Abstract
The transcription factor BCL11B plays essential roles during development of the immune, nervous, and cutaneous systems. Here we show that BCL11B is expressed in both osteogenic and sutural mesenchyme of the developing craniofacial complex. Bcl11b−/− mice exhibit increased proliferation of osteoprogenitors, premature osteoblast differentiation, and enhanced skull mineralization leading to synostoses of facial and calvarial sutures. Ectopic expression of Fgfr2c, a gene implicated in craniosynostosis in mice and humans, and that of Runx2 was detected within the affected sutures of Bcl11b−/− mice. These data suggest that ectopic expression of Fgfr2c in the sutural mesenchyme, without concomitant changes in the expression of FGF ligands, appears to induce the RUNX2-dependent osteogenic program and craniosynostosis in Bcl11b−/− mice.
Keywords: BCL11B/CTIP2, craniofacial development, craniosynostosis, midfacial hypoplasia, Fgfr2c, Runx2, Twist1
Introduction
Flat bones of the mammalian skull and face develop by intramembranous ossification of mesenchyme derived from the mesoderm and neural crest cells (NCCs) (Jiang et al., 2002). In this process, mesenchymal cells condense, differentiate, and form ossification centers (Opperman, 2000; Rice, 2008). Craniofacial sutures are formed within the margins between developing bones. While human sutures remain patent through early life stages, these tissues harbor proliferating osteoprogenitors that give rise to osteoblasts at the osteogenic fronts of skull bones. Craniofacial sutures serve as important sites of skull growth during fetal development and into young adulthood, when sutures undergo ossification (Rice, 2008).
Suture development is precisely controlled and synchronized with brain growth. Premature osteoblast differentiation and suture ossification underlie the condition known as craniosynostosis, which occurs in 1 out of 2,200 live births. Craniosynostosis is associated with restricted skull expansion, midfacial hypoplasia (MFH), increased intracranial pressure, and craniofacial dysmorphologies, all of which may negatively impact respiration, vision, hearing, and cognition (Nie, 2005; Purushothaman et al., 2011; Rosenberg et al., 1997).
Activating mutations within the fibroblast growth factor receptor 2 isoform IIIc (FGFR2c) locus are the most common cause of syndromic craniosynostosis, of which the most prevalent forms are Crouzon and Apert syndromes (Cunningham et al., 2007; Purushothaman et al., 2011). FGFRs act through protein kinase C (PKC) and mitogen-activated protein kinase (MAPK) pathways to stimulate or de-repress the function of transcription factor RUNX2, a master regulator of osteogenesis (Fitzpatrick, 2013; Kim et al., 2003a; Park et al., 2010). As a result, constitutive activation of FGFR2c leads to induction of the RUNX2-dependent osteogenic program and premature ossification of sutures. However, little is known about molecular control of FGFR2c expression during craniofacial development or in craniosynostosis.
We demonstrated previously that the transcription factor BCL11B regulates expression of components of the FGF signaling pathway during mouse incisor development (Kyrylkova et al., 2012a). Our present analyses of NCC-specific and germline Bcl11b mutants (Bcl11bncc−/− and Bcl11b−/−, respectively) reveal that BCL11B also plays an essential role in mouse craniofacial development and maintenance of sutural patency.
Materials and Methods
Mouse Lines
Mice carrying floxed allele of Bcl11b (Bcl11fl/fl; same as Bcl11L2/L2) and germ-specific deletion of Bcl11b (Bcl11b−/−) have been described previously (Golonzhka et al., 2009). Neural crest-specific deletion of Bcl11b (Bcl11bncc−/−; same as Bcl11bmes−/−) was achieved by crossing Bcl11fl/fl mice with the Wnt1-cre deleter line (Danielian et al., 1998; Kyrylkova et al., 2012a). Animal experiments were approved by the Oregon State University Institutional Animal Care and Use Committee, protocol 4279.
Micro-CT Analysis
Mouse skulls were scanned using a Scanco μCT40 scanner (Scanco Medical AG, Basserdorf, Switzerland) at a voxel size of 12×12×12 µm (for E16.5-P5) or 16×16×16 µm (for P10-P21), 55 kVp X-ray voltage, 145 µA intensity, and 200 ms integration time. Filtering parameters sigma and support were set to 0.8 and 1, respectively. For evaluation of total skull bone volume (BV) and visualization of sutures, bone segmentation was conducted at a threshold (scale, 0–1000) of 115 (for E16.5-P0), 135 (for P5-P14), or 145 (for P21). Three to six skulls were scanned for each genotype within each age group. For evaluation of BV in frontal and parietal bones in E16.5, E18.5, and P0 mice, whole skull scan data were segmented at a threshold of 115 (scale, 0–1000) and the frontal and parietal bones were selected and quantified. An unpaired, two-tailed Student's t-test was performed using GraphPad Prism software to determine statistical significance.
Histological Analyses
Von Kossa staining was performed according to standard protocols. RNA in situ hybridization (ISH) and immunohistochemistry using anti-BCL11B (Abcam, 1:300) were performed on 16 µm-thick sagittal (for coronal suture) and horizontal (for facial sutures) sections using anti-rat biotin and HRP-streptavidin conjugate (Jackson) as described previously (Kyryachenko et al., 2012; Kyrylkova et al., 2012b). At least three mice were used for each genotype and experiment. At least ten serial sections across the facial and coronal sutures were analyzed for each experiment.
BrdU labeling and detection (2 hours after injection) was performed using anti-BrdU antibody (Accurate Chemical, 1:100) on 16 µm-thick horizontal sections as described previously (Kyryachenko et al., 2012; Kyrylkova et al., 2012b). Images of serial sections from both control and mutant heads were aligned across the entire face in order to compare sections from the same sectioning plane. Images with the premaxillary osteogenic mesenchyme (the tissue with the most striking difference) were used for the subsequent analysis. In order to distinguish the premaxillary osteogenic mesenchyme on the sections stained for BrdU, adjacent sections were stained using Runx2 probe and superimposed to outline the borders of future premaxilla. At least five serial sections across the facial and coronal sutures from three mice were quantified per each genotype and experiment. The BrdU index was calculated as a mean relative amount of BrdU-positive cells as a fraction of total, hematoxylin-positive cells. An unpaired, two-tailed Student's t-test was performed using GraphPad Prism software to determine statistical significance.
Quantitative reverse transcription (qRT)-PCR analyses
Frontal and parietal bones of P21 mice and facial tissue of E14.5 embryos were dissected and stored in RNAlater reagent (Qiagen) at 4°C until processed. Total RNA was extracted using TRIzol reagent (Invitrogen) followed by purification using RNeasy spin columns (Qiagen). Synthesis of cDNA was carried out using random hexamer primers and SuperScript III kit (Invitrogen). The resulting cDNA was amplified with gene-specific primers using QuantiTect SYBR Green PCR kit (Qiagen) on a 7500 Real-Time PCR System (Applied Biosystems). Amplification of all targets was normalized to that of the housekeeping gene Hprt. All reactions were performed in triplicate; at least three mice were used for each genotype and experiment. An unpaired, two-tailed Student's t-test was performed using GraphPad Prism software to determine statistical significance. Primer sequences for amplification reactions are shown in Table 1.
Table 1.
Primer sequences used for quantitative PCR
| Gene | Forward primer (5'…3') | Reverse primer (5'…3') |
|---|---|---|
| Bsp | GGGAGGCAGTGACTCTTCAG | CTGGTCTTCATTCCCCTCAG |
| Fgfr2c | GTGCTTGGCGGGTAATTCTA | GGAAGCCGTGATCTCCTTCT |
| Hprt | GTTAAGCAGTACAGCCCC | AGGGCATATCCAACAACA |
| Il1b | AAGATGAAGGGCTGCTTCCAA | TGAAGGAAAAGAAGGTGCTCATG |
| Il6 | GAGGATACCACTCCCAACAGACC | AAGTGCATCATCGTTGTTCATACA |
| Runx2 | CGGTAACCACAGTCCCATCT | ACTGGCGGTGCAACAAGAC |
| Tnf | CTGTAGCCCACGTCGTAGCA | GTGTGGGTGAGGAGCACGTA |
Results and Discussion
NCC and Germline Deletion of Bcl11b Leads to Craniofacial Synostoses
We previously generated a mouse line conditionally null for Bcl11b expression in NCC-derived mesenchyme (Bcl11bncc−/− in current manuscript) using Wnt1-cre deleter strain (Danielian et al., 1998; Kyrylkova et al., 2012a). Within the skull, NCCs give rise to the facial skeleton, frontal, and squamosal bones as well as contribute to the interparietal bone and base of the skull (Jiang et al., 2002; Pietri et al., 2003).
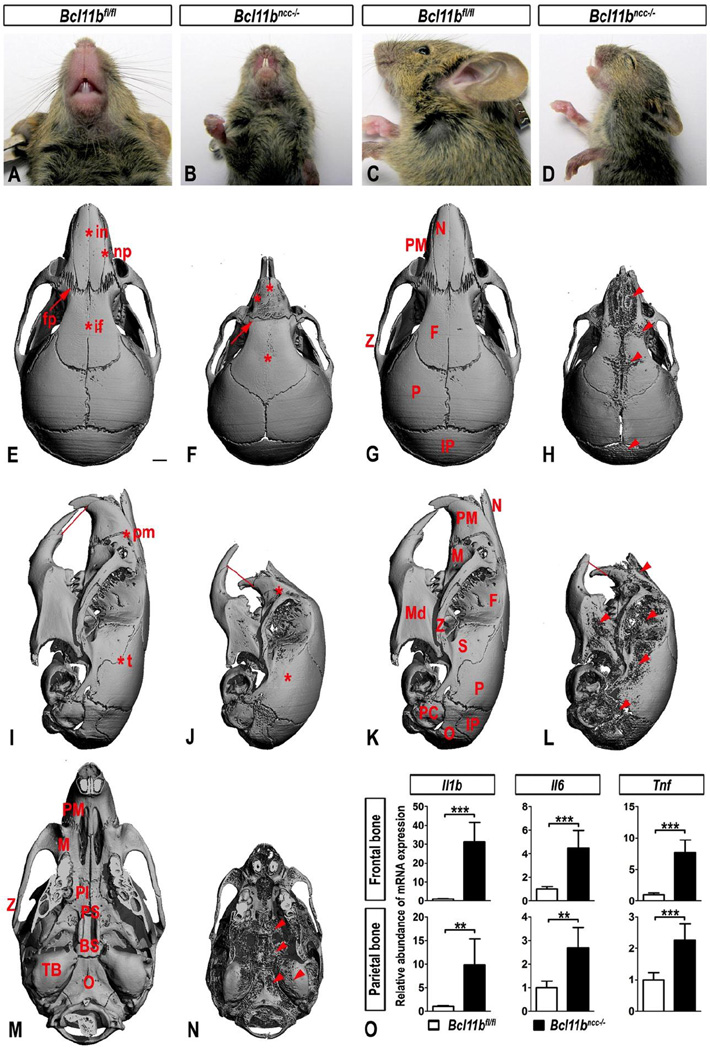
Bcl11bncc−/− mice (unlike Bcl11b−/− mice) survived after birth for approximately three weeks (post-natal day 21; P21). At this age, Bcl11bncc−/− mice exhibited abnormally small and misshapen heads, severe MFH, and malocclusion (Fig. 1A–D). We observed fusion of multiple craniofacial sutures, including the internasal, naso-premaxillary, interfrontal, premaxillary-maxillary, and temporal sutures. In addition, the Bcl11bncc−/− premaxillary-frontal suture exhibited abnormal morphology and lacked its canonical, fractal interdigitation (Fig. 1E, F, I, J). Some Bcl11bncc−/− skulls at P21 were characterized by a high degree of porosity, which was generalized and not limited to NCC-derived bones, (Fig. 1G, H, K, L, M, N). The bone porosity phenotype of Bcl11bncc−/− mice was accompanied by and possibly secondary to synostosis-induced inflammation, as expression of pro-inflammatory cytokines Il1b, Il6, and Tnf, increased dramatically in both frontal and parietal bones of the mutants at P21 (Fig. 1O).
Fig. 1. Bcl11bncc−/− mice exhibit craniosynostosis and variable degree of bone porosity at three weeks of age.
(A–D) Ventral and lateral views of mouse heads show abnormal shape, short snouts, and severe malocclusion in Bcl11bncc−/− mice at P21. (E–N) Dorsal, lateral, and ventral views of the micro-CT images of control (Bcl11bfl/fl2) and Bcl11bncc−/− mouse skulls at P21. Red lines indicate dental occlusion. Asterisks denote sutures affected by synostosis in Bcl11bncc−/− skulls and corresponding patent sutures in the control mice. Arrow points to the lack of fractal interdigitation in Bcl11bncc−/− fronto-premaxillary suture. Arrowheads point to increased porosity in multiple Bcl11bncc−/− skull bones. Bones affected by porosity: BS, basisphenoid; F, frontal; IP, interparietal; M, maxillary; Md, mandible; N, nasal; O, occipital; P, parietal; Pl, palatine; PC, periotic capsule; PM, premaxillary; PS, presphenoid; S, squamosal; TB, tympanic bulla; Z, zygomatic. Sutures affected by synostosis: fp, fronto-premaxillary; if, interfrontal; in, internasal; np, naso-premaxillary; pm, premaxillary-maxillary; t, temporal. Scale bars: 1 mm. (O) qRT-PCR analysis indicates up-regulated expression of pro-inflammatory cytokines in parietal and frontal bones of Bcl11bncc−/− skulls. Error bars indicate standard deviation. ** denotes statistical significance at p ≤ 0.01; *** denotes statistical significance at p ≤ 0.001 (n = 4 for each genotype).
Postnatal Bcl11bncc−/− mice exhibit severe MFH, a condition that often accompanies syndromic craniosynostosis in humans. Until recently, the most common theory posited that MFH arose from premature ossification of one or more sutures at the cranial base (Rosenberg et al., 1997; Stewart et al., 1977). However, analyses of mice harboring constitutively active mutations in Fgfr1 and Fgfr2 genes, which underlie Pfeiffer and Apert syndromes, respectively, revealed that MFH is a consequence of synostoses of facial sutures (Purushothaman et al., 2011). Consistent with this, Bcl11bncc−/− mice did not exhibit premature ossification of the cranial base (Figs. 1M, N and S1), but did exhibit synostoses of facial skeleton, including premaxillary-maxillary and naso-premaxillary sutures (Fig. 1E, F, I, J), further supporting the role of facial skeletal pathology in etiology of MFH. In comparison to the known biology of cranial sutures, development of facial sutures is poorly understood. Growth at the osteogenic fronts of the facial bones is similar to that of the cranial vault bones, but facial bones differ in that they overlie the cartilaginous elements of the facial complex rather than the dura mater. Moreover, development of some facial sutures, such as fronto-premaxillary, is characterized by complex interdigitation patterns (Fig. 1E) that are not observed within any sutures of the cranial vault (Nelson and Williams, 2004). Therefore, Bcl11bncc−/− mice may serve as a valuable model to study pathogenesis of facial synostoses and development of MFH.
We traced back the developmental dynamics of the conditional knockout phenotype by conducting micro-CT analyses of multiple Bcl11bncc−/− skulls at different postnatal stages. We observed development of four major craniofacial phenotypes from P5 to P14 in Bcl11bncc−/− skulls (see legend of Fig. S1): (1) synostoses of facial sutures, which were completely fused throughout post-natal development, and progressive fusion of temporal, fronto-maxillary, internasal, and interfrontal sutures (middle row of Fig. S1); (2) expansions of sagittal, lambdoid, and coronal sutures, which appeared to be of a compensatory nature (see brackets in Fig. S1A, B, G, H); (3) complete obliteration of the interdigitated pattern of the naso-premaxillary suture in the Bcl11bncc−/− skulls (Fig. S1G, H, M, N); and (4) bone porosity that began in the anterior region of frontal bones and at the base of Bcl11bncc−/− skulls and progressively worsened throughout the postnatal period to affect nearly every bone of the mutant head (Fig. S1). Cranial bone porosity in Bcl11bncc−/− mice did not appear to be the result of bone malformation because the morphologies of Bcl11bncc−/− mutant and wild-type bones were largely indistinguishable at birth (Fig. 2).
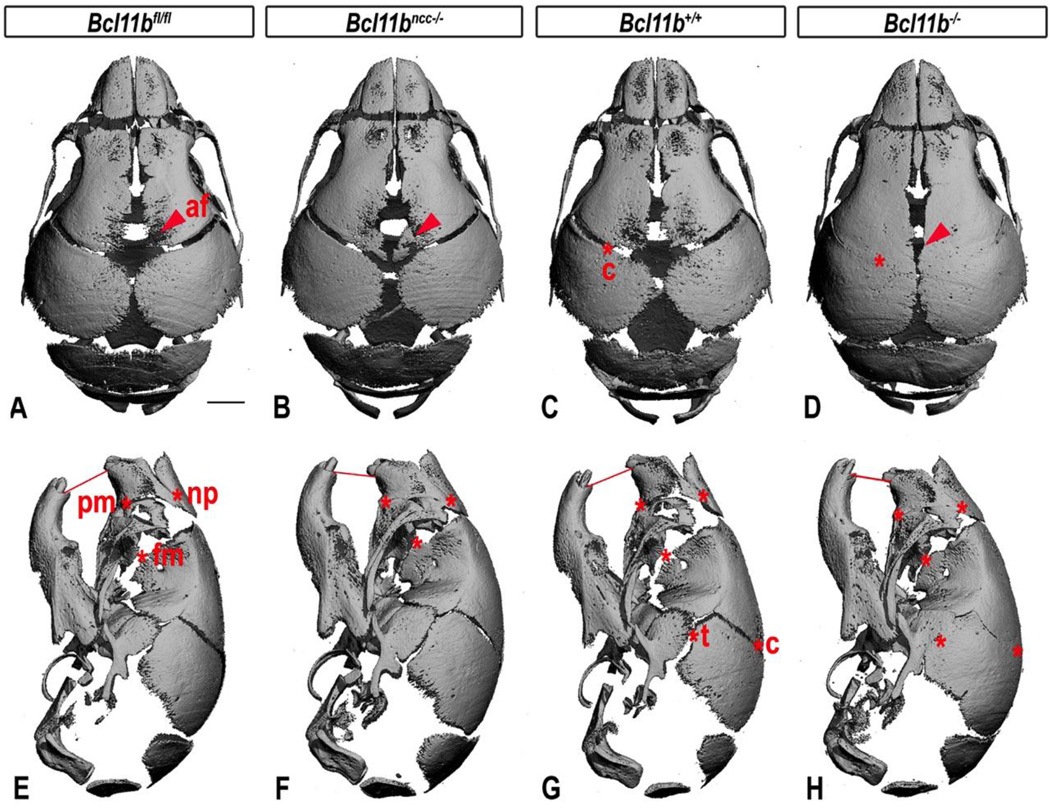
Fig. 2. Craniofacial defects of Bcl11bncc−/− and Bcl11b−/− mice at birth.
Dorsal and lateral views of the micro-CT images of control (Bcl11bfl/fl and Bcl11b+/+), Bcl11bncc−/−, and Bcl11b−/− mouse skulls at P0. Red lines indicate dental occlusion. Asterisks denote sutures affected by synostosis in Bcl11bncc−/− and Bcl11b−/− skulls and corresponding patent sutures in the control mice. Arrowheads indicate increased mineralization and reduced anterior fontanelle (af) within Bcl11bncc−/− and Bcl11b−/− calvaria. Affected sutures: c, coronal; fm, fronto-maxillary; np, naso-premaxillary; pm, premaxillary-maxillary; t, temporal. Scale bars: 1 mm.
Premaxillary-maxillary and naso-premaxillary sutures in Bcl11bncc−/− mice were already fused at birth, at which time mild MFH and a reduced sutural field between frontal and maxillary bones were also evident (Fig. 2A, B, E, F). The early onset of the craniofacial phenotype of Bcl11bncc−/− mice led us to examine Bcl11b−/− mice, which harbor a germline deletion of Bcl11b and die shortly after birth (Golonzhka et al., 2009). Bcl11b−/− skulls exhibited facial synostoses at P0 that were indistinguishable from those of Bcl11bncc−/− mice (Fig. 2B, F, D, H). However, premature fusion of the temporal and coronal sutures was also evident in Bcl11b−/− mice at P0, a phenotype that has not been observed in Bcl11bncc−/− mice (Fig. 2).
The coronal suture, a major growth center of the skull vault, is formed at the interface of the NCC-derived frontal bone and mesoderm-derived parietal bone. However, the non-osteogenic mesenchyme of the coronal suture is derived from the mesoderm (Yoshida et al., 2008), and this likely explains our observation of coronal suture synostosis in Bcl11b−/−, but not in Bcl11bncc−/−, mice.
Bcl11b−/− mice exhibited enhanced mineralization of the facial skeleton at P0, as demonstrated by von Kossa staining (Fig. S2). Enhanced ossification was also noted within the anterior fontanelle of Bcl11b−/− mice, affecting both frontal and parietal bones, and selectively within frontal bones of Bcl11bncc−/− mice (Fig. 1A–D). The latter was reflected in a significantly increased ratio of frontal/parietal bone volume (BV) in Bcl11bncc−/− mice (2.04 ± 0.02; n = 5) at P0 relative to the Bcl11fl/fl controls (1.76 ± 0.03; n = 5, P < 0.0001).
Collectively, these findings demonstrate that mice lacking BCL11B exhibit multiple facial and cranial synostoses at birth, implying that the observed craniofacial pathology likely initiated during embryonic development.
BCL11B Is Expressed in Osteogenic and Sutural Mesenchyme during Embryonic Development
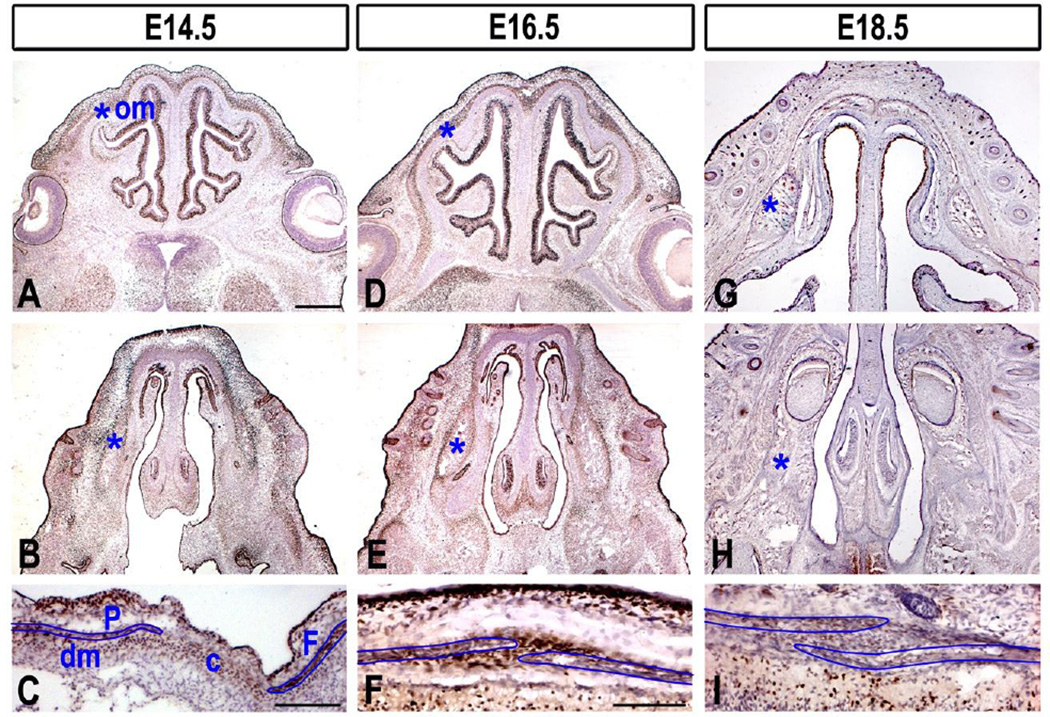
Expression of BCL11B within the osteogenic mesenchyme of the face was first detected at embryonic day (E)13.5 (Fig. S3C). High levels of BCL11B expression persisted at E14.5 and E16.5 within the facial skeleton, osteogenic fronts of frontal and parietal bones, in the coronal suture, and dura mater (Fig. 3A–F), consistent with previous report (Holmes et al., 2015). BCL11B expression appeared somewhat down-regulated within the facial and cranial skeleton by E18.5 (Fig. 3G–I) and remained so to at least P5 (Fig. S4), suggesting that this transcription factor functions primarily during a finite window of embryogenesis.
Fig. 3. BCL11B expression in osteogenic and sutural mesenchyme.
BCL11B immunostaining in sections of embryonic faces (two upper rows represent sections from two different planes) and calvaria (lower row) at indicated stages. Osteogenic mesenchyme (om) is denoted by asterisk in the facial sections and outlined by blue line in the sections of calvaria. c, coronal suture; dm, dura mater; F, frontal bone; P, parietal bone. Scale bars: (two upper rows) 500 µm; (C), (F,I) 200 µm.
In Bcl11bncc−/− mice, expression of BCL11B was ablated specifically in NCC-derived facial mesenchyme, including dermal and tooth mesenchyme at E12.5 and osteogenic mesenchyme at E13.5 (Fig. S3). This suggests that the Bcl11b locus had likely been excised before the onset of BCL11B expression within the osteogenic mesenchyme of the facial skeleton. Levels of BCL11B protein were unaltered in all epithelial structures of Bcl11bncc−/− heads (Fig. S3).
Embryonic Skulls of Bcl11b−/− Mice Exhibit Premature Mineralization and Craniofacial Synostoses
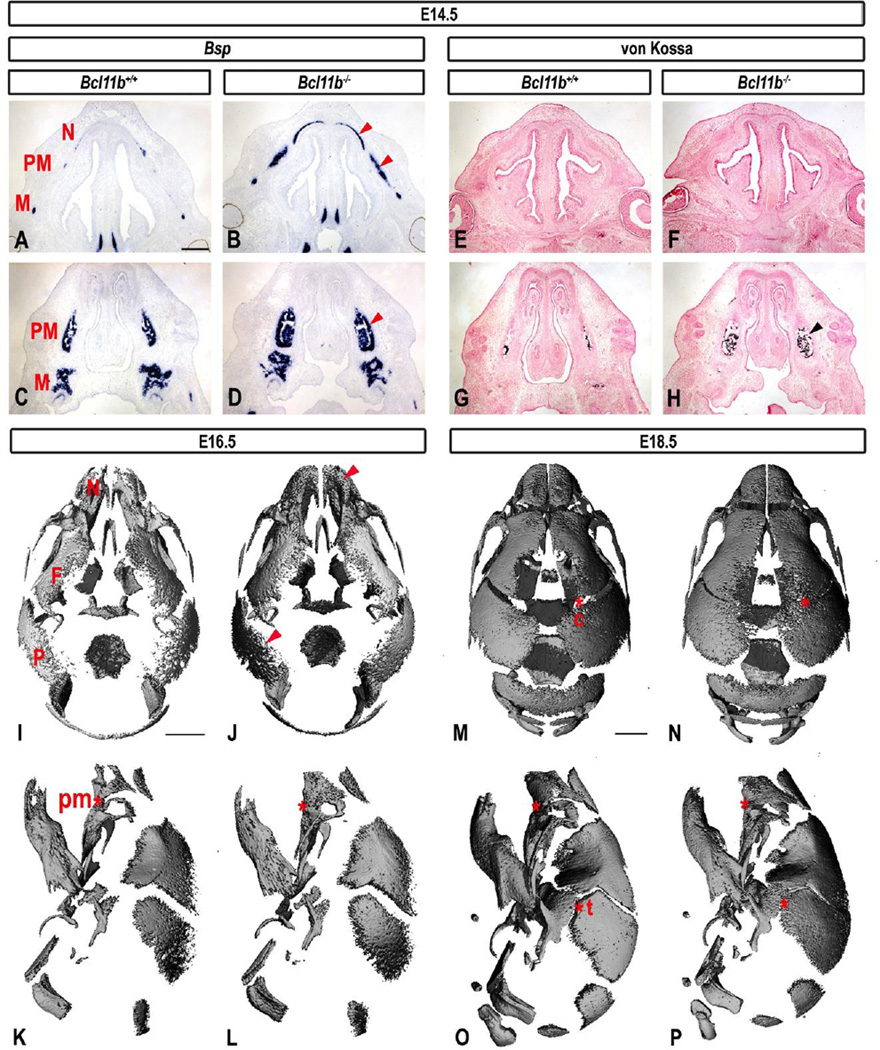
Expression of bone sialoprotein (Bsp), a marker of osteoblast maturation, was up-regulated within the facial skeleton of Bcl11b−/− mice at E14.5 (Figs. 4A–D). Moreover, Bcl11b−/− mice exhibited increased mineralization within the facial skeleton relative to control mice at this stage, as determined by von Kossa staining (Fig. 4E–H).
Fig. 4. Increased osteoblast maturation, mineralization, and craniofacial synostoses in Bcl11b−/− embryonic skulls.
(A–D) RNA ISH using a Bsp probe and (E–H) von Kossa staining in sections of control and Bcl11b−/− heads at two different planes in E14.5 embryos. Arrowheads point to enhanced osteoblast maturation and increased mineralization, respectively, in Bcl11b−/− facial mesenchyme. (I–P) Dorsal and lateral views of the micro-CT images of control and Bcl11b−/− mouse skulls at indicated stages. Arrowheads point to increased mineralization in craniofacial bones of Bcl11b−/− skulls. Asterisks denote sutures affected by synostosis in Bcl11b−/− skulls and corresponding patent sutures in the control mice. Affected bones: F, frontal; M, maxillary; N, nasal; P, parietal; PM, premaxillary. Affected sutures: c, coronal; pm, premaxillary-maxillary; t, temporal. Scale bars: (A–H) 500 µm; (I–L), (M–P) 1 mm.
Micro-CT analyses of Bcl11b−/− skulls at E16.5 and E18.5 also revealed increased mineralization within the facial skeleton and calvaria of mutant skulls (Fig. 4I–P). This was confirmed by determination of BV in frontal and parietal bones, as well as of the entire head. Bcl11b−/− mice exhibited a 32% and a 44% increase in frontal and parietal BVs, respectively, at E16.5, and these differences were statistically significant (Table 2). Analysis of the entire head also indicated that Bcl11b−/− mice exhibited greater bone volume at E16.5 (30% higher). However, these differences in BVs between mutant and wild-type mice were not maintained in older embryos (E18.5; Table 2).
Table 2.
Bone volumes (in mm3) at E16.5 in wild-type and Bcl11b−/− mice
| Embryonic age | ||||
|---|---|---|---|---|
| E16.5 | E18.5 | |||
| Bcl11b+/+ | Bcl11b−/− | Bcl11b+/+ | Bcl11b−/− | |
| Frontal | 0.34 ± 0.05 | 0.45 ± 0.02** | 1.14 ± 0.23 | 1.29 ± 0.25 |
| Parietal | 0.14 ± 0.01 | 0.2 ± 0.02** | 0.58 ± 0.13 | 0.71 ± 0.21 |
| Total | 2.26 ± 0.18 | 2.94 ± 0.32* | 8.8 ± 2.3 | 8.9 ± 2.2 |
p < 0.05
p < 0.01
The Bcl11b−/− premaxillary-maxillary suture was already fused at E16.5 (Fig. 4I–L); however, initiation of coronal and temporal synostoses was not observed until E18.5 in the Bcl11b−/− skulls (Fig. 4M–P). These data demonstrate that BCL11B normally limits premature bone mineralization and suppresses ossification of facial and coronal sutures.
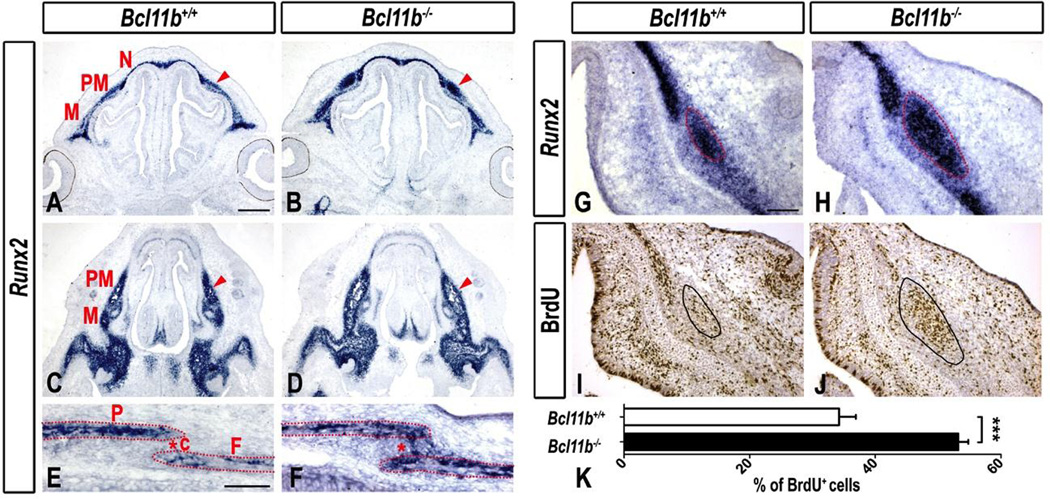
Increased Osteoblast Differentiation and Ectopic Expression of Runx2 and Fgfr2c in Coronal and Facial Sutures of Bcl11b−/− Mice
The transcription factor RUNX2 is a master regulator of osteogenesis required for determination of the osteoblast lineage (Komori, 2010; Komori et al., 1997; Otto et al., 1997). Runx2 was strongly expressed in differentiating osteoblasts of the facial and calvarial osteogenic mesenchyme at E14.5 and E16.5, respectively, in control mice (Fig. 5A, C, E, G). Low and undetectable levels of Runx2 transcripts were observed in the premaxillary-maxillary and coronal sutures, respectively, of control skulls (Figs. 5A, C, E, G). An expansion of Runx2-positive cells, accompanied by an increase in Runx2 expression, was observed in the osteogenic mesenchyme of the Bcl11b−/− facial skeleton at E14.5 (Figs. 5B, D, H), and ectopic Runx2 transcripts were detected within coronal suture of these mutants at E16.5 (Fig. 5F). In addition, osteoblasts (defined by expression of Runx2) of Bcl11b−/− premaxillary mesenchyme exhibited 60% increase in proliferation compared to the osteoblasts of control mice at E14.5 (Figs. 5I–K). This observation suggests that BCL11B limits osteoblast expansion and represses Runx2 expression within sutural mesenchyme of the facial skeleton.
Fig. 5. Increased Runx2 expression and proliferation in the Bcl11b−/− embryonic faces and ectopic Runx2 expression within the Bcl11b−/− coronal suture.
(A–H) RNA ISH using a Runx2 probe in sections of embryonic faces at E14.5 at two different planes (A–D) and at higher magnification (G, H) and in coronal suture at E16.5 (E, F). Arrowheads denote up-regulation and expansion of Runx2 expression. Asterisks indicate ectopic expression of Runx2 in Bcl11b−/− sutures. (I, J) BrdU immunostaining in sections of control and Bcl11bncc−/− heads at E14.5. Bones: F, frontal; M, maxillary; N, nasal; P, parietal; PM, premaxillary. Sutures: c, coronal. Scale bars: (A–D) 500 µm; (E, F) 200 µm; (G, J) 100 µm. (K) Quantification of the BrdU index (BrdU+ cells as a fraction of hematoxylin+ cells). Error bars indicate standard deviation. *** denotes statistical significance at p ≤ 0.001 (n = 3).
Runx2 expression, as well as the stability, DNA-binding, and transcriptional activity of this transcription factor are induced by FGF signaling pathways, which play important roles in regulation of craniofacial sutural patency (Kim et al., 2003a; Park et al., 2010; Twigg et al., 2013; Xiao et al., 2002). Activating mutations within FGFR receptors are associated with syndromic and non-syndromic craniosynostosis in mice and humans (Nie et al., 2006). The FGF family is comprised of 22 genes encoding structurally related growth factors (Ornitz and Itoh, 2001). Fgf2, Fgf9, Fgf10 and Fgf18 are expressed in developing bones and/or sutural mesenchyme and have been associated with enhanced ossification and skeletal defects including craniosynostosis (Behr et al., 2010; Hajihosseini et al., 2009; Harada et al., 2009; Kim et al., 1998; Liu et al., 2002; Montero et al., 2000; Ohbayashi et al., 2002; Rice et al., 2000). Although Fgf4 is not expressed in the developing facial or cranial skeleton, direct application of FGF4 in the vicinity of sutures promotes osteoblast proliferation and/or differentiation and subsequent sutural closure (Greenwald et al., 2001; Kim et al., 2003b; Kim et al., 1998; Nagayama et al., 2013).
FGFs bind four high-affinity receptor tyrosine kinases (FGFR1 to FGFR4). Mutations in genes encoding FGFR1-3, but not FGFR4, are associated with craniosynostosis in mice and humans (Chim et al., 2011). In general, all of these mutations result in receptors with enhanced and/or constitutive activity (Cunningham et al., 2007). Expression of Fgfr2c, as well as low levels of Fgfr1c, Fgfr2b, and Fgfr3c, has been reported in developing calvarial bones and particularly at the osteogenic fronts of these bones (Rice et al., 2003).
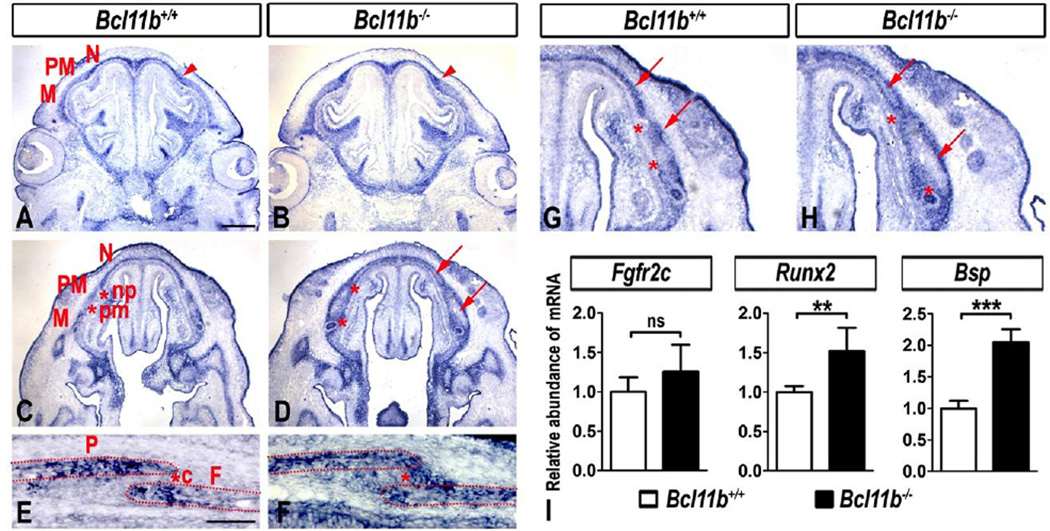
We showed previously that BCL11B regulates FGF signaling during incisor development (Kyrylkova et al., 2012a). Therefore, we analyzed Bcl11b−/− mice for expression of multiple FGF ligands and receptors that play roles during craniofacial development. We did not detect alterations in expression of genes encoding FGF or FGFR family members in Bcl11b−/− mice (Figs. S5, S6), with the exception of Fgfr2c (Figs. 6A–H). Fgfr2c expression was detected widely within differentiating osteoblasts of the face and head of control mice but was generally excluded from sutural mesenchyme (Fig. 6A, C, E, G), as reported previously (Iseki et al., 1997; Rice et al., 2003). Fgfr2c transcripts were ectopically expressed within facial and coronal sutures of Bcl11b−/− mice at E14.5 and E16.5, respectively (Fig. 6B, D, F, H). These results suggest that BCL11B normally represses Fgfr2c expression within craniofacial sutures.
Fig. 6. Altered Fgfr2c expression in the Bcl11b−/− embryonic faces and coronal suture.
(A–H) RNA ISH using an Fgfr2c probe in sections of embryonic faces at E14.5 at two different planes (A–D) and at higher magnification (G, H) and in coronal suture at E16.5 (E, F). Arrowheads denote up-regulation and expansion of Fgfr2c expression. Arrows point to the lack of Fgfr2c expression within the central osteoid of facial bones in Bcl11b−/− mice. Asterisks indicate ectopic expression of Fgfr2c in Bcl11b−/− sutures. Bones: F, frontal; M, maxillary; N, nasal; P, parietal; PM, premaxillary. Sutures: c, coronal; np, naso-premaxillary; pm, premaxillary-maxillary. Scale bars: (A–D) 500 µm; (E, F) 200 µm; (G, J) 100 µm. (I) qRT-PCR analysis indicates quantitatively non-significant change in Fgfr2c expression and an increase in Runx2 and Bsp expression in Bcl11bncc−/− faces at E14.5. Error bars indicate standard deviation. ns, not significant; ** denotes statistical significance at p ≤ 0.01; *** denotes statistical significance at p ≤ 0.001 (n = 3).
Fgfr2c transcripts were generally excluded from the more differentiated mesenchyme within the central osteoid of Bcl11b−/− facial bones (Fig. 6D, H). Fgfr2 isoforms are expressed early in the osteoblast lineage and are down-regulated as osteoprogenitors mature toward terminal differentiation (Iseki et al., 1997). Therefore, decreased Fgfr2c expression in the central osteoid of facial bones of the Bcl11b−/− mice is consistent with enhanced osteoblast differentiation in these areas of the craniofacial skeleton.
Analysis of facial gene expression by qRT-PCR did not reveal significant, quantitative changes in expression of Fgfr2c between Bcl11b−/− and control samples (Fig. 6I), consistent with the fact that Fgfr2c expression was decreased in the central osteoid and increased in the facial sutures of Bcl11b−/− mice (Fig. 6A–D, G, H). However, expression of both Runx2 and Bsp was significantly up-regulated in the facial tissue of Bcl11b−/− mice by 1.6- and 2-fold, respectively (Fig. 6I), supporting the findings of histological analyses (Fig 4A–D, 5A–D, G, H).
We observed increased osteoprogenitor proliferation and Runx2 expression in Bcl11b−/− skulls, both of which phenocopy defects of mice harboring gain-of-function mutation of Fgfr2c (Fgfr2cc342y/+) (Eswarakumar et al., 2004). Bcl11b−/− mice over- and ectopically-express a wild-type form of Fgfr2c, yet exhibit a craniosynostosis phenotype that is much more severe than that of mice heterozygously expressing a constitutive active form of the receptor, e.g. Fgfr2cc342y/+. This may be explained by the fact that Fgfr2 overexpression results in ligand-independent activation of the receptor, as was shown in other cell systems (Turner et al., 2010). Moreover, overexpression of Fgfr1, driven by transient transfection, results in FGF2-mediated proliferation in neonatal cardiac myocyte cultures (Sheikh et al., 1999). In the latter case, overexpressed FGFR1 would appear to be stimulated by an FGF ligand(s) that is in the vicinity. Therefore, we suggest that overexpression of Fgfr2c in the sutural mesenchyme, without concomitant changes in the expression of FGF ligands, contributes to craniosynostosis in Bcl11b−/− mice.
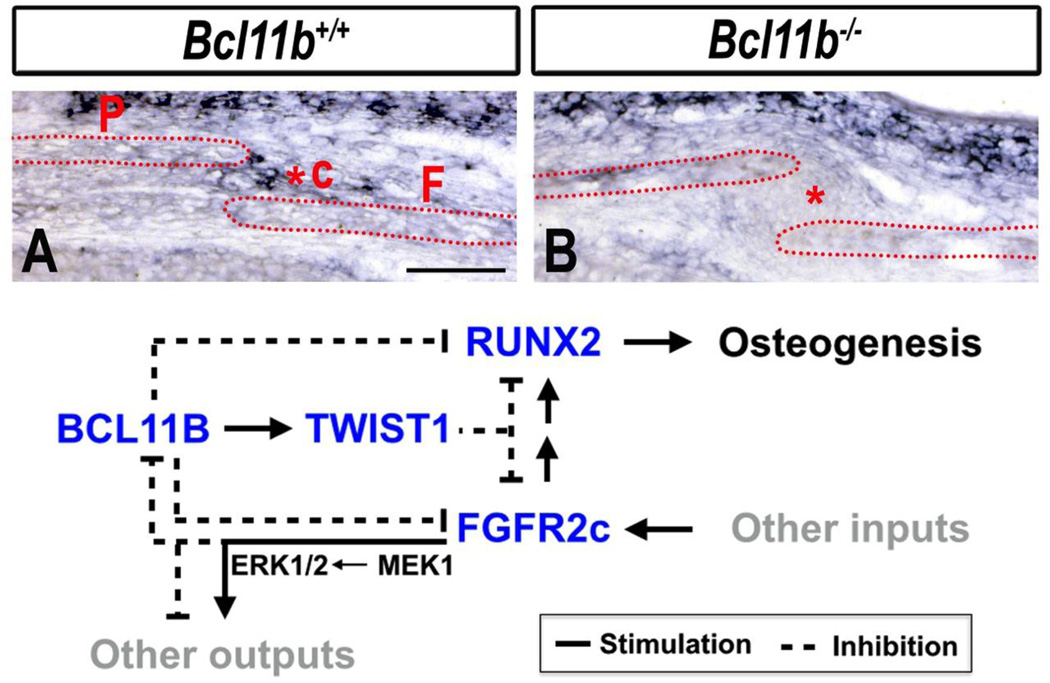
Finally, BCL11B may play an additional or alternative role in establishing and/or maintaining the boundary between osteogenic and sutural mesenchyme, as previously described for Twist1+/− mice (Yen et al., 2010), a model of Saethre–Chotzen syndrome (el Ghouzzi et al., 1997; Paznekas et al., 1998). In this regard, it may be of interest that expression of Twist1 was down-regulated in the coronal suture of Bcl11b−/− mice (Fig. 7A–B). These findings suggest that the Twist1 gene, like the genes encoding RUNX2 and FGFR2, appears to be downstream of BCL11B in the craniofacial skeleton.
Fig. 7. Loss of expression of Twist1 in coronal suture of Bcl11b−/− mice at E16.5 and model for role of BCL11B in maintaining patency of the coronal suture.
(A, B) RNA ISH using a Twist1 probe in sections of control and Bcl11b−/− embryonic heads at E16.5. c, coronal suture; F, frontal bone; P, parietal bone. Scale bar: 200 µm. (C) Working model of BCL11B action in non-osteogenic mesenchyme of coronal suture. We propose that BCL11B induces or maintains expression of Twist1 while repressing expression of both Fgfr2c and Runx2. TWIST1 may also repress expression of Runx2 and Fgfr2c. This model posits that activation of the FGF pathways, via FGFR2c, leads to down-regulation of Bcl11b expression or BCL11B protein levels, which attenuates BCL11B-mediated maintenance of Twist1 expression and relieves BCL11B-mediated repression of expression of both Fgfr2c and Runx2, and facilitating osteogenesis.
Conclusions
Data presented herein demonstrate that BCL11B plays an important role in maintaining sutural patency from the earliest stages of mouse craniofacial development. BCL11B appears to control a gene network that regulates timely proliferation and differentiation of osteoprogenitors, limits premature bone mineralization, and suppresses ossification of facial and coronal sutures. We speculate that the principal mechanistic basis of these actions of BCL11B is the activation and/or maintenance of Twist1 expression, coupled with repression of Fgfr2c expression and the RUNX2-dependent osteogenic program within osteogenic and sutural mesenchyme. The model shown in Fig. 7C depicts placement of BCL11B in osteogenic pathway within the coronal suture. This model is based on current data and our previous demonstration that BCL11B and FGF signaling participate in a reciprocal, inhibitory circuit during incisor development (Kyrylkova et al, 2012). Because BCL11B appears to regulate expression of Twist1 positively in the coronal suture but negatively regulates expression of Fgfr2c and Runx2, BCL11B is depicted upstream of all three signaling pathways.
Craniosynostosis may be isolated (non-syndromic) or associated with other clinical signs as part of a syndrome. Plausible causative mutations have been identified only in ~30% of the cohort that comprised over 300 cases of craniosynostosis (Wilkie et al., 2010). Genes that are predominantly affected in craniosynostosis are FGFR2, FGFR3, TWIST1, EFNB1, TCF12 and ERF (Fitzpatrick, 2013; Sharma et al., 2013; Twigg et al., 2004; Twigg et al., 2013; Wilkie et al., 2010). However, ~70% of craniosynostosis cases still have unknown genetic etiology (Fitzpatrick, 2013). We do not presently know if mutations at the BCL11B locus contribute to craniosynostosis in humans, as suboptimal exome coverage of this locus hinders identification of possible mutations in humans (A. Wilkie, personal communication). However, we have observed that disruption of a single Bcl11b allele in mice in some cases results in synostoses within the facial skeleton at P20 (Fig. S7), indicating that heterozygous mutation of the Bcl11b locus is sufficient to produce facial synostoses. Moreover, Potter and colleagues recently reported that expression of BCL11B was down-regulated 5-fold in the ossified, compared to non-ossified, sutures of craniosynostosis patients (Potter et al., 2015). The latter finding is consistent with in vivo data from both mouse models described herein. These findings, together with our data, establish plausibility that dysregulated expression of BCL11B may be implicated in craniosynostosis in humans. Further understanding of the underpinnings of BCL11B action in the non-osteogenic mesenchyme of craniofacial sutures should provide insight into transcriptional mechanisms controlling maintenance of sutural patency in embryonic and neonatal life.
Supplementary Material
Highlights.
Bcl11bncc−/− and Bcl11b−/− mice exhibit severe cranial and facial synostoses.
BCL11B is expressed in developing craniofacial osteogenic and sutural mesenchyme.
BCL11B restricts premature osteoblast differentiation and mineralization of the skull.
BCL11B prevents ectopic expression of Fgfr2c and Runx2 in sutures.
BCL11B maintains expression of Twist1 in sutures.
Acknowledgements
We thank David Rice, Irma Thesleff, Ophir Klein, Chrissa Kioussi, and Carmen Wong for providing plasmids, antibodies, and primers. We also thank Dawn Olson and Jody Gordon for assisting in micro-CT analysis. This work was supported by NIH/NIDCR grant DE021879 (to ML).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Behr B, Leucht P, Longaker MT, Quarto N. Fgf-9 is required for angiogenesis and osteogenesis in long bone repair. Proc Natl Acad Sci U S A. 2010;107:11853–11858. doi: 10.1073/pnas.1003317107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chim H, Manjila S, Cohen AR, Gosain AK. Molecular signaling in pathogenesis of craniosynostosis: the role of fibroblast growth factor and transforming growth factor-beta. Neurosurg Focus. 2011;31:E7. doi: 10.3171/2011.5.FOCUS1197. [DOI] [PubMed] [Google Scholar]
- Cunningham ML, Seto ML, Ratisoontorn C, Heike CL, Hing AV. Syndromic craniosynostosis: from history to hydrogen bonds. Orthod Craniofac Res. 2007;10:67–81. doi: 10.1111/j.1601-6343.2007.00389.x. [DOI] [PubMed] [Google Scholar]
- Danielian PS, Muccino D, Rowitch DH, Michael SK, McMahon AP. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr Biol. 1998;8:1323–1326. doi: 10.1016/s0960-9822(07)00562-3. [DOI] [PubMed] [Google Scholar]
- el Ghouzzi V, Le Merrer M, Perrin-Schmitt F, Lajeunie E, Benit P, Renier D, Bourgeois P, Bolcato- Bellemin AL, Munnich A, Bonaventure J. Mutations of the TWIST gene in the Saethre-Chotzen syndrome. Nat Genet. 1997;15:42–46. doi: 10.1038/ng0197-42. [DOI] [PubMed] [Google Scholar]
- Eswarakumar VP, Horowitz MC, Locklin R, Morriss-Kay GM, Lonai P. A gain-of-function mutation of Fgfr2c demonstrates the roles of this receptor variant in osteogenesis. Proc Natl Acad Sci U S A. 2004;101:12555–12560. doi: 10.1073/pnas.0405031101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick DR. Filling in the gaps in cranial suture biology. Nat Genet. 2013;45:231–232. doi: 10.1038/ng.2557. [DOI] [PubMed] [Google Scholar]
- Golonzhka O, Liang X, Messaddeq N, Bornert JM, Campbell AL, Metzger D, Chambon P, Ganguli-Indra G, Leid M, Indra AK. Dual role of COUP-TF-interacting protein 2 in epidermal homeostasis and permeability barrier formation. J Invest Dermatol. 2009;129:1459–1470. doi: 10.1038/jid.2008.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald JA, Mehrara BJ, Spector JA, Warren SM, Fagenholz PJ, Smith LE, Bouletreau PJ, Crisera FE, Ueno H, Longaker MT. In vivo modulation of FGF biological activity alters cranial suture fate. Am J Pathol. 2001;158:441–452. doi: 10.1016/s0002-9440(10)63987-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajihosseini MK, Duarte R, Pegrum J, Donjacour A, Lana-Elola E, Rice DP, Sharpe J, Dickson C. Evidence that Fgf10 contributes to the skeletal and visceral defects of an Apert syndrome mouse model. Dev Dyn. 2009;238:376–385. doi: 10.1002/dvdy.21648. [DOI] [PubMed] [Google Scholar]
- Harada M, Murakami H, Okawa A, Okimoto N, Hiraoka S, Nakahara T, Akasaka R, Shiraishi Y, Futatsugi N, Mizutani-Koseki Y, Kuroiwa A, Shirouzu M, Yokoyama S, Taiji M, Iseki S, Ornitz DM, Koseki H. FGF9 monomer-dimer equilibrium regulates extracellular matrix affinity and tissue diffusion. Nat Genet. 2009;41:289–298. doi: 10.1038/ng.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes G, van Bakel H, Zhou X, Losic B, Jabs EW. BCL11B expression in intramembranous osteogenesis during murine craniofacial suture development. Gene Expr Patterns. 2015;17:16–25. doi: 10.1016/j.gep.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iseki S, Wilkie AO, Heath JK, Ishimaru T, Eto K, Morriss-Kay GM. Fgfr2 and osteopontin domains in the developing skull vault are mutually exclusive and can be altered by locally applied FGF2. Development. 1997;124:3375–3384. doi: 10.1242/dev.124.17.3375. [DOI] [PubMed] [Google Scholar]
- Jiang X, Iseki S, Maxson RE, Sucov HM, Morriss-Kay GM. Tissue origins and interactions in the mammalian skull vault. Dev Biol. 2002;241:106–116. doi: 10.1006/dbio.2001.0487. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Kim JH, Bae SC, Choi JY, Ryoo HM. The protein kinase C pathway plays a central role in the fibroblast growth factor-stimulated expression and transactivation activity of Runx2. J Biol Chem. 2003a;278:319–326. doi: 10.1074/jbc.M203750200. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Lee MH, Park HS, Park MH, Lee SW, Kim SY, Choi JY, Shin HI, Ryoo HM. Erk pathway and activator protein 1 play crucial roles in FGF2-stimulated premature cranial suture closure. Dev Dyn. 2003b;227:335–346. doi: 10.1002/dvdy.10319. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Rice DP, Kettunen PJ, Thesleff I. FGF-, BMP- and Shh-mediated signalling pathways in the regulation of cranial suture morphogenesis and calvarial bone development. Development. 1998;125:1241–1251. doi: 10.1242/dev.125.7.1241. [DOI] [PubMed] [Google Scholar]
- Komori T. Regulation of osteoblast differentiation by Runx2. Adv Exp Med Biol. 2010;658:43–49. doi: 10.1007/978-1-4419-1050-9_5. [DOI] [PubMed] [Google Scholar]
- Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, Sato M, Okamoto R, Kitamura Y, Yoshiki S, Kishimoto T. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- Kyryachenko S, Kyrylkova K, Leid M, Kioussi C. Immunohistochemistry and detection of proliferating cells by BrdU. Methods Mol Biol. 2012;887:33–39. doi: 10.1007/978-1-61779-860-3_4. [DOI] [PubMed] [Google Scholar]
- Kyrylkova K, Kyryachenko S, Biehs B, Klein O, Kioussi C, Leid M. BCL11B regulates epithelial proliferation and asymmetric development of the mouse mandibular incisor. PLoS One. 2012a;7:e37670. doi: 10.1371/journal.pone.0037670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrylkova K, Kyryachenko S, Kioussi C, Leid M. Determination of gene expression patterns by in situ hybridization in sections. Methods Mol Biol. 2012b;887:23–31. doi: 10.1007/978-1-61779-860-3_3. [DOI] [PubMed] [Google Scholar]
- Liu Z, Xu J, Colvin JS, Ornitz DM. Coordination of chondrogenesis and osteogenesis by fibroblast growth factor 18. Genes Dev. 2002;16:859–869. doi: 10.1101/gad.965602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero A, Okada Y, Tomita M, Ito M, Tsurukami H, Nakamura T, Doetschman T, Coffin JD, Hurley MM. Disruption of the fibroblast growth factor-2 gene results in decreased bone mass and bone formation. J Clin Invest. 2000;105:1085–1093. doi: 10.1172/JCI8641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagayama T, Okuhara S, Ota MS, Tachikawa N, Kasugai S, Iseki S. FGF18 accelerates osteoblast differentiation by upregulating Bmp2 expression. Congenit Anom (Kyoto) 2013;53:83–88. doi: 10.1111/cga.12012. [DOI] [PubMed] [Google Scholar]
- Nelson DK, Williams T. Frontonasal process-specific disruption of AP-2alpha results in postnatal midfacial hypoplasia, vascular anomalies, and nasal cavity defects. Dev Biol. 2004;267:72–92. doi: 10.1016/j.ydbio.2003.10.033. [DOI] [PubMed] [Google Scholar]
- Nie X. Cranial base in craniofacial development: developmental features, influence on facial growth, anomaly, and molecular basis. Acta Odontol Scand. 2005;63:127–135. doi: 10.1080/00016350510019847. [DOI] [PubMed] [Google Scholar]
- Nie X, Luukko K, Kettunen P. FGF signalling in craniofacial development and developmental disorders. Oral Dis. 2006;12:102–111. doi: 10.1111/j.1601-0825.2005.01176.x. [DOI] [PubMed] [Google Scholar]
- Ohbayashi N, Shibayama M, Kurotaki Y, Imanishi M, Fujimori T, Itoh N, Takada S. FGF18 is required for normal cell proliferation and differentiation during osteogenesis and chondrogenesis. Genes Dev. 2002;16:870–879. doi: 10.1101/gad.965702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opperman LA. Cranial sutures as intramembranous bone growth sites. Dev Dyn. 2000;219:472–485. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1073>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Ornitz DM, Itoh N. Fibroblast growth factors. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-3-reviews3005. REVIEWS3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GW, Beddington RS, Mundlos S, Olsen BR, Selby PB, Owen MJ. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- Park OJ, Kim HJ, Woo KM, Baek JH, Ryoo HM. FGF2-activated ERK mitogen-activated protein kinase enhances Runx2 acetylation and stabilization. J Biol Chem. 2010;285:3568–3574. doi: 10.1074/jbc.M109.055053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paznekas WA, Cunningham ML, Howard TD, Korf BR, Lipson MH, Grix AW, Feingold M, Goldberg R, Borochowitz Z, Aleck K, Mulliken J, Yin M, Jabs EW. Genetic heterogeneity of Saethre-Chotzen syndrome, due to TWIST and FGFR mutations. Am J Hum Genet. 1998;62:1370–1380. doi: 10.1086/301855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietri T, Eder O, Blanche M, Thiery JP, Dufour S. The human tissue plasminogen activator- Cre mouse: a new tool for targeting specifically neural crest cells and their derivatives in vivo. Dev Biol. 2003;259:176–187. doi: 10.1016/s0012-1606(03)00175-1. [DOI] [PubMed] [Google Scholar]
- Potter AB, Rhodes JL, Vega RA, Ridder T, Shiang R. Gene expression changes between patent and fused cranial sutures in a nonsyndromic craniosynostosis population. Eplasty. 2015;15:e12. [PMC free article] [PubMed] [Google Scholar]
- Purushothaman R, Cox TC, Maga AM, Cunningham ML. Facial suture synostosis of newborn Fgfr1(P250R/+) and Fgfr2(S252W/+) mouse models of Pfeiffer and Apert syndromes. Birth Defects Res A Clin Mol Teratol. 2011;91:603–609. doi: 10.1002/bdra.20811. [DOI] [PubMed] [Google Scholar]
- Rice DP. Developmental anatomy of craniofacial sutures. Front Oral Biol. 2008;12:1–21. doi: 10.1159/000115028. [DOI] [PubMed] [Google Scholar]
- Rice DP, Aberg T, Chan Y, Tang Z, Kettunen PJ, Pakarinen L, Maxson RE, Thesleff I. Integration of FGF and TWIST in calvarial bone and suture development. Development. 2000;127:1845–1855. doi: 10.1242/dev.127.9.1845. [DOI] [PubMed] [Google Scholar]
- Rice DP, Rice R, Thesleff I. Fgfr mRNA isoforms in craniofacial bone development. Bone. 2003;33:14–27. doi: 10.1016/s8756-3282(03)00163-7. [DOI] [PubMed] [Google Scholar]
- Rosenberg P, Arlis HR, Haworth RD, Heier L, Hoffman L, LaTrenta G. The role of the cranial base in facial growth: experimental craniofacial synostosis in the rabbit. Plast Reconstr Surg. 1997;99:1396–1407. doi: 10.1097/00006534-199704001-00030. [DOI] [PubMed] [Google Scholar]
- Sharma VP, Fenwick AL, Brockop MS, McGowan SJ, Goos JA, Hoogeboom AJ, Brady AF, Jeelani NO, Lynch SA, Mulliken JB, Murray DJ, Phipps JM, Sweeney E, Tomkins SE, Wilson LC, Bennett S, Cornall RJ, Broxholme J, Kanapin A, Johnson D, Wall SA, van der Spek PJ, Mathijssen IM, Maxson RE, Twigg SR, Wilkie AO. Mutations in TCF12, encoding a basic helix-loop-helix partner of TWIST1, are a frequent cause of coronal craniosynostosis. Nat Genet. 2013;45:304–307. doi: 10.1038/ng.2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikh F, Fandrich RR, Kardami E, Cattini PA. Overexpression of long or short FGFR-1 results in FGF-2-mediated proliferation in neonatal cardiac myocyte cultures. Cardiovasc Res. 1999;42:696–705. doi: 10.1016/s0008-6363(99)00008-5. [DOI] [PubMed] [Google Scholar]
- Stewart RE, Dixon G, Cohen A. The pathogenesis of premature craniosynostosis in acrocephalosyndactyly (Apert's syndrome). A reconsideration. Plast Reconstr Surg. 1977;59:699–707. doi: 10.1097/00006534-197705000-00013. [DOI] [PubMed] [Google Scholar]
- Turner N, Lambros MB, Horlings HM, Pearson A, Sharpe R, Natrajan R, Geyer FC, van Kouwenhove M, Kreike B, Mackay A, Ashworth A, van de Vijver MJ, Reis-Filho JS. Integrative molecular profiling of triple negative breast cancers identifies amplicon drivers and potential therapeutic targets. Oncogene. 2010;29:2013–2023. doi: 10.1038/onc.2009.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twigg SR, Kan R, Babbs C, Bochukova EG, Robertson SP, Wall SA, Morriss-Kay GM, Wilkie AO. Mutations of ephrin-B1 (EFNB1), a marker of tissue boundary formation, cause craniofrontonasal syndrome. Proc Natl Acad Sci U S A. 2004;101:8652–8657. doi: 10.1073/pnas.0402819101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twigg SR, Vorgia E, McGowan SJ, Peraki I, Fenwick AL, Sharma VP, Allegra M, Zaragkoulias A, Sadighi Akha E, Knight SJ, Lord H, Lester T, Izatt L, Lampe AK, Mohammed SN, Stewart FJ, Verloes A, Wilson LC, Healy C, Sharpe PT, Hammond P, Hughes J, Taylor S, Johnson D, Wall SA, Mavrothalassitis G, Wilkie AO. Reduced dosage of ERF causes complex craniosynostosis in humans and mice and links ERK1/2 signaling to regulation of osteogenesis. Nat Genet. 2013;45:308–313. doi: 10.1038/ng.2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie AO, Byren JC, Hurst JA, Jayamohan J, Johnson D, Knight SJ, Lester T, Richards PG, Twigg SR, Wall SA. Prevalence and complications of single-gene and chromosomal disorders in craniosynostosis. Pediatrics. 2010;126:e391–e400. doi: 10.1542/peds.2009-3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao G, Jiang D, Gopalakrishnan R, Franceschi RT. Fibroblast growth factor 2 induction of the osteocalcin gene requires MAPK activity and phosphorylation of the osteoblast transcription factor, Cbfa1/Runx2. J Biol Chem. 2002;277:36181–36187. doi: 10.1074/jbc.M206057200. [DOI] [PubMed] [Google Scholar]
- Yen HY, Ting MC, Maxson RE. Jagged1 functions downstream of Twist1 in the specification of the coronal suture and the formation of a boundary between osteogenic and non-osteogenic cells. Dev Biol. 2010;347:258–270. doi: 10.1016/j.ydbio.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Vivatbutsiri P, Morriss-Kay G, Saga Y, Iseki S. Cell lineage in mammalian craniofacial mesenchyme. Mech Dev. 2008;125:797–808. doi: 10.1016/j.mod.2008.06.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.