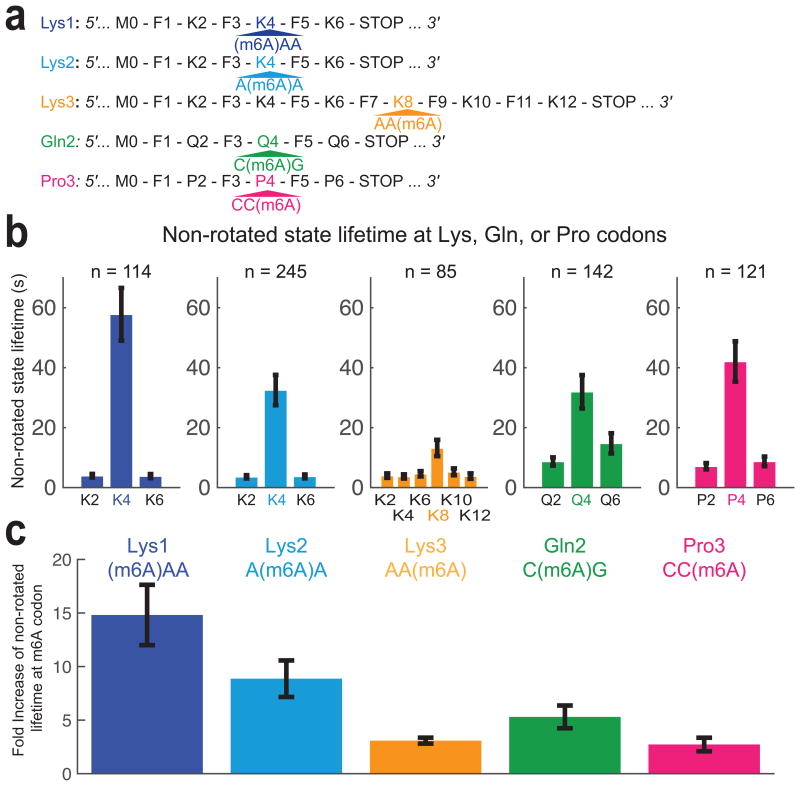
Using this approach, we measured the rotated and non-rotated lifetimes for each codon during translation of a twelve-codon mRNA sequence with repeating phenylalanine (Phe) and lysine (Lys) codons, containing an m6A modification at the 3rd base of the 8th codon (Lys with AA(m6A) codon), which we called Lys3 mRNA (Fig. 2a). In the presence of EF-Tu–GTP–Lys- (Cy5)tRNALys ternary complex (TC) and EF-Tu–GTP–Phe-tRNAPhe TC, we observed a 3-fold increase in non-rotated state lifetime for a modified Lys codon relative to non-rotated state lifetime for non-modified Lys codons in the same mRNA (Fig. 2b, c). These dynamic effects were specific to A site occupancy by the modified codon; we did not observe other effects on translational dynamics as m6A enters the ribosomal entry channel (corresponds to translational dynamics on codons 4-6), enters the ribosomal A site (codons 7), or leaves the ribosome (codons 9-12) (Supplementary Fig. 1), consistent with a model that ascribes the observed perturbation to A site codon:anticodon interaction. Furthermore, we did not observe any effect on rotated state lifetimes, suggesting that m6A does not affect the rates of translocation (Supplementary Fig. 1).
Figure 2.
Single-base m6A-modification of codon delays tRNA accommodation. (a) mRNA constructs used in single-molecule assay. All mRNA constructs have six codons in the coding region with m6A-modified codon in the fourth codon, except Lys3, where twelve-codon long construct was used to test the effect of m6A entering and leaving ribosome completely. (b) The non-rotated state lifetimes at Lys, Gln or Pro codons present in the mRNA used. The error bars represent s.e.m. (95% confidence interval) from fitting a single-exponential distribution to number of molecule specified. (c) Fold-increases of m6A-modified codon rotated state lifetime are compared across experimental conditions and mRNAs used. Error bars are calculated using the propagation of error method from data shown in (b).
Our x-ray crystal structures of translational decoding complexes, containing m6A-modified short RNA oligonucleotides, further support the observations above. We purified and crystallized Thermus thermophilus 30S ribosomal subunits, and soaked them with an oligonucleotide corresponding to the modified anticodon stem loop (ASL) of human tRNALys3 and with four different short RNA26–28 ((m6A)AAUUU, A(m6A)AUUU, AA(m6A)UUU, and AAAUUU, written from 5′ to 3′). From our four complete x-ray diffraction data sets with resolution ranging from 3.35 Å to 3.45 Å for each crystallized sample (Data Table 1), we observed a well-defined difference electron density in the decoding site, showing canonical Watson-Crick base pairing interaction for both A-U and m6A-U in the ribosomal A site (Supplementary Fig. 2). At the current resolution ranges, we could not obtain well-ordered electron density for the 6-methyl group of m6A, perhaps due to rotational movement about the C-N bond. Our result agreed well with results from NMR experiments on m6A base pairing18, which exclusively showed canonical base pairing between m6A and U, despite steric clash between added methyl group and N7 of adenosine. The structures obtained by x-ray crystallography indicated that the m6A does not perturb canonical base paring in the final state of tRNA accommodation, while steric effects observed by NMR and manifested by decreased thermodynamic stability of modified A-U pairs could underlie perturbed tRNA selection dynamics observed in our single-molecule assays.
Data Table 1. Data collection and refinement statistics (molecular replacement).
| Unmodified (4X62) |
m6A +1 (4X64) |
m6A +2 (4X65) |
m6A +3 (4X66) |
|
|---|---|---|---|---|
| Data collection | ||||
| Space group | P41212 | P41212 | P41212 | P41212 |
| Cell dimensions | ||||
| a, b, c (Å) | 400.8, 400.8, 175.1 | 400.5, 400.5, 175.5 | 401.4, 401.4, 175.9 | 401.3, 401.3, 175.6 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 35.0-3.45 (3.51-3.45) | 35.0-3.35 (3.41-3.35) | 35.0-3.35 (3.41-3.35) | 55.0-3.45 (3.51-3.45) |
| Rmerge | 0.168 (0.984) | 0.130 (0.993) | 0.141 (0.929) | 0.210 (0.961) |
| I / σI | 15.10 (2.76) | 12.29 (1.25) | 19.08 (2.83) | 9.04 (1.77) |
| Completeness (%) | 99.2 (100.0 | 96.4 (91.4) | 100.0 (100.0) | 96.6 (98.7) |
| Redundancy | 10.5 (10.7) | 5,8 (2.7) | 11.9 (9.2) | 5.1 (4.9) |
| Refinement | ||||
| Resolution (Å) | 35.0-3.45 (3.49-3.45) | 35.0-3.35 (3.38-3.35) | 35.0-3.35 (3.38-3.35) | 50.0-3.45 (3.49-3.45) |
| No. reflections | 183968 (5548) | 171102 (1033) | 204105 (5914) | 180755 (5622) |
| Rwork / Rfree | 0.181/0.214 (0.272/0.296) | 0.165/0.204 (0.291/0.314) | 0.185/0.211) (0.261/0.303) | 0.194/0.226 (0.297/0.354) |
| No. atoms | ||||
| Protein | 19121 | 19121 | 19121 | 19121 |
| RNA | 32874 | 32875 | 32875 | 32875 |
| Ligand/Ion/Water | 782 | 829 | 844 | 826 |
| B factors | ||||
| Protein | 110.3 | 94.8 | 77.9 | 99.4 |
| RNA | 97.8 | 80.2 | 66.1 | 86.5 |
| Ligand/Ion/Water | 103.4 | 83.6 | 74.6 | 87.3 |
| r.m.s. deviations | ||||
| Bond lengths (Å) | 0.004 | 0.004 | 0.005 | 0.005 |
| Bond angles (°) | 0.790 | 0.859 | 0.819 | 0.801 |
One crystal was used for each dataset. aValues in parentheses are for highest-resolution shell.
[AU: Equations defining various R values are standard and hence are no longer defined in the footnotes.]
[AU: Ramachandran statistics should be in methods section at the end of the refinement sub-section.]
[AU: Wavelength of data collection, temperature, beamline should all be in methods section.]
Codon context of m6A affects decoding dynamics
To determine the positional effects of m6A modification within the codon on tRNA decoding, we monitored translation of two six-codon long mRNA constructs, containing an m6A modification at either the 1st or 2nd base of the 4th codon, which we called Lys1 and Lys2 mRNA constructs respectively (Fig. 2a). Using these short mRNAs with an m6A-modified codon close to the start codon allowed the modified codon to enter the A site earlier in the experiment, increasing the probability of observing its translation prior to reporter dye photobleaching. Comparing the effect of m6A in the different mRNAs, the observed prolonged non-rotated state lifetimes were greatest for m6A at the first codon position (15-fold delay in accommodation compared to unmodified Lys codons), second longest for the second position (9-fold) and smallest for m6A in the third position (3-fold) (Fig. 2b, c). Prolonged non-rotated state lifetimes on m6A-modified codons indicated that the m6A modification has an effect on ternary complex initial selection and/or the kinetic proofreading step during decoding prior to peptide bond formation. In comparison, the greatest decreases in tRNALys selection rates were usually observed for single-base changes at position 2, with smallest effects at position 3 of near-cognate codon22.
We next probed the role for sequence context of m6A modifications on decoding dynamics. For m6A in E. coli, a consensus sequence of GGCC(m6A)G is reported2. To test this more native codon context in which m6A would most likely occur within E. coli, we used mRNA constructs with a C(m6A)G codon (Glutamine) or a CC(m6A) codon (Proline), which we named Gln2 and Pro3 respectively (Fig. 2a). We replaced tRNAs with Phe-(Cy5)tRNAPhe and Phe amino-acid depleted total amino-acylated tRNA mix to observe correlation between inter-subunit FRET signal and Phe-(Cy5)tRNAPhe transit during translation of these mRNAs, allowing us to score for actively translating complexes. Modifications in both the Gln or Pro slowed translation elongation: We observed a 5-fold increase in non-rotated state lifetime for the C(m6A)G Gln codon compared with the other unmodified Gln codons, and a 3-fold increase in non-rotated state lifetime for the CC(m6A) Pro codon compared to other Pro codons. Our results showed that while position of m6A within codon still delays tRNA decoding dynamics, the other bases within m6A-modified codon modulate the magnitude of effect. Particularly, replacing nearby A-U pairs with G-C pairs mitigates the m6A-induced delay in tRNA incorporation.
m6A delays distinct steps of tRNA incorporation differently
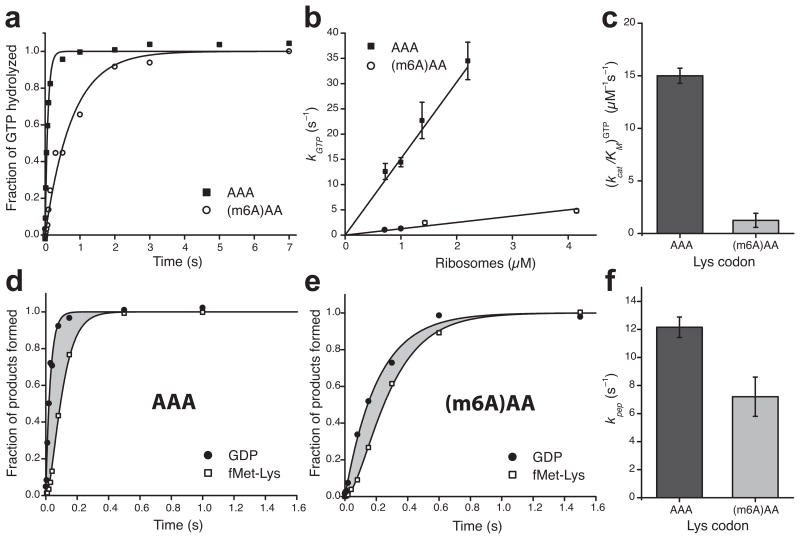
We next used bulk kinetic assays with quench-flow instrumentation in order to measure the effect of m6A on different decoding steps. Bulk kinetic assays were used to measure the ribosome-dependent rate of GTP-hydrolysis by elongation factor Tu (EF-Tu) (kGTP) and the rate of cognate tRNA accommodation plus peptidyl transfer (kpep), which can reveal the effect of m6A-modification on these processes. We measured the rate of [3H]-GTP-hydrolysis by EF-Tu after mixing EF-Tu–GTP–Lys-tRNALys TC with the 70S decoding complex in excess over TC and programmed with f[3H]Met-tRNAfMet in the P site and an AAA or (m6A)AA codon in the A site. This showed slower GTP-hydrolysis in decoding the (m6A)AA codon than decoding the AAA codon (Fig. 3a). By measuring kGTP at varying ribosome concentrations, we observed for both codons a linear increase in kGTP with respect to ribosome concentration and estimated the Michaelis-Menten parameter (kcat/KM)GTP for GTP-hydrolysis from a linear regression22, as shown in Figure 3b. The (kcat/KM)GTP-value for decoding (m6A)AA codon was 12-fold smaller than that for decoding AAA codon (Fig. 3c). In this experiment, the m6A-induced GTP-hydrolysis delay primarily prolonged the time the TC remained unbound to the ribosome, while it does not indicate a detectable difference in GTP-hydrolysis after TC bound to the ribosome.
Figure 3.
Single-base m6A-modification slows down binding of ternary complexes to the A site of ribosomes during decoding and has minor effect on the subsequent steps. (a) Kinetics of GTP hydrolysis after binding of Lys-tRNALys ternary complexes (0.3 μM) to 70S (1 μM) initiation complexes programmed with AAA or (m6A)AA in the A site. (b) Dependence of the rate of GTP-hydrolysis, kGTP, on ribosome concentration. (c) Estimates of kcat/KM-values for GTP-hydrolysis from b. (d) and (e) Kinetics of GTP-hydrolysis and dipeptide fMet-Lys formation measured simultaneously in the same experiment. The grey areas represent the total time for all subsequent steps after GTP-hydrolysis up to and including peptidyl transfer. (f) Estimates of the compounded rate constant, kpep, for the steps after GTP-hydrolysis on EF-Tu up to and including peptidyl transfer, from experiments shown in d and e. See Supplementary Data Table 1 for data in c and f. Kinetic data in a, d and e are representative of three independent experiments. Error bars in b, c, and f represent s.d. (n = 3, technical replicates) as calculated from the fitting procedure32.
To investigate whether m6A codon modification affects steps subsequent to GTP-hydrolysis, we measured the kinetics of GTP-hydrolysis and accommodation plus dipeptide fMet-Lys formation in the very same experiment (Fig. 3d, e). From the difference in the times for GTP-hydrolysis and dipeptide formation, we calculated the compounded rate constant kpep for all steps subsequent to GTP-hydrolysis up to and including peptidyl transfer22. At standard conditions kpep was slightly less than twofold smaller for (m6A)AA compared to AAA codon (Fig. 3f), revealing a defect also on tRNA accommodation plus peptidyl transfer caused by m6A. Another remarkable m6A defect was the 1.5 fold excess hydrolysis of GTP per peptidyl transfer reaction (Supplementary Fig. 3a), showing that m6A induces substantial proofreading (f = 1.5) of the cognate tRNALys compared to the unmodified case where f = 1.0. This means that under the high accuracy condition, the Michaelis-Menten parameter (kcat/KM)pep for peptide bond formation at (m6A)AA codon was reduced in relation to at the AAA codon by a factor of 12 due to reduced efficiency of GTP-hydrolysis (Fig. 3c) and by another factor of 1.5 due to m6A induced proofreading of tRNALys (Supplementary Data Table 1), amounting to an overall kinetic loss of 18.
The magnitude of the perturbation caused by m6A in the Lys codon correlated positively with the decoding accuracy during translation. In experiments with free Mg2+ concentration of 7.5 mM, which greatly reduces the accuracy of decoding compared to that at 1.3 mM Mg2+ concentration22, the Michaelis-Menten parameter (kcat/KM)pep for peptide bond formation at (m6A)AA codon was reduced in relation to at the AAA codon by a factor of 4.5 due to reduced efficiency of GTP-hydrolysis (Supplementary Fig. 4b) and by another factor of 1.3 due to m6A induced proofreading of tRNALys (Supplementary Fig. 4e), amounting to an overall kinetic loss of 6 (Supplementary Data Table 1).
Comparing methods used to probe translational dynamics
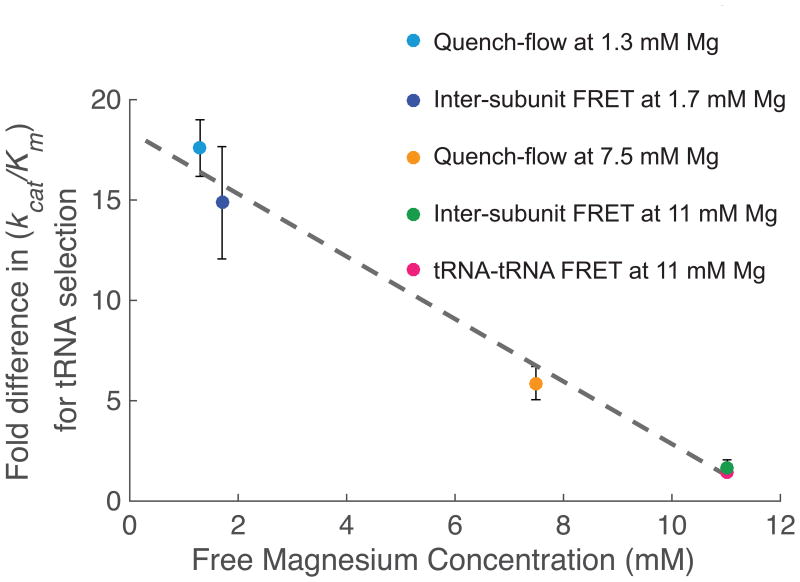
We then sought to compare the effect of m6A to tRNA incorporation measured by two different methods. Our single-molecule assay measures the time between translocation to next codon, correlated with back-rotation of ribosomal subunits and tRNA departure from E-site of ribosome, and stable tRNA accommodation correlated with rotation of ribosomal subunits, while our bulk kinetic assay measures the rate of GTP-hydrolysis and dipeptide formation. Assuming dipeptide formation occurs concurrent with the rotation of ribosomal subunits, we can compare the measured lengthening of non-rotated state due to m6A in single-molecule assays directly with the overall kinetic loss measured in bulk kinetic assays. By accounting for the difference in free magnesium concentration, which affects the accuracy and efficiency of translation, we observed linear relationship between magnesium concentration and the effect of m6A to tRNA selection (Fig. 4), analogous to relationships seen in near-cognate tRNA selection22,29.
Figure 4.
The effect of m6A in delaying tRNA incorporation scales measured across methods. Comparing fold difference in m6A-induced delay in tRNA shows agreement across different methods. Inter-subunit FRET at 1.7mM Mg data point corresponds to data from Figure 2, while Quench-flow at 1.3 mM and 7.5 mM Mg data points corresponds to data from Figure 3 and Figure 4, respectively. Inter-subunit FRET at 11 mM magnesium concentration was measured taking ratio between two fittings of single-exponential distributions to time between 50S joining event and Lys-(Cy5)tRNALys arrival event happening near simultaneously with increase in inter-subunit FRET efficiency, for both AAA and (m6A)AA (number of molecule analyzed = 113 and 80, for AAA and (m6A)AA, respectively). The error for this measurement is calculated by the propagation of error method from s.e.m calculated in these two experiments. tRNA-tRNA FRET at 11mM Mg data point has been measured as indicated in the text.
Probing the effect of m6A during early stages of decoding
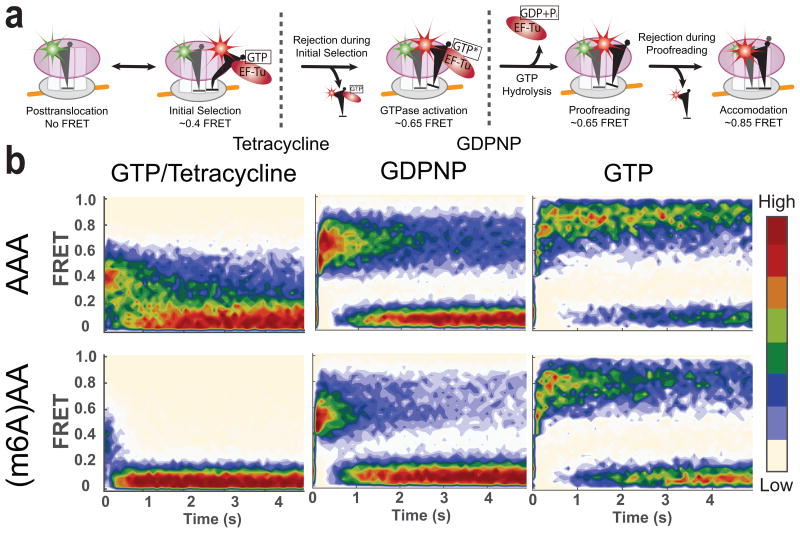
To investigate in depth the effect of m6A at distinct stages during tRNA accommodation to the ribosome, we performed single-molecule experiments tracking FRET between fluorescently labeled tRNAs23. Previously, we have identified two different intermediate stages of translational decoding prior to accommodation using chemical agents known to inhibit translation at specific steps23,30. Without any perturbation to translation, we observed a full accommodation of tRNA to the A site, indicated by high FRET state (FRET efficiency near 0.85) between the P site tRNA and the A site tRNA. By substituting GTP with GDPNP, a non-hydrolyzable analog of GTP, we observed a mid-FRET state (FRET efficiency near 0.65), which corresponds to the GTPase-active state prior to a full accommodation. Further, in the presence of tetracycline, an antibiotic known to block progression from initial selection stage, FRET values were reduced to a low-FRET state (FRET efficiency near 0.4) (Fig. 5a). By comparing evolution of tRNA-tRNA FRET signals from ribosomes programmed with either unmodified or m6A-modified Lys codon in the A site at different conditions, we tracked the effect of m6A to codon:anticodon interaction at distinct stages of translational decoding.
Figure 5.
Effect of m6A on different stages of decoding observed using tRNA-tRNA FRET. (a) A model of tRNA selection steps and corresponding approximate FRET values. Monitoring FRET between fMet-(Cy3)tRNAfMet and Lys-(Cy5)tRNALys can be used to track the tRNA selection process. (b) Contour plots of the time evolution of population FRET at various conditions, generated by superimposing all the observed FRET events (in the case of the GTP experiment (left panels), first observed event per immobilized molecule) synchronized to the start of the event defined by the FRET efficiency larger than 0.35. Contours are plotted from tan (lowest population) to red (highest population). The number of FRET events post-synchronized for each experiment is 350, 350, 948, 1573, 520 and 396 for experimental conditions with GTP–and–AAA, GTP–and–(m6A)AA, GDPNP–and–AAA, GDPNP–and–(m6A)AA, GTP–and–tetracycline–and–AAA and GTP–and–tetracycline–and–(m6A)AA, respectively.
In order to validate our tRNA-tRNA FRET assay, we first measured the effects of m6A in tRNA accommodation stage and compared the results to those obtained above by bulk kinetics and single-molecule inter-subunit FRET experiments. Using the same mRNAs as in the bulk kinetic experiments, we measured the Lys-(Cy5)tRNALys selection time to either AAA or (m6A)AA codon in the A site after mixing EF-Tu–GTP–Lys-(Cy5)tRNALys TC and the 70S decoding complex with fMet-(Cy3)tRNAfMet in the P site, where the Lys-(Cy5)tRNALys selection time was defined between the time of mixing and the time of first occurrence of the FRET value to a high-FRET state with FRET efficiency near 0.85 (ref. 23). At 11 mM free Mg2+ concentration, the average Lys-(Cy5)tRNALys selection time to (m6A)AA codon was 10.4 second compared to 7.6 second for AAA codon, an almost 1.4-fold increase. This delay in tRNA selection agrees well with the 1.6-fold delay in tRNA selection measured using the inter-subunit FRET and Lys-(Cy5)tRNALys-selection assay on the same mRNA constructs at similar Mg2+ concentration. Furthermore, the magnitude of the delay in tRNA selection also agreed with extrapolation from bulk measurements and single-molecule inter-subunit FRET and Lys-(Cy5)tRNALys-selection assay as shown in Figure 4.
After observing the agreement on the effect of m6A among tRNA-tRNA FRET, bulk kinetic assays, and inter-subunit FRET assays, we perturbed tRNA selection by either substituting GTP with GDPNP or including antibiotic tetracycline (Fig. 5a). In all three experimental conditions at high free Mg2+ concentration (11 mM), average FRET lifetimes decreased by 1.4- to 3.0-fold for (m6A)AA compared to average FRET lifetimes for AAA, which indicated that the effect of m6A precedes initial selection inhibited by tetracycline (Fig. 5b). Furthermore, m6A-modification in the A site codon slightly lowered the average FRET efficiency especially in the presence of GDPNP (Supplementary Fig. 5), mirroring a previous result for selection of near-cognate tRNA in the A site30. At low free Mg2+ concentrations (1 mM), the FRET lifetime decreases severely (Supplementary Fig. 6).
Discussion
Here, we have investigated the effect of m6A occurring within mRNA on translation using biochemical, structural and single-molecule methods. Our data show that the presence of an m6A within a codon alters cognate tRNA selection to be kinetically unfavorable. The structural data from x-ray crystallography show that a stable codon:anticodon interaction containing m6A can form within the A site with little perturbation in local or global structure, but minor steric constrains due to the modification may lead to kinetic and thermodynamic destabilization of the m6A-U pairing, as shown previously18. The greatest effect of m6A on tRNA selection occurs at the most thermodynamically unstable steps of initial selection from codon:anticodon recognition through GTP-hydrolysis by EF-Tu. m6A modification has a smaller but significant (1.5-fold) effect on proofreading after initial selection, as the ribosome evolves to the canonical accommodated codon:anticodon complex observed in our structures.
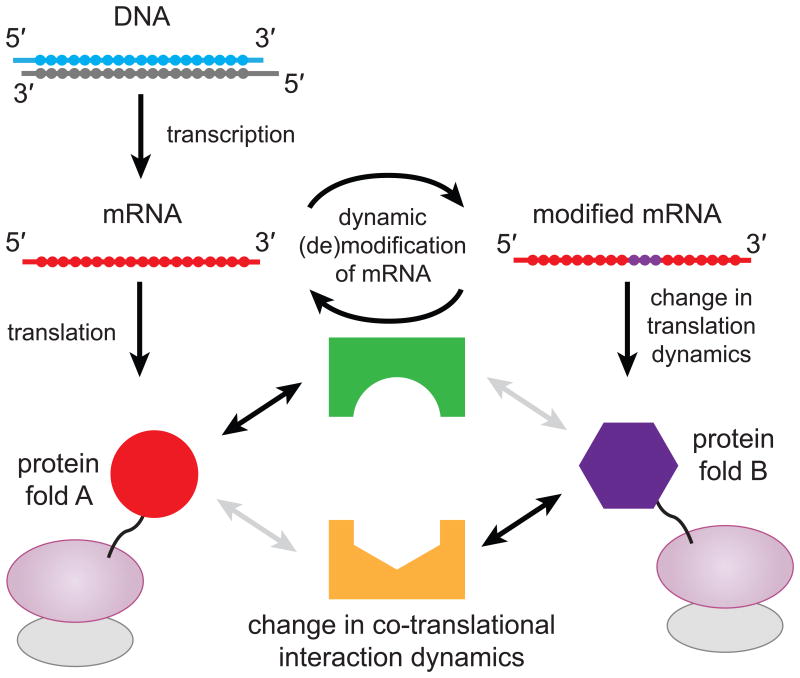
Our observations suggest that dynamic mRNA modifications within coding regions could be an important factor in controlling translation elongation dynamics locally and promptly at a post-transcriptional level. Regulating local translation elongation rate could in turn modulate coupled co-translational process such as protein folding31 and recognition of nascent peptide chain by chaperones and/or signal recognition particles (Fig. 6), possibly resulting in different functional forms or localizations of protein products from a single gene.
Figure 6.
Proposed model for protein synthesis regulation via dynamic modification of mRNA. Regulation of mRNA modification could act as a localized toggle switch between two translation elongation dynamics for a single gene. Each set of elongation dynamic could influence folding of proteins or interaction between nascent protein and other factors.
Accession Codes
Coordinates and structure factors have been deposited in the Protein Data Base under accession codes 4X62, 4X64, 4X65 and 4X66 for data sets unmodified mRNA, mRNA-1 ((m6A)AAUUU), mRNA-2 (A(m6A)AUUU) and mRNA-3 (AA(m6A)UUU) respectively.
Online Methods
Purification and crystallization of 30S ribosomal subunits
We purified and crystallized 30S ribosomal subunits from T. thermophilus HB8 strain essentially as described27,28. The m6A modified mRNA fragments (with codon sequences underlined), 5′(m6A)AAUUU3′, 5′A(m6A)AUUU3′, 5′AA(m6A)UUU3′ and native mRNA 5′AAAUUU3′ were purchased from Dharmacon. The presence of m6A modification was confirmed by mass spectrometry. The ASL , (with anticodon sequence underlined GCAGACU(mcm5s2U)UU(ms2t6A)AΨCUGC), is a generous gift of Paul Agris (University of Albany). 30S crystals were sequentially transferred to the final buffer with 26% v/v 2-methyl-2,4-pentanediol (MPD) for cryoprotection and soaks were performed in the final buffer supplemented with 200μM of each mRNA oligos, ASL and 80μM paromomycin for 48hours. Crystals were flash-frozen for data collection by plunging them directly into liquid nitrogen.
Data collection and refinement of structure obtained by x-ray crystallography
X-ray diffraction data were collected from a single crystal for all m6A modified mRNA datasets, and two crystals were used for unmodified mRNA dataset. All datasets were collected with a Pilatus 6M detector at beamline BL12-2 at the Stanford Synchrotron Radiation Lightsource (SSRL) in Menlo Park, CA. Diffraction data sets were processed with the HKL2000 package33. Coordinates of the 30S subunit structure excluding mRNA and ASL (PDB accession code 3T1H26 with additional 30S rRNA and protein modifications were used for initial rigid body refinement with Phenix34 for all data sets. After simulated-annealing refinement, individual coordinates, three group B factors per residue, and TLS parameters were refined. Potential positions of magnesium or potassium ions were compared with those in a high-resolution (2.5 Å) 30S subunit structure (PDB accession code 2VQE35) in program COOT36, and positions with strong difference density were retained. All magnesium atoms were replaced with magnesium hexahydrate. Water molecules located outside of significant electron density were manually removed. A similar refinement protocol was used for all datasets. Structure alignments were performed using the alignment algorithm of PyMOL with the default 2σ rejection criterion and five iterative alignment cycles. All X-ray crystal structure figures were produced with PyMOL37.
Reagents and buffers for single-molecule experiments
Reagents and buffers for inter-subunit FRET single-molecule experiment, including modified E. coli ribosomal subunits capable of incorporation of fluorescence dye labeled DNA oligonucleotides, translation factors (IF2, EF-Tu, EF-G, EF-Ts), S1, and aminoacyl-tRNA total tRNA mix (purchased from Sigma-Aldrich), Cy3 labeled tRNAfMet (purchased from Chemical Block Ltd, labeled at s4U8 with Cy3 maleimide purchased from GE Healthcare), Cy5 labeled tRNAPhe (purchased from Sigma-Aldrich, labeled at the elbow position (acp3U47) with Cy5 NHS ester purchased from GE Healthcare) and tRNALys (purchased from Chemical Block Ltd, labeled at the elbow position (acp3U47) with Cy5 NHS ester purchased from GE Healthcare)) were prepared and purified as described before38,39. Synthetic biotinylated mRNA used were purchased from GE Dharmacon, with m6A modification at desired base. All mRNAs had 5′-biotin followed by a 5′-UTR and Shine–Dalgarno sequence derived from gene 32 of the T4 phage (sequence described previously40), an AUG start codon, followed by the sequence of interest. All experiments were conducted in a Tris-based polymix buffer consisting of 50 mM Tris-acetate (pH 7.5), 100 mM potassium chloride, 5 mM ammonium acetate, 0.5 mM calcium acetate, 5 mM magnesium acetate, 0.5 mM EDTA, 5 mM putrescine-HCl, and1 mM spermidine. For single-molecule FRET experiments, 4 mM GTP (or 4 mM GDPNP, where applicable) were added and were performed at 20 °C.
Single-molecule inter-subunit FRET / tRNA-transit experiment on ZMW instrument
The 3′-dye labeled DNA oligonucleotides (labeled with Cy3B or tandem BHQ-2-BHQ-2) complementary to the mutant ribosome hairpins were ordered from Trilink. Immediately before each experiment, purified 30S and 50S ribosomal subunits (final concentration, 1 mM) were mixed in a 1:1 ratio with the 39 dye-labeled oligonucleotides specific for the hairpin extensions in each subunit for 37 °C for 10 min and then at 30 °C for 20 min in Polymix buffer system. The 30S pre-initiation complexes (PICs) were formed as described by incubating the following at 37 °C for 5 min: 0.25 mM Cy3B-30S, pre-incubated with stoichiometric S1, 1 mM IF2, 1 mM fMet-tRNAfMet, 1 mM mRNA, and 4 mM GTP to form 30S PICs in the polymix buffer.
Before use, we pre-incubate a SMRT Cell V3 from Pacific Biosciences (Menlo Park, CA, USA), a zero-mode waveguide (ZMW) chip, with a 1 mg/ml Neutravidin solution in 50 mM Tris-acetate pH 7.5 and 50 mM KCl at room temperature for 5 min. The cell is then washed with Buffer 6 (50 mM Tris-acetate pH 7.5, 100 mM potassium chloride, 5 mM ammonium acetate, 0.5 mM calcium acetate, 5 mM magnesium acetate, and 0.5 mM EDTA). We then dilute the 30S PICs with our Tris-based polymix buffer containing 1 mM IF2 and 4 mM GTP down to 10 nM PIC concentration. The diluted PICs are then loaded into the SMRT cell at room temperature for 3 min to immobilize the 30S PICs into the ZMW wells. We wash away excessive unbound material with our Tris-based polymix buffer containing 1 mM IF2, 4 mM GTP, 1 mM Trolox, and a PCA/PCD oxygen scavenging system (2.5 mM 3,4-dihydroxybenzoic acid and 250 nM protocatechuate deoxygenase)41. We formed ternary complexes between total charged E. coli tRNAs and EF-Tu–GTP as previously described. Cy5-labeled Lys and Phe aminoacyl-tRNA–EF-Tu–GTP ternary complexes for experiments with Lys1, Lys2 and Lys3 mRNA constructs and Cy5-labeled Phe and total tRNA charged Phe-depleted amino-acids for experiments with Gln2 and Pro3 mRNA constructs were pre-formed by incubating (2 min at 37 °C) the amicoacyl-tRNA with five-fold excess of EF-Tu, GTP (1 mM), PEP (3 mM) and EF-Ts (40 mM) in Polymix. The ternary complexes (final concentration 10.8nM for Lys-(Cy5)tRNALys, 17.5nM for Phe-tRNAPhe, 27nM for Phe-(Cy5)tRNAPhe and 750nM for Phe-depleted total amico-acylated tRNA) were added to BHQ-2-50S (200 nM), EF-G (120 nM), IF2 (1 mM), GTP (4 mM), 2.5 mM Trolox, and the oxygen scavenging system (PCA/PCD) to form a delivery mix in polymix buffer. Before starting an experiment, the SMRT Cell is loaded into a modified PacBio RS sequencer. At the start of the elongation experiment, the instrument illuminates the SMRT cell with a green laser and then delivers 20 μl of a delivery mixture onto the cell surface at t = 10 second.
Single-molecule tRNA-tRNA FRET experiments on TIRFM instrument
The preparation for tRNA-tRNA FRET experiments was the same as ZMW experiments with the following exception: Wash and delivery mixture contains extra 10 mM Mg(OAc)2 (nearly 11mM free magnesium concentration, where applicable), 1 mM IF2, 10.8 nM Lys-(Cy5)tRNALys ternary complexes, 4mM of GDPNP (instead of GTP, where applicable) and 100 μM of tetracycline (where applicable).
TIRF and ZMW instrumentation and data analysis
Single-molecule inter-subunit FRET and Lys-(Cy5)tRNALys-transit experiment were conducted using a commercial PacBio RS sequencer that we modified to allow the collection of single-molecule fluorescence intensities from individual ZMW wells about 130 nm in diameter in 4 different dye channels corresponding to Cy3, Cy3.5, Cy5, and Cy5.5. The RS sequencer has two lasers for dye excitation at 532 nm and 632 nm. In all experiments, data was collected at 10 frames per second (100 ms exposure time) for 8 min. The energy flux of the green laser is 0.60 mW per mm2 and the red laser is at 0.10 mW per mm2.
The instrumental setup for TIRFM was same as previously described38,40. For excitation of Cy3 (or Cy3B), a diode-pumped solid-state 532 nm laser at 1 kW cm-2 was used. A Quad-View device (Photometrics) separated fluorescence emissions into four channels where two of the channels were filtered to correspond to emission spectrum of Cy3B and Cy5 respectively. The signal was then projected onto two 512×512 pixel quadrants of an EMCCD camera (Andor Technology). Videos were recorded (typically at 10 frames per second with a total of 1500 frames collected, but for experiments with tetracycline, we used 20 frames per second with a total 2400 frames) using the MetaMorph software package (Molecular Devices).
Data analyses for all experiments are conducted with MATLAB (MathWorks) scripts written in-house21,24. Briefly, traces from either the ZMW wells or immobilized complexes on TIRFM slide are initially selected based on fluorescence intensity, fluorescence lifetime and the changes in intensity. Exhaustively filtered traces exhibiting intersubunit FRET or single-molecule binding signals are then selected for further data analysis. The FRET states are assigned as previously described based on a hidden Markov model based approach and visually corrected. For tRNA-tRNA FRET data, FRET value less than 0.35 was assigned to no-FRET state, as before23. For tetracycline experiments, FRET events with FRET value higher than 0.5 were rejected to account for less than complete binding of tetracycline to ribosomes. n in single-molecule experiments was determined by the number of molecules analyzed. All lifetimes were fitted to a single-exponential distribution using maximum-likelihood parameter estimation in MATLAB.
Bulk kinetic experiments
Low Mg2+ buffer was Polymix buffer42, containing 95 mM KCl, 5 mM NH4Cl, 0.5 mM CaCl2, 8 mM putrescine, 1 mM spermidine, 5 mM potassium phosphate, 1 mM DTE and 5 mM Mg(OAc)2, supplemented with energy regeneration components containing 1 mM ATP + 1 mM GTP for the ribosome mixture or 2 mM ATP for the ternary complex mixture, 10 mM phosphoenolpyruvate (PEP), 50 μg/ml pyruvate kinase (PK), and 2 μg/ml myokinase (MK). High Mg2+ buffer contained 10 mM extra Mg(OAc)2 in addition to low Mg2+ buffer. Assuming PEP chelates Mg2+ with a Kd of 6 mM and one ATP or GTP molecule chelates one Mg2+ (ref. 27), the free Mg2+ concentrations were calculated as 1.3 and 7.5 mM for low and high Mg2+ buffers, respectively.
Fast kinetic measurements were performed at 20 °C in a temperature controlled quench-flow instrument (RQF-3; Kintek Corp.), in which ribosome and ternary complex mixtures were rapidly mixed and the reactions were stop by quenching with 17 % (final) formic acid at different incubation times. Ribosome and ternary complex mixtures were prepared as described22 except here [3H]GTP instead of [3H]GDP was used for preparing ternary complexes. Ribosome mixture contained 70S ribosomes (variable concentrations as indicated in the experiments), f[3H]Met-tRNAfMet (1.2 × ribosomes), mRNA (encoding AUG-AAA-Stop or AUG-(m6A)AA-Stop; 1.5 × ribosomes), IF1 (1.5 × ribosomes), IF2 (0.5 × ribosomes) and IF3 (1.5 × ribosomes). Ternary complex mixture contained E. coli tRNALys (4 μM), 0.5 μM EF-Tu, 0.5μM [3H]GTP, 0.2 mM Lys and 1.5 units/μL Lys-tRNA synthetase. The extend of [3H]GDP and [3H]fMet-Lys formation was quantified by HPLC equipped with a β-RAM model 4 radioactivity detector (IN/US Systems) as described22. Rates of GTP-hydrolysis (kGTP) and dipeptide formation (kdip) were estimated by fitting the data into a single exponential model and a 2-step exponential model, respectively, as described32. Ribosomes were always in excess over ternary complexes so that rates were limited by ribosome concentration.
To measure proofreading during tRNA selection, two ribosome mixtures (1 μM), one displaying AAA and the other one displaying (m6A)AA codon in A site, were mixed with the same ternary complex mixture (0.5 μM) in parallel. The lower plateau of dipeptide fMet-Lys formation for (m6A)AA indicates the rejection of tRNALys after GTP hydrolysis. Proofreading factor f was given by the ratio of maximal yield of fMet-Lys formed between reactions with AAA and (m6A)AA codons.
To estimate the compound rate constant, kpep, for all subsequent steps after GTP hydrolysis on EF-Tu up to and including peptidyl transfer, we measured GTP hydrolysis and dipeptide formation in the very same experiment. At low Mg2+ condition, experiments were done at 2 μM of ribosomes programmed with AAA or 4 μM of ribosomes programmed with (m6A)AA, and 0.5 μM of ternary complexes. At high Mg2+ condition, experiments were done at 0.7 μM ribosomes programmed with AAA or (m6A)AA, and 0.2 μM ternary complexes. Taking the proofreading factor f into account29, kpep was calculated as 1/kpep = f · (1/kdip − 1/kGPT).
Supplementary Material
Acknowledgments
This work was supported by US National Institutes of Health (NIH) grants GM51266 and GM099687 to J. D. Puglisi; by grants from the Knut and Alice Wallenberg Foundation (RiboCORE) and the Swedish Research Council and the Human Frontier Science Program to M. Ehrenberg; by US NIH grants GM111858 to S. E. O'Leary; by grants from the Israel Science Foundation (ISF grant no. 1667/12), the Israeli Centers of Excellence (I-CORE) Program (ISF grants no. 41/11 and no. 1796/12), and the Ernest and Bonnie Beutler Research Program to G. Rechavi; by a Human Frontier Science Program long-term fellowship to D. Dominissini; by a Stanford Bio-X fellowship to J. Choi. Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource (SSRL), a national user facility operated by Stanford University on behalf of the US Department of Energy, US Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the US Department of Energy, US Office of Biological and Environmental Research, US National Institutes of Health, US National Center for Research Resources, US Biomedical Technology Program, and the US National Institute of General Medical Sciences. G. Rechavi is a supported as a member of the Sagol Neuroscience Network and by the Kahn Family Foundation support. We thank Paul Agris in University of Albany for a human ASL reagent, and members of Puglisi lab for discussion. J. Choi thanks J. B. Choi for support.
Footnotes
Author Contributions: J. Choi, K. Ieong, and H. Demirci performed all the experiments and the data analysis; J. Choi performed single-molecule experiments; K. Ieong performed bulk-kinetic experiments; H. Demirci performed x-ray crystallography. D. Dominissini and G. Rechavi provided reagents and conceived this project with J. Choi, K. Ieong, H. Demirci, J. Chen, M. Ehrenberg and J. D. Puglisi. J. Chen, A. Petrov, and A. Prabhakar helped reagent preparations. J. Choi, K. Ieong, H. Demirci, S. E. O'Leary, M. Ehrenberg and J. D. Puglisi wrote manuscript.
References
- 1.Moore M. From birth to death: the complex lives of eukaryotic mRNAs. Science (80- ) 2005;309:1514. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- 2.Deng X, et al. Widespread occurrence of N6-methyladenosine in bacterial mRNA. Nucleic Acids Res. 2015:1–11. doi: 10.1093/nar/gkv596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dominissini D, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–6. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 4.Meyer KD, et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–46. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang X, et al. N6-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell. 2015;161:1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bokar JA. The biosynthesis and functional roles of methylated nucleosides in eukaryotic mRNA. Top Curr Genet. 2005;12:141–178. [Google Scholar]
- 7.Karikó K, Buckstein M, Ni H, Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–75. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 8.Zheng G, Dahl J, Niu Y, Fedorcsak P. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Batista PJ, et al. m6A RNA Modification Controls Cell Fate Transition in Mammalian Embryonic Stem Cells. Cell Stem Cell. 2014;15:707–719. doi: 10.1016/j.stem.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fustin JM, et al. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell. 2013;155:793–806. doi: 10.1016/j.cell.2013.10.026. [DOI] [PubMed] [Google Scholar]
- 11.Jia G, Fu Y, Zhao X, Dai Q, Zheng G. N6-Methyladenosine in Nuclear RNA is a Major Substrate of the Obesity-Associated FTO. Nat Chem Biol. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–20. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, et al. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol. 2014;16:191–8. doi: 10.1038/ncb2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao X, et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014;24:1403–19. doi: 10.1038/cr.2014.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Niu Y, et al. N6-methyl-adenosine (m6A) in RNA: an old modification with a novel epigenetic function. Genomics Proteomics Bioinformatics. 2013;11:8–17. doi: 10.1016/j.gpb.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jia G, Fu Y, He C. Reversible RNA adenosine methylation in biological regulation. Trends Genet. 2013;29:108–15. doi: 10.1016/j.tig.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwartz S, et al. High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell. 2013;155:1409–1421. doi: 10.1016/j.cell.2013.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roost C, et al. Structure and thermodynamics of N6-methyladenosine in RNA: a spring-loaded base modification. J Am Chem Soc. 2015;137:2107–15. doi: 10.1021/ja513080v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu N, et al. N6-methyladenosine-dependent RNA structural switches regulate RNA–protein interactions. Nature. 2015;518:560–564. doi: 10.1038/nature14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spitale RC, et al. Structural imprints in vivo decode RNA regulatory mechanisms. Nature. 2015;519:486–490. doi: 10.1038/nature14263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen J, et al. High-throughput platform for real-time monitoring of biological processes by multicolor single-molecule fluorescence. Proc Natl Acad Sci U S A. 2014;111:664–9. doi: 10.1073/pnas.1315735111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johansson M, Zhang J, Ehrenberg M. Genetic code translation displays a linear trade-off between efficiency and accuracy of tRNA selection. Proc Natl Acad Sci U S A. 2012;109:131–6. doi: 10.1073/pnas.1116480109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blanchard SC, Jr, R LG, Kim HD, Chu S, Puglisi JD. tRNA selection and kinetic proofreading in translation. Nat Struct Mol Biol. 2004;11:1008–1014. doi: 10.1038/nsmb831. [DOI] [PubMed] [Google Scholar]
- 24.Chen J, Petrov A, Tsai A, O'Leary SE, Puglisi JD. Coordinated conformational and compositional dynamics drive ribosome translocation. Nat Struct Mol Biol. 2013;20:718–27. doi: 10.1038/nsmb.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uemura S, et al. Real-time tRNA transit on single translating ribosomes at codon resolution. Nature. 2010;464:1012–7. doi: 10.1038/nature08925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vendeix FaP, et al. Human tRNA(Lys3)(UUU) is pre-structured by natural modifications for cognate and wobble codon binding through keto-enol tautomerism. J Mol Biol. 2012;416:467–85. doi: 10.1016/j.jmb.2011.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Demirci H, et al. Modification of 16S ribosomal RNA by the KsgA methyltransferase restructures the 30S subunit to optimize ribosome function. RNA. 2010;16:2319–2324. doi: 10.1261/rna.2357210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wimberly BT, et al. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 29.Johansson M, Bouakaz E, Lovmar M, Ehrenberg M. The Kinetics of Ribosomal Peptidyl Transfer Revisited. Mol Cell. 2008;30:589–598. doi: 10.1016/j.molcel.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 30.Lee T, Blanchard SC, Kim HD, Puglisi JD, Chu S. The role of fluctuations in tRNA selection by the ribosome. Proc Natl Acad Sci. 2007;104:13661–13665. doi: 10.1073/pnas.0705988104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim SJ, et al. Translational tuning optimizes nascent protein folding in cells. Science (80- ) 2015;348:1–5. doi: 10.1126/science.aaa3974. [DOI] [PubMed] [Google Scholar]
- 32.Johansson M, et al. pH-sensitivity of the ribosomal peptidyl transfer reaction dependent on the identity of the A-site aminoacyl-tRNA. Proc Natl Acad Sci U S A. 2011;108:79–84. doi: 10.1073/pnas.1012612107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 34.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr Sect D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kurata S, et al. Modified uridines with C5-methylene substituents at the first position of the tRNA anticodon stabilize U??G wobble pairing during decoding. J Biol Chem. 2008;283:18801–18811. doi: 10.1074/jbc.M800233200. [DOI] [PubMed] [Google Scholar]
- 36.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr Sect D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schrödinger. The PyMOL Molecular Graphics System, Version 1.7.4. Schrödinger, LLC; 2015. [Google Scholar]
- 38.Marshall RA, Dorywalska M, Puglisi JD. Irreversible chemical steps control intersubunit dynamics during translation. Proc Natl Acad Sci U S A. 2008;105:15364–15369. doi: 10.1073/pnas.0805299105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dorywalska M, et al. Site-specific labeling of the ribosome for single-molecule spectroscopy. Nucleic Acids Res. 2005;33:182–189. doi: 10.1093/nar/gki151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aitken CE, Puglisi JD. Following the intersubunit conformation of the ribosome during translation in real time. Nat Struct Mol Biol. 2010;17:793–800. doi: 10.1038/nsmb.1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aitken CE, Marshall RA, Puglisi JD. An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments. Biophys J. 2008;94:1826–1835. doi: 10.1529/biophysj.107.117689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kurland C, Ehrenberg M. Optimization of translation accuracy. Prog Nucleic Acid Res Mol Biol. 1984;31:191–219. doi: 10.1016/s0079-6603(08)60378-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.