Abstract
Cone snails comprise approximately 500 species of venomous molluscs which have evolved the ability to generate multiple toxins with varied and exquisite selectivity. α-Conotoxin is a powerful tool for defining the composition and function of nicotinic acetylcholine receptors which play a crucial role in excitatory neurotransmission and are important targets for drugs and insecticides. An α4/7 conotoxin, Lp1.1, originally identified by cDNA and genomic DNA cloning from Conus leopardus, was found devoid of the highly conserved Pro residue in the first intercysteine loop. To further study this toxin, α-Lp1.1 was chemically synthesized and refolded into its globular disulfide isomer. The synthetic Lp1.1 induced seizure and paralysis on freshwater goldfish and selectively reversibly inhibited ACh-evoked currents in Xenopus oocytes expressing rat α3β2 and α6α3β2 nAChRs. Comparing the distinct primary structure with other functionally related α-conotoxins could indicate structural features in Lp1.1 that may be associated with its unique receptor recognition profile.
Keywords: Conus, α-conotoxins, neuronal nicotinic acetylcholine receptor subtypes, Xenopus oocytes
1. Introduction
Venomous marine molluscs belonging to the genus Conus utilize a unique neurochemical strategy for prey capture, defense, and competitor deterrence [45]. The venom of this large genus of predatory snails that feed on fish, other molluscs, and marine worms is composed of a complex mixture of different bioactive and highly modified peptides which are notable for their small size, potency, and target receptor subtype selectivity [45]. Many of these peptides are disulfide-rich neurotoxins, termed conotoxins, that selectively inhibit ligand- and voltage-gated ion channels. This high selectivity of conotoxins has lead to a great interest in the use of these molecules as pharmacological tools and the design of novel therapeutics [1, 37].
The primary structures of more than 100 conotoxins have been determined and classified into gene superfamilies on the basis of the amino acid sequences of the signal peptides of their precursors [37]. In general, the members of each superfamily have a characteristic arrangement of cysteine residues. Each gene superfamily comprises one or more pharmacologic families, i.e., the A superfamily, containing α-conotoxins, αA-conotoxins, and κ;A-conotoxins; the O superfamily, containing ω-conotoxins, κ;-conotoxins, δ-conotoxins, and μO-conotoxins; and the M superfamily, containing μ-conotoxins, ψ-conotoxins, and κ;M-conotoxins [45]. Most conotoxin families have a corresponding ion channel family target (i.e., ω-conotoxins towards calcium channels, α-conotoxins towards nicotinic receptors), and different families may have different ligand binding sites on the same ion channel target. The individual peptides in a conotoxin family are each selectively targeted to a diverse set of molecular isoforms within the same ion channel family. It has been postulated that divergence within a single superfamily to produce functionally different families is one of the strategies employed by cone snails, and may account, in part, for their success in nature [37, 39].
Nicotinic acetylcholine receptors (nAChRs) mediate synaptic transmission at neuromuscular and/or neuronal-neuronal synapses as well as extrasynaptical locations [33]. They belong to a superfamily of ligand-gated ion channels which include glycine and GABA-A receptors and are found across the whole phylum of bilateria from nematodes to mammals [35]. Their prominent functions are reflected in a variety of defense mechanisms and prey capture strategies in which many toxins secreted by predatory animals target this receptor [33]. Different nAChR subunits may combine in a variety of ways to form receptors which have been shown to be involved in learning, antinociception, nicotine addiction and neurological disorders such as Parkinson’s and Alzheimer’s disease [35]. Muscle nAChRs are pentamers containing four different subunits, α, β, γ (or ε), δ. Neuronal nAChRs are homologous to muscle nAChRs and fall into two different pharmacological classes: one binds α-bungarotoxin with high affinity and is predominantly composed of only α7 subunits, and the other binds agonists with high-affinity and is composed of α (α2-α6) and β (β2-β4) subunits [24]. The combinatorial diversity of nAChRs presents an opportunity to develop selective nAChR agonists and modulators for the specific treatment of neurological disorders. However, a prerequisite for the development of selective drugs is the identification and pharmacological characterization of the various receptor subtypes, and the determination of their precise subunit composition and physiological functions. Since it is difficult to distinguish subtly divergent molecular forms of these receptors with most available ligands, a considerable effort should be made to develop further and more subtype-specific nicotinic agonists and antagonists to probe nAChRs.
A remarkable variety of Conus peptides have been identified to date that alter the function of various nAChRs. A rationale for the presence of several different nicotinic ligands in Conus venoms follows. Previously characterized conotoxin nAChR antagonists have been members of the A, C, J, L, M and S superfamily of conotoxins [18, 38]. They are powerful tools for the purification, subtype differentiation and histologic labelling of nAChRs.
α-Conotoxins, a series of structurally and functionally related peptides, are between 11 and 16 amino acids in length, and are competitive antagonists of the nAChRs [35]. Thus far, these peptides have been divided into four groups based on the number of residues between the second and third cysteines and on the spacing between the third and fourth cysteines in the mature toxins [24]. These groups have different degrees of antagonistic effect on distinct nAChRs. The α4/7 subfamily, as well as the α4/3 and α4/4 subfamilies, targets neuronal nAChRs with one exception of α-EI [24, 31]. Such a characteristic of the α4/7 subfamily differs from that of the α3/5 subfamily that mostly blocks neuromuscular α1β1γδ subtypes [24]. It has been pointed out that α-conotoxins specific for neuronal subtypes of nAChR are neutral or negatively charged, whereas α-conotoxins that target muscle receptors have a net positive charge [19]. Even more surprising was the finding that in contrast to all previously studied α4/7-conotoxins, peptides SrIA and SrIB have a nAChR-potentiating activity [21]. In addition to being remarkable probes for structural studies, α-conotoxins also have therapeutic potential. For example, Vc1.1, the first α-conotoxin being developed to treat neuropathic pain causes an accelerated recovery of injured neurons [37, 41].
In this article, we describe the synthesis, oxidative folding and functional characterization by in vivo bioassay and electrophysiology of α-conotoxin Lp1.1 identified from the worm-hunting species Conus leopardus collected from the South China Sea. The detailed functional analyses of Lp1.1 will provide a basis for establishing structure/function relationships and may facilitate structure-based design studies to produce more potent and selective molecules.
2. Materials and methods
2.1. Materials
ZORBAX 300SB-C18 semipreparative column was purchased from Agilent Technologies (Santa Clara, CA, USA), and trifluoroacetic acid (TFA) and acetonitrile (ACN) for HPLC were from Merck (Darmstadt, Germany). Other reagents were of analytical grade.
2.2. Peptide synthesis and refolding
Lp1.1 was assembled by the solid-phase method on an ABI 433A peptide synthesizer (ABI, Foster City, USA) using standard Fmoc [N-(9-fluorenyl)methoxycarbonyl] chemistry and side-chain protection. Orthogonal protection was used on Cys residues: Cys2 and Cys8 were protected as the stable Cys (S-acetamidomethyl); Cys3 and Cys16 were protected as the acid-labile Cys (S-trityl). Linear peptide was cleaved from the resin by treatment with TFA/H2O/ethanedithiol/phenol/thioanisole (90:5:2.5:7.5:5 by volume), side chains of the acid-labile Cys and other non-Cys residues were de-protected during cleavage of the peptide from resin. The released peptide was precipitated and washed several times with cold methyl-t-butyl ether (MTBE). The crude peptide was then purified by RP-HPLC on a ZORBAX 300SB-C18 semi-preparative column (9.4 × 250 mm, Agilent Technologies) using a linear gradient of ACN at a flow rate of 2 mL/min and characterized by ESI-MS. The product was lyophilized and dissolved in 50 mM NH4HCO3 buffer.
The disulfide bond between Cys3 and Cys16 was formed by air oxidation. The linear peptide was stirred at room temperature for 8 hr, and the single disulfide bonded product was purified on a semi-preparative C18 column. The S-acetamidomethyl group was then removed from Cys2 and Cys8 in 0.02 mg/mL iodine, 4% TFA and 10% ACN, and a disulfide bond was formed simultaneously. The reaction was quenched by adding ascorbic acid. The predominant form of peptide obtained after oxidation was purified on a semi-preparative C18 reverse-phase column. The final product was applied to an analytical C18 column to verify its identity, using the following gradient of ACN at a flow rate of 0.5 mL/min: 0–5 min 10–15% buffer B, 5–45 min 15–35% buffer B. Buffer B is 0.1% TFA in 100% ACN.
2.3. In vivo bioassay
Gold fish (Gambusia affinis) were injected intramuscularly (i.m.) using 29-gauge insulin syringes with 20 μL of different doses of lyophilized peptide dissolved in normal saline solution (PBS) and placed in an aquarium for observation. The paralytic state of the fish was deduced from its inability to resist to a weak vortex current in a large beaker. The control fish were similarly injected with PBS.
2.4. Electrophysiology
The mouse muscle and rat neuronal nAChR expression clones were used to make capped RNA (cRNA) for injection into oocytes of Xenopus laevis frogs. cRNA was prepared using mMESSAGE mMACHINE (ABI, Foster City, USA) in vitro RNA transcription kits with either T7 or SP6 promoter and recovered by lithium chloride precipitation, according to the manufacturer’s protocol. Xenopus oocytes were harvested and dissociated with collagenase I at room temperature for 1 hr. The defolliculated oocytes were injected with 50 nL of cRNA 1–2 days after being harvested [40]. For expression of skeletal muscle nAChRs, 1 ng of each subunit cRNA was injected into the nucleus of each oocyte. For expression of neuronal nAChRs, typically 5 ng of each subunit cRNA was injected per oocyte. The oocytes were incubated at 15ºC in ND96 buffer (96 mM NaCl, 2 mM KCl, 1.8 mM CaCl2, 1 mM MgCl2, and 5 mM HEPES at pH 7.5) with 1 mM glucose-6-phosphate, 50 mg/L gentamycin, and 5 mM pyruvic acid to increase their survival for up to 7 days after injection [20, 36]. The oocyte recordings were obtained 1–6 days post-injection.
Two-electrode voltage clamp recording of Xenopus oocyte nAChR currents was conducted on an OC-725C amplifier (Warner Instruments, Hamden, USA). Oocytes were placed in a Warner RC-3Z recording chamber and attached with an OC-725 bath clamp. Electrodes with the resistance between 0.05 and 0.2 MΩ were filled with 3 M KCl. Oocytes were voltage clamped at a holding potential of −60 mV while perfused with OR2 buffer at 5 mL/min by gravity flow controlled by a Warner BPS-8 controller [42]. One micromolar atropine was added to OR2 buffer to block endogenous muscarinic acetylcholine receptors for all nAChR recordings, with the exception of α7 nAChRs, which atropine inhibits [12]. Currents were elicited by 5-s pulses of gravity-perfused agonist solution applied every 2 min to obtain the baseline activity: 10 μM ACh for muscle constructs and 100 μM ACh for neuronal constructs. All data recordings were carried out at room temperature. To identify the effect of the peptide on a particular receptor subtype, a predetermined concentration of Lp1.1 was applied to an oocyte in a static bath to allow the peptide to reach equilibrium with receptors for 10 min prior to restoration of the perfusion and ACh pulses. ACh was reapplied to assess the fractional response and recovery from the toxin exposure. Data were sampled at 500 Hz and filtered at 200 Hz. Peak current amplitude was measured before and following incubation of the peptide. The amplitude of the elicited current, following the peptide application and equilibration period, was calculated as a percentage of the amplitude of the elicited current prior to toxin application. All data were pooled (n = 3–5 for each receptor subtype) and represented as arithmetic means ± S.D.
3. Results
3.1. Unique primary structure
The conservation of the signal peptide and the UTR of each conotoxin superfamily has often been used to clone novel conotoxins from venom duct cDNAs. Recent research indicates that cloning from genomic DNA is a better alternative to acquiring more conotoxins than cDNA cloning. The two strategies have been used to clone a novel α-conotoxin Lp1.1 [46]. It shares the highly conserved signal peptide and the unique cysteine pattern in the mature toxins (Fig. 1). The pattern and the spacing of cysteines indicate that Lp1.1 belongs to the α4/7 subfamily of conotoxins. All known neuronally active α4/7 conotoxins have a conserved Ser in the first position of loop 1 and a Pro residue in the same loop. However, these conserved Ser and Pro residues are absent in Lp1.1. This toxin has a very unique primary structure with three Ala residues and a basic Arg residue in its first loop.
Figure 1.
Comparison of conotoxin prepropeptides belonging to the α family. The identical and similar amino acids are outlined in dark and gray background, respectively. Gaps are inserted to maximize similarity.
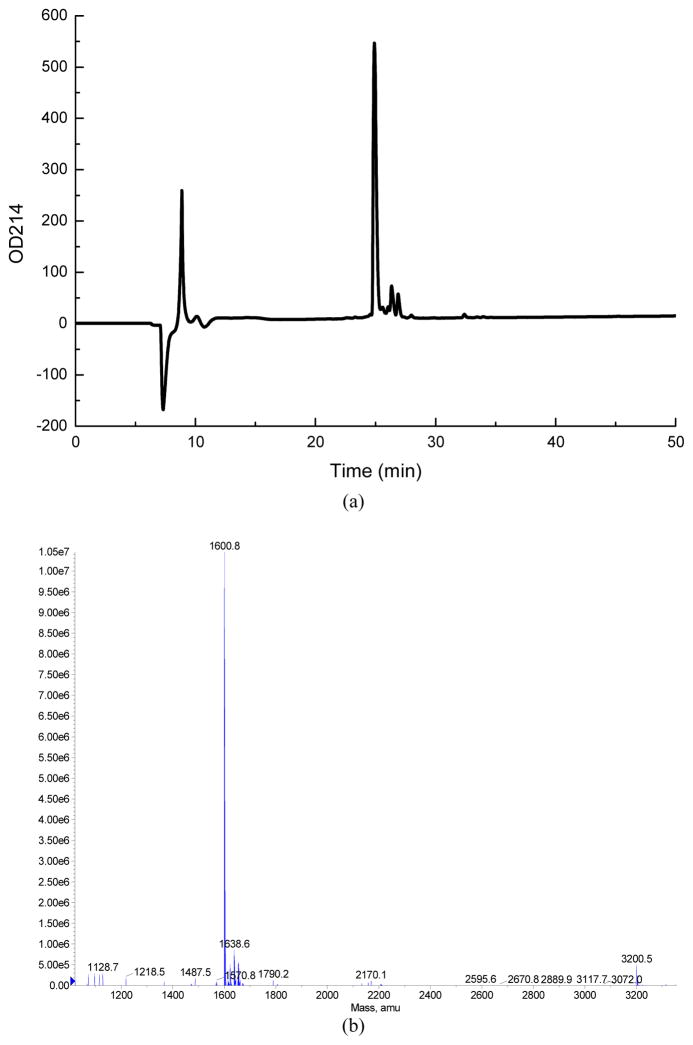
According to some rules emerging from matching the sequences of the mature peptides with the nucleotide sequences of the cDNAs encoding conotoxins [3], Lp1.1 is assumed to have 16 residues with the amidated C-terminus. The disulfide connectivity of Lp1.1 is assumed to be Cys2-Cys8 and Cys3-Cys16, consistent with the conserved α-conotoxin framework in which the four cysteines in their natural conformation form disulfide bridges giving the molecule a globular two-loop configuration with side chains projecting from a rigid backbone [7]. Lp1.1 was successfully assembled by solid phase peptide synthesis and the disulfide bonds were formed using a two-step oxidative folding approach in aqueous buffer at basic pH. The synthetic Lp1.1 has at least 95% purity as analysed by HPLC (Fig. 2). The mass of the synthetic Lp1.1 was consistent with the amidated sequence (average [M + H]+: calculated, 1600.88Da; observed, 1600.8Da).
Figure 2.
HPLC chromatogram (a) and MS profile (b) of the synthetic Lp1.1. In (a), analysis of the synthetic Lp1.1 by HPLC was performed on a PepMap C18 analytical column (4.6mm×250mm) (LC Packings). The peptide was loaded onto the column in 90% buffer A and eluted in a gradient of 15–35% buffer B with a flow rate of 0.5 mL/min over 40 min. Buffer A= 0.1% TFA; buffer B=0.1% TFA in 100% ACN. Absorbance was measured at 214nm.
3.2. Physiologic effect and inhibitory action
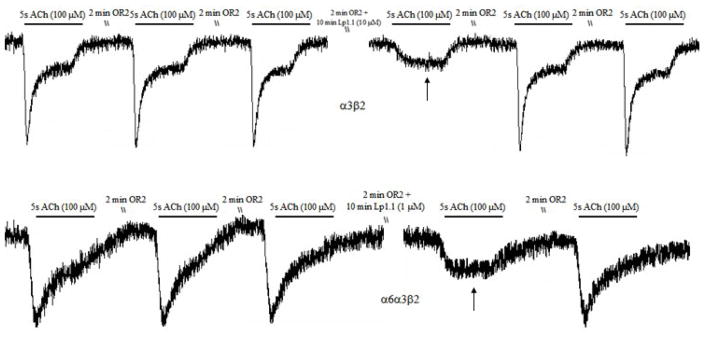
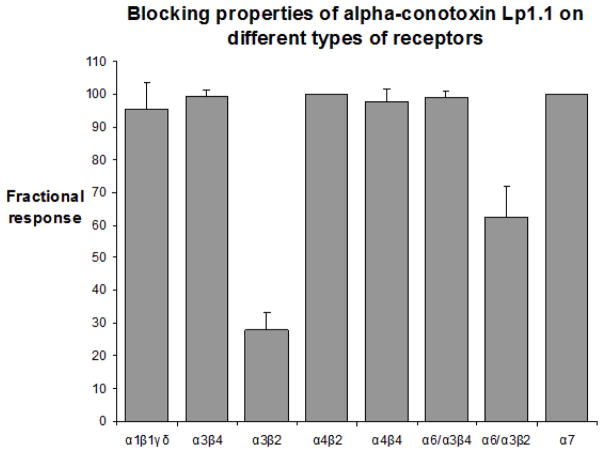
The in vivo bioassay and electrophysiological studies have been carried out to determine the activity of Lp1.1 with the unusual sequence. When Lp1.1 was administered i.m. in goldfish, 5 μg caused uncoordinated movement. Similar symptom was observed for 10 μg synthetic peptide and 20 μg of Lp1.1 caused seizure and paralysis after injection (Table 1). In parallel, the effect of Lp1.1 on ACh-evoked currents in Xenopus oocytes expressing the mouse and rat nAChR subtypes was investigated. At 10 μM, Lp1.1 blocked the ACh (100 μM)-elicited currents about 72% in recombinant α3β2 nAChRs, and the toxin dissociated rapidly from the receptor. A block of approximately 38% of the elicited current was obtained in recombinant α6α3β2, with a similar dissociation rate (Fig. 3). However, Lp1.1 exhibited no inhibition at concentrations up to 10 μM on other receptor subtypes tested (Fig. 4).
Table 1.
Biological activity of peptide Lp1.1 from C. leopardus
| Normal saline solution (20 μl) (n=3) | Lp1.1 (5 μg/20 μl) (n=3) | Lp1.1 (10 μg/20 μl) (n=3) | Lp1.1 (20 μg/20 μl) (n=3) |
| normal activity | fast breathing, unbalanced body, sporadic rolling swimming | fast breathing, moderate body shaking behavior | fast breathing, seizure and paralysis |
Figure 3.
Representative current traces for α-Lp1.1 at 10 μM and 1 μM on the rat α3β2 and α6α3β2 nAChRs, respectively. Control traces are shown prior to the application of peptide. The arrow marks the first current trace elicited after a 10-minute application of peptide. Subsequent current traces show peptide dissociation and washout.
Figure 4.
Lp1.1 is highly selective for rat neuronal α3β2 nAChR. Each bar indicates the average percent response (± S.D.) after the application of 10 μM Lp1.1 to Xenopus oocytes expressing a variety of nAChRs. To determine the average percent response, the peptide was tested ≥3 times against each receptor subtype. In contrast to the low nanomolar affinity of other α-conotoxins for a certain nAChR subtype, Lp1.1 demonstrated » 10 μM IC50 values for most of the receptors tested. Due to the limited amount of the toxin, only 1 μM of Lp1.1 was administered on rat neuronal α6α3β2 and α7 nAChRs, and mouse muscle α1β1γδ nAChR.
4. Discussion
Due to their slow mobility and a wide variety of prey (five different phyla), cone snails produce hypervariable toxins with the ability to target different subtypes of the same membrane receptor or ion channel with high affinity. α-Conotoxins from marine snails are known to be selective and potent competitive antagonists of nicotinic acetylcholine receptors (Table 2). Here we describe the total synthesis, oxidative folding, and activity of a novel toxin, Lp1.1, cloned from C. leopardus, a species that preys exclusively on hemichordates. As determined by cDNA cloning, the toxin is supposed to be a typical α4/7 conotoxin containing 16 amino acid residues. Indeed, the regiospecifically synthesized globular disulfide isomer of Lp1.1 showed blocking effects on α3β2 and α6α3β2 rat nAChR subtypes (Fig. 3 and 4).
Table 2.
Physical-chemical characteristics and biological activities of peptides belonging to α family identified from piscivorous (p), molluscivorous (m) and vermivorous (v) Conus species
| Peptide | Sequence | Species | Prey | Selectivity | References |
|---|---|---|---|---|---|
| 3/5 subfamily of α-conotoxins | |||||
| CnIA | GRCCHPACGKYYSC* | C. consors | p | α1β1γδ » α7 | [10] |
| GI | ECCNPACGRHYSC* | C. geographus | p | α1β1εδ | [13] |
| GIA | ECCNPACGRHYSCGK* | C. geographus | p | α1β1γδ | [13] |
| GII | ECCNPACGKHFSC* | C. geographus | p | α1β1γδ | [13] |
| MI | GRCCHPACGKNYSC* | C. magus | p | α1β1γδ | [4] |
| SI | ICCNPACGPKYSC* | C. striatus | p | α1β1γδ | [47] |
| 4/3 subfamily of α-conotoxins | |||||
| ImI | GCCSDPRCAWRC* | C. imperialis | v | α7 | [32] |
| ImIIA | YCCHRGPCMVWC* | C. imperialis | v | N/A | Genbank Q9U619 |
| ImII | ACCSDRRCRWRC | C. imperialis | v | α7 | [9] |
| Reg1b | GCCSDORCKHQC* | C. regius | v | N/A | [11] |
| Reg1c | GCCSDPRCKHQC* | C. regius | v | N/A | [11] |
| RgIA | GCCSDPRCRYRCR | C. regius | v | α9α10 | [8] |
| 4/4 subfamily of α-conotoxins | |||||
| BuIA | GCCSTPPCAVLYC* | C. bullatus | p | α6/α3 β4/β2 | [2] |
| PIB | ZSOGCCWNPACVKNRC* | C. purpurascens | p | α1β1δ (γ/ε) | [22] |
| 4/5 subfamily of α-conotoxins | |||||
| Ca1.1 | QNCCSIPSCWEKYKCS | C. caracteristicus | v | N/A | [43] |
| 4/6 subfamily of α-conotoxins | |||||
| AuIB | GCCSYPPCFATNPDC* | C. aulicus | m | α3/β4 | [26] |
| 4/7 subfamily of α-conotoxins | |||||
| AnIA | CCSHPACAANNQDYC* | C. anemone | v | α3/β2, α7 | |
| AnIB | GGCCSHPACAANNQDYC* | C. anemone | v | α3/β2, α7 | [25] |
| AnIC | GGCCSHPACFASNPDYC* | C. anemone | v | α3/β2, α7 | |
| AuIA | GCCSYPPCFATNSDYC* | C. aulicus | m | (less active) | [26] |
| AuIC | GCCSYPPCFATNSGYC* | C. aulicus | m | (less active) | |
| EI | RDOCCYHPTCNMSNPQIC* | C. ermineus | p | α1β1γδ, α3β4, α4β2 | [28] |
| EpI | GCCSDPRCNMNNPDYC* | C. episcopatus | m | α3β2/α3β4, α7 | [23] |
| GIC | GCCSHPACAGNNQHIC* | C. geographus | P | α3β2 | [29] |
| GID | IRDγCCSNPACRVNNOHVC | C. geographus | p | α3β2 | [34] |
| MII | GCCSNPVCHLEHSNLC* | C. magus | p | α3β2, α6β2β3 | [4] |
| OmIA | GCCSHPACNVNNPHICG* | C. omaria | m | α3β2, α7 | [5, 44] |
| PeIA | GCCSHPACSVNHPELC* | C. pergrandis | unknown | α9α10 | [30] |
| PIA | RDPCCSNPVCTVHNPQIC* | C. purpurascens | p | α6β2β3 | [6] |
| PnIA | GCCSLPPCAANNPDYC* | C. pennaceus | m | α3β2 | [17, 27] |
| [A10L]PnIA | GCCSLPPCALNNPDYC* | C. pennaceus | m | α7 | [15, 27] |
| PnIB | GCCSLPPCALSNPDYC* | C. pennaceus | m | α7 | [16, 27] |
| SrIA | RTCCSROTCRMγYPγLCG* | C. spurius | v | α4β2, α1β1γδ | |
| SrIB | RTCCSROTCRMEYPγLCG* | C. spurius | v | α4β2, α1β1γδ | [21] |
| [γ15E]SrIB | RTCCSROTCRMEYPELCG* | C. spurius | v | α4β2, α1β1γδ | |
| Vc1.1 | GCCSDORCNYDHPγIC* | C. victoriae | m | α3α7β4/α3α5β4 | [41] |
| Lp1.1 | GCCARAACAGIHQELC* | C. leopardus | v | α3β2, α6α3β2 | This work |
Conserved cysteine residues and the highly conserved serine and proline in the first loop are highlighted in bold-face. For post-translational modifications: γ, γ-carboxyglutamate; Z, pyroglutamic acid; O, hydroxyproline;
, amidated C-terminus;
Y, sulfotyrosine. N/A, not available.
However, the activity of Lp1.1 is considerably weaker than those of other α4/7-conotoxins, with the IC50 of Lp1.1 being higher than 10 μM. Previous results have suggested that the activities of α-conotoxins are related with the sequences of the second intercysteine loop [24, 35], as this loop exhibits higher sequence diversity (Table 2) and can give different molecular surface on the conserved backbone conformation. The NMR study of Lp1.1 is now in progress to see whether Lp1.1 has any unique structural feature that confers its specificity and low potency. Another possible reason for the low activity of Lp1.1 is the possible post-translational modification in the native Lp1.1 toxin which can not be predicted based on the cDNA sequence. On the other hand, we can not rule out the possibility that Lp1.1 may have species specificity and show higher activity on AChR of other species which are not determined in this work.
The most striking point of Lp1.1 sequence is the absence of the conserved proline or hydroxyproline residue in the first intercysteine loop (Table 2). This highly conserved Pro residue, as well as the C-terminal amidation, has recently been reported to be essential structural determinant on folding patterns and conformational switches in α- and χ/λ-conotoxins [19]. Substitution of proline in the first loop of α-conotoxin ImI switches the folding pattern from the globular form (C1–C3, C2–C4) to the ribbon form (C1–C4, C2–C3), while substitution of Lys6 in the first loop of conotoxin CMrVIA with Pro shifts the folding preference from its native ribbon conformation to the globular conformation. The AChBP binding activity of ImI is also dramatically affected by the substitution and the consequent conformational change [19]. It has been reported that the same cysteine pattern may correlate with a different connectivity of disulfide bridges [14, 48]. Thus it would be very intriguing to see, without the conserved Pro residue, how the native Lp1.1 toxin folds and acts on AChR.
In conclusion, we have described the synthesis, folding and functional characterization of Lp1.1, a selective antagonist of the rat α3β2 and α6α3β2 nAChRs. Lp1.1 has also been shown to be potent in goldfish model of i.m. injection. These initial functional data on Lp1.1 will facilitate further structure/function studies to understand the molecular mechanism of this molecule via a range of approaches including peptide and receptor mutagenesis, molecular modeling studies and peptide engineering.
Acknowledgments
This work was supported by National Basic Research Program of China (2004CB719904) and by the Chinese Academy of Sciences for Key Topics in Innovation Engineering (KSCX2-YW-R-104). Chunguang Wang is supported by Program for young excellent talents in Tongji University (2006KJ063) and Dawn Program of Shanghai Education Commission (06SG26).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Refercences
- 1.Armishaw CJ, Alewood PF. Conotoxins as research tools and drug leads. Curr protein Pept Sci. 2005;6:221–40. doi: 10.2174/1389203054065437. [DOI] [PubMed] [Google Scholar]
- 2.Azam L, Dowell C, Watkins M, Stitzel JA, Olivera BM, McIntosh JM. α-Conotoxin BuIA, a novel peptide from Conus bullatus, distinguishes among neuronal nicotinic acetylcholine receptors. J Biol Chem. 2005;280:80–7. doi: 10.1074/jbc.M406281200. [DOI] [PubMed] [Google Scholar]
- 3.Buczek O, Bulaj G, Olivera BM. Conotoxins and the posttranslational modification of secreted gene products. Cell Mol Life Sci. 2005;62:3067–79. doi: 10.1007/s00018-005-5283-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cartier GE, Yoshikami D, Gray WR, Luo S, Olivera BM, McIntosh JM. A new α-conotoxin which targets α3β2 nicotinic acetylcholine receptors. J Biol Chem. 1996;271:7522–8. doi: 10.1074/jbc.271.13.7522. [DOI] [PubMed] [Google Scholar]
- 5.Chi SW, Kim DH, Olivera BM, McIntosh JM, Han KH. Solution conformation of a neuronal nicotinic acetylcholine receptor antagonist α-conotoxin OmIA that discriminates α3 vsα6 nAChR subtypes. Biochem Biophys Res Commun. 2006;345:248–54. doi: 10.1016/j.bbrc.2006.04.099. [DOI] [PubMed] [Google Scholar]
- 6.Dowell C, Olivera BM, Garrett JE, Staheli ST, Watkins M, Kuryatov A, et al. α-Conotoxin PIA is selective for α6 subunit-containing nicotinic acetylcholine receptors. J Neurosci. 2003;23:8445–52. doi: 10.1523/JNEUROSCI.23-24-08445.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dutton JL, Craik DJ. alpha-Conotoxins: nicotinic acetylcholine receptor antagonists as pharmacological tools and potential drug leads. Curr Med Chem. 2001;8:327–44. doi: 10.2174/0929867013373453. [DOI] [PubMed] [Google Scholar]
- 8.Ellison M, Haberlandt C, Gomez-Casati ME, Watkins M, Elgoyhen AB, McIntosh JM, et al. α-RgIA: a novel conotoxin that specifically and potently blocks the α9α10 nAChR. Biochemistry. 2006;45:1511–7. doi: 10.1021/bi0520129. [DOI] [PubMed] [Google Scholar]
- 9.Ellison M, McIntosh JM, Olivera BM. α-Conotoxins ImI and ImII. Similar α7 nicotinic receptor antagonists act at different sites. J Biol Chem. 2003;278:757–64. doi: 10.1074/jbc.M204565200. [DOI] [PubMed] [Google Scholar]
- 10.Favreau P, Krimm I, Le Gall F, Bobenrieth MJ, Lamthanh H, Bouet F, et al. Biochemical characterization and nuclear magnetic resonance structure of novel α-conotoxins isolated from the venom of Conus consors. Biochemistry. 1999;38:6317–26. doi: 10.1021/bi982817z. [DOI] [PubMed] [Google Scholar]
- 11.Franco A, Pisarewicz K, Moller C, Mora D, Fields GB, Marì F. Hyperhydroxylation: a new strategy for neuronal targeting by venomous marine molluscs. Prog Mol Subcell Biol. 2006;43:83–103. doi: 10.1007/978-3-540-30880-5_4. [DOI] [PubMed] [Google Scholar]
- 12.Gerzanich V, Anand R, Lindstrom J. Homomers of α8 and α7 subunits of nicotinic acetylcholine receptors exhibit similar channel but contrasting binding site properties. Mol Pharmacol. 1994;45:212–20. [PubMed] [Google Scholar]
- 13.Gray WR, Luque A, Olivera BM, Barrett J, Cruz LJ. Peptide toxins from Conus geographus venom. J Biol Chem. 1981;256:4734–40. [PubMed] [Google Scholar]
- 14.Han YH, Wang Q, Jiang H, Liu L, Xiao C, Yuan DD, et al. Characterization of novel M-superfamily conotoxins with new disulfide linkage. FEBS J. 2006;273:4972–82. doi: 10.1111/j.1742-4658.2006.05493.x. [DOI] [PubMed] [Google Scholar]
- 15.Hogg RC, Miranda LP, Craik DJ, Lewis RJ, Alewood PF, Adams DJ. Single amino acid substitutions in α-conotoxin PnIA shift selectivity for subtypes of the mammalian neuronal nicotinic acetylcholine receptor. J Biol Chem. 1999;274:36559–64. doi: 10.1074/jbc.274.51.36559. [DOI] [PubMed] [Google Scholar]
- 16.Hu SH, Gehrmann J, Alewood PF, Craik DJ, Martin JL. Crystal structure at 1. 1 angstrom resolution of α-conotoxin PnIB: Comparison with α-conotoxins PnIA and GI. Biochemistry. 1997;36:11323–30. doi: 10.1021/bi9713052. [DOI] [PubMed] [Google Scholar]
- 17.Hu SH, Gehrmann J, Guddat LW, Alewood PF, Craik DJ, Martin JL. The 1. 1 angstrom crystal structure of the neuronal acetylcholine receptor antagonist, α-conotoxin PnIA from Conus pennaceus. Structure. 1996;4:417–23. doi: 10.1016/s0969-2126(96)00047-0. [DOI] [PubMed] [Google Scholar]
- 18.Jimenez EC, Olivera BM, Teichert RW. αC-conotoxin PrXA: a new family of nicotinic acetylcholine receptor antagonists. Biochemistry. 2007;46:8717–24. doi: 10.1021/bi700582m. [DOI] [PubMed] [Google Scholar]
- 19.Kang TS, Radić Z, Talley TT, Jois SD, Taylor P, Kini RM. Protein folding determinants: structural features determining alternative disulfide pairing in α- and χ/λ-conotoxins. Biochemistry. 2007;46:3338–55. doi: 10.1021/bi061969o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levandoski MM, Lin Y, Moise L, McLaughlin JT, Cooper E, Hawrot E. Chimeric analysis of a neuronal nicotinic acetylcholine receptor reveals amino acids conferring sensitivity to α-bungarotoxin. J Biol Chem. 1999;274:26113–9. doi: 10.1074/jbc.274.37.26113. [DOI] [PubMed] [Google Scholar]
- 21.López-Vera E, Aguilar MB, Schiavon E, Marinzi C, Ortiz E, Restano Cassulini R, et al. Novel α-conotoxins from Conus spurius and the α-conotoxin EI share high-affinity potentiation and low-affinity inhibition of nicotinic acetylcholine receptors. FEBS J. 2007;274:3972–85. doi: 10.1111/j.1742-4658.2007.05931.x. [DOI] [PubMed] [Google Scholar]
- 22.López-Vera E, Jacobsen RB, Ellison M, Olivera BM, Teichert RW. A novel α-conotoxin (α-PIB) isolated from C. purpurascens is selective for skeletal muscle nicotinic acetylcholine receptors. Toxicon. 2007;49:1193–9. doi: 10.1016/j.toxicon.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 23.Loughnan M, Bond T, Atkins A, Cuevas J, Adams DJ, Broxton NM, et al. α-Conotoxin EpI, a novel sulfated peptide from Conus episcopatus that selectively targets neuronal nicotinic acetylcholine receptors. J Biol Chem. 1998;273:15667–74. doi: 10.1074/jbc.273.25.15667. [DOI] [PubMed] [Google Scholar]
- 24.Loughnan ML, Alewood PF. Physico-chemical characterization and synthesis of neuronally active α-conotoxins. Eur J Biochem. 2004;271:2294–304. doi: 10.1111/j.1432-1033.2004.04146.x. [DOI] [PubMed] [Google Scholar]
- 25.Loughnan ML, Nicke A, Jones A, Adams DJ, Alewood PF, Lewis RJ. Chemical and functional identification and characterization of novel sulfated α-conotoxins from the cone snail Conus anemone. J Med Chem. 2004;47:1234–41. doi: 10.1021/jm031010o. [DOI] [PubMed] [Google Scholar]
- 26.Luo S, Kulak JM, Cartier GE, Jacobsen RB, Yoshikami D, Olivera BM, et al. α-Conotoxin AuIB selectively blocks α3β4 nicotinic acetylcholine receptors and nicotine-evoked norepinephrine release. J Neurosci. 1998;18:8571–9. doi: 10.1523/JNEUROSCI.18-21-08571.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo S, Nguyen TA, Cartier GE, Olivera BM, Yoshikami D, McIntosh JM. Single-residue alteration in α-conotoxin PnIA switches its nAChR subtype selectivity. Biochemistry. 1999;38:14542–8. doi: 10.1021/bi991252j. [DOI] [PubMed] [Google Scholar]
- 28.Martinez JS, Olivera BM, Gray WR, Craig AG, Groebe DR, Abramson SN, et al. α-Conotoxin EI, a new nicotinic acetylcholine receptor antagonist with novel selectivity. Biochemistry. 1995;34:14519–26. doi: 10.1021/bi00044a030. [DOI] [PubMed] [Google Scholar]
- 29.McIntosh JM, Dowell C, Watkins M, Garrett JE, Yoshikami D, Olivera BM. α-Conotoxin GIC from Conus geographus, a novel peptide antagonist of nicotinic acetylcholine receptors. J Biol Chem. 2002;277:33610–5. doi: 10.1074/jbc.M205102200. [DOI] [PubMed] [Google Scholar]
- 30.McIntosh JM, Plazas PV, Watkins M, Gomez-Casati ME, Olivera BM, Elgoyhen AB. A novel α-conotoxin, PeIA, cloned from Conus pergrandis, discriminates between rat α9α10 and α7 nicotinic cholinergic receptors. J Biol Chem. 2005;280:30107–12. doi: 10.1074/jbc.M504102200. [DOI] [PubMed] [Google Scholar]
- 31.McIntosh JM, Santos AD, Olivera BM. Conus peptides targeted to specific nicotinic acetylcholine receptors subtypes. Annu Rev Biochem. 1999;68:59–88. doi: 10.1146/annurev.biochem.68.1.59. [DOI] [PubMed] [Google Scholar]
- 32.McIntosh JM, Yoshikami D, Mahe E, Nielsen DB, Rivier JE, Gray WR, et al. A nicotinic acetylcholine receptor ligand of unique specificity, α-conotoxin ImI. J Biol Chem. 1994;269:16733–9. [PubMed] [Google Scholar]
- 33.Nicke A. Learning about structure and function of neuronal nicotinic acetylcholine receptors. Lessons from snails. Eur J Biochem. 2004;271:2293. doi: 10.1111/j.1432-1033.2004.04171.x. [DOI] [PubMed] [Google Scholar]
- 34.Nicke A, Loughnan ML, Millard EL, Alewood PF, Adams DJ, Daly NL, et al. Isolation, structure, and activity of GID, a novel α4/7-conotoxin with an extended N-terminal sequence. J Biol Chem. 2003;278:3137–44. doi: 10.1074/jbc.M210280200. [DOI] [PubMed] [Google Scholar]
- 35.Nicke A, Wonnacott S, Lewis RJ. α-Conotoxins as tools for the elucidation of structure and function of neuronal nicotinic acetylcholine receptor subtypes. Eur J Biochem. 2004;271:2305–19. doi: 10.1111/j.1432-1033.2004.04145.x. [DOI] [PubMed] [Google Scholar]
- 36.Nutt LK, Margolis SS, Jensen M, Herman CE, Dunphy WG, Rathmell JC, et al. Metabolic regulation of oocyte cell death through the CaMKII-mediated phosphorylation of caspase-2. Cell. 2005;123:89–103. doi: 10.1016/j.cell.2005.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Olivera BM. Conus peptides: biodiversity-based discovery and exogenomics. J Biol Chem. 2006;281:31173–7. doi: 10.1074/jbc.R600020200. [DOI] [PubMed] [Google Scholar]
- 38.Peng C, Tang S, Pi C, Liu J, Wang F, Wang L, et al. Discovery of a novel class of conotoxin from Conus litteratus, lt14a, with a unique cysteine pattern. Peptides. 2006;27:2174–81. doi: 10.1016/j.peptides.2006.04.016. [DOI] [PubMed] [Google Scholar]
- 39.Remigio EA, Duda TF., Jr Evolution of ecological specialization and venom of a predatory marine gastropod. Mol Ecol. 2008;17:1156–62. doi: 10.1111/j.1365-294X.2007.03627.x. [DOI] [PubMed] [Google Scholar]
- 40.Rosenthal JA, Levandoski MM, Chang B, Potts JF, Shi QL, Hawrot E. The functional role of positively charged amino acid side chains in α-bungarotoxin revealed by site-directed mutagenesis of a His-tagged recombinant α-bungarotoxin. Biochemistry. 1999;38:7847–55. doi: 10.1021/bi990045g. [DOI] [PubMed] [Google Scholar]
- 41.Sandall DW, Satkunanathan N, Keays DA, Polidano MA, Liping X, Pham V, et al. A novel α-conotoxin identified by gene sequencing is active in suppressing the vascular response to selective stimulation of sensory nerves in vivo. Biochemistry. 2003;42:6904–11. doi: 10.1021/bi034043e. [DOI] [PubMed] [Google Scholar]
- 42.Sanders T, Hawrot E. A novel pharmatope tag inserted into the beta4 subunit confers allosteric modulation to neuronal nicotinic receptors. J Biol Chem. 2004;279:51460–5. doi: 10.1074/jbc.M409533200. [DOI] [PubMed] [Google Scholar]
- 43.Santos AD, McIntosh JM, Hillyard DR, Cruz LJ, Olivera BM. The A-superfamily of conotoxins: structural and functional divergence. J Biol Chem. 2004;279:17596–606. doi: 10.1074/jbc.M309654200. [DOI] [PubMed] [Google Scholar]
- 44.Talley TT, Olivera BM, Han KH, Christensen SB, Dowell C, Tsigelny I, et al. α-Conotoxin OmIA is a potent ligand for the acetylcholine-binding protein as well as α3β2 and α7 nicotinic acetylcholine receptors. J Biol Chem. 2006;281:24678–86. doi: 10.1074/jbc.M602969200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Terlau H, Olivera BM. Conus venoms: a rich source of novel ion channel-targeted peptides. Physiol Rev. 2004;84:41–68. doi: 10.1152/physrev.00020.2003. [DOI] [PubMed] [Google Scholar]
- 46.Yuan DD, Han YH, Wang CG, Chi CW. From the identification of gene organization of α-conotoxins to the cloning of novel toxins. Toxicon. 2007;49:1135–49. doi: 10.1016/j.toxicon.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 47.Zafaralla GC, Ramilo C, Gray WR, Karlstrom R, Olivera BM, Cruz LJ. Phylogenetic specificity of cholinergic ligands: α-conotoxin SI. Biochemistry. 1988;27:7102–5. doi: 10.1021/bi00418a065. [DOI] [PubMed] [Google Scholar]
- 48.Zugasti-Cruz A, Aguilar MB, Falcón A, Olivera BM, Heimer de la Cotera EP. Two new 4-Cys conotoxins (framework 14) of the vermivorous snail Conus austini from the Gulf of Mexico with activity in the central nervous system of mice. Peptides. 2008;29:179–85. doi: 10.1016/j.peptides.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]