SUMMARY
The interconversion between naïve and primed pluripotent states is accompanied by drastic epigenetic rearrangements. However, it is unclear whether intrinsic epigenetic events can drive reprogramming to naïve pluripotency, or if distinct chromatin states are instead simply a reflection of discrete pluripotent states. Here, we show that blocking histone H3K4 methyltransferase MLL1 activity with the small molecule inhibitor MM-401 reprograms mouse epiblast stem cells (EpiSCs) to naïve pluripotency. This reversion is highly efficient and synchronized, with over 50% of treated EpiSCs exhibiting features of naïve embryonic stem cells (ESCs) within three days. Reverted ESCs (rESCs) reactivate silenced X chromosomes and contribute to embryos following blastocyst injection, generating germline-competent chimeras. Importantly, blocking MLL1 leads to global redistribution of H3K4me1 at enhancers and represses lineage determinant factors and EpiSC markers, which indirectly regulate ESC transcription circuitry. These findings show that discrete perturbation of H3K4 methylation is sufficient to drive reprogramming to naïve pluripotency.
INTRODUCTION
Several metastable pluripotent states arise from either developing embryos in vivo or cell cultures in vitro (Cahan and Daley, 2013; Nichols and Smith, 2009). Mouse embryonic stem cells (ESCs) derived from the embryonic inner cell mass (ICM) represent the naïve pluripotency. Naïve ESCs harbor the requisite developmental potency and flexibility to produce all embryonic lineages when injected into the blastocyst embryo. Upon implantation, epiblast precursors differentiate to a ‘primed’ pluripotent state in the post-implantation epiblast (EpiSCs). This primed state can be recapitulated by culturing ESCs in medium containing bFGF (also called FGF2) and Activin in vitro (Brons et al., 2007; Tesar et al., 2007). ESCs and EpiSCs represent two distinct pluripotent states and have different characteristics with regard to morphology, growth factor dependency, epigenetic states and the ability to integrate into ICM and contribute to the germ line (Nichols and Smith, 2009). EpiSCs and ESCs are interconvertible. Transition from ESCs to EpiSCs is relatively straightforward and can be achieved by adapting culture conditions (Buecker et al., 2014; Schulz et al., 2014). In comparison, EpiSC reversion to ESCs, either spontaneously or through 2i treatment, is extremely inefficient (Bao et al., 2009; Han et al., 2010). EpiSC reversion can be facilitated by overexpression of specific factors such as Klf4 (Guo et al., 2009), Klf2 and Nanog (Stuart et al., 2014), Esrrb (Festuccia et al., 2012), Tfcp2l1 (Ye et al., 2013) and Nr5a (Guo and Smith, 2010) or by deletion of Mbd3 (Rais et al., 2013). With the exception of Mbd3 deletion, whose role in reprogramming is still being debated (dos Santos et al., 2014), these manipulations shunt EpiSCs back to ESCs at a conversion rate of ~1–5%, even in the presence of 2i and LIF (Nichols and Smith, 2009). The low efficiency associated with EpiSC reprogramming suggests that there could be an unknown transcriptional/epigenetic barrier that prevents reversion of developmental commitments.
Mechanistic studies show that conversions between ESCs and EpiSCs are accompanied by dramatic reorganization of the epigenetic landscape (Buecker et al., 2014; Factor et al., 2014; Gafni et al., 2013). The transition of naïve ESCs to EpiSCs is accompanied by global up regulation of H3K27me3 and DNA methylation (Theunissen et al., 2014), concomitant with the rise of heterochromatin in EpiSCs (Orkin and Hochedlinger, 2011). Consistently, X-chromosome inactivation in female cells is a hallmark that differentiate the primed vs. naïve pluripotent state (De Los Angeles et al., 2015). In contrast to repressive chromatin marks, there is no global change in the level of H3K4me between ESCs and EpiSCs (Li et al., 2012; Marks et al., 2012). However, dynamic modulation of H3K4me at key regulatory loci has been described (Papp and Plath, 2013; Voigt et al., 2013). Emergence of ‘poised’ enhancers that are critical for differentiation and decommissioning of ‘seed’ enhancers that are important for naïve pluripotent state have also been reported (Buecker et al., 2014; Factor et al., 2014). Notably, despite extensive studies depicting epigenetic changes, it is not clear whether they are merely the consequence of rewiring of the regulatory/transcription circuitry during cell fate alteration. Causative epigenetic modifications that are able to initiate EpiSC reversion and reset the naïve pluripotent state remain largely unknown.
In metazoans, H3K4me is mainly deposited by the MLL family histone methyltransferases (HMTs) (Rao and Dou, 2015). The activities of the MLL family HMTs are tightly regulated by a core complex containing several conserved interacting proteins (i.e. WDR5, RbBP5, and ASH2L) (Dou et al., 2006). Given the highly accessible and hyperactive chromatin structures in ESCs, it is generally assumed that H3K4me plays an important ’housekeeping’ role in ESCs and is necessary for ESCs to maintain self-renewal and unlimited differentiation potential (De Los Angeles et al., 2015). However, genetic studies show that depletion of core components of the MLL complexes, WDR5 or ASH2L/DPY30, leads to distinct outcomes (Ang et al., 2011; Jiang et al., 2011). Furthermore, deletion of Mll1, Mll2 or Set1a gene in ESCs has no major effects on ESC self-renewal despite their essential functions during embryonic development in vivo (Bledau et al., 2014; Ernst et al., 2004; Glaser et al., 2009). These studies raise the question of whether H3K4me plays important roles in the pluripotent stem cells and, if so, what is its function? In this study, we use a small molecule inhibitor MM-401 that specifically targets MLL1, but not other MLL family HMTs (Cao et al., 2014; Karatas et al., 2013), and demonstrate that MLL1-mediated H3K4me is an intrinsic epigenetic determinant that regulates acquisition of differentiated pluripotent identity. Blocking MLL1 function is sufficient to reprogram EpiSCs to naïve pluripotency.
RESULTS
MLL1 is up regulated during ESC differentiation to EpiSCs
Upon examination of expression of the MLL family HMTs (through FUNGENE database) (Fish et al., 2013), we found that while most MLL family HMTs (i.e. Mll1-4, Set1a, Set1b) as well as components of their residing complexes (e.g. Wdr5, Rbbp5) were expressed in ESCs, their respective expression pattern during spontaneous ESC differentiation was different (Figure S1A). Notably, Mll1 was dynamically regulated at different time points during ESC differentiation, being up regulated at day 2–3, day 6 and day10.5 respectively (Figure S1A). Consistently, up regulation of Mll1 at both early and late stages of embryonic development (Kojima et al., 2014) was also found in vivo (Figure S1B). Interestingly, early up regulation of Mll1 coincided with that of the epiblast marker Fgf5 and was reversely correlated with naïve pluripotent cell maker Rex1 (Figure S1B). Examination of Mll1 expression in multiple ESC and EpiSC lines confirmed that Mll1 expression was correlated more with that of EpiSC markers (e.g., Fgf5 and Cer1) than naïve stem cell markers (e.g., Nanog and Rex1) (Figure S1C and S1D). As a control, expression of Wdr5 and Rbbp5, two common components of the MLL family complexes, did not show correlation with EpiSC markers (Figure S1C). The RNA-seq results were validated by real-time RT-PCR for expression of Mll1, Mll3 and Wdr5 as well as selected pluripotent stem cell markers in ESCs and EpiSCs (Figure S1D).
MLL1 regulates ESC differentiation to EpiSCs
To test whether MLL1 and its H3K4 methyltransferase activity play roles in ESC differentiation to EpiSCs, we used our recently developed inhibitor MM-401, which inhibits MLL1 activity by blocking the MLL1-WDR5 interaction (Cao et al., 2014). To determine MM-401 dose, we first treated Mll1f/f and Mll1−/− ESCs with increasing concentration of MM-401 for either three or six days. Enantiomer MM-NC-401 or DMSO was used as the controls. As shown in Figure S2A, both MM-NC-401 and MM-401 showed toxicity at concentration above 100µM after 6-day treatment, inhibiting growth of both Mll1f/f and Mll1−/− ESCs (Figure S2A and data not shown). Notably, MM-401 had no effects on Mll1−/− ESCs at concentration of 100µM or lower despite modest growth inhibition on Mll1f/f ESCs. Based on these results, we decided to use 50µM or 100µM MM-401 in our assays to avoid toxicity or off-target effects. At these concentrations, MM-401 had no effects on EpiSC growth (Figure S2B), cell attachments after passaging (Figure S2C) and embryoid body (EB) formation (data not shown).
Whereas both MM-401 treatment and Mll1 deletion had no effects on ESC self-renewal as demonstrated by strong alkaline phosphatase (AKP) staining (Figure S2D), they led to reduced expression of epiblast markers, i.e. Mixl1, Wnt3 and Evx1 and Fgf5, at early stage of EB differentiation (Figure S2E). To directly test whether MLL1 inhibition or deletion affects EpiSC differentiation, ESCs treated with either 4-hydroxytamoxifen (4-OHT, for Mll1 deletion) or MM-401 (for Mll1 inhibition) were cultured in bFGF/Activin A/KSR media. It has been reported that bFGF/Activin A leads to efficient ESC to EpiLC differentiation in vitro, as demonstrated by weakened AKP staining and loss of Rex1 expression (Schulz et al., 2014). Interestingly, this process was delayed by Mll1 deletion or MM-401 treatment. After 72-hour culture in EpiLCpromoting conditions, a significant number of Mll1−/− and MM-401 treated colonies retained strong AKP staining as compared to untreated Mll1f/f ESCs that had attenuated AKP staining (Figure S2F). Taken together, these data show that genetic deletion or pharmacologic MLL1 inhibition impairs ESC differentiation to EpiLCs.
Inhibition of MLL1 promotes reversion of EpiSCs to ESCs
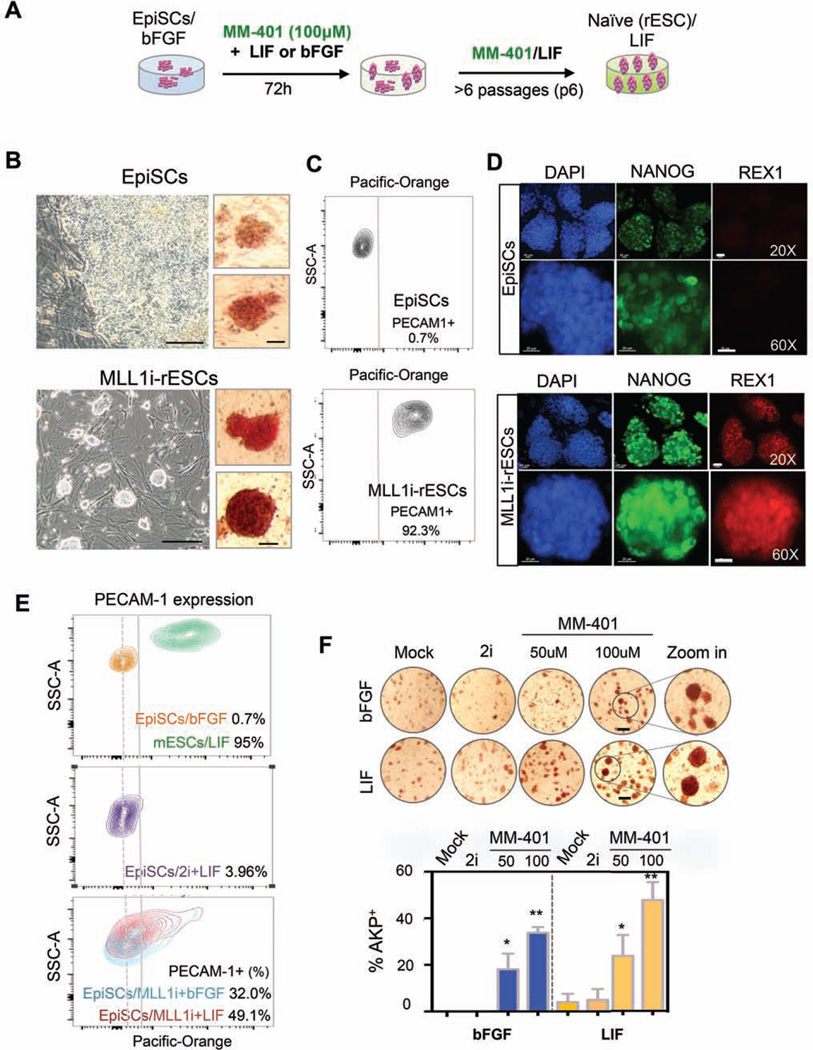
We next considered the possibility that MLL1 inhibition might promote naïve pluripotent state. To test this, we treated the EpiSC line (#9F) with MM-401 in LIF/KSR or bFGF/KSR media for 72 hours and continued to culture the cells in the presence of MM-401 and LIF/KSR beyond 6 passages (Figure 1A). To our surprise, EpiSC clones that had flat morphology, weak AKP staining and lacked expression of REX1 (Factor et al., 2013; Tesar et al., 2007), changed dramatically upon MM-401 treatment. The clones became dome-shaped with compact cells in the center and exhibited intense AKP staining (Figure 1B). Continued culturing of the ESC-like clones in MM-401 and LIF/KSR medium for 6 passages led to establishment of stably reverted ESC lines (MLL1i-rESC). The MLL1irESCs had high expression of naïve ESC markers PECAM1 and REX1 (Figure 1C and 1D) as well as homogenous expression of Nanog (Figure 1D). These naïve ESC characteristics (dos Santos et al., 2014; Stuart et al., 2014; Takashima et al., 2014) were stably maintained even after MM-401 was withdrawn for at least 30 passages (Figure S3A). To our knowledge, reversion of EpiSCs to ESCs by targeting a discrete histone modification has not yet been described and warrants further analyses.
Figure 1. MM-401 efficiently reprograms EpiSCs to ESCs.
A. Schematics of EpiSC reprogramming experiments. B. Representative images of EpiSCs with (bottom) or without MM-401 (top) at passage 6. Left, phase contrast; scale bar, 100µm. Right, AKP+ staining; scale bar, 20µm). C. FACS for PECAM1 in EpiSCs (top) and MLL1i-rESCs (bottom). Gray line, gating for PECAM1+ cells. Dash line, high frequency peak of EpiSCs distribution in PECAM-1 staining. D. Immunofluoresence staining for EpiSCs and MLL1i-rESCs as indicated. Scale bar, 20µm. E. Contour plots of PECAM1 FACS signal in ESCs, EpiSCs and MM-401 treated EpiSCs. Dash line indicated gating for PECAM1+ cells. F. AKP staining of cells as indicated. bFGF/KSR or LIF/KSR media were used as indicated on left. 100–210 clones were quantified for AKP++ and morphology change under each condition. Experiments were repeated at least twice. Scale bar, 200µm. This Figure is related to Supplemental Figure S1, S2 and S3.
To quantify EpiSC reversion efficiency en masse, we performed FACS analysis on PECAM1 following treatment with MM-401. Interestingly, 49.1% and 32.0% EpiSCs that were cultured with LIF/KSR or bFGF/KSR, respectively showed increased PECAM1 expression as early as 72hr after MM-401 (100µM) treatment (Figure 1E). Despite initial expression of PECAM1, the naïve pluripotency characteristics could not be stably maintained in cells cultured with bFGF/KSR and MM-401. The cells gradually lost PECAM1 expression after passaging (Figure S3B). Similar to EpiSC reversion en masse, MLL1 inhibition also led to rapid clonal reversion in a dose-dependent manner. About 25% and 48% EpiSC clones gained strong AKP staining after 72hrs of 50 or 100µM MM-401 treatment, respectively (Figure 1F). Similarly, strong AKP staining was detected for 33.8% clones in media containing bFGF/KSR and MM-401 (Figure 1F). Notably, MM-401 mediated reversion was much more efficient than spontaneous or 2i-induced EpiSC conversion (Figure 1E and 1F) (Greber et al., 2010; Lanner and Rossant, 2010).
MLL1 inhibition reactivates silenced X-chromosome in EpiSCs
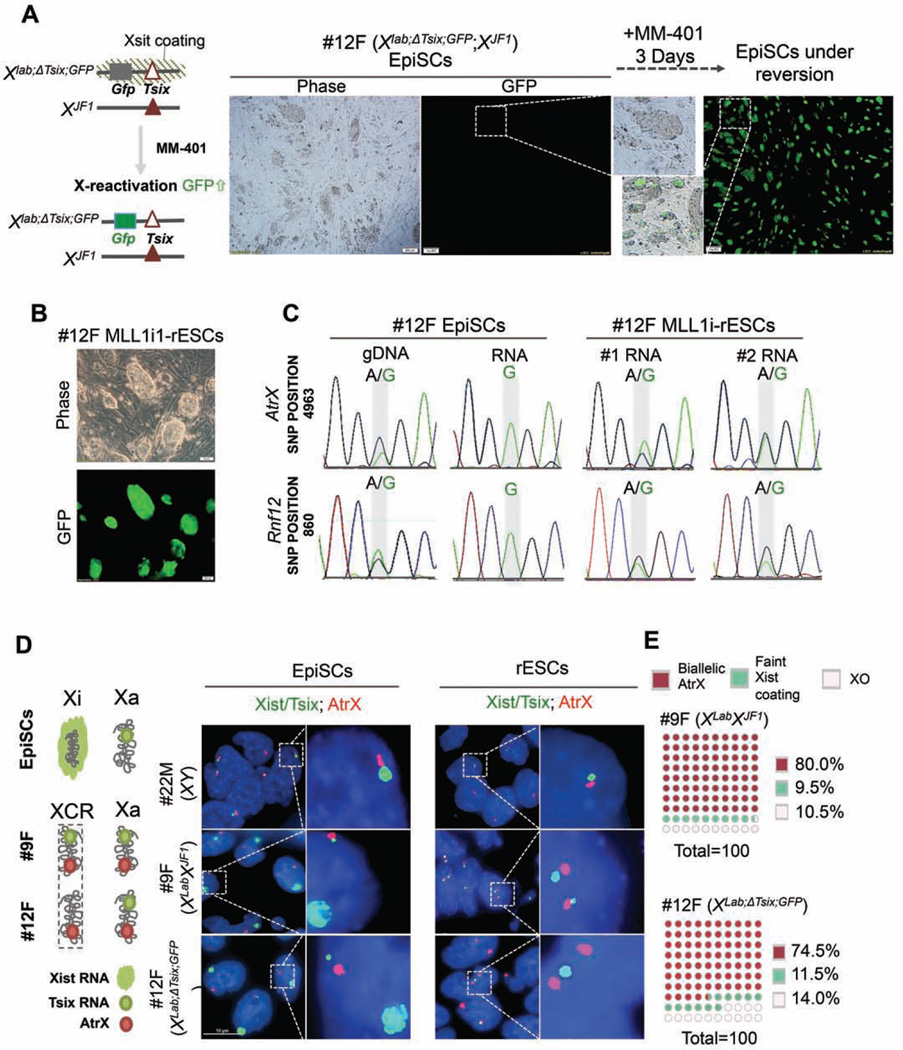
One hallmark of the naïve ESCs as compared to the primed EpiSCs is the lack of inactive Xchromosome (Xi) in female cells (De Los Angeles et al., 2015). Therefore, reactivation of Xi has been used as a bona fide marker for successful EpiSC reprogramming in vitro (Han et al., 2011). To examine whether MLL1 inhibition reactivates Xi in EpiSCs, we used a female EpiSC line (12F) derived from F1 hybrid embryos that carry polymorphic X-chromosomes (Mus musculus-derived XLab and Mus molossinus-derived XJF1) (see Methods). As shown in Figure S3C, the XLab Xchromosome harbors a Gfp transgene and a small deletion of the Tsix gene (ΔTsix) while the XJF1 Xchromosome is wild type (Gayen et al., 2015). In this cell line, the XLab X-chromosome exclusively expresses the Xist RNA undergoes X-inactivation (Gayen et al., 2015). Reactivation of XLab allele in this cell line could be monitored via re-expression of Gfp transgene. Strikingly, MM-401 treatment led to GFP expression from the inactivated XLab allele after three days (Figure 2A, right panel). Half of the cells (~51.6%) in culture were GFP+ at day 3 with concomitant morphological changes and PECAM1 expression (Figure 2A and data not shown). After 2 passages (~ 8 days), almost all clones in culture showed homogeneous GFP expression (Figure 2B). To test clonal conversion efficiency by reactivation of the Xi chromosomes, we seeded 300 EpiSCs in 32 wells, which led to ~90 clones in each well with ~31% plating rate (Figure S3D). MM401 treatment did not affect EpiSCs seeding/attachment (Figure S3E). After 3 days of MM-401 treatment, GFP+ clones emerged in all 32 wells in the presence of either LIF or bFGF (Figure S3F). About 45% (LIF) and 30% (bFGF) of clones in each well were GFP+ (Figure S3D, bottom panel). No GFP+ clones were observed in wells seeded with untreated EpiCSs (Figure S3D and S3F). These results suggest that initiation of EpiSC reprogramming to ESC by MM-401 is efficient, synchronized and independent of exogenous LIF or bFGF signaling.
Figure 2. Reactivation of Xi-chromosome during EpiSC reversion.
A. Left, X-chromosome allele information for 12F female EpiSCs. Right, representative phase/ GFP images of 12F EpiSCs before and after MM-401 treatment. B. Representative images of phase/GFP for Mll1i-rESC after passages 2. C. SNP sequencing for Atrx and Rnf12. The divergent nucleotides in SNP position were highlighted in grey. D. Left, schematics for X-chromosome status in EpiSCs and ESCs. Right, RNA-FISH for Xist/Tsix (green) and Atrx (red) in EpiSCs and Mll1irESCs. Scale bar, 10µm. E. Quantification of biallelic expression of Atrx (red) or incomplete Xi reactivation in Mll1i-rESCs. XO, cells with only one detected X-chromosome.100 nuclei were counted for each cell line from two independent experiments. This Figure is related to Figure S3.
To test whether Xi-reactivation was stably maintained in MLL1i-rESCs, we examined expression of the X-linked genes from XLab allele after multiple passages. Transcripts from genes on XLab can be distinguished from those on XJF1 by single nucleotide polymorphisms (SNPs) (Gayen et al., 2015; Maclary et al., 2014). We analyzed two transcripts associated with SNPs at AtrX (SNP 4393) and Rnf12 (SNP 860) genes in the 12F EpiSC line and reverted MLL1i-rESCs. In EpiSCs, all AtrX and Rnf12 transcripts were from the XJF1 chromosome that had adenosine at SNP 4693 and SNP 860, respectively (Figure 2C, left panel). In MLL1i-rESCs, however, AtrX and Rnf12 transcripts were detected from both X chromosomes (Figure 2C, right two panels). The comparable Sanger chromatogram peaks of the two alleles indicated that XLab in EpiSCs was stably reactivated, confirming stable EpiSC reversion.
MLL1 inhibition robustly reprograms multiple EpiSC lines in vitro
To rule out that the reactivation of the Xi-chromosome after MM-401 treatment was affected by the Tsix mutation in the 12F EpiSC line, we used fluorescence in situ hybridization (RNA-FISH) to examine X-chromosome reactivation in another female EpiSC line (9F) that carried two wild type Xchromosomes. RNA-FISH experiments for the 12F EpiSC line and a male EpiSC line (22M) were included as the controls. RNA-FISH probes against Xist/Tsix (green) and AtrX (red) were used to differentiate inactive vs. active X chromosomes as previously described (Gayen et al., 2015; Maclary et al., 2014). As shown in Figure 2D, both 9F and 12F EpiSCs showed Xist ‘cloud’ on the Xi-chromosome and AtrX expression on the active X chromosome. However, the MM-401-treated 9F MLL1i-rESCs showed bi-allelic expression of AtrX and Tsix with simultaneous loss of Xistcoating (Figure 2D, middle right panel), which indicated activation of both X chromosomes. The control 12F MLL1i-rESCs showed bi-allelic expression of AtrX and single-allelic expression of Tsix due to Tsix mutation on the XLab allele (Figure 2D, bottom right panel). The percentage of 9F and 12F MLL1i-rESCs cells with bi-allelic expression of Atrx was 80% and 75%, respectively (Figure 2E). Tsix and Atrx expression were also detected from the only X-chromosome in male 22M EpiSCs and its derived MLL1i-rESCs (Figure 2D, top panels).
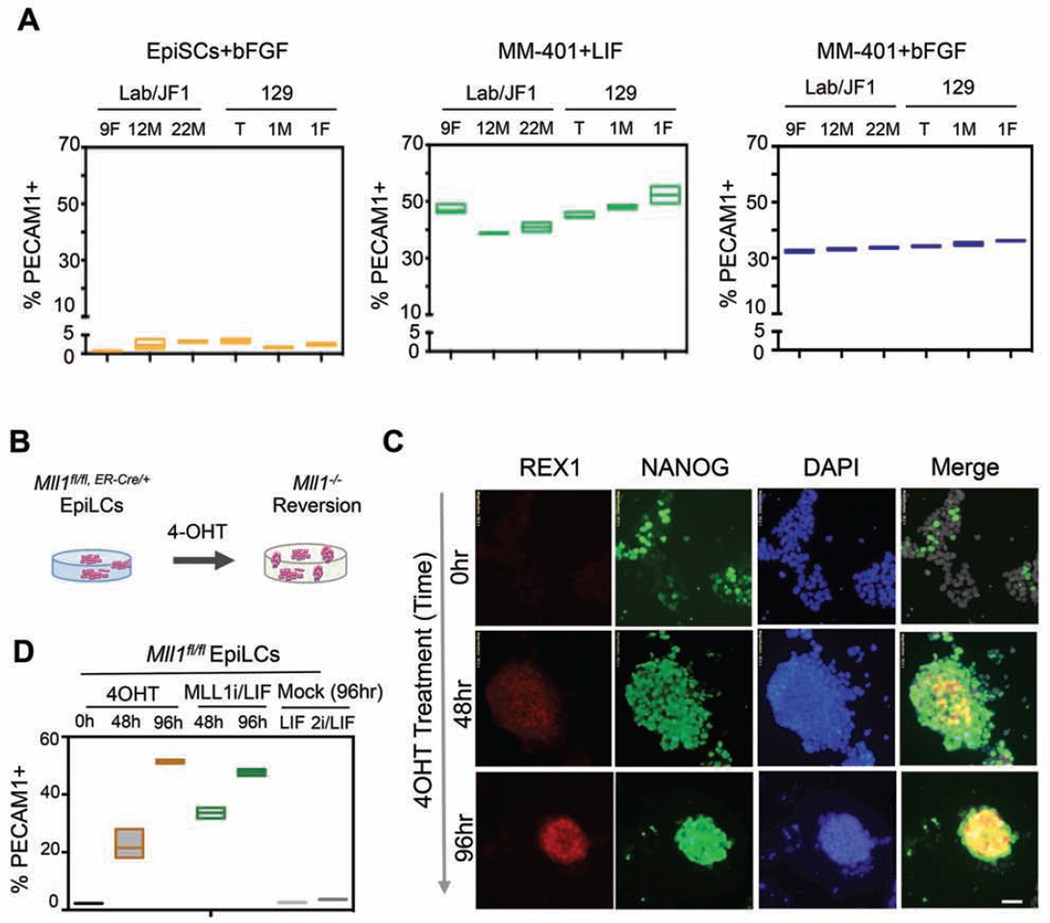
To assess whether pharmacologic MLL1 inhibition could revert EpiSC to naïve ESCs across diverse genetic backgrounds and/or gender, we treated MM-401 on separately derived, additional EpiSC cell lines including inbred 129.1M (male) and 129.1F (female) from the 129/Sv mice and F1 hybrid 12M (male) and 22M (male) from a cross of Mus musculus and Mus molossinus mice as previously described (Gayen et al., 2015). We similarly treated a previous female EpiSC line 129.T (Tesar et al, 2007). As shown in Figure 3A, 3-day MM-401 treatment of the EpiSC lines led to a substantial increase in reversion regardless of genetic background. Similar to 9F cells (Figure 1E), MM-401 treatment led to an increase of PECAM1+ cells from initial 2.3±1.1% to 45.4±4.8% and 34.2±1.3% in LIF/KSR or bFGF/KSR, respectively (Figure 3A, middle and right panels). Homogenous expression of naïve pluripotent markers REX1 and NANOG in these cells further confirmed that these cells had undergone reversion (Figure S3G). Taken together, our studies showed that MLL1 inhibition robustly reprogrammed multiple EpiSC lines to naïve ESCs in vitro.
Figure 3. MLL1 deletion is sufficient to reprogram EpiLCs.
A. FACS for PECAM1 in 6 EpiSC lines that were mock or MM-401 treated as indicated. Y-axis, percent of PECAM1+ cells. B. Schematics for 4-OHT-induced Mll1 deletion. C. Immunofluorescence for NANOG and REX1 after 4-OHT treatments. DAPI was used as counterstain. Scale bar, 20µm. D. FACS for PECAM1 in cells as indicated on top. For A and D, top and bottom edges of the box represents maximum to minimum changes, respectively. Middle line represents mean of three independent experiments. This Figure is related to Figure S3.
MLL1 deletion induces reversion from the primed state in vitro
To confirm that MM-401 acted through MLL1 inhibition during reversion, we tested whether genetic deletion of Mll1 gene promoted reversion of the primed state to the naïve pluripotent state. To this end, we derived the EpiLCs from the Mll1fl/fl;ER-cre/+ ESCs as previously described (Schulz et al., 2014). Mll1 deletion in the cells could be efficiently induced by 4-hydroxytamoxifen (4-OHT) treatments (Figure S3H). Mock-treated EpiLCs were stable under our assay conditions as demonstrated by lack of expression of PECAM1+ and REX1 at all time points (Figure 3 C and D). Similar to MM-401 treatment, Mll1 deletion led to REX1 expression as early as 48 hours after 4-OHT treatments (Figure 3B). Marked and homogenous NANOG expression was also observed upon Mll1 deletion (Figure 3B). The reversion efficiency of Mll1−/− EpiLCs to ESCs was quantified by FACS analyses on PECAM1+ (Figure 3D). About 50% of cells after Mll1 deletion became PECAM1+ (Figure 3D). To further demonstrate that loss of MLL1 indeed led to EpiSC reversion, we depleted MLL1 by shRNA in the EpiSC line 12F (Xlab;ΔTsix;GFP;XJF1) (Figure 2). MLL1 shRNA was confirmed by real time PCR and immunoblot (Figure S3I). After 72hrs of shRNA treatment, 55% of Mll1 shRNA transfected EpiSCs (RFP+) showed Xi-reactivation, as indicated by GFP fluorescence (Figure S3J). No cells that were transfected with scrambled-shRNA had GFP expression (Figure S3J). Taken together, these results strongly argue that blocking MLL1 is the causal epigenetic change that promotes EpiSC reprogramming. These results also confirm that MM-401 promotes EpiSC reversion via MLL1 inhibition.
MLLi-rESCs is developmentally competent in vivo
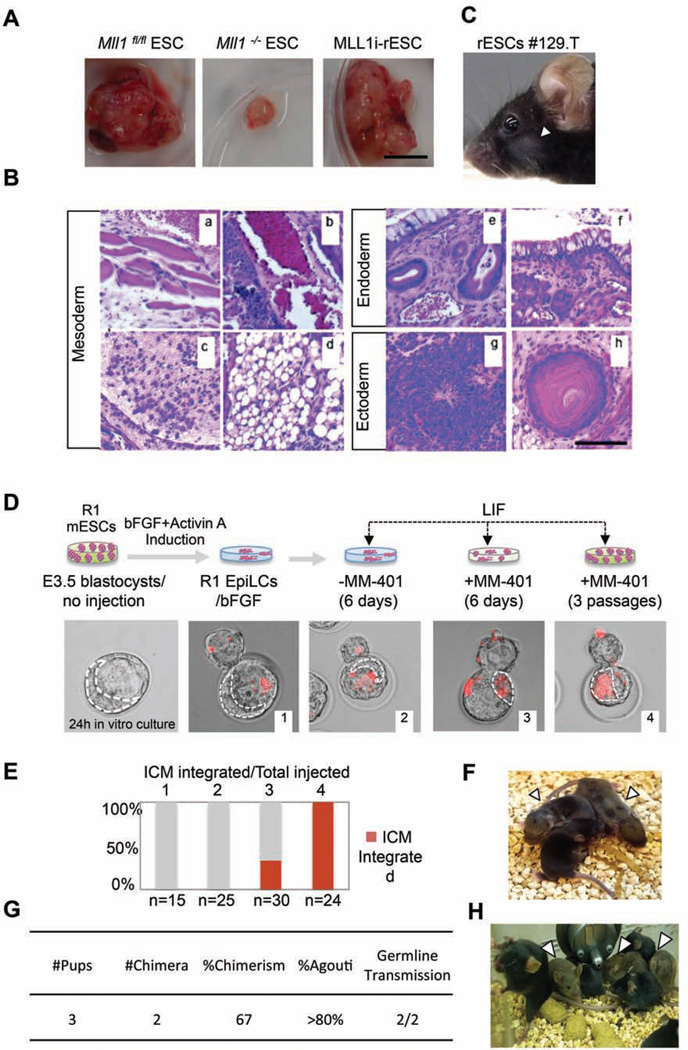
To determine whether rESCs derived by MLL1 inhibition are pluripotent in vivo, we tested the ability of MLL1i-rESCs in developing teratomas in vivo. To this end, we engrafted 12F MLL1i-rESCs to severe combined immunodeficiency (SCID) mice and monitored growth of teratomas. We also engrafted the Mll1flox/flox and Mll1−/− ESCs into SCID mice as the control. After 6 weeks, Mll1−/− rESC-derived teratomas were much smaller in size (Figure 4A) and showed defects in ectoderm and mesoderm differentiation (e.g. red blood cells and blood vessel formation) (Figure S4B), consistent with previous reports on Mll1 null ESCs (Ernst et al., 2004; Katada and Sassone-Corsi, 2010; Yu et al., 1995). In contrast, the stably established MLL1i-rESCs showed no defects in teratoma development (Figure 4A). MLL1i-rESCs-derived teratomas contained well-differentiated endoderm, ectoderm and mesoderm tissues indistinguishable from those of Mll1flox/flox ESCs (Figure 4B and Figure S4A). The differences between MLL1i-rESCs and Mll1−/− rESCs are due to reversible effects of pharmacological inhibitor since MM-401 was not present in vivo after engraftment. The histology results were further confirmed by expression of lineage specific markers in Mll1flox/flox ESCs, Mll1−/− ESCs and MLL1i-rESCs (Figure S4C). These results suggest that MLL1i-rESCs are able to fully differentiate into tissues from all three germ layers. They also highlight a dynamic requirement of MLL1 (or MLL1 inhibition) in stem cell compartment that is different from its function at late gestational stages (see discussion).
Figure 4. MLL1i-rESCs are developmentally competent in vivo.
A. Teratomas generated from engrafted cells as indicated on top. Scale bar, 1cm. B. Hematoxylin and Eosin (H&E) staining of MLL1i-rESC teratoma sections. (a) Muscle; (b) Blood vessel; (c) Cartilage; (d) Adipose-like tissue; (e) respiratory-like epithelium; (f) Gastrointestinal-like epithelium; (g) Neural epithelium; (h) Hair follicle. Scale bar, 500µm. C. F1 chimeric mouse from 129.T MLL1irESCs injected blastocyst. D. Top, schematic of ICM incorporation experiments. Bottom, representative merged fluorescent/phase contrast images of blastocysts after microinjection. Red, dye-labeled R1-EpiLCs. Dash line, inner cell mass (ICM) analyzed by CY5.5 filter. E. Quantification of ICM incorporation in D. 1, R1-EpiLCs/bFGF; 2, R1-EpiLCs/LIF; 3, R1-EpiLCs treated with MM- 401/LIF for 6 days; 4, R1-EpiLCs treated with MM-401/LIF for 3 passages (12 days). Y-axis, % of ICM with labeled rESCs. The number of microinjected blastocysts was indicated on bottom. F. F1 chimera from host mice engrafted with Mll1i-R1-rESC containing blastocyst. G. Summary of chimeric contribution in F1. H. F2 progenies from chimeric F1 mice. In C, F and G, arrowheads show agouti coat color. This Figure is related to Figure S4.
To determine whether MLL1i-rESCs could contribute to chimeras, we injected the 129.T-rESCs (Figure 3A and Figure S3G) into C57BL/6 blastocysts. MM-401 was withdrawn from the cell culture prior to the injection. The injected blastocysts were then transferred into the C57BL/6 host mice. Chimeric animals were recovered at ~10% frequency (3 out of 33) among live pups from the injected embryos (Figure 4C). The ability of MLL1i-rESCs to integrate into the blastocyst and give rise to chimeric mice unequivocally established MLL1i-rESCs as bona fide naïve pluripotent cells.
To assess the developmental potential of ESCs and MLL1i-rESCs of isogenic origin without genetic variability/diversity, we compared the abilities of well-established R1 ESCs and our reprogrammed R1 EpiLCs to contribute into the ICM integration and chimera generation. To this end, we treated R1-EpiLCs, confirmed by immunofluorescence for NANOG and REX1 (Figure S4C), with MM-401 for either 6 days or three passages (12 Days) in LIF/KSR media (Schematics in Figure S4D). Mock-treated R1-EpiLCs in bFGF/KSR or LIF/KSR media were included as the controls. About 10hrs prior to blastocyst injection, the cells were labeled by cell-permeable dye (Vybrant® DyeCycle™ Ruby) for visualization. As shown in Figure 4D and 4E, the reprogrammed R1-EpiLCs were incorporated into ICM and the incorporation efficiency correlated with duration of MM-401 treatment. At 20hrs post-injection, 100% of blastocysts (i.e. 24/24) injected with MM-401 treated passage 3 R1-EpiLCs had labeled cells in the ICM. Additionally, 33% of blastocysts (i.e. 10/30) injected with 6-day MM-401 treated R1-EpiLCs harbored labeled cells in the ICM. As a control, mock-treated R1-EpiLCs did not contribute to the ICM (0/15 and 0/25, Figure 4D and 4E), confirming that R1-EpiLCs maintained a stable primed state in the absence of MM-401 treatment. The reprogrammed R1-rESCs contributed to live-born chimeras that developed into healthy adults (Figure 4F). MLL1i-R1-rESCs injected blastocysts resulted in 67% chimeric animals with 80% coat color chimerism (% agouti), similar to that of naïve R1-ESCs (Nagy et al., 1993). Importantly, the reprogrammed R1-rESCs could be transmitted to germline and gave rise to healthy F2 progenies (Figure 4H). Taken together, the results showed that MLL1 inhibition efficiently reprogrammed EpiSCs/EpiLCs to an authentic naïve pluripotent state.
MLL1 inhibition rapidly leads to transcriptional changes during EpiSC reprogramming
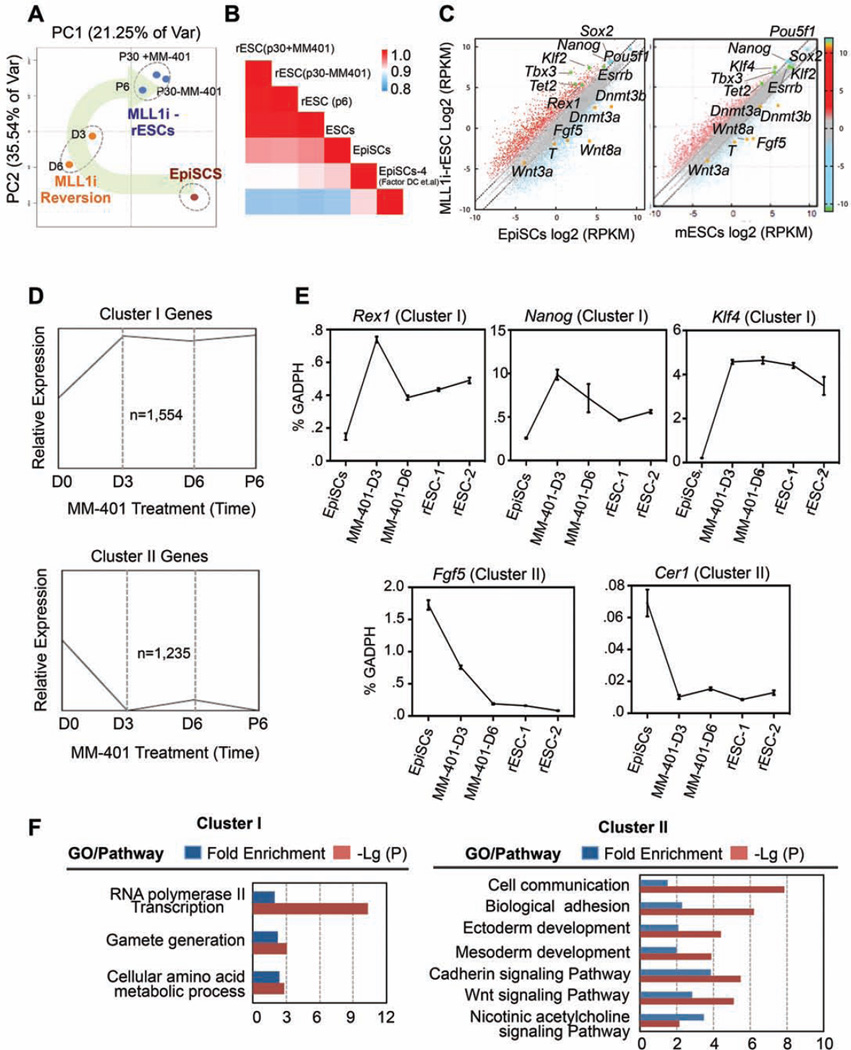
To understand the underlying mechanisms, we examined transcriptome changes by RNA-sequencing (RNA-seq) at day 0, 3, 6, as well as passage 6 (P6) and 30 (P30) during EpiSC reversion. We also analyzed the transcriptome of P30 MLL1i-rESC cells that were cultured without MM-401 since P6. The obtained transcriptomes were subjected to principal component analysis (PCA). Interestingly, transcription profiles of EpiSCs after 3–6 days of MM-401 treatment were distinct from both untreated EpiSCs and final MLL1i-rESCs (Figure 5A). The rapid transcriptome changes after MM-401 treatment were consistent with the phenotypic changes in these cells. Furthermore, global reprogramming of the transcriptome was completed at P6 after MM-401 treatment. Transcriptome at this time point clustered together with that of P30 MLL1i-rESC cells. Interestingly, P30 MLL1i-rESCs that were cultured without MM-401 after P6 had similar transcription profiles as those P6 and P30 cells cultured with continuous presence of MM-401 (Figure 5A). These results suggested that MM-401 induced reversion resulted in a metastable pluripotent state at P6, which could be sustained independent of Mll1. An unsupervised Pearson correlation analysis showed that this metastable pluripotent state bore a more resemblance to naïve ESCs than EpiSCs at the transcriptional level (Figure 5B). In support of these global analyses, P6 MLL1i-rESCs exhibited higher expression of naïve specific markers (e.g., Klf4/2, Tbx3, Essrb, Tet2 and Rex1) and lower expression of epiblast markers (e.g., Fgf5, Wnt8a, Dnmt3a/b and T) as compared to initial untreated EpiSCs (Figure 5C). Higher expression of Sox2 and Nanog was also observed in these cells (Figure 5C, left panel). Furthermore, a comparison between MLL1i-rESC and authentic ESCs (LIF/serum) showed that MLL1i-rESCs had higher expression of naïve markers (e.g. Klf4, Tbx3) and lower expression of primed cell markers (e.g., Wnt3a, Fgf5, Dnmt3b) (Figure 5C, right panel), a feature reminiscent of the ground state ESCs.
Figure 5. Characterization of the MLL1-dependent transcriptome.
A. Principle component analyses on transcriptome of EpiSCs at different time points after MM-401 treatment. The reprogramming process is highlighted. B. Pearson correlation coefficient for pair-wise comparison as indicated. RNA-seq for EpiSCs-4 was from (Factor et al., 2014). C. Scatter plots of global gene expression by RNA-seq in log2(RPKM)(reads per kilobase per million) in MLL1i-rESCs vs. EpiSCs (left) and MLL1i rESCs vs. ESCs/LIF/Serum (right). The grey dash lines delineate the boundaries of 2-fold difference in gene expression. Pluripotent markers were highlighted in blue or green and EpiSC makers in orange. D. Gene population clustered by common expression changes (RPKM>1) during reprogramming (K-means=3). E. Real time PCR for selected cluster I and cluster II genes. Gene expression (mean ± s.d.) was presented relative to Gapdh in each sample. F. Panther statistical overrepresentation test of cluster I and II genes. This Figure is related to Figure S6 and Table S1, S6 and S7
We categorized characteristic gene expression patterns after MM-401 treatment. Among ~8,000 genes commonly identified at all time points by RNA-seq (RPKM>1), ~2,800 genes were grouped into two clusters that shared similar expression kinetics during EpiSC reversion. Specifically, they were either abruptly up regulated (1554 genes, cluster I) or down regulated (1235 genes, cluster II) at day 3 of MM-401 treatment and maintained relative expression afterwards (Figure 5D and Figure S6A). Naïve ESC markers Rex1, Nanog, and Klf4 as well as EpiSC markers Fgf5 and Cer1 were found in clusters I and II, respectively. Their expression was confirmed by real-time PCR (Figure 5E). Complete lists of cluster I and II genes were included in Table S1. These results support a highly synchronized EpiSC reversion at the molecular level. Gene Ontology (GO) term analyses showed that cluster I genes were mostly involved RNA polymerase II transcription and amino acid metabolism (Figure 5F). In contrast, cluster II genes were enriched for several pathways important for epiblast development in vivo or ex vivo (Gadue et al., 2006; ten Berge et al., 2008). They include cell communication, ectoderm/mesoderm development, biological adhesion as well as Cadherin/Wnt signaling pathways (Figure 5F).
MM-401 treatment alters MLL1 binding, H3K4me1 distribution and gene expression in EpiSCs
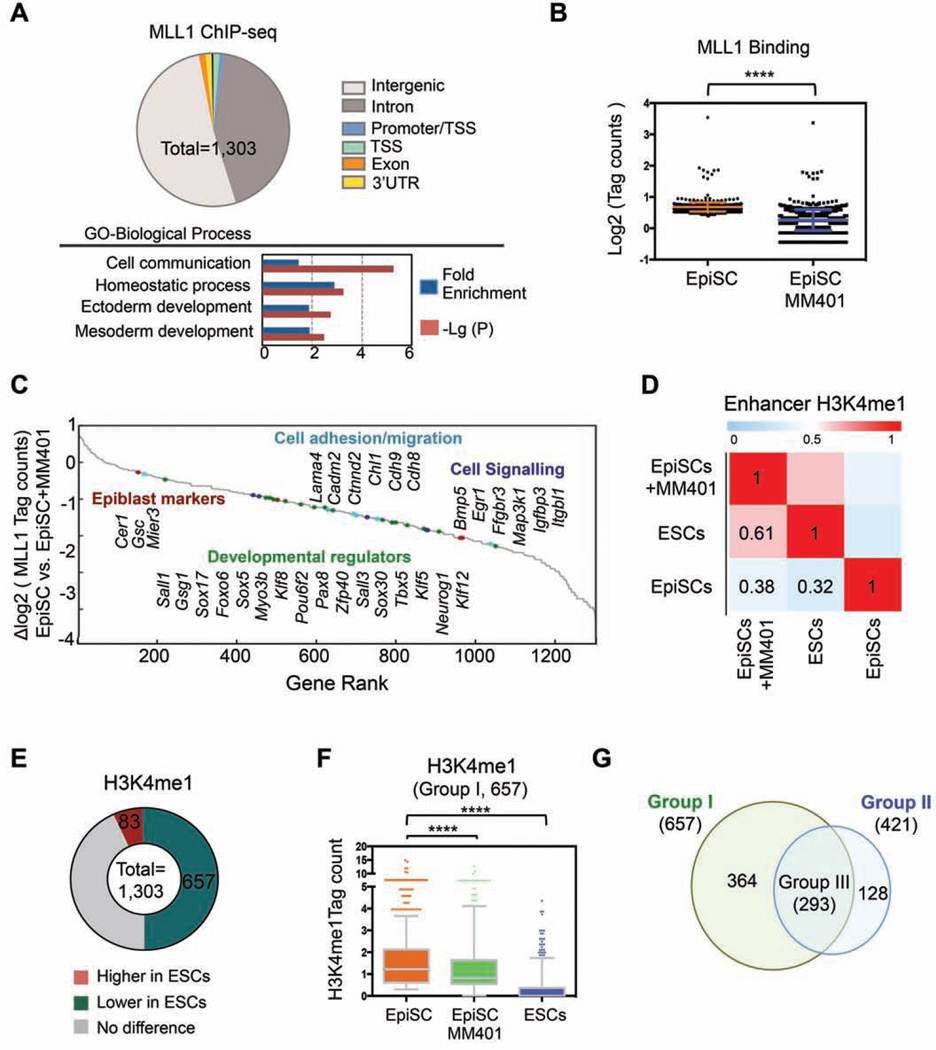
To identify MLL1 direct targets in EpiSCs and to characterize changes in H3K4me upon MM-401 treatment, we performed chromatin immunoprecipitation followed by Illumina-based, next generation sequencing (ChIP-seq) for MLL1, H3K4me1 and H3K4me3 in EpiSCs with or without 4-day MM-401 treatment. ChIP-seq identified 1,303 MLL1 peaks in EpiSCs. As shown in Figure 6A, majority of MLL1 binding sites in EpiSCs were at either intergenic regions (51.8%) or introns (42.7%) (complete list see Table S2), suggesting that MLL1 probably functions at regulatory enhancers (defined by H3K4me1) in EpiSCs. About 8.3% of cluster II genes in Figure 5D were MLL1 direct targets. Importantly, MLL1 binding was reduced at ~62% of MLL1 direct targets after MM-401 treatment (Figure 6B, log2 (tag ratio) <−1)), consistent with disruption of the MLL1 complex by MM-401 (Cao et al., 2014). The gene rank highlighted lineage specific transcription factors (e.g. Sox5, Sox17, Klf5), epiblast markers (e.g. Cer1 and Gsc), cadherins (e.g. Cdh8, Cdh9) and cell signaling genes (e.g. Bmp5, Egr1) as the MLL1 direct targets (Figure 6C). Notably, MLL1 did not bind to naïve ESC markers (e.g Nanog and Pou5f1), consistent with lack of self-renewal defects in Mll1−/− ESCs (Ernst et al., 2004). ChIP-seq results were confirmed by ChIP-qPCR at selected genes (Figure S5).
Figure 6. MLL1 regulates dynamic H3K4me during pluripotent stem cells conversion.
A. Top, MLL1 distribution in EpiSCs relative to gene structure. Bottom, Panther pathway analyses on the annotated MLL1 direct targets. B. MLL1 binding in EpiSCs treated with or without MM-401. Y-axis shows the compiled log2 tag counts within MLL1 peak center ± 200bp. Data is presented as mean ± s.d. ****, p<0.0001 in Mann-Whitney test. C. Gene rank based on changes in MLL1 binding after MM-401 treatment. D. Pearson correlation of H3K4me1 in two pluripotent states (n=103,748). H3K4me1 in ESCs (GSE47949) and EpiSCs (GSE57407) were from public database. E. Changes of H3K4me1 at MLL1 binding sites in ESCs vs. EpiSCs. F. Box plots for H3K4me1 level in EpiSCs, ESCs and EpiSCs treated with MM-401. Y-axis, H3K4me1 tag counts within MLL1 peak center ± 200bp. 657 genes defined in E were included in this analyses. In Box plot, central mark represents median value and edges represent 25th and 75th percentiles of H3K4me1 level. The whiskers extended to 5th to 95th percentile and outliers are plotted individually. ****, p<0.0001 in Mann- Whitney test. G. Venn diagram for genes that had lower H3K4me1 in ESCs (Group I) or lower H3K4me1 in EpiSCs after MM-401 treatment (Group II). This Figure is related to Figure S5, S6 and Table S2–5.
ChIP-seq for H3K4me1 and H3K4me3 in EpiSCs identified 99,356 and 26,543 peaks in EpiSCs, respectively (Table S3 and S4). Consistent with previous studies (Factor et al., 2014), H3K4me1, but not H3K4me3, was dramatically different between ESCs and EpiSCs with Pearson correlation coefficient of 0.32 (Figure 6D and Figure S6B). Interestingly, MM-401 treatment led to genome-wide change of H3K4me1 in EpiSCs (Figure 6D). The enhancer H3K4me1 profile in EpiSCs treated with MM-401 for 3 days bore a more resemblance to that of ESCs than EpiSCs, with Pearson correlation coefficients of 0.61 and 0.32, respectively (Figure 6D). In comparison, H3K4me3 at gene promoters was little affected by MM-401 treatment (Figure S6B).
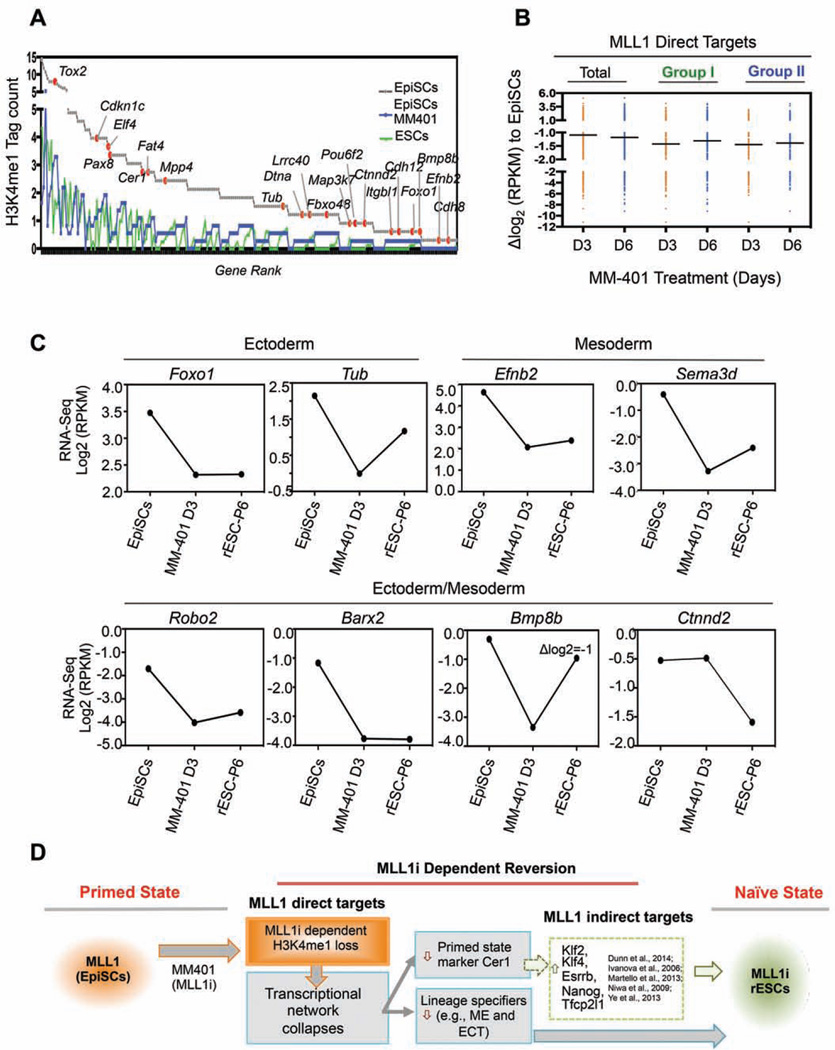
We next examined H3K4me1 changes at MLL1 direct targets. We found two typical H3K4me1 distribution patterns relative to MLL1 peaks in EpiSCs (Figure S6C, blue and purple lines). MM-401 treatment led to significant down regulation of H3K4me1 surrounding both classes of MLL1 targets (Figure S6C). We further examined H3K4me1 within 1 nucleosome (+/− 200bp) of the MLL1 peak centers in EpiSCs and ESCs. Strikingly, significant difference in H3K4me1 (Δlog2 H3K4me1 tag count <−1 or >1) was found at 740 out of 1,303 MLL1 peaks between EpiSCs and ESCs (Figure 6E). Among them, 657 MLL1 targets (Group I, 50.5% of total) had drastically lower H3K4me1 in ESCs than EpiSCs (Figure 6E). More importantly, H3K4me1 at these physiologically relevant sites was significantly down regulated by MM-401 during EpiSC reprogramming (Figure 6F). In fact, vast majority of MLL1 targets that had lower H3K4me1 (Δ log2 H3K4me1 tag count < −0.5, Group II) after MM-401 treatment overlapped with Group I genes above (Figure 6G and complete list see Table S5). As the result, we identified 293 genes (Group III) that had significant H3K4me1 reduction in both ESCs and MM401-treated EpiSCs. Gene rank based on H3K4me1 tag count in EpiSCs was shown in Figure 7A. Representative genes with known functions at the stage of epiblast development were highlighted (Figure 7A, grey line). ChIP-qPCR confirmation for H3K4me1 at selected genes was shown in Figure S5. Gene expression analyses showed that majority of the MLL1 direct targets were rapidly down regulated after MM-401 treatment (Figure 7B). Median reductions for Group I and Group II genes were ~2.8 fold (Δ log2 RPKM = −1.5) (Figure 7B and Figure S6D). Real-time qPCR confirmation of selected genes was shown in Figure S6E. These results suggest that MM-401 promotes EpiSC reversion by blocking developmentally up-regulated H3K4me1 and consequent expression of MLL1 direct targets.
Figure 7. Direct MLL1 gene network in EpiSCs.
A. Gene rank of H3K4me1 tag counts at Group III genes in EpiSCs and their levels in ESCs and MM401-treated EpiSCs. B. Scattered plot for expression of Group I and Group II genes after MM-401 treatment at day 3 (D3) and day 6 (D6). Genes that had log2(fold change)>1 or <−1 were included in this analyses. For A and B, Group I and II are defined in Figure 6G. C. Expression of selected MLL1 direct targets during reversion. Average Log2(RPKM) from RNA-seq duplicates was presented after normalization. D. The model for the molecular roadmap of MM-401-induced EpiSC reversion. This Figure is related to Figure S7.
MLL1 regulates a gene network in EpiSCs
Analysis of the MLL1 direct targets in EpiSCs showed that 222 out of 421 Group II genes (Figure 6G) belong to a gene network (Figure S7A). They distributed widely in cells (Figure S7A), suggesting that MLL1 is probably a master regulator and influences multifaceted functions in EpiSCs. Ninety-seven genes (44% of total) in this network had significant and rapid reduction in expression (log2 fold change <−1) upon MM-401 treatment (Figure S7A, yellow). Interestingly, two pathways were especially enriched in this MLL1 network: 1) the biological adhesion pathway as exemplified by multiple membrane-bound proteins (e.g. Cacna2d4, Stx18, Pappa, Dpp4) (Figure S7A, green circle) and 2) the developmental pathway as exemplified by prominent regulators of early cell lineage specification (e.g. Nerrog1, Tub, Cdh9, and Efna5) (Figure 7C and S7A, red circle) (Brennan et al., 2001; Puigserver et al., 2003). TGFβ/Cadherin pathway genes were also identified in the MLL1 network. The prominence of lineage specification and cell adhesion pathways in the MLL1 network is stark contrast to the lack of naïve ESC factors (e.g. Pou5f1, Nanog and Sox2) (Figure 5F and Figure 7C). These results suggest that MLL1 does not regulate the ESC core transcription circuitry. Instead, MLL1 inhibition seems to initiate reprogramming by repressing characteristic EpiSC features (see discussion).
DISCUSSION
Here we show that MLL1 inhibition by small molecule inhibitor MM-401 or Mll1 deletion is sufficient to reprogram the primed EpiSCs to naïve pluripotency. The EpiSC reprogramming occurs with high efficiency, with 50% cells exhibiting naïve ESC features after 3-days. The rESCs have full development potential and give rise to live chimeras. Mechanistic studies identify an MLL1 network in EpiSCs and show that direct modulation of a discrete histone mark is sufficient to promote the naïve pluripotent state. Rather than a passive epigenetic mark, MLL1-mediated H3K4me plays a causal role in the acquisition of naïve pluripotency.
Our study shows that MM-401 is a powerful tool to study the role of Mll1 in cell fate determination. MM-401 disrupts MLL1 chromatin binding at a significant subset of MLL1 targets in EpiSCs (Figure 6B) and induces EpiSC reprogramming in a fashion similar to Mll1 gene deletion (Figure 3B). Previous genetic studies have established that Mll1 deletion does not affect ESC self-renewal (Ernst et al., 2004; Glaser et al., 2009). Instead, it impairs ESC differentiation into neural or hematopoietic lineages, consistent with in vivo studies (Jude et al., 2007; Lim et al., 2009; McMahon et al., 2007). Here we take advantage of the reversibility of the pharmacological inhibitor MM-401 to reveal a dynamic requirement of MLL1 in defining early pluripotent states. Unlike Mll1−/− ESCs, MLL1i-rESCs have full developmental potential upon MM-401 withdrawal in vivo. Our study indicates that MLL1 probably acts as a one-way directional valve in preventing course reversal at key junctions of early epiblast differentiation, which is consistent with its dynamic expression during development (Figure S1). Notably, Mll1−/− embryos are able to develop till mid-to-late gestation (Yu et al., 1995), suggesting that loss of MLL1 function is not sufficient to entirely block developmental signals that drive early embryogenesis. This is consistent with our observation that MM-401 cannot stably sustain reverted naïve ESCs in the presence of continuous bFGF signaling (Figure S3B). We also would like to point out that the effects of MM-401 or Mll1 deletion in ESCs are distinct from those of Wdr5 depletion (Ang et al., 2011), which probably disrupts ESC transcription circuitry independent of MLL (Li et al., 2012).
The EpiSC reprogramming by MLL1 inhibition or deletion is a surprise given the lack of apparent functions for MLL1 in ESC self-renewal and maintenance. This is a departure from previously reported reprogramming methods that involve either overexpression of ESC transcription factors and/or blockade of cell signaling (Cahan and Daley, 2013; Nichols and Smith, 2009). Instead, MLL1 inhibition directly affects lineage commitments towards ectoderm, mesoderm or both germ layers (Figure 7C and S7A). It also represses the EpiSC markers (Figure 7C). These results raise an interesting possibility that MM-401 restores naïve pluripotency by blocking pathways that lead to the primed pluripotent state. It supports the notion that the naïve pluripotent state represents a ‘passive’ or ‘uninstructed’ state (Silva and Smith, 2008) that can be captured by blocking alternative cell identity. In this context, we envision that blocking MLL1 serves to revert stem cells to the ‘uninstructed’ naïve pluripotent state by erasing developmental ‘scripts’. Alternatively, since the naïve pluripotent state is actively maintained by the network of core transcription factors and requires network reorganization for both entry and exit of the naïve state (Factor et al., 2014; Gafni et al., 2013), it is also possible that MLL1 inhibition perturbs lineage determinant factors, which indirectly reorganize the naïve transcription network through extensive feedback controls or crossregulations (De Los Angeles et al., 2015). Indeed, transcriptome analyses show that MLL1 inhibition indirectly up-regulates ESC markers during the reprogramming process (Figure 5F and Figure 7). Interestingly, they include previously reported EpiSC reprogramming factors such as Klf4 (Guo et al., 2009), Klf2 and Nanog (Stuart et al., 2014), Esrrb (Festuccia et al., 2012), Tfcp2l1 (Ye et al., 2013) and orphan nuclear receptor Nr5a2 (Guo and Smith, 2010) (Figure 5E and S7B). We also see a modest reduction of Mbd3 (Rais et al., 2013) (Figure S7B). Simultaneous regulations of parallel reprogramming pathways probably underlie the highly efficient and synchronized EpiSC reversion by MM-401. Specifically, MM-401-induced EpiSC reprogramming changes clone morphology within 24hrs (data not shown). About 50% cells have up regulation of PECAM1 and REX1 as well as Xireactivation occurring within 72hrs (Figure 1 and Figure 2). We also would like to point out that MLL1i-rESCs, once reverted, stably maintain naïve characteristics even in the absence of continuous MLL1 inhibition (Figure 4 and 5A).
Dynamic reorganization of H3K4me1-defined enhancer landscapes has been reported when ESCs differentiate into EpiSCs (Buecker et al., 2014; Factor et al., 2014). However, the causal link between epigenetic modifications and initiation of cell fate conversion has not been established. In fact, epigenetic changes are often depicted as consequences of changed transcription network or cell signaling (De Los Angeles et al., 2015). Here we show that epigenetic change itself may be sufficient to trigger EpiSC reprogramming and blocking MLL1 function constitutes a key rate-limiting step or barrier in EpiSC reversion. MLL1 inhibition in EpiSCs drastically shifts the enhancer landscape to that of naïve ESCs (Figure 6D). Moreover, most of MLL1 binding sites undergo reversal of otherwise developmentally up regulated H3K4me1 upon MM-401 treatment (Figure 6F). Notably, MLL1 direct targets represent only a small fraction of H3K4me1 alteration during EpiSC reversion. It is likely that MLL1 inhibition leads to subsequent changes by other H3K4 methyltransferases that warrant future studies. One caveat is that although non-histone substrates of MLL1 have yet to be identified, we cannot completely rule out the possibility that other MLL1-dependent methylation event(s) drive the collapse of EpiSCs network.
Finally, our study supports that erasure, rather than deposition, of key epigenetic marks is associated with restoration of naïve pluripotent state. Factors, such as MLL1, whose inhibition or deletion does not inhibit epiblast development in vivo or affect the ESC self-renewal in vitro, may still play a critical role to regulate acquisition of naïve pluripotency. In this context, it would be interesting to examine whether blocking H3K27me3 and DNA methylation, which are dynamically up regulated, affects EpiSC reprogramming (Gafni et al., 2013; Nora et al., 2012). Future interrogation of the roles of epigenetic regulators at crucial developmental windows is key to furthering our understanding of epigenetic regulation in cell fate determination.
EXPERIMENTAL PROCEDURE
Establish and culture of pluripotent cell lines
Mll1f/f; Cre-ERTM ESCs and Mll1−/− ESCs were derived from ICM of 3.5 d.p.c blastocysts and cultured on top of MEF feeder cells in GMEM containing 15% FBS and LIF. EpiSCs were derived from periimplantation stage mouse embryos and cultured in in K15F5 medium containing Knockout DMEM with standard supplements. EpiLC induction was performed in N2B27-based medium containing 15% KSR, 10ng/ml bFGF and 20 ng/ml Activin A.
EpiSC reprogramming experiment
EpiSCs were passaged in single cell or small clumps on MEF feeder cells. MM-401 (50–100uM final concentration) was added to culture medium immediately or 2–3 days after EpiSCs forming clones. MM-401 is replenished to reach 50–100µM concentration during each passage. rESC lines can be established by clone picking or en masse. rESCs for chimera test were derived clonally.
Immunofluoresence (IF), AKP staining, RNA-FISH
IF was carried out for NANOG, OCT4 and REX1 at 1:200, 1:100 and 1:200 dilution, respectively. The Vector™ Alkaline Phosphatase (AP) Staining Kit was used for AKP staining. RNA-FISH was performed as described in Maclary et al., 2014. Details see Supplemental Information.
In vivo characterization of rESCs
The in vivo experiments were performed at Transgenic Core Facility of University of Michigan. For ICM integration, rESCs were labeled with Vybrant® DyeCycle™ Ruby dye 10 hours prior to blastocyst injection. The cells were visualized at 20 hours after injection.
ChIP-Seq and RNA-Seq Analyses
H3K4me1 and H3K4me3 ChIP-seq data for untreated cells were from GSE47949 (ESCs) and GSE57407 (EpiSCs). ChIP-Seq reads were aligned to UCSC mm9 using Bowtie2 and analyzed by HOMER. RNA-seq was analyzed by Tophat (version 2.0.3). Details on pathway, PCA and network analyses as well as antibody info see Supplemental Information. Anti-MLL1 antibody was described in (Dou et al., 2005).
Supplementary Material
HIGHLIGHTS.
MLL1 inhibition by MM-401 promotes reversion of EpiSCs to naïve pluripotency
MLL1 controls the rate-limiting step in EpiSC reprogramming
Identification of an MLL1-dependent gene network in pluripotent stem cells
Epigenetic perturbation is sufficient to initiate EpiSC-to-ESC reversion
Acknowledgments
This work is supported by NIGMS (GM082856), Leukemia and Lymphoma Society to Y.D, an NIH Director’s New Innovator Award (DP2-OD-008646-01) to S.K, NIGMS (5R01GM094231) to AN and NCI (CA117307-04) to Y.D and S.W. We are grateful to Dr. Saunders and Ms. Hughes at University of Michigan for blastocyst injections and generation of chimeras. We are grateful to Drs. Brady for Mll1flox/flox mice and Tesar for 129.T EpiSCs.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
H.Z designed, performed experiments and wrote the manuscript. S.G derived EpiSCs and performed RNA-FISH and SNP analyses. J.X performed bioinformatics analyses. S.G and J.X contribute equally to this work. B.Z generated the Mllf/f ESCs and conducted the teratoma assays. Y.S performed MLL1 ChIP-seq on EpiSCs. A.S, supervised by A.N, generated the MLL1 network. H.K, L.L and S.W developed MM-401. S.K, R.R and Y.D contributed to experimental designs and manuscript editing.
Accession numbers
The data are accessible through NCBI's Gene Expression Omnibus with accession number GSE66112.
References
- Ang YS, Tsai SY, Lee DF, Monk J, Su J, Ratnakumar K, Ding J, Ge Y, Darr H, Chang B, et al. Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell. 2011;145:183–197. doi: 10.1016/j.cell.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S, Tang F, Li X, Hayashi K, Gillich A, Lao K, Surani MA. Epigenetic reversion of post-implantation epiblast to pluripotent embryonic stem cells. Nature. 2009;461:1292–1295. doi: 10.1038/nature08534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bledau AS, Schmidt K, Neumann K, Hill U, Ciotta G, Gupta A, Torres DC, Fu J, Kranz A, Stewart AF, et al. The H3K4 methyltransferase Setd1a is first required at the epiblast stage, whereas Setd1b becomes essential after gastrulation. Development. 2014;141:1022–1035. doi: 10.1242/dev.098152. [DOI] [PubMed] [Google Scholar]
- Brennan J, Lu CC, Norris DP, Rodriguez TA, Beddington RS, Robertson EJ. Nodal signalling in the epiblast patterns the early mouse embryo. Nature. 2001;411:965–969. doi: 10.1038/35082103. [DOI] [PubMed] [Google Scholar]
- Brons IG, Smithers LE, Trotter MW, Rugg-Gunn P, Sun B, Chuva de Sousa Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA, et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- Buecker C, Srinivasan R, Wu Z, Calo E, Acampora D, Faial T, Simeone A, Tan M, Swigut T, Wysocka J. Reorganization of enhancer patterns in transition from naive to primed pluripotency. Cell Stem Cell. 2014;14:838–853. doi: 10.1016/j.stem.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahan P, Daley GQ. Origins and implications of pluripotent stem cell variability and heterogeneity. Nat Rev Mol Cell Biol. 2013;14:357–368. doi: 10.1038/nrm3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao F, Townsend EC, Karatas H, Xu J, Li L, Lee S, Liu L, Chen Y, Ouillette P, Zhu J, et al. Targeting MLL1 H3K4 methyltransferase activity in mixed-lineage leukemia. Mol Cell. 2014;53:247–261. doi: 10.1016/j.molcel.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Los Angeles A, Ferrari F, Xi R, Fujiwara Y, Benvenisty N, Deng H, Hochedlinger K, Jaenisch R, Lee S, Leitch HG, et al. Hallmarks of pluripotency. Nature. 2015;525:469–478. doi: 10.1038/nature15515. [DOI] [PubMed] [Google Scholar]
- dos Santos RL, Tosti L, Radzisheuskaya A, Caballero IM, Kaji K, Hendrich B, Silva JC. MBD3/NuRD facilitates induction of pluripotency in a context-dependent manner. Cell Stem Cell. 2014;15:102–110. doi: 10.1016/j.stem.2014.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Y, Milne TA, Ruthenburg AJ, Lee S, Lee JW, Verdine GL, Allis CD, Roeder RG. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat Struct Mol Biol. 2006;13:713–719. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- Dou Y, Milne TA, Tackett AJ, Smith ER, Fukuda A, Wysocka J, Allis CD, Chait BT, Hess JL, Roeder RG. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 2005;121:873–885. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- Ernst P, Mabon M, Davidson AJ, Zon LI, Korsmeyer SJ. An Mll-dependent Hox program drives hematopoietic progenitor expansion. Current biology : CB. 2004;14:2063–2069. doi: 10.1016/j.cub.2004.11.012. [DOI] [PubMed] [Google Scholar]
- Factor DC, Corradin O, Zentner GE, Saiakhova A, Song L, Chenoweth JG, McKay RD, Crawford GE, Scacheri PC, Tesar PJ. Epigenomic comparison reveals activation of "seed" enhancers during transition from naive to primed pluripotency. Cell Stem Cell. 2014;14:854–863. doi: 10.1016/j.stem.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Factor DC, Najm FJ, Tesar PJ. Generation and characterization of epiblast stem cells from blastocyst-stage mouse embryos. Methods Mol Biol. 2013;1074:1–13. doi: 10.1007/978-1-62703-628-3_1. [DOI] [PubMed] [Google Scholar]
- Festuccia N, Osorno R, Halbritter F, Karwacki-Neisius V, Navarro P, Colby D, Wong F, Yates A, Tomlinson SR, Chambers I. Esrrb is a direct Nanog target gene that can substitute for Nanog function in pluripotent cells. Cell Stem Cell. 2012;11:477–490. doi: 10.1016/j.stem.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish JA, Chai B, Wang Q, Sun Y, Brown CT, Tiedje JM, Cole JR. FunGene: the functional gene pipeline and repository. Front Microbiol. 2013;4:291. doi: 10.3389/fmicb.2013.00291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadue P, Huber TL, Paddison PJ, Keller GM. Wnt and TGF-beta signaling are required for the induction of an in vitro model of primitive streak formation using embryonic stem cells. Proc Natl Acad Sci U S A. 2006;103:16806–16811. doi: 10.1073/pnas.0603916103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gafni O, Weinberger L, Mansour AA, Manor YS, Chomsky E, Ben-Yosef D, Kalma Y, Viukov S, Maza I, Zviran A, et al. Derivation of novel human ground state naive pluripotent stem cells. Nature. 2013;504:282–286. doi: 10.1038/nature12745. [DOI] [PubMed] [Google Scholar]
- Gayen S, Maclary E, Buttigieg E, Hinten M, Kalantry S. A Primary Role for the Tsix lncRNA in Maintaining Random X-Chromosome Inactivation. Cell Rep. 2015;11:1251–1265. doi: 10.1016/j.celrep.2015.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser S, Lubitz S, Loveland KL, Ohbo K, Robb L, Schwenk F, Seibler J, Roellig D, Kranz A, Anastassiadis K, et al. The histone 3 lysine 4 methyltransferase, Mll2, is only required briefly in development and spermatogenesis. Epigenetics Chromatin. 2009;2:5. doi: 10.1186/1756-8935-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greber B, Wu G, Bernemann C, Joo JY, Han DW, Ko K, Tapia N, Sabour D, Sterneckert J, Tesar P, et al. Conserved and divergent roles of FGF signaling in mouse epiblast stem cells and human embryonic stem cells. Cell Stem Cell. 2010;6:215–226. doi: 10.1016/j.stem.2010.01.003. [DOI] [PubMed] [Google Scholar]
- Guo G, Smith A. A genome-wide screen in EpiSCs identifies Nr5a nuclear receptors as potent inducers of ground state pluripotency. Development. 2010;137:3185–3192. doi: 10.1242/dev.052753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G, Yang J, Nichols J, Hall JS, Eyres I, Mansfield W, Smith A. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development. 2009;136:1063–1069. doi: 10.1242/dev.030957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han DW, Greber B, Wu G, Tapia N, Arauzo-Bravo MJ, Ko K, Bernemann C, Stehling M, Scholer HR. Direct reprogramming of fibroblasts into epiblast stem cells. Nat Cell Biol. 2011;13:66–71. doi: 10.1038/ncb2136. [DOI] [PubMed] [Google Scholar]
- Han DW, Tapia N, Joo JY, Greber B, Arauzo-Bravo MJ, Bernemann C, Ko K, Wu G, Stehling M, Do JT, et al. Epiblast stem cell subpopulations represent mouse embryos of distinct pregastrulation stages. Cell. 2010;143:617–627. doi: 10.1016/j.cell.2010.10.015. [DOI] [PubMed] [Google Scholar]
- Jiang H, Shukla A, Wang X, Chen WY, Bernstein BE, Roeder RG. Role for Dpy-30 in ES cell-fate specification by regulation of H3K4 methylation within bivalent domains. Cell. 2011;144:513–525. doi: 10.1016/j.cell.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jude CD, Climer L, Xu D, Artinger E, Fisher JK, Ernst P. Unique and independent roles for MLL in adult hematopoietic stem cells and progenitors. Cell stem cell. 2007;1:324–337. doi: 10.1016/j.stem.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karatas H, Townsend EC, Cao F, Chen Y, Bernard D, Liu L, Lei M, Dou Y, Wang S. High-affinity, small-molecule peptidomimetic inhibitors of MLL1/WDR5 protein-protein interaction. Journal of the American Chemical Society. 2013;135:669–682. doi: 10.1021/ja306028q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katada S, Sassone-Corsi P. The histone methyltransferase MLL1 permits the oscillation of circadian gene expression. Nat Struct Mol Biol. 2010;17:1414–1421. doi: 10.1038/nsmb.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima Y, Kaufman-Francis K, Studdert JB, Steiner KA, Power MD, Loebel DA, Jones V, Hor A, de Alencastro G, Logan GJ, et al. The transcriptional and functional properties of mouse epiblast stem cells resemble the anterior primitive streak. Cell Stem Cell. 2014;14:107–120. doi: 10.1016/j.stem.2013.09.014. [DOI] [PubMed] [Google Scholar]
- Lanner F, Rossant J. The role of FGF/Erk signaling in pluripotent cells. Development. 2010;137:3351–3360. doi: 10.1242/dev.050146. [DOI] [PubMed] [Google Scholar]
- Li X, Li L, Pandey R, Byun JS, Gardner K, Qin Z, Dou Y. The histone acetyltransferase MOF is a key regulator of the embryonic stem cell core transcriptional network. Cell stem cell. 2012;11:163–178. doi: 10.1016/j.stem.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim DA, Huang YC, Swigut T, Mirick AL, Garcia-Verdugo JM, Wysocka J, Ernst P, Alvarez-Buylla A. Chromatin remodelling factor Mll1 is essential for neurogenesis from postnatal neural stem cells. Nature. 2009;458:529–533. doi: 10.1038/nature07726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maclary E, Buttigieg E, Hinten M, Gayen S, Harris C, Sarkar MK, Purushothaman S, Kalantry S. Differentiation-dependent requirement of Tsix long non-coding RNA in imprinted X-chromosome inactivation. Nat Commun. 2014;5:4209. doi: 10.1038/ncomms5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks H, Kalkan T, Menafra R, Denissov S, Jones K, Hofemeister H, Nichols J, Kranz A, Stewart AF, Smith A, et al. The transcriptional and epigenomic foundations of ground state pluripotency. Cell. 2012;149:590–604. doi: 10.1016/j.cell.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon KA, Hiew SY, Hadjur S, Veiga-Fernandes H, Menzel U, Price AJ, Kioussis D, Williams O, Brady HJ. Mll has a critical role in fetal and adult hematopoietic stem cell self-renewal. Cell stem cell. 2007;1:338–345. doi: 10.1016/j.stem.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder JC. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc Natl Acad Sci U S A. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najm FJ, Chenoweth JG, Anderson PD, Nadeau JH, Redline RW, McKay RD, Tesar PJ. Isolation of epiblast stem cells from preimplantation mouse embryos. Cell Stem Cell. 2011;8:318–325. doi: 10.1016/j.stem.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J, Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin SH, Hochedlinger K. Chromatin connections to pluripotency and cellular reprogramming. Cell. 2011;145:835–850. doi: 10.1016/j.cell.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp B, Plath K. Epigenetics of reprogramming to induced pluripotency. Cell. 2013;152:1324–1343. doi: 10.1016/j.cell.2013.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigserver P, Rhee J, Donovan J, Walkey CJ, Yoon JC, Oriente F, Kitamura Y, Altomonte J, Dong H, Accili D, et al. Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1alpha interaction. Nature. 2003;423:550–555. doi: 10.1038/nature01667. [DOI] [PubMed] [Google Scholar]
- Rais Y, Zviran A, Geula S, Gafni O, Chomsky E, Viukov S, Mansour AA, Caspi I, Krupalnik V, Zerbib M, et al. Deterministic direct reprogramming of somatic cells to pluripotency. Nature. 2013;502:65–70. doi: 10.1038/nature12587. [DOI] [PubMed] [Google Scholar]
- Rao RC, Dou Y. Hijacked in cancer: the KMT2 (MLL) family of methyltransferases. Nat Rev Cancer. 2015;15:334–346. doi: 10.1038/nrc3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz EG, Meisig J, Nakamura T, Okamoto I, Sieber A, Picard C, Borensztein M, Saitou M, Bluthgen N, Heard E. The two active X chromosomes in female ESCs block exit from the pluripotent state by modulating the ESC signaling network. Cell Stem Cell. 2014;14:203–216. doi: 10.1016/j.stem.2013.11.022. [DOI] [PubMed] [Google Scholar]
- Silva J, Smith A. Capturing pluripotency. Cell. 2008;132:532–536. doi: 10.1016/j.cell.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart HT, van Oosten AL, Radzisheuskaya A, Martello G, Miller A, Dietmann S, Nichols J, Silva JC. NANOG amplifies STAT3 activation and they synergistically induce the naive pluripotent program. Curr Biol. 2014;24:340–346. doi: 10.1016/j.cub.2013.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima Y, Guo G, Loos R, Nichols J, Ficz G, Krueger F, Oxley D, Santos F, Clarke J, Mansfield W, et al. Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell. 2014;158:1254–1269. doi: 10.1016/j.cell.2014.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Berge D, Koole W, Fuerer C, Fish M, Eroglu E, Nusse R. Wnt signaling mediates self-organization and axis formation in embryoid bodies. Cell Stem Cell. 2008;3:508–518. doi: 10.1016/j.stem.2008.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RD. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- Theunissen TW, Powell BE, Wang H, Mitalipova M, Faddah DA, Reddy J, Fan ZP, Maetzel D, Ganz K, Shi L, et al. Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell. 2014;15:471–487. doi: 10.1016/j.stem.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt P, Tee WW, Reinberg D. A double take on bivalent promoters. Genes Dev. 2013;27:1318–1338. doi: 10.1101/gad.219626.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye S, Li P, Tong C, Ying QL. Embryonic stem cell self-renewal pathways converge on the transcription factor Tfcp2l1. EMBO J. 2013;32:2548–2560. doi: 10.1038/emboj.2013.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu BD, Hess JL, Horning SE, Brown GA, Korsmeyer SJ. Altered Hox expression and segmental identity in Mll-mutant mice. Nature. 1995;378:505–508. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.