Abstract
Extracellular vesicles (EVs) hold immense promise for utilization as biotherapeutics and drug delivery vehicles due to their nature as biological nanoparticles that facilitate intercellular molecular transport. Specifically, EVs have been identified as natural carriers of nucleic acids, sparking interest in their use for gene therapy and RNA interference applications. So far, small RNAs (siRNA and miRNA) have been successfully loaded into EVs for a variety of delivery applications, but the potential use of EVs for DNA delivery has scarcely been explored. Here, we report that exogenous linear DNA can be associated with EVs via electroporation in quantities sufficient to yield an average of hundreds of DNA molecules per vesicle. We determined that loading efficiency and capacity of DNA in EVs is dependent on DNA size, with linear DNA molecules less than 1000 bp in length being more efficiently associated with EVs compared to larger linear DNAs and plasmid DNAs using this approach. We further showed that EV size is also determinant with regard to DNA loading, as larger microvesicles encapsulated more linear and plasmid DNA than smaller, exosome-like EVs. Additionally, we confirmed the ability of EVs to transfer foreign DNA loaded via electroporation into recipient cells, although functional gene delivery was not observed. These results establish critical parameters that inform the potential use of EVs for gene therapy and, in agreement with other recent results, suggest that substantial barriers must be overcome to establish EVs as broadly applicable DNA delivery vehicles.
Keywords: extracellular vesicles (EVs), exosomes, microvesicles, electroporation, DNA delivery, gene therapy
Graphical abstract
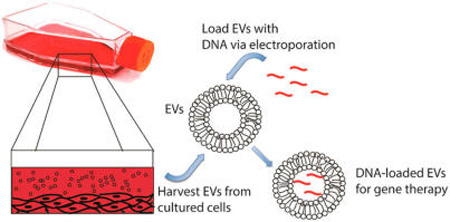
INTRODUCTION
Extracellular vesicles (EVs) are natural nanoscopic particles produced by most cells that hold immense promise for utilization as drug carriers in personalized medicine, as they can theoretically be procured from a patient’s own cells thus limiting potential immunogenicity.1–4 EVs comprise a heterogeneous population of phospholipid-based particles that are produced via multiple mechanisms and are difficult to completely separate using conventional methods. EV subsets include exosomes (typically 30–120 nm), which are formed inside multivesicular endosomes and released to the extracellular environment upon fusion with the plasma membrane; as well as microvesicles (typically 50–1000 nm), which are produced by the outward budding and fission of membrane vesicles from the cell surface.5–7 EVs are thought to play a significant role in intercellular communication, especially via horizontal transfer of various nucleic acids between cells, including microRNAs and long noncoding RNAs (lncRNAs).8,9 The discovery of RNA in EVs8 led to studies examining their therapeutic potential for exogenous RNA delivery, with promising initial results10,11 that have spawned heightened interest in the field. Specifically, a seminal study by Wood and colleagues showed that exosomes could be loaded with siRNA via electroporation, resulting in knockdown of a target protein in mice.10 However, other groups have reported an inability to load EVs efficiently with small RNAs via electroporation,11,12 potentially due to electric field-induced molecular aggregation,12 and alternative methods such as producer–cell transfection have been employed.11,13,14 Transfection-based approaches offer potential advantages in loading efficiency and molecular stability compared to electroporation. Yet, as elegantly explained by Kooijmans et al.,12 transfection-based approaches are limited by toxicity and safety concerns associated with the potential for transfection reagents to alter producer cell gene expression, which could in turn result in undesirable changes in EV cargo and bioactivity. Cell transfection efficiency is also inherently variable and is thus unlikely to produce EVs with consistent levels of a desired therapeutic molecule. Further, sequence-specific variation inherent in cell transfection would likely mandate process reformulation for each potential therapeutic cargo. However, electroporation has long been associated with functional nucleic acid transfer and intracellular delivery,15–18 and there are potential avenues to reduce nucleic acid aggregation during EV loading, such as by lipid complexation.19 Ultimately, the issue of how to most effectively load EVs with therapeutic nucleic acids has not been settled, and further explorations of the potential methods are needed.20
In addition to RNA, DNAs have also been detected in EVs by several groups.21–24 Yet, to date there have been few reported attempts to harness EVs for exogenous DNA delivery. Contag and colleagues recently reported that plasmid DNA incorporated into EVs via a transfection-based approach could be transferred to recipient cells by both exosomes and microvesicles, with only the microvesicle-associated DNA resulting in functional protein expression.14 However, nontransfection-based methods for DNA loading into EVs have not been reported, and the biological parameters and therapeutic potential of EVs for DNA delivery remain almost completely undefined. Given that the promise of gene therapy25,26 continues to be hampered by a lack of appropriate delivery systems, further exploration of the potential of EVs for this purpose is warranted.
In this study, we demonstrate that nucleic acids larger than siRNA or miRNA can be loaded into EVs via electroporation at quantities of, on average, hundreds of molecules per vesicle. Also, we establish that loading efficiency of DNAs using electroporation is dependent on both DNA size and EV size. We further show that EVs loaded via electroporation can transfer exogenous DNA to recipient cells, but functional gene transfer was not observed. Overall, these results identify biological parameters that inform future consideration of EV-based gene therapy approaches and highlight that a diversity of methods must be explored to broaden and enhance the therapeutic potential of EVs.
MATERIALS AND METHODS
Cell Culture
EVs were isolated from a variety of cell types in these studies. HEK293T cells (obtained from American Type Culture Collection (ATCC)) were cultured in DMEM High Glucose with sodium pyruvate (110 mg/mL), l-glutamine (6 mM), penicillin/streptomycin (100 units/mL), and 10% FBS. Human umbilical vein endothelial cells (HUVEC; Promocell) and retrovirally telomerized HUVEC (HRVT; a kind gift of Dr. Antonino Passaniti) were cultured in EGM-2 media (Lonza). Human mesenchymal stem cells (hMSC; Lonza) were cultured in DMEM high glucose supplemented with l-glutamine (4 mM), penicillin/streptomycin (100 units/mL), nonessential amino acids (0.1 mM), and 10% FBS. All media were filter sterilized and depleted of serum-derived EVs prior to use by centrifuging complete media at 100,000g for 16 h at 4 °C.
EV Isolation
To isolate EVs, ~2 × 106 cells were seeded into T150 flasks and allowed to incubate for 24 h or until ~50% confluent. Media was then aspirated, cells were washed once with 20 mL of prewarmed 1× PBS, and 35 mL of prewarmed EV-depleted media were added to the flask. Cells were incubated until ~90% confluent, media was collected, and differential centrifugation was employed to isolate EVs. Media were initially centrifuged at 300g for 10 min to remove cellular debris. Supernatant was then carefully transferred into new tubes and two subsequent centrifugation steps were carried out at 2000g for 20 min and 10,000g for 30 min, respectively, to remove larger sized vesicles. Finally, supernatant was centrifuged for 2 h at 100,000g to pellet EVs using Optiseal tubes (Beckman Coulter) and a T70i ultracentrifuge rotor (Beckman Coulter). After discarding the supernatant, the EV pellet was resuspended into 1 mL of 1× PBS, an additional 29 mL of cold 1× PBS was added to same tube and 70 min of ultracentrifugation was carried out at 100,000g to remove residual media components from EVs. In the last step, EVs were resuspended into 1 mL of 1× PBS and placed at −80 °C for long-term storage. All centrifugation steps were carried out at 4 °C.
EV Characterization
The concentration and size of vesicles were measured by Nanotracking Analysis (NTA) using a NanoSight LM10 (Malvern). EVs were diluted 40× in 1× PBS prior to measurement. NTA measures EV size and concentration based on light scattering and Brownian motion of colloids in liquid suspension, using the Stokes–Einstein equation to calculate hydrodynamic diameter.
RNA was isolated from EVs by addition of 700 µL of QIAzol reagent (Qiagen) directly into Optiseal tubes prior to resuspending the EV pellet. RNA was then purified using the miRNeasy mini kit (Qiagen #217004) according to the manufacturer’s instructions. The amount of RNA was quantified using a nanospectrophotometer, and RNA integrity was assessed by running samples in 15% TBE-Urea gels.
Total protein present in EV samples was determined using the BCA assay (Pierce #23225). To assess specific protein content via immunoblotting, EVs were mixed with 2× protein sample buffer [125 mM Tris×HCl pH 6.8, 4% (wt/vol) SDS, 20% (v/v) glycerol, 100 mM DTT, 0.01% bromophenol blue] and were heated at 90 °C for 5 min. Samples were immediately quenched on ice and were loaded onto 10% Mini-PROTEAN TGX gels (Bio-Rad). Protein was transferred onto nitrocellulose membranes using a Trans-Blot Turbo apparatus (Bio-Rad 170-4270), and standard immunoblotting techniques were employed. Alix and GAPDH antibodies were purchased from Cell Signaling. Odyssey blocking buffer from LI-COR (catalog # 927-40000) was used for blocking. Primary and secondary antibodies were diluted 1000–10,000-fold in 0.5× blocking buffer. Final gel images were obtained by scanning with a LI-COR Odyssey CLX.
DNA Loading into EVs by Electroporation
HEK293T derived EVs were mixed with dsDNA in electroporation buffer (1.15 mM potassium phosphate, pH = 7.2, 25 mM potassium chloride, 21% Optiprep; as described previously10,27). Electroporation was carried out using Gene Pulser/Micropulser Cuvettes (Bio-Rad #165-2089) in a GenePulser Xcell electroporator (Bio-Rad). Subsequently, all samples were filtered through Nanosep centrifugal devices with Omega membranes (300 kDa MWCO; Pall #OD300C33) to remove free DNA and buffer components.28,29 Unless otherwise indicated, 10 µg of EVs, corresponding to ~3 × 108 EVs as measured by NTA, were mixed with 5 µg of dsDNA in a final volume of 50 µL electroporation buffer (note that 10 µg refers to total protein content as measured by BCA assay). Electroporation was carried out at 400 V and 125 µF with two pulses. Electroporated samples were transferred into 0.5 mL tubes, and 1 mM EDTA was added to alleviate nucleic acid aggregation based on the results of Kooijmans et al.12 Comparison of the effect of EDTA addition before and after electroporation revealed similar efficiency in reduction of DNA aggregation. Samples were then incubated at room temperature for 15 min and subsequently transferred into Nanosep tubes and centrifuged at 5000g at 4 °C for 5 min to remove buffer and unincorporated dsDNA. To remove excess unbound dsDNA, samples were digested in the same tube by adding DNase I enzyme (1.5 Kunitz units/µL, Qiagen #79254) and incubating for 20 min. DNase I activity was confirmed by digesting an equivalent amount of loaded DNA in solution. After digestion, DNase I was inactivated by adding EDTA to a final concentration of 20 mM. Five hundread microliters of 1× TE was then added, and the solution was again centrifuged at 5000g at 4 °C for 5 min to remove digested DNA, inactivated enzyme, and other buffer components. This washing step was repeated two more times (total of three washes). Finally, EVs were resuspended in 100 µL of 1× TE by pipetting up and down several times. DNA quantification was performed using Quant-it PicoGreen Assay kit (Life Technologies, catalog #P7589) following the manufacturer’s protocol. Known quantities of DNA were labeled in the same fashion to establish a standard curve for quantification. The number of DNA molecules (copy number) was calculated using an average molecular weight value for 250 bp dsDNA and the amount of 250 bp DNA associated with EVs after electroporation.
EV-Associated DNA Transfer to Cells
S. cerevisiae dsDNA encoding the tRNA Ser(CGA) gene, including 300 nucleotides upstream and downstream sequences (750 bp total), was amplified by PCR using S. cerevisiae total DNA as a template. Ten micrograms of purified DNA was mixed with 10 µg of EVs, and electroporation was carried out as described. After electroporation, samples were filtered, digested with DNase I, and resuspended with 500 µL of EV-depleted media for HEK293T cells. DNA-loaded EVs were then exposed to HEK293Ts in culture at ~50% confluency in 6 well plates. After 24 h, cells were washed 3× with PBS and lysed, and PCR amplification was carried out using cellular DNA as a template. Primers that specifically recognize the S. cerevisiae tRNA Ser(CGA) gene were used for amplification and amplified PCR products were run on agarose gels.
Statistical Analysis
Parametric statistical tests (one-way analysis of variance (ANOVA), 2-sample t test) were used as appropriate, and statistical significance level is indicated for each figure where it was calculated.
RESULTS AND DISCUSSION
Selection of EV Producing Cells for DNA Delivery
Numerous cell types have been employed for potential generation of EVs for drug delivery applications.10,11,14,27,30–36 36 It was hypothesized that EVs with minimal intrinsic biological cargo might be best suited for therapeutic delivery applications in that nonspecific molecular delivery would be minimized in such EVs. Since the primary biological cargo of EVs has been identified to be protein and RNAs,4,20 total protein and RNA contents of EVs from several common cell sources, including HEK293T cells, HUVEC, HRVT, and hMSC, were analyzed.
EV isolation was initially verified via NTA. A representative profile of HEK293T EVs is shown in Figure 1A; mean EV diameter for these EVs was assessed to be 85 ± 41 nm. No significant differences in EV size distribution were observed for any of the other cells used for EV production (mean EV diameters: HUVEC 96 ± 55 nm; HRVT 124 ± 86 nm; hMSC 93 ± 50 nm). Further validation was demonstrated by visualization of enrichment of Alix, a marker associated with the exosomal subset of EVs,3,37 via immunoblot in HEK293T EVs (Figures 1B,C). Equal amounts of EVs from four different cell lines (HUVEC, hMSC, HRVT, and HEK293T) were used to analyze protein and RNA content. SDS-PAGE analysis of EV protein did not reveal any significant differences among HEK293T, MSC, and HUVEC; however, HRVT EVs showed a broad distribution of proteins (Figures 1D,F), indicating that the immortalization process involved in HRVT creation from HUVEC38 imparts changes in cellular protein content that are retained in EVs. This same trend was evident in total EV RNA (Figures 1E,G), leading to the conclusion that HRVTs are not ideal generators of EVs for therapeutic applications. Interestingly, RNA analysis also revealed that HEK293T-derived EVs contain low levels of RNA compared to other cell types assessed (Figures 1E,G), suggesting that these EVs may be suitable for loading of exogenous cargo for therapeutic delivery with reduced potential for nonspecific intrinsic cargo delivery. Therefore, HEK293T-derived EVs were used exclusively for additional experiments in this study.
Figure 1.
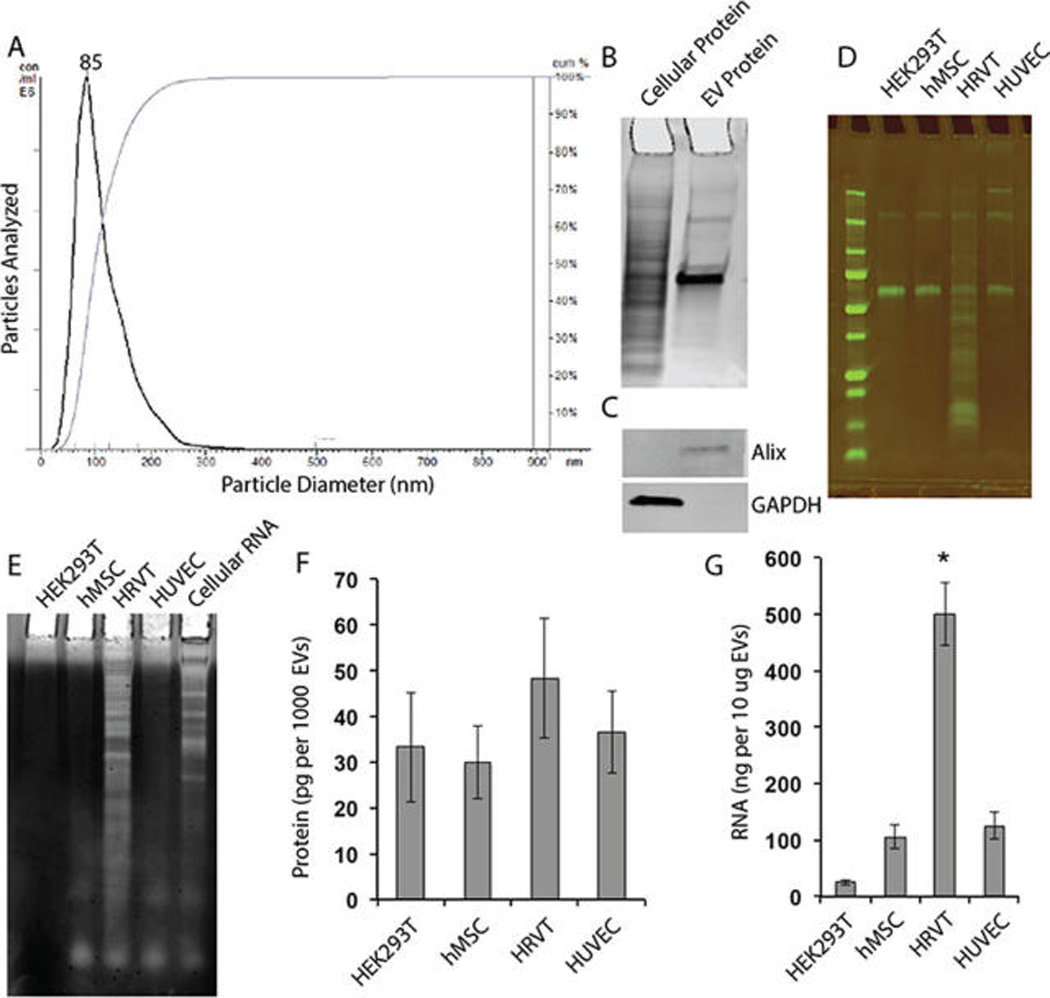
Characterization of EVs isolated from HEK293T and other cell lines. (A) Isolated EVs were characterized by NTA. A representative profile of HEK293T EVs is shown; mean particle diameter was found to be ~85 nm. (B) Representative coomassie blue stained SDS-PAGE gel of cellular and EV protein fractions from HEK293T cells. (C) Immunoblots for Alix, an exosomal marker, and GAPDH, a cellular marker, in the cellular and EV protein fractions from HEK293T cells. (D) SDS-PAGE of protein isolated from EVs from the following cells: Human Embryonic Kidney 293T (HEK293T); human Mesenchymal Stem Cells (hMSC); retrovirally telomerized human umbilical vein endothelial cells (HRVT); human umbilical vein endothelial cells (HUVEC). (E) TBE-Urea gel analysis of EV RNA content from HEK293T, hMSC, HRVT, and HUVEC. (F) Relative EV protein level and (G) relative EV RNA level in different cell types. Each panel is representative of at least three independent experiments. *P < 0.05 compared to all other groups (one-way ANOVA).
Assessment of Electroporation for Short Linear DNA Loading into EVs
Although small nucleic acids such as siRNAs have been successfully loaded into EVs via electroporation,10 DNA loading into EVs has so far been limited primarily to transfection-based approaches.14 To determine if electroporation could be adapted for loading of EVs with DNA, a 250 bp dsDNA from the VA1 gene (adenovirus virus associated gene 1) was prepared by PCR amplification from a pVA1 plasmid, based on the hypothesis that short linear DNAs are most likely to be efficiently loaded into EVs by electroporation based on prior success of this technique with loading 20–25 bp siRNA. Indeed, DNA associated with HEK293T-derived EVs was significantly increased by electroporation (Figure 2A). Exposure to DNase I revealed that a majority of DNA associated with EVs during electroporation was not protected from degradation; however, the remaining DNA still constituted an amount, on average, of hundreds of copies per EV (Figure 2B).
Figure 2.
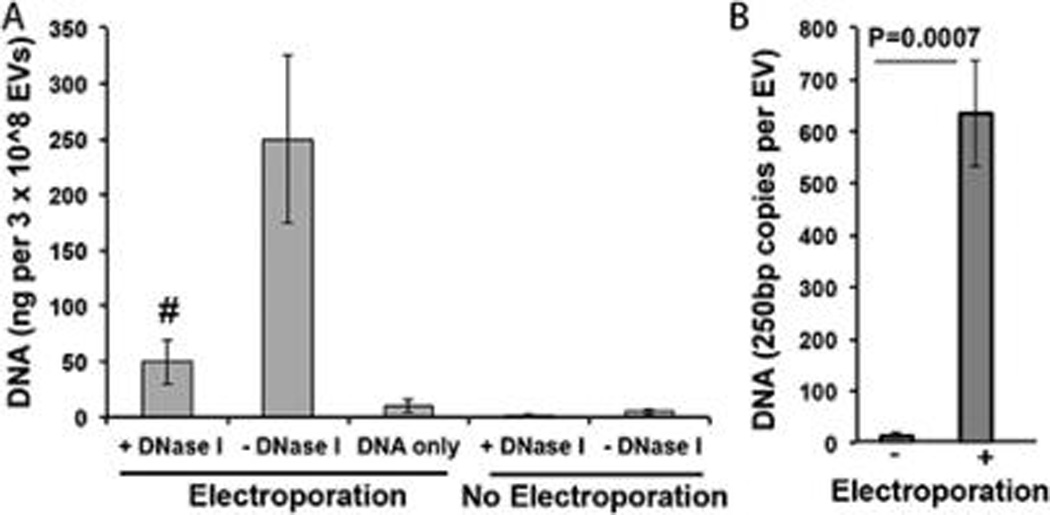
DNA loading into EVs by electroporation. (A) HEK293T-derived EVs were loaded with 250 bp dsDNA via electroporation. DNA amounts as detected by PicoGreen assay after extensive washing as described in Methods are shown for the following groups (bars from left to right): (1) EVs electroporated in the presence of DNA and subsequently treated with DNase I; (2) EVs electroporated in the presence of DNA and not treated with DNase I; (3) DNA electroporated without EVs present (and not treated with DNase I); (4) EVs incubated, but not electroporated, in the presence of DNA and subsequently treated with DNase I; and (5) EVs incubated, but not electroporated, in the presence of DNA and not treated with DNase I. n = 3 for all groups. #P < 0.01 for 1 compared to both 3 and 4. (B) The number of DNA copies per vesicle was calculated on an average basis from bulk data using an estimated weight associated with a 250 bp dsDNA sequence.
Notably, electroporation of 250 bp linear dsDNA alone did induce a small positive signal as measured by PicoGreen assay, similar to the phenomenon noted for electroporation of siRNA.12 However, in this case, DNA association with EVs still appeared to be significant compared to this background signal (Figure 2A). Additionally, electroporation induced only a slight change in average size of EVs (Figure S1), indicating that potential vesicle aggregation was minimized.39 It is also notable that the average number of DNA molecules per vesicle is significantly higher than what would be expected based on purely diffusion-mediated molecular distribution and loading. This could indicate specific interactions, likely electrostatically driven,40 between DNA and lipids (or proteins) that facilitate more efficient association with EVs. This concept is similar to the previously characterized effect of electrophoretic DNA transfer into cells during electroporation41,42 and is reinforced by the higher association of DNA with the external layer of EVs than with the internal compartment (Figure 2A).
Examination of Electroporation Parameters
Efficiency of loading is a critical parameter in determining efficacy as well as commercial viability of drug delivery systems. Thus, DNA loading amount and electroporation parameters were modulated in an attempt to increase DNA incorporation into EVs. Upon increasing the initial DNA quantity for loading, a saturation effect was observed, and maximal loading efficiency was achieved at approximately 2.5 µg of 250 bp linear dsDNA (Figure 3A). This result is significant in that it demonstrated a ~2-fold increase in efficiency over the initial method, which utilized a starting amount of 5 µg of this DNA. However, overall DNA loading efficiency in order to achieve maximal DNA loading in these experiments was limited to ~2% (~50 ng of DNA incorporated into EVs/ ~2500 ng of DNA required to achieve maximum loading × 100%). In all experiments, measurements were corrected for background signal generated by DNA alone.
Figure 3.
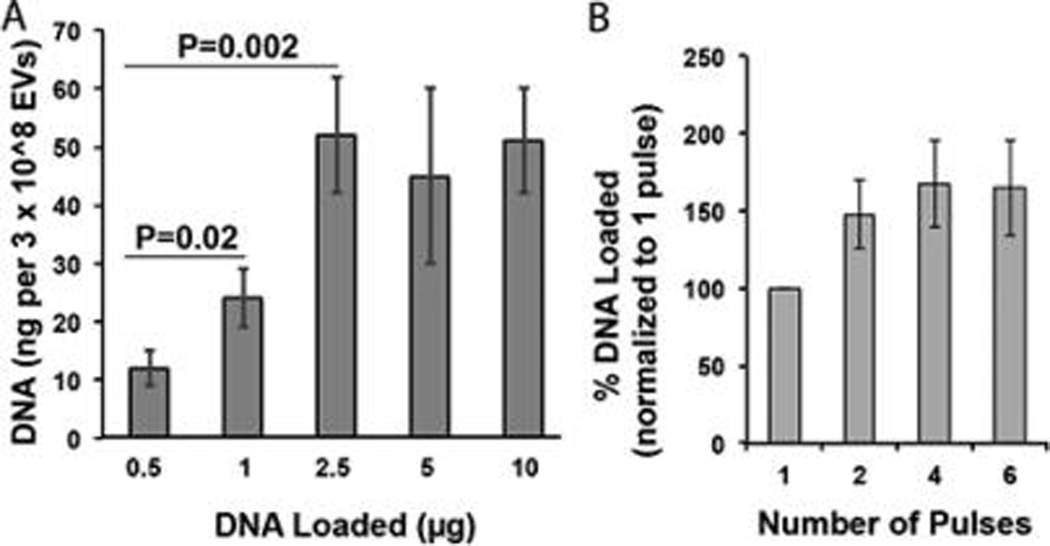
Evaluation of DNA loading efficiency into EVs via electroporation. (A) HEK293T-derived EVs were electroporated in the presence of various initial amounts of 250 bp linear dsDNA (DNA Loaded) and DNA incorporated into EVs was assessed by PicoGreen assay. All data were normalized to background signal generated by electroporated DNA only at each loading amount. (B) Similarly, HEK293T-derived EVs were electroporated in the presence of 5 µg of this DNA, and the number of pulses was varied as indicated. DNA incorporated into EVs was assessed by PicoGreen assay. For both panels, n = 3.
Electroporation parameters were also examined to see if DNA loading could be improved. Decreasing pulse number from the initial procedure of 2 down to 1 pulse did lead to a decrease in DNA incorporation into EVs (Figure 3B). However, increasing the pulses above 2 did not result in any significant increase in DNA incorporation (Figure 3B). Modulation of electroporation voltage (up to 500 V) and pulse type (exponential vs square-wave) did not affect DNA loading. As before, all measurements were corrected for background signal generated by DNA alone at each appropriate concentration.
Effect of DNA Size on DNA Loading into EVs
Since increasing lengths of DNA have increased associated volumes, it was hypothesized that loading of DNA molecules into EVs might be size dependent. To test this hypothesis, linear dsDNA fragments of 250 bp, 500 bp, 750 bp, 1000 bp, 1500 bp, and 4000 bp in length were prepared via PCR amplification and incorporation into EVs following electroporation was assessed. Strikingly, a significant decrease in DNA incorporation was observed between the 750 bp DNA and the 1000 bp DNA (Figure 4). All DNAs above 750 bp tested exhibited very low levels of DNA incorporation, suggesting a size limitation cutoff in the range of 750–1000 bp (Figure 4). This size limitation was also apparent for plasmid DNA molecules ranging in size from ~4.5 to ~10 kb (Figure 5). Plasmid DNA exhibited similar responsiveness to changes in initial loading amount and electroporation pulses as linear DNA. However, overall, plasmid DNA was incorporated into EVs at very low efficiency (<0.2%).
Figure 4.
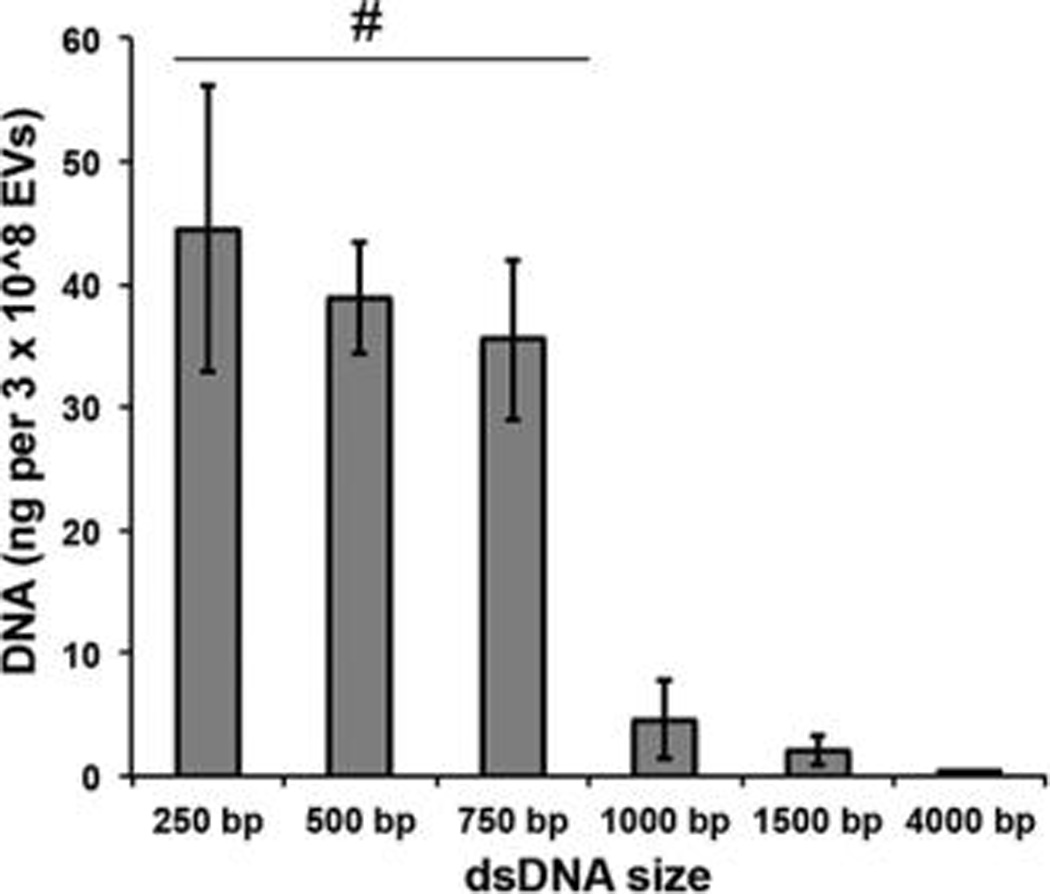
Linear DNA loading into EVs by electroporation is size limited. HEK293T-derived EVs were electroporated in the presence of linear dsDNAs of the indicated sizes, and DNA loading was assessed after DNase I digestion via the PicoGreen assay. For all groups, n = 3, and all data were normalized to background signal generated by electroporated DNA only for each size. #P < 0.01 for each group compared to 1000 bp, 1500 bp, and 4000 bp groups.
Figure 5.
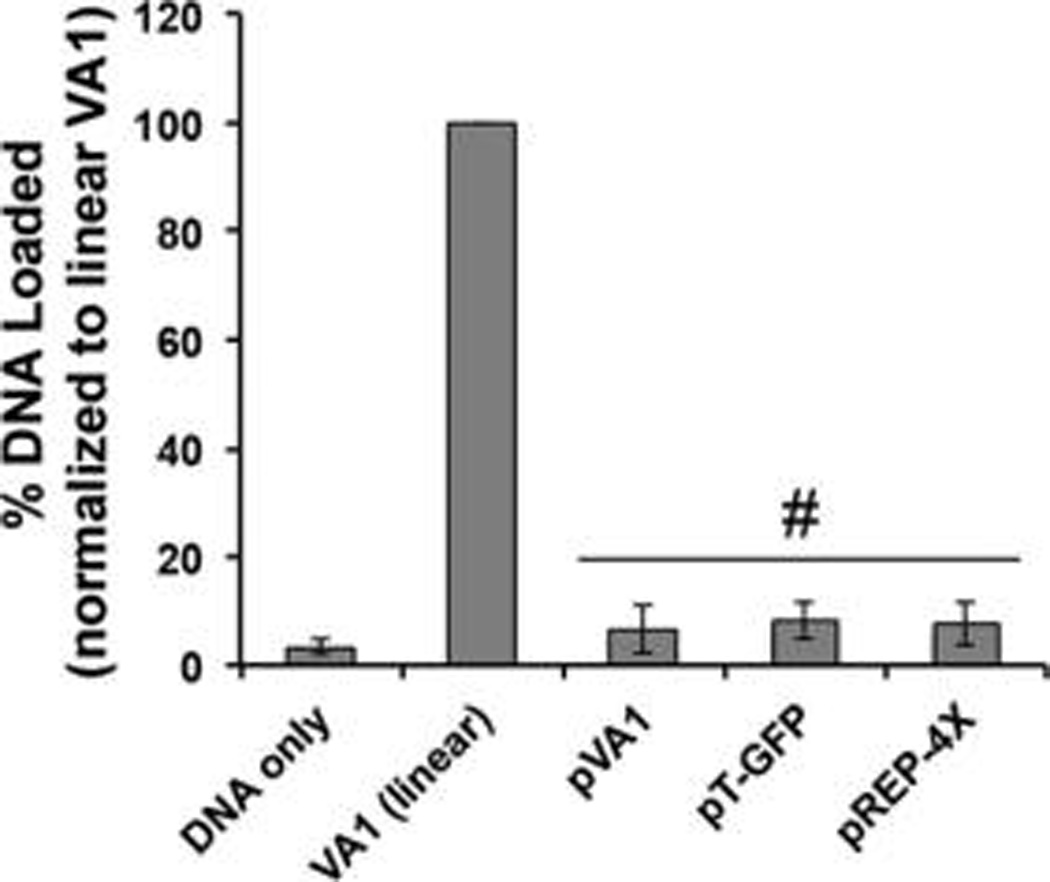
Plasmid DNA loading into EVs by electroporation is less efficient than linear DNA loading. HEK293T-derived EVs were electroporated in the presence of the indicated plasmid DNAs, and DNA loading was assessed after DNase I digestion via the PicoGreen assay. For all groups, n = 3. #P < 0.01 for each group compared to linear VA1.
The apparent size limitation for DNA loading observed in this study could be caused by a number of factors. Pore size restrictions could prevent larger molecules from entering the luminal space of EVs with the same efficiency as smaller DNAs. Additionally, diffusion limitations associated with larger molecules could reduce their efficiency of migration through transient pores generated by electroporation, decreasing overall loading compared to smaller molecules. These findings are not without precedent, as when electroporation is carried out to load DNA plasmids into bacteria, transformation efficiency decreases with increasing plasmid size.43,44
Effect of EV Size on DNA Loading into EVs
A recent report by Kanada et al. highlighted the distinctions between different EV subsets with regard to their potential for DNA delivery, showing that one EV subset, microvesicles (MVs), could enable functional transfer of DNA to cells, while another EV subset, exosomes, could not.14 To examine whether MVs might have greater potential for electroporation-mediated DNA loading and delivery, MVs were isolated from the 10,000g centrifugation supernatant during EV purification (Figure 6A) and were electroporated using identical conditions as reported for EVs. Upon normalization to the total number of vesicles, the DNA loading capacity of MVs was observed to be ~3-fold that of EVs for linear 250 bp DNA and ~4-fold that of EVs for ~6 kb plasmid DNA (Figure 6B).
Figure 6.
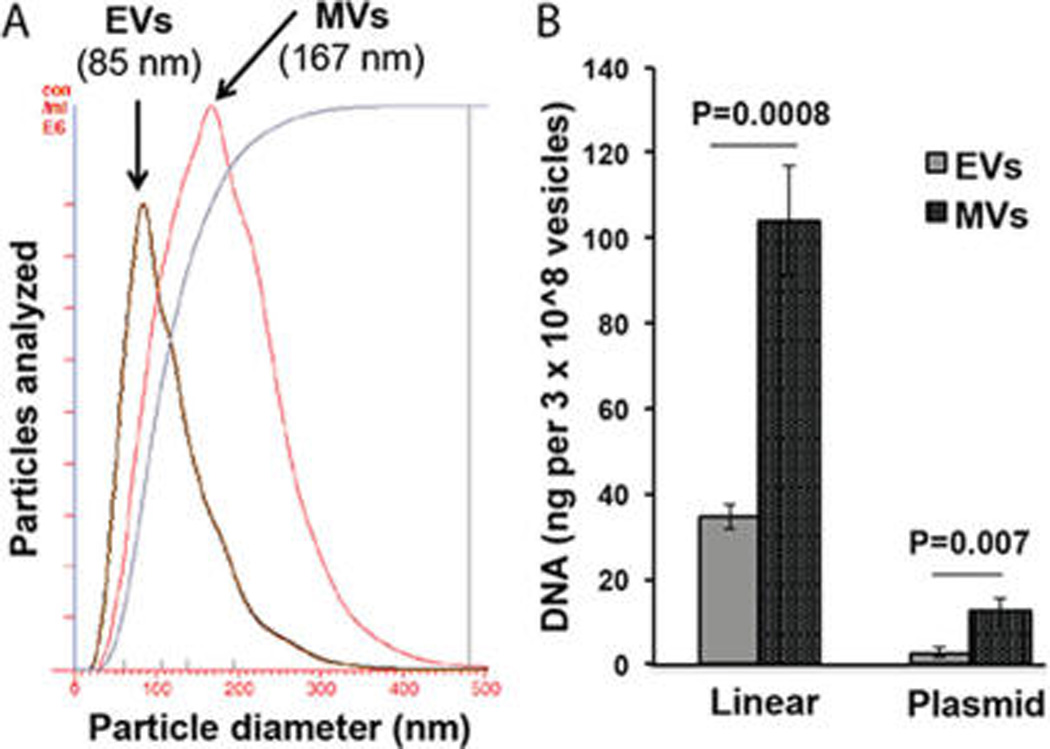
DNA loading by electroporation varies across EV subsets. (A) EV and MV populations were characterized by NTA; mean diameters are reported. Data are representative of at least 3 EV isolations. (B) Loading capacities of linear 250 bp DNA and ~6 kb plasmid DNA in EVs and MVs were compared normalizing for total number of vesicles in each population. In each case, 4 µg of DNA was electroporated with 10 µg of vesicles as described in Methods, and DNA was quantified after DNase I digestion via the PicoGreen assay. For all groups, n = 3.
While it is difficult to accurately project the theoretical maximum DNA loading into EVs due to the presence of intrinsic biological cargo, these data could indicate that the diffusion-driven DNA migration into electroporated EVs has reached saturation, and thus only larger areas can result in enhanced DNA loading via this method. Given that MVs are plasma membrane-derived vesicles while exosome-like EVs originate from multivesicular bodies inside cells,4 it is also possible that the MVs have different lipid compositions or responses to electroporation that result in enhanced permeability to DNAs compared to EVs. It should be noted that the population of isolated EVs is thought to contain both exosomes and microvesicles, as these subsets cannot be completely separated using standard methods. In any case, these data are generally supportive of the findings of Kanada et al.14 and suggest that MVs may hold enhanced potential for DNA delivery compared to smaller, exosome-like EVs.
EV-Mediated DNA Transfer to Cells
The capacity of electroporation-loaded EVs to transfer DNA to recipient cells was assessed to provide insight into the ultimate potential of EVs as DNA delivery vehicles. HEK293T-derived EVs were loaded with a 750 bp linear dsDNA of the S. cerevisiae tRNA Ser(CGA) gene via electroporation and treated with DNase I. After washing and filter sterilization, the DNA-loaded EVs incubated with cultured HEK293T cells at ~50% confluency in EV-depleted media for 24 h. After this time, cells were washed extensively to remove any unincorporated EVs, and DNA was harvested from cells using standard molecular biology methods. The 750 bp S. cerevisiae DNA, which is not normally present in HEK293T cells, was detected by PCR analysis in cells exposed to EVs loaded with this DNA, but not in cells exposed to unloaded EVs alone (Figure 7), indicating that EVs can transfer DNA to recipient cells
Figure 7.
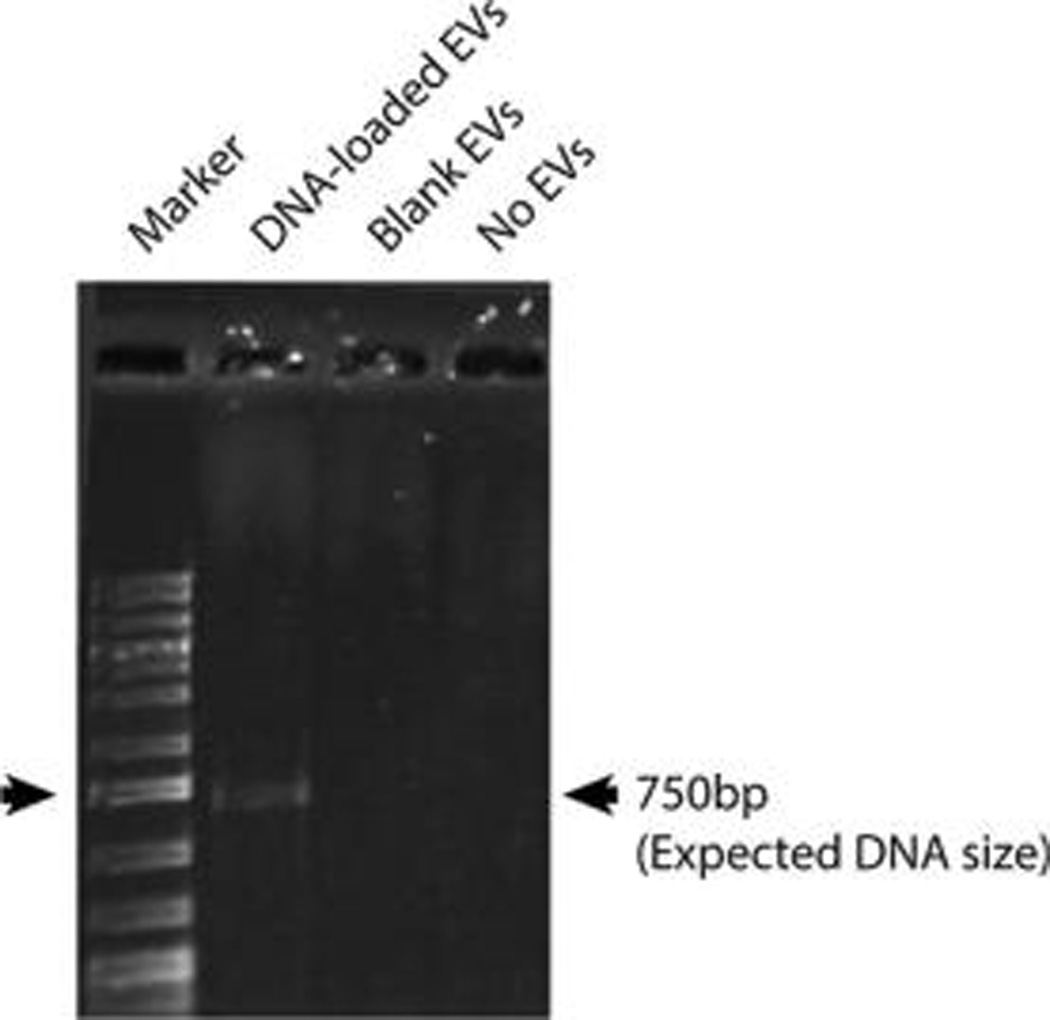
EVs can transfer DNA loaded by electroporation to recipient cells. HEK293T-derived EVs were loaded with a 750 bp linear dsDNA of the S. cerevisiae tRNA Ser(CGA) gene via electroporation, treated with DNase I, washed, and incubated with HEK293T cells at ~50% confluency in EV-depleted media for 24 h. Unloaded HEK293T-derived EVs (Blank EVs) media without exogenous EVs (No EVs) were also incubated with cells under the same conditions. The presence of the 750 bp DNA was probed for with specific primers by PCR with putative amplicons run on an agarose gel. Data are representative of three independent experiments.
Potential transfer of plasmid DNA to recipient cells was also assessed using the same experimental set up. However, no PCR product was observed following incubation of pT-GFP-loaded EVs with HEK293T cells. Expectedly, GFP protein expression was undetectable by immunoblot in these cells, while cells that were transfected with the same plasmid via lipofectamine 3000 showed GFP expression by Western blotting (Figure S2). Increasing the amount of EVs exposed to cells did not result in any measurable DNA transfer or protein expression in recipient HEK293Ts, nor did use of MVs. Thus, functional DNA transfer was not achieved in these studies, reflecting the findings of others12,14 and highlighting current limitations of this method for gene delivery.
In total, these data support several conclusions. DNA can be encapsulated into EVs via electroporation, and this encapsulation is dependent on DNA size. Linear DNA incorporation into EVs via electroporation is much more efficient than plasmid DNA incorporation, but overall efficiency of DNA encapsulation is low. Additionally, MVs are able to incorporate higher amounts of DNA via electroporation than exosome-like EVs. Finally, while electroporation can be employed to successfully deliver DNA to cells, functional protein expression following DNA transfer was not observed in these studies.
Although this work does not directly address the controversy in the field about electroporation-mediated loading of siRNAs and miRNAs into EVs, some observations may be relevant to this issue. These results share some similarities with findings of inefficient siRNA incorporation into EVs via electroporation;12 however, in this case it appears that electroporation-induced aggregation of DNA is less than that of siRNA, and thus, encapsulation of some useful amount of DNA into EVs using this method is still possible. Additionally, these data suggest that small RNAs are of the appropriate size to be effectively loaded using this method. However, the reduced stability of RNA compared to DNA may result in relatively higher aggregation and degradation of these molecules and thus ineffective loading. Overall, these data support the conclusions of others12 that electroporation as currently applied has limited utility for nucleic acid loading into EVs.
The therapeutic potential of EVs for gene therapy has recently been broadened,14 and there is significant potential benefit in avoiding transfection-based approaches to incorporating DNA constructs into EVs. However, these studies suggest that loading of EVs with DNA via electroporation is unlikely to have widespread utility for gene delivery applications unless efficiency and capacity can be greatly improved. While not as broadly applicable as plasmid DNA for gene therapy, linear DNA delivery, for which EVs loaded by electroporation could be used, has therapeutic potential. PCR-generated linear DNA fragments, similar to those used in this study, can be used as antiviral vaccines45 and as producers of antiviral miRNA shuttles.46 Additionally, further development of microlinear vectors for gene delivery47 may allow enhanced utility of electroporation-mediated DNA loading of EVs for therapeutic applications in the future.
CONCLUSIONS
In conclusion, these results emphasize the need for continued exploration of EV loading approaches in order to enhance the utility of EVs as drug delivery vehicles. These data establish the concept that EVs loaded with DNA using a nontransfection-based method can transfer that DNA to a recipient cell and also define parameters of biological cargo that impact EV encapsulation efficiency. Additionally, these data support the concept that different subsets of EVs have different potentials for DNA delivery, with microvesicles emerging as a promising vehicle for further exploration. However, overall this study highlights the critical nature of further discovery of fundamental biological properties that dictate the therapeutic functionality of engineered EVs for drug delivery.
Supplementary Material
Acknowledgments
This work was supported by NIH grant R00 HL112905, a University of Maryland Tier 1 seed grant, a University of Maryland-Baltimore–University of Maryland-College Park seed grant, and a Ralph E. Powe Junior Faculty Enhancement Award from the Oak Ridge Associated Universities (all to S.M.J.). The authors thank Prof. John P. Fisher and lab members for use of instruments in their lab.
Footnotes
ASSOCIATED CONTENT
Supporting Information
- Supporting Information (PDF)
The authors declare no competing financial interest.
REFERENCES
- 1.Lamichhane TN, Sokic S, Schardt JS, Raiker RS, Lin JW, Jay SM. Emerging roles for extracellular vesicles in tissue engineering and regenerative medicine. Tissue Eng., Part B. 2015;21(1):45–54. doi: 10.1089/ten.teb.2014.0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Katsuda T, Kosaka N, Takeshita F, Ochiya T. The therapeutic potential of mesenchymal stem cell-derived extracellular vesicles. Proteomics. 2013;13(10–11):1637–1653. doi: 10.1002/pmic.201200373. [DOI] [PubMed] [Google Scholar]
- 3.Lee Y, El Andaloussi S, Wood MJ. Exosomes and microvesicles: extracellular vesicles for genetic information transfer and gene therapy. Hum. Mol. Genet. 2012;21(R1):R125–R134. doi: 10.1093/hmg/dds317. [DOI] [PubMed] [Google Scholar]
- 4.El Andaloussi S, Mager I, Breakefield XO, Wood MJ. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat. Rev. Drug Discovery. 2013;12(5):347–357. doi: 10.1038/nrd3978. [DOI] [PubMed] [Google Scholar]
- 5.Gyorgy B, Szabo TG, Pasztoi M, Pal Z, Misjak P, Aradi B, Laszlo V, Pallinger E, Pap E, Kittel A, Nagy G, Falus A, Buzas EI. Membrane vesicles, current state-of-the-art: emerging role of extracellular vesicles. Cell. Mol. Life Sci. 2011;68(16):2667–2688. doi: 10.1007/s00018-011-0689-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colombo M, Raposo G, Thery C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014;30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 7.Vlassov AV, Magdaleno S, Setterquist R, Conrad R. Exosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim. Biophys. Acta, Gen. Subj. 2012;1820(7):940–948. doi: 10.1016/j.bbagen.2012.03.017. [DOI] [PubMed] [Google Scholar]
- 8.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007;9(6):654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 9.Ludwig AK, Giebel B. Exosomes: small vesicles participating in intercellular communication. Int. J. Biochem. Cell Biol. 2012;44(1):11–15. doi: 10.1016/j.biocel.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 10.Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 2011;29(4):341–345. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 11.Ohno S, Takanashi M, Sudo K, Ueda S, Ishikawa A, Matsuyama N, Fujita K, Mizutani T, Ohgi T, Ochiya T, Gotoh N, Kuroda M. Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells. Mol. Ther. 2013;21(1):185–191. doi: 10.1038/mt.2012.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kooijmans SA, Stremersch S, Braeckmans K, de Smedt SC, Hendrix A, Wood MJ, Schiffelers RM, Raemdonck K, Vader P. Electroporation-induced siRNA precipitation obscures the efficiency of siRNA loading into extracellular vesicles. J. Controlled Release. 2013;172(1):229–238. doi: 10.1016/j.jconrel.2013.08.014. [DOI] [PubMed] [Google Scholar]
- 13.Akao Y, Iio A, Itoh T, Noguchi S, Itoh Y, Ohtsuki Y, Naoe T. Microvesicle-mediated RNA molecule delivery system using monocytes/macrophages. Mol. Ther. 2011;19(2):395–399. doi: 10.1038/mt.2010.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanada M, Bachmann MH, Hardy JW, Frimannson DO, Bronsart L, Wang A, Sylvester MD, Schmidt TL, Kaspar RL, Butte MJ, Matin AC, Contag CH. Differential fates of biomolecules delivered to target cells via extracellular vesicles. Proc. Natl. Acad. Sci. U. S. A. 2015;112(12):E1433–E1442. doi: 10.1073/pnas.1418401112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neumann E, Schaefer-Ridder M, Wang Y, Hofschneider PH. Gene transfer into mouse lyoma cells by electroporation in high electric fields. EMBO J. 1982;1(7):841–845. doi: 10.1002/j.1460-2075.1982.tb01257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chu G, Hayakawa H, Berg P. Electroporation for the efficient transfection of mammalian cells with DNA. Nucleic Acids Res. 1987;15(3):1311–1326. doi: 10.1093/nar/15.3.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Broderick KE, Chan A, Lin F, Shen X, Kichaev G, Khan AS, Aubin J, Zimmermann TS, Sardesai NY. Optimized in vivo transfer of small interfering RNA targeting dermal tissue using in vivo surface electroporation. Mol. Ther.–Nucleic Acids. 2012;1:e11. doi: 10.1038/mtna.2012.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mir LM. Electroporation-based gene therapy: recent evolution in the mechanism description and technology developments. Methods Mol. Biol. 2014;1121:3–23. doi: 10.1007/978-1-4614-9632-8_1. [DOI] [PubMed] [Google Scholar]
- 19.Bagai S, Sun C, Tang T. Lipid-modified polyethylenimine-mediated DNA attraction evaluated by molecular dynamics simulations. J. Phys. Chem. B. 2014;118(25):7070–7076. doi: 10.1021/jp503381r. [DOI] [PubMed] [Google Scholar]
- 20.Gyorgy B, Hung ME, Breakefield XO, Leonard JN. Therapeutic applications of extracellular vesicles: clinical promise and open questions. Annu. Rev. Pharmacol. Toxicol. 2015;55:439–464. doi: 10.1146/annurev-pharmtox-010814-124630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thakur BK, Zhang H, Becker A, Matei I, Huang Y, Costa-Silva B, Zheng Y, Hoshino A, Brazier H, Xiang J, Williams C, Rodriguez-Barrueco R, Silva JM, Zhang W, Hearn S, Elemento O, Paknejad N, Manova-Todorova K, Welte K, Bromberg J, Peinado H, Lyden D. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res. 2014;24(6):766–769. doi: 10.1038/cr.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT, Jr, Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008;10(12):1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kahlert C, Melo SA, Protopopov A, Tang J, Seth S, Koch M, Zhang J, Weitz J, Chin L, Futreal A, Kalluri R. Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J. Biol. Chem. 2014;289(7):3869–3875. doi: 10.1074/jbc.C113.532267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balaj L, Lessard R, Dai L, Cho YJ, Pomeroy SL, Breakefield XO, Skog J. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat. Commun. 2011;2:180. doi: 10.1038/ncomms1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morgan RA, Dudley ME, Wunderlich JR, Hughes MS, Yang JC, Sherry RM, Royal RE, Topalian SL, Kammula US, Restifo NP, Zheng Z, Nahvi A, de Vries CR, Rogers-Freezer LJ, Mavroukakis SA, Rosenberg SA. Cancer regression in patients after transfer of genetically engineered lymphocytes. Science. 2006;314(5796):126–129. doi: 10.1126/science.1129003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mancuso K, Hauswirth WW, Li Q, Connor TB, Kuchenbecker JA, Mauck MC, Neitz J, Neitz M. Gene therapy for red-green colour blindness in adult primates. Nature. 2009;461(7265):784–787. doi: 10.1038/nature08401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.El-Andaloussi S, Lee Y, Lakhal-Littleton S, Li J, Seow Y, Gardiner C, Alvarez-Erviti L, Sargent IL, Wood MJ. Exosome-mediated delivery of siRNA in vitro and in vivo. Nat. Protoc. 2012;7(12):2112–2126. doi: 10.1038/nprot.2012.131. [DOI] [PubMed] [Google Scholar]
- 28.Lasser C, Alikhani VS, Ekstrom K, Eldh M, Paredes PT, Bossios A, Sjostrand M, Gabrielsson S, Lotvall J, Valadi H. Human saliva, plasma and breast milk exosomes contain RNA: uptake by macrophages. J. Transl. Med. 2011;9:9. doi: 10.1186/1479-5876-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ekstrom K, Valadi H, Sjostrand M, Malmhall C, Bossios A, Eldh M, Lotvall J. Characterization of mRNA and microRNA in human mast cell-derived exosomes and their transfer to other mast cells and blood CD34 progenitor cells. J. Extracell. Vesicles. 2012;1:1. doi: 10.3402/jev.v1i0.18389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhuang X, Xiang X, Grizzle W, Sun D, Zhang S, Axtell RC, Ju S, Mu J, Zhang L, Steinman L, Miller D, Zhang HG. Treatment of Brain Inflammatory Diseases by Delivering Exosome Encapsulated Anti-inflammatory Drugs From the Nasal Region to the Brain. Mol. Ther. 2011;19:1769. doi: 10.1038/mt.2011.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wahlgren J, De LKT, Brisslert M, Vaziri Sani F, Telemo E, Sunnerhagen P, Valadi H. Plasma exosomes can deliver exogenous short interfering RNA to monocytes and lymphocytes. Nucleic Acids Res. 2012;40(17):e130. doi: 10.1093/nar/gks463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lai RC, Yeo RW, Tan KH, Lim SK. Exosomes for drug delivery - a novel application for the mesenchymal stem cell. Biotechnol. Adv. 2013;31(5):543–551. doi: 10.1016/j.biotechadv.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 33.Munoz JL, Bliss SA, Greco SJ, Ramkissoon SH, Ligon KL, Rameshwar P. Delivery of Functional Anti-miR-9 by Mesenchymal Stem Cell-derived Exosomes to Glioblastoma Multiforme Cells Conferred Chemosensitivity. Mol. Ther.–Nucleic Acids. 2013;2:e126. doi: 10.1038/mtna.2013.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tian Y, Li S, Song J, Ji T, Zhu M, Anderson GJ, Wei J, Nie G. A doxorubicin delivery platform using engineered natural membrane vesicle exosomes for targeted tumor therapy. Biomaterials. 2014;35(7):2383–2390. doi: 10.1016/j.biomaterials.2013.11.083. [DOI] [PubMed] [Google Scholar]
- 35.Banizs AB, Huang T, Dryden K, Berr SS, Stone JR, Nakamoto RK, Shi W, He J. In vitro evaluation of endothelial exosomes as carriers for small interfering ribonucleic acid delivery. Int. J. Nanomed. 2014;9:4223–4230. doi: 10.2147/IJN.S64267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cooper JM, Wiklander PB, Nordin JZ, Al-Shawi R, Wood MJ, Vithlani M, Schapira AH, Simons JP, El-Andaloussi S, Alvarez-Erviti L. Systemic exosomal siRNA delivery reduced alpha-synuclein aggregates in brains of transgenic mice. Mov. Disord. 2014;29(12):1476–1485. doi: 10.1002/mds.25978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baietti MF, Zhang Z, Mortier E, Melchior A, Degeest G, Geeraerts A, Ivarsson Y, Depoortere F, Coomans C, Vermeiren E, Zimmermann P, David G. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat. Cell Biol. 2012;14(7):677–685. doi: 10.1038/ncb2502. [DOI] [PubMed] [Google Scholar]
- 38.Yang J, Nagavarapu U, Relloma K, Sjaastad MD, Moss WC, Passaniti A, Herron GS. Telomerized human microvasculature is functional in vivo. Nat. Biotechnol. 2001;19(3):219–224. doi: 10.1038/85655. [DOI] [PubMed] [Google Scholar]
- 39.Hood JL, Scott MJ, Wickline SA. Maximizing exosome colloidal stability following electroporation. Anal. Biochem. 2014;448:41–49. doi: 10.1016/j.ab.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuvichkin VV. DNA-lipid interactions in vitro and in vivo. Bioelectrochemistry. 2002;58(1):3–12. doi: 10.1016/s1567-5394(02)00123-8. [DOI] [PubMed] [Google Scholar]
- 41.Titomirov AV, Sukharev S, Kistanova E. In vivo electroporation and stable transformation of skin cells of newborn mice by plasmid DNA. Biochim. Biophys. Acta, Gene Struct. Expression. 1991;1088(1):131–134. doi: 10.1016/0167-4781(91)90162-f. [DOI] [PubMed] [Google Scholar]
- 42.Sukharev SI, Klenchin VA, Serov SM, Chernomordik LV, Chizmadzhev, Yu A. Electroporation and electrophoretic DNA transfer into cells. The effect of DNA interaction with electropores. Biophys. J. 1992;63(5):1320–1327. doi: 10.1016/S0006-3495(92)81709-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ohse M, Takahashi K, Kadowaki Y, Kusaoke H. Effects of plasmid DNA sizes and several other factors on transformation of Bacillus subtilis ISW1214 with plasmid DNA by electroporation. Biosci., Biotechnol., Biochem. 1995;59(8):1433–1437. doi: 10.1271/bbb.59.1433. [DOI] [PubMed] [Google Scholar]
- 44.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 45.Johansson P, Lindgren T, Lundstrom M, Holmstrom A, Elgh F, Bucht G. PCR-generated linear DNA fragments utilized as a hantavirus DNA vaccine. Vaccine. 2002;20(27–28):3379–3388. doi: 10.1016/s0264-410x(02)00265-7. [DOI] [PubMed] [Google Scholar]
- 46.Chattopadhyay S, Ely A, Bloom K, Weinberg MS, Arbuthnot P. Inhibition of hepatitis B virus replication with linear DNA sequences expressing antiviral micro-RNA shuttles. Biochem. Biophys. Res. Commun. 2009;389(3):484–489. doi: 10.1016/j.bbrc.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 47.Wang HS, Chen ZJ, Zhang G, Ou XL, Yang XL, Wong CK, Giesy JP, Du J, Chen SY. A novel micro-linear vector for in vitro and in vivo gene delivery and its application for EBV positive tumors. PLoS One. 2012;7(10):e47159. doi: 10.1371/journal.pone.0047159. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


