Abstract
Nε-acetylation is emerging as an abundant post-translational modification of bacterial proteins. Two mechanisms have been identified: one is enzymatic, dependent on an acetyltransferase and acetyl coenzyme A; the other is non-enzymatic and depends on the reactivity of acetyl phosphate. Some, but not most, of those acetylations are reversed by deacetylases. This review will briefly describe the current status of the field and raise questions that need answering.
Keywords: acetyl phosphate, acetyltransferase, CRP, deacetylase, metabolism, post-translational modification
Nε-acetylation is abundant in bacteria
Post-translational modifications control protein structure, stability and function. In eukaryotes, Nε-lysine acetylation is an abundant post-translational modification affecting thousands of proteins in diverse processes (Glozak and Seto, 2007; Rardin et al., 2013; Verdin and Ott, 2015; Yang and Seto, 2008a). Compelling evidence exists that Nε-acetylation is also abundant in bacteria. Global surveys have revealed acetylation in diverse bacterial phyla (Baeza et al., 2014; Kim et al., 2013; Kosono et al., 2015; Kuhn et al., 2014; Lee et al., 2013; Wang et al., 2010; Weinert et al., 2013; Wu et al., 2013; Yu et al., 2008; Zhang et al., 2009; Zhang et al., 2013). Recent studies have detected acetylation of 15–20% of the Escherichia coli and Bacillus subtilis proteomes (Kosono et al., 2015; Kuhn et al., 2014; Weinert et al., 2013; Zhang et al., 2013). Here, I will provide a brief review and raise questions I consider to be most important.
Nε-acetylation can occur by two different mechanisms
The conventional mechanism for Nε-acetylation is enzymatic (Fig. 1i), relying on a lysine acetyltransferase (KAT) to catalyze donation of the acetyl group from acetyl-coenzyme A (acCoA) to the ε-amino group of a deprotonated lysine. This is the mechanism used by KATs to acetylate lysines in the unstructured N-termini of eukaryotic histones. KATs from diverse bacteria have been identified and some characterized. (For reviews, see (Bernal et al., 2014; Hentchel and Escalante-Semerena, 2015; Hu et al., 2010; Jones and O'Connor, 2011; Kim and Yang, 2011; Soppa, 2010; Thao and Escalante-Semerena, 2011b)).
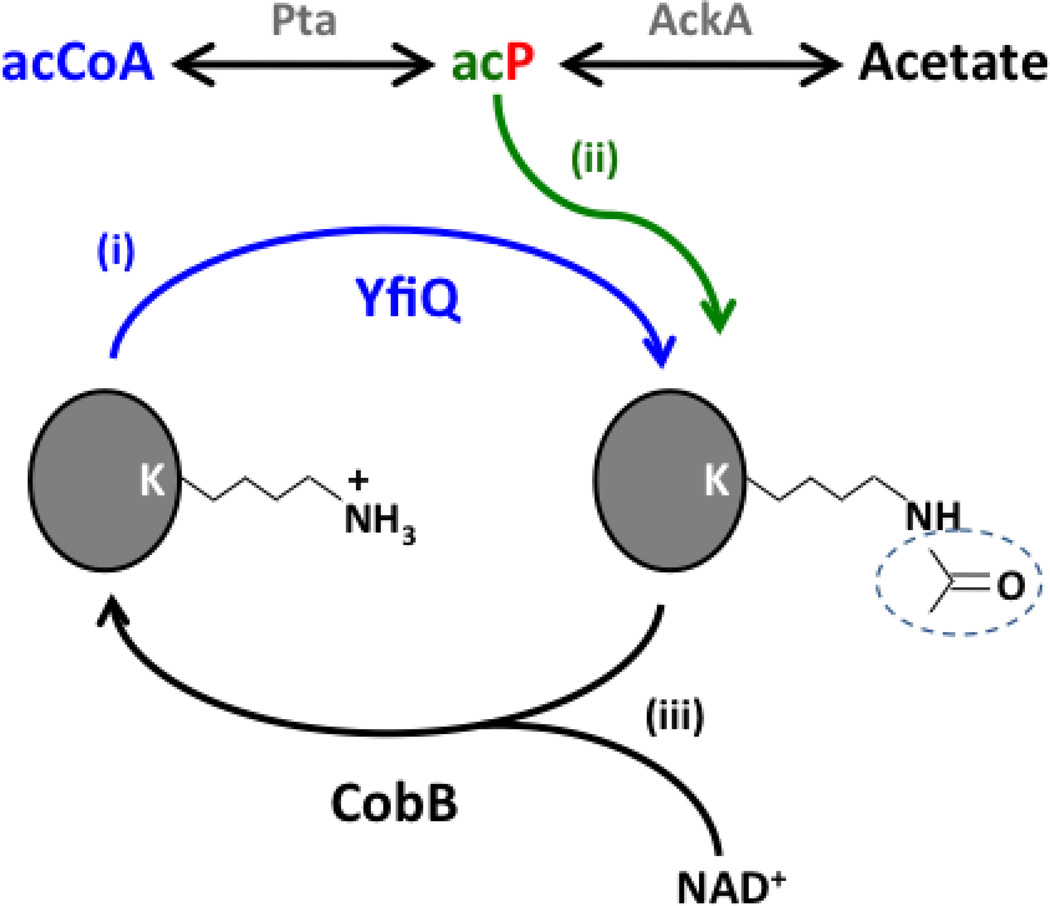
Fig. 1. Mechanisms for Nε-lysine acetylation and deacetylation.
(i) Acetyltransferase (YfiQ)- and acCoA-dependent acetylation (blue). (ii) Non-enzymatic acP-dependent acetylation (green). (iii) The NAD+-dependent deacetylase CobB can remove acetyl groups from some acetyllysines. acP is the high-energy intermediate of the phosphotransacetylase (Pta) - acetate kinase (AckA) pathway.
In E. coli, researchers have identified a single KAT: a member of the GCN5-like acetyltransferase (GNAT) family called YfiQ. It was first identified and characterized in Salmonella enterica (Hentchel and Escalante-Semerena, 2015; Starai and Escalante-Semerena, 2004b; Thao and Escalante-Semerena, 2011a), where it is called Protein AcetylTransferase or Pat. To avoid confusion, use of this acronym in E. coli should be discouraged, as it was previously assigned to Putrescine AminoTransferase. For a similar reason, an alternative acronym (Pka) should also be discouraged, as it is the widely used acronym for Protein Kinase A, which modifies eukaryotic proteins by phosphorylation.
It is possible that E. coli expresses KATs in addition to YfiQ, as several E. coli GNATs have not yet been assigned a function. However, previous attempts to identify additional KATs have failed. New approaches should be applied.
Recent studies reveal a novel non-enzymatic mechanism, in which acetyl phosphate (acP) directly donates its acetyl group to the ε-amino group of a deprotonated lysine (Kuhn et al., 2014; Weinert et al., 2013). The result is the same as the enzymatic mechanism, acetylation of the Nε-amino group of a lysine (Fig. 1ii). In sheer numbers of sensitive lysines, acP-dependent acetylation dwarfs YfiQ-dependent acetylation (Kuhn et al., 2014; Weinert et al., 2013). A novel, label-free quantitative mass spectrometric (MS) method called Skyline ‘MS1 Filtering’ (Rardin et al., 2013; Schilling et al., 2012) was used to determine, with statistical significance, that 592 lysines from 292 proteins were sensitive to acP levels. In contrast, many fewer lysines (~6×) were sensitive to the KAT YfiQ (Kuhn et al., 2014). Using more conventional SILAC methods, others obtained similar results (Weinert et al., 2013). Thus, in terms of sheer numbers of acetylated lysines, the non-enzymatic, acP-dependent mechanism predominates in E. coli. Non-enzymatic, acP-dependent acetylation is not confined to E. coli; a similar story is unfolding in B. subtilis (Kosono et al., 2015).
The impact of acetylation is predicted to depend upon its stoichiometry. A highly regulated and common acetylation likely has more impact than a highly regulated but uncommon acetylation. To address this issue, John Denu and co-workers developed a method to calculate global acetyl-stoichiometry in cell lysates. They modified an approach previously developed by Brad Gibson’s group to determine relative lysine reactivities in an isolated protein (Guo et al., 2008). Denu’s team detected substantial acetyl-stoichiometry in E. coli; hundreds of quantified peptides had stoichiometries greater than 10%, with 150 peptides acetylated at stoichiometries >20% (Baeza et al., 2014). Since both acetylating mechanisms were operable in the tested cells, it is unclear whether the detected acetyllysines resulted from the enzymatic, acCoA-dependent mechanism or from the non-enzymatic, acP-dependent mechanism. Further analyses of stoichiometry should be pursued under a variety of physiologically relevant conditions. Such studies will help prioritize the study of acetyllysines predicted to have the largest impact.
The deacetylase CobB can reverse acetylations from either mechanism
Acetyllysines are quite stable; however, they can be enzymatically reversed by lysine deacetylases (KDACs) (Fig. 1iii). Two major families of KDACs have been identified: the zinc-dependent Rpd3/Hda1 family (Yang and Seto, 2008b) and the NAD+-dependent sirtuin family (Blander and Guarente, 2004). For each HDAC family, putative bacterial homologues have been identified [reviewed by (Hildmann et al., 2007)], but only a few have been shown to serve as protein deacetylases [for a review, see (Hentchel and Escalante-Semerena, 2015)]. The best example of a bacterial sirtuin is CobB, first identified and characterized in S. enterica (Starai et al., 2002; Starai et al., 2003). In E. coli, the sirtuin CobB appears to be the sole (or predominant) KDAC (AbouElfetouh et al., 2015). It shows no preference for acetyl donors (AbouElfetouh et al., 2015); it can deacetylate both YfiQ-catalyzed acCoA-dependent acetyllysines (e.g., K609 of acetyl-CoA synthetase) and non-enzymatic acP-dependent acetyllysines (e.g. K154 of RcsB) (AbouElfetouh et al., 2015). Thus, it is not a member of a system designed specifically to reversibly acetylated a few key lysines, as suggested (Hentchel and Escalante-Semerena, 2015), but instead appears to operate independently, at least in E. coli. How CobB recognizes its substrates remains to be determined; however, we know that specificity depends on accessibility and three-dimensional microenvironment of the target acetyllysine (AbouElfetouh et al., 2015). In contrast to dogma, most acetyllysines appear to be insensitive to CobB (Kuhn et al., 2014; Weinert et al., 2013). MS1 Filtering identified and quantified only 69 CobB-sensitive lysines. This contrasts with the 592 acP-dependent acetyllysines (Kuhn et al., 2014). Only 24 were sensitive to both acP and CobB (AbouElfetouh et al., 2015). Thus, the vast majority of acetyllysines are not reversed and a major unanswered question is how does the cell cope with acetylations that are not reversed?
acP-dependent acetylation is specific
Most lysines are not acetylated, or only at very low undetectable levels. Some (e.g. K609 of Acs) are specifically acetylated by the enzymatic reaction (Hentchel and Escalante-Semerena, 2015). Those that are acetylated in an acP-dependent manner exhibit different acetylation rates with different fold changes over time (Schilling et al., 2015). Most acetylated lysines reside in a three-dimensional environment conducive to non-enzymatic acetylation. This environment contains residues that bind the phosphoryl group; these residues are either positively charged or tend towards H-bond formation (Baeza et al., 2015; Kuhn et al., 2014). Also, the lysine must be deprotonated. In the enzymatic, acCoA-dependent mechanism, often a negatively charged residue on the enzyme surface acts as a general base catalyst to perform this function. In the non-enzymatic, acP-dependent mechanism, the catalyst appears to be a negatively charged residue adjacent to the target lysine (Kuhn et al., 2014). This model should be rigorously tested, but independent evidence supports it. A prime example is the cAMP receptor protein (CRP), the major carbon regulator in δ-proteobacteria, e.g. Enterobactericaeae (Shimada et al., 2011) and Vibrionaeae (Colton and Stabb, 2015). CRP has two adjacent lysines (K100 and K101) that exhibit strikingly different behaviors with respect to acetylation. AcP-dependent acetylation of K100 is almost always detected, whereas acetyl-K101 has never been detected (Baeza et al., 2014; Kuhn et al., 2014; Weinert et al., 2013; Zhang et al., 2013). The former includes all the aforementioned molecular features; the latter does not (Davis et al., in preparation).
acP-dependent acetylation is a consequence of acetate fermentation
Enzyme-dependent acetylation is regulated at several levels. As mentioned previously, CobB deacetylates a subset of YfiQ-dependent acetyllysines in S. enterica and E. coli (AbouElfetouh et al., 2015; Garrity et al., 2007; Li et al., 2010; Starai et al., 2002; Zhao et al., 2004). In S. enterica, the myo-inositol catabolism repressor LolR activates transcription of pat and cobB, as well as that of acs, which encodes their substrate acetyl-CoA synthetase (Hentchel et al., 2015). Transcription of pat, but not cobB, decreases under acid stress. This might be due to reduced cAMP-CRP complex as acid stress also reduced cya and crp transcription (Ren et al., 2015) and E. coli yfiQ transcription depends on the cAMP-CRP complex (Castano-Cerezo et al., 2011). In E. coli, acP-dependent acetylation also requires cAMP-CRP (see below), but not YfiQ, and CobB can deacetylate a subset of acP-dependent acetyllysines (AbouElfetouh et al., 2015). Also, acP-dependent acetylation seems to result from overflow metabolism, which occurs if the CoA pool is overwhelmed; for example, when carbon exceeds nitrogen and/or oxygen. Cells solve this metabolic bottleneck by recycling CoA via mixed acid fermentation (Wolfe, 2005). Species that perform mixed acid fermentation are found in the genera Escherichia, Neisseria, Bacillus, and Vibrio, among others (Wolfe, 2015). The major products of mixed acid fermentation are ATP and acetate. The intermediate of this pathway (Pta-AckA) is acP (Wolfe, 2005). With millimolar levels of acP (Klein et al., 2007), many lysines become acetylated (Kuhn et al., 2014; Weinert et al., 2013). We propose a simple model: overflow metabolism causes acP accumulation and acP accumulation causes acetylation. This scenario is supported by a recent study that showed global acetylation increasing over time and correlating closely with glucose consumption and acetate excretion (Schilling et al., 2015). Attempts to further test this model are underway. If it is correct, then genetic or environmental manipulations that alter the timing of glucose consumption should simultaneously influence acetylation.
AcP-dependent acetylation is enriched in central metabolic pathways
If acetylation is a response to overflow metabolism, then acetylation might feed back to regulate central metabolism. If so, then central metabolic enzymes should be enriched for acetylation. Indeed, MS1 Filtering revealed that proteins with glucose-regulated acetyl sites are overwhelmingly involved in central metabolism, notably glycolysis, gluconeogenesis, the TCA cycle, the glyoxylate bypass, fatty acid biosynthesis, and pyruvate metabolism, which links them all. Our group and others had previously shown that acetylation is enriched in central metabolic pathways (Kuhn et al., 2014; Wang et al., 2010; Weinert et al., 2013; Zhang et al., 2013), but this was the first demonstration that central metabolic enzymes become acetylated over time in response to glucose consumption. It also revealed that most glucose-induced acetylation was acP-dependent (Schilling et al., 2015). Moreover, this effect is not restricted to glucose, as similar results were obtained with other carbon sources (e.g. lactate) that result in acetate fermentation [(Schilling et al., 2015) and unpublished data].
cAMP-CRP regulates glucose-induced acP-dependent acetylation
Since glucose-regulated acetylation was enriched in central metabolism, we tested the hypothesis that some central metabolic regulator would regulate acetylation. Glucose consumption and acetate excretion were delayed and global acetylation was essentially eliminated in cya and crp mutants, but not in mutants lacking other central metabolic regulators, including arcA, cra and csrA (Schilling et al., 2015). Most CRP-dependent acetylation requires acP synthesis: acetylation was low in mutants that fail to synthesize acP, even when CRP was overexpressed. In contrast, acetylation was restored in crp mutants that accumulated acP. These results are consistent with a model in which acP is downstream of CRP. The mechanism by which cAMP-CRP-dependent transcription facilitates acP synthesis remains unknown, but it likely involves transcriptional control of carbon transport and flux through glycolytic pathways (Schilling et al., 2015).
The impact of acetylation on protein function and cellular physiology
We now know that protein acetylation occurs in diverse bacteria. It is abundant, stoichiometrically relevant, and regulated at multiple levels. As such, bacterial protein acetylation resembles eukaryotic protein acetylation, including non-enzymatic acetylation, which occurs at low levels in mitochondria (Lombard et al., 2015; Wagner and Payne, 2013; Weinert et al., 2015). The inference is that acetylation in bacteria works much like it does in eukaryotes and that it exerts a similar impact on protein function and cellular physiology.
Most of what we know about the impact of Nε-acetylation of bacterial protein function comes from the study of a small subset of proteins. For acetyl-CoA synthetase (Acs), reversible acetylation serves as a simple on-off switch. A KAT (Pat in S. enterica, YfiQ in E. coli) inhibits Acs activity by acetylating a lysine (K609) located in Acs’ active site (Gardner et al., 2006; Starai et al., 2002). The mechanism by which acetylation inhibits Acs activity is unknown; however, reactivation occurs upon deacetylation (Starai et al., 2002), catalyzed by the sirtuin CobB (Starai et al., 2002; Starai et al., 2003). For the response regulator CheY, acetylation of several lysines influences (i) reversible phosphorylation of an aspartyl residue, (ii) rotational direction of the flagellar motor, and (iii) CheY’s ability to form complexes with 3 protein targets (Barak and Eisenbach, 2001; Barak et al., 2004; Barak et al., 1992; Barak et al., 2006; Fraiberg et al., 2014; Li et al., 2010; Liarzi et al., 2010; Yan et al., 2008). For RNase R, acetylation controls protein turnover: acetylation and thus neutralization of a lysine residue is proposed to break a salt bridge that sequesters the docking site for a chaperone that recruits proteases (Liang et al., 2011). For RcsB, it appears that acetylation of one lysine (K154) inhibits its function (Castano-Cerezo et al., 2014; Hu et al., 2013), but the mechanism remains unknown. For the alpha subunit of RNA polymerase, acetylation of one lysine (K298) is proposed to enhance glucose-induced CpxR-dependent transcription of cpxP (Lima et al., 2011), whereas acetylation of a second lysine (K291) inhibits that transcription (Lima et al., 2012). Thus, acetylation of two different lysines on the same protein can differentially regulate its function.
As with protein function, the impact of acetylation on cellular physiology is poorly understood. Some patterns, however, have emerged. First, there is the obvious connection to central metabolism. The acetyl donors (acCoA and acP) are central metabolites as is the necessary substrate for deacetylation (NAD+). The major carbon regulator in E. coli and other enteric bacteria CRP regulates both non-enzymatic acP-dependent acetylation and enzymatic acCoA-dependent acetylation and fermentation results in acP-dependent acetylation. Furthermore, reversible acetylation regulates Acs activity (Gardner et al., 2006; Starai et al., 2002; Starai and Escalante-Semerena, 2004a; Starai and Escalante-Semerena, 2004b). Because this activity involves both acCoA and ATP, acetylation modulates energy status with its attendant consequences (Chan et al., 2011). AcP-dependent and CobB-sensitive acetylation of K154 inhibits RcsB function (Castano-Cerezo et al., 2014; Hu et al., 2013), which responds to peptidoglycan stress (Cambre et al., 2015; Laubacher and Ades, 2008), controls cell shape (Ranjit and Young, 2013) and contributes to inherent antibiotic resistance (Laubacher and Ades, 2008). RcsB also inhibits flagellar expression (Fredericks et al., 2006), activates extracellular polysaccharide biosynthesis (Fredericks et al., 2006), enhances the acid stress response (Castanie-Cornet et al., 2010) and controls transcription of a small RNA (rprA) that modulates the response to stationary phase stress (Hu et al., 2013; Majdalani et al., 2002). The link between acetylation and stress is supported by several other observations. Glucose-induced acetylation of alpha regulates transcription of cpxP (Lima et al., 2011; Lima et al., 2012), which encodes a chaperone involved the periplasmic unfolded protein response (Danese and Silhavy, 1998). Furthermore, YfiQ/Pat and CobB have been associated with stresses associated with low pH (Ren et al., 2015), high temperature (Ma and Wood, 2011) and reactive oxidative species (Ma and Wood, 2011). Finally, acP has been associated with ATP-dependent proteolysis (Mizrahi et al., 2006) and both protein folding and aggregation (Mizrahi et al., 2009). ATP-dependent proteases are linked to carbon metabolism (Yang and Lan, 2015). Most of these proteases and their chaperones are acetylated in an acP-dependent manner (Kuhn et al., 2014; Schilling et al., 2015). Since it is associated with stresses of multiple types, could acetylation be a response to stresses associated with rapid carbon flux through central metabolism?
Consequences for pharmaceutical production
Lysine acetylation has been detected following production in E. coli of recombinant therapeutic proteins, including the chemokine RANTES (d'Alayer et al., 2007), human basic fibroblast growth factor mutein (Suenaga et al., 1996), human carbonic anhydrase (Mahon et al., 2015), insulin lispro (Szewczak et al., 2015), interleukin-10 (Pflumm et al., 1997), interleukin-2 (Moya et al., 2002), interferon alpha (Takao et al., 1987), neurotropin-3 (Ross et al., 1996), and somatotropin (Violand et al., 1994). Nearly 30% of currently approved recombinant therapeutic proteins are produced in E. coli (Huang et al., 2012). All post-translational modifications in a recombinant therapeutic protein could potentially influence its safety and efficacy and should be assessed for toxicity, immunogenicity and biological activity (Mahon et al., 2015). Thus, it is critical to identify the acetylating mechanism, understand its regulation, and determine the effect that acetylation exerts on the function of these heterologously expressed proteins.
Acknowledgments
This work was supported by grants from the NIGMS (R01 GM066130) and the DOE (DE-SC00124430. I would like to acknowledge my collaborators, Bradford Gibson, Wayne Anderson and Christopher Rao, as well as present and former members of my laboratory.
References
- AbouElfetouh A, Kuhn ML, Hu LI, Scholle MD, Sorensen DJ, Sahu AK, Becher D, Antelmann H, Mrksich M, Anderson WF, et al. The E. coli sirtuin CobB shows no preference for enzymatic and nonenzymatic lysine acetylation substrate sites. Microbiologyopen. 2015;4:66–83. doi: 10.1002/mbo3.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeza J, Dowell JA, Smallegan MJ, Fan J, Amador-Noguez D, Khan Z, Denu JM. Stoichiometry of site-specific lysine acetylation in an entire proteome. J Biol Chem. 2014;289:21326–21338. doi: 10.1074/jbc.M114.581843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeza J, Smallegan MJ, Denu JM. Site-specific reactivity of nonenzymatic lysine acetylation. ACS Chem Biol. 2015;10:122–128. doi: 10.1021/cb500848p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak R, Eisenbach M. Acetylation of the response regulator, CheY, is involved in bacterial chemotaxis. Molecular Microbiology. 2001;40:731–743. doi: 10.1046/j.1365-2958.2001.02425.x. [DOI] [PubMed] [Google Scholar]
- Barak R, Prasad K, Shainskaya A, Wolfe AJ, Eisenbach M. Acetylation of the Chemotaxis Response Regulator CheY by Acetyl-CoA Synthetase Purified from Escherichia coli. Journal of Molecular Biology. 2004;342:383–401. doi: 10.1016/j.jmb.2004.07.020. [DOI] [PubMed] [Google Scholar]
- Barak R, Welch M, Yanovsky A, Oosawa K, Eisenbach M. Acetyladenylate or its derivative acetylates the chemotaxis protein CheY in vitro and increases its activity at the flagellar switch. Biochemistry. 1992;31:10099–10107. doi: 10.1021/bi00156a033. [DOI] [PubMed] [Google Scholar]
- Barak R, Yan J, Shainskaya A, Eisenbach M. The Chemotaxis Response Regulator CheY Can Catalyze its Own Acetylation. Journal of Molecular Biology. 2006;359:251–265. doi: 10.1016/j.jmb.2006.03.033. [DOI] [PubMed] [Google Scholar]
- Bernal V, Castano-Cerezo S, Gallego-Jara J, Ecija-Conesa A, de Diego T, Iborra JL, Canovas M. Regulation of bacterial physiology by lysine acetylation of proteins. N Biotechnol. 2014;31:586–595. doi: 10.1016/j.nbt.2014.03.002. [DOI] [PubMed] [Google Scholar]
- Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu Rev Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- Cambre A, Zimmermann M, Sauer U, Vivijs B, Cenens W, Michiels CW, Aertsen A, Loessner MJ, Noben JP, Ayala JA, et al. Metabolite profiling and peptidoglycan analysis of transient cell wall-deficient bacteria in a new Escherichia coli model system. Environ Microbiol. 2015;17:1586–1599. doi: 10.1111/1462-2920.12594. [DOI] [PubMed] [Google Scholar]
- Castanie-Cornet MP, Cam K, Bastiat B, Cros A, Bordes P, Gutierrez C. Acid stress response in Escherichia coli: mechanism of regulation of gadA transcription by RcsB and GadE. Nucleic Acids Res. 2010;38:3546–3554. doi: 10.1093/nar/gkq097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castano-Cerezo S, Bernal V, Blanco-Catala J, Iborra J, Canovas M. cAMP-CRP co-ordinates the expression of the protein acetylation pathway with central metabolism in Escherichia coli. Molecular Microbiology. 2011;82:1110–1128. doi: 10.1111/j.1365-2958.2011.07873.x. [DOI] [PubMed] [Google Scholar]
- Castano-Cerezo S, Bernal V, Post H, Fuhrer T, Cappadona S, Sanchez-Diaz NC, Sauer U, Heck AJ, Altelaar AF, Canovas M. Protein acetylation affects acetate metabolism, motility and acid stress response in Escherichia coli. Mol Syst Biol. 2014;10:762. doi: 10.15252/msb.20145227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CH, Garrity J, Crosby HA, Escalante-Semerena JC. In Salmonella enterica, the sirtuin-dependent protein acylation/deacylation system (SDPADS) maintains energy homeostasis during growth on low concentrations of acetate. Molecular Microbiology. 2011;80:168–183. doi: 10.1111/j.1365-2958.2011.07566.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colton DM, Stabb EV. Rethinking the roles of CRP, cAMP, and sugar-mediated global regulation in the Vibrionaceae. Curr Genet. 2015 Jul 28; doi: 10.1007/s00294-015-0508-8. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- d'Alayer J, Expert-Bezancon N, Beguin P. Time- and temperature-dependent acetylation of the chemokine RANTES produced in recombinant Escherichia coli. Protein Expr Purif. 2007;55:9–16. doi: 10.1016/j.pep.2007.04.022. [DOI] [PubMed] [Google Scholar]
- Danese PN, Silhavy TJ. CpxP, a stress-combative member of the Cpx regulon. J Bacteriol. 1998;180:831–839. doi: 10.1128/jb.180.4.831-839.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraiberg M, Afanzar O, Cassidy CK, Gabashvili A, Schulten K, Levin Y, Eisenbach M. CheY's acetylation sites responsible for generating clockwise flagellar rotation in Escherichia coli. Mol Microbiol. 2014 doi: 10.1111/mmi.12858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredericks CE, Shibata S, Aizawa S-I, Reimann SA, Wolfe AJ. Acetyl phosphate-sensitive regulation of flagellar biogenesis and capsular biosynthesis depends on the Rcs phosphorelay. Molecular Microbiology. 2006;61:734–747. doi: 10.1111/j.1365-2958.2006.05260.x. [DOI] [PubMed] [Google Scholar]
- Gardner JG, Grundy FJ, Henkin TM, Escalante-Semerena JC. Control of Acetyl-Coenzyme A Synthetase (AcsA) Activity by Acetylation/Deacetylation without NAD+ Involvement in Bacillus subtilis. Journal of Bacteriology. 2006;188:5460–5468. doi: 10.1128/JB.00215-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrity J, Gardner JG, Hawse W, Wolberger C, Escalante-Semerena JC. N-lysine propionylation controls the activity of propionyl-CoA synthetase. J Biol Chem. 2007;282:30239–30245. doi: 10.1074/jbc.M704409200. [DOI] [PubMed] [Google Scholar]
- Glozak MA, Seto E. Histone deacetylases and cancer. Oncogene. 2007;26:5420–5432. doi: 10.1038/sj.onc.1210610. [DOI] [PubMed] [Google Scholar]
- Guo X, Bandyopadhyay P, Schilling B, Young MM, Fujii N, Aynechi T, Guy RK, Kuntz ID, Gibson BW. Partial Acetylation of Lysine Residues Improves Intraprotein Cross-Linking. Analytical Chemistry. 2008;80:951–960. doi: 10.1021/ac701636w. [DOI] [PubMed] [Google Scholar]
- Hentchel KL, Escalante-Semerena JC. Acylation of Biomolecules in Prokaryotes: a Widespread Strategy for the Control of Biological Function and Metabolic Stress. MMBR. 2015;79:321–346. doi: 10.1128/MMBR.00020-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentchel KL, Thao S, Intile PJ, Escalante-Semerena JC. Deciphering the Regulatory Circuitry That Controls Reversible Lysine Acetylation in Salmonella enterica. MBio. 2015;6 doi: 10.1128/mBio.00891-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildmann C, Riester D, Schwienhorst A. Histone deacetylases--an important class of cellular regulators with a variety of functions. Appl Microbiol Biotechnol. 2007;75:487–497. doi: 10.1007/s00253-007-0911-2. [DOI] [PubMed] [Google Scholar]
- Hu LI, Chi BK, Kuhn ML, Filippova EV, Walker-Peddakotla AJ, Basell K, Becher D, Anderson WF, Antelmann H, Wolfe AJ. Acetylation of the Response Regulator RcsB Controls Transcription from a Small RNA Promoter. Journal of Bacteriology. 2013;195:4174–4186. doi: 10.1128/JB.00383-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu LI, Lima BP, Wolfe AJ. Bacterial protein acetylation: the dawning of a new age. Molecular Microbiology. 2010;77:15–21. doi: 10.1111/j.1365-2958.2010.07204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CJ, Lin H, Yang X. Industrial production of recombinant therapeutics in Escherichia coli and its recent advancements. J Ind Microbiol Biotechnol. 2012;39:383–399. doi: 10.1007/s10295-011-1082-9. [DOI] [PubMed] [Google Scholar]
- Jones JD, O'Connor CD. Protein acetylation in prokaryotes. Proteomics. 2011;11:3012–3022. doi: 10.1002/pmic.201000812. [DOI] [PubMed] [Google Scholar]
- Kim D, Yu BJ, Kim JA, Lee Y-J, Choi S-G, Kang S, Pan J-G. The acetylproteome of Gram-positive model bacterium Bacillus subtilis. Proteomics. 2013;13:1726–1736. doi: 10.1002/pmic.201200001. [DOI] [PubMed] [Google Scholar]
- Kim GW, Yang XJ. Comprehensive lysine acetylomes emerging from bacteria to humans. Trends Biochem Sci. 2011;36:211–220. doi: 10.1016/j.tibs.2010.10.001. [DOI] [PubMed] [Google Scholar]
- Klein AH, Shulla A, Reimann SA, Keating DH, Wolfe AJ. The Intracellular Concentration of Acetyl Phosphate in Escherichia coli Is Sufficient for Direct Phosphorylation of Two-Component Response Regulators. Journal of Bacteriology. 2007;189:5574–5581. doi: 10.1128/JB.00564-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosono S, Tamura M, Suzuki S, Kawamura Y, Yoshida A, Nishiyama M, Yoshida M. Changes in the Acetylome and Succinylome of Bacillus subtilis in Response to Carbon Source. PLoS One. 2015;10:e0131169. doi: 10.1371/journal.pone.0131169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn ML, Zemaitaitis B, Hu LI, Sahu A, Sorensen D, Minasov G, Lima BP, Scholle M, Mrksich M, Anderson WF, et al. Structural, kinetic and proteomic characterization of acetyl phosphate-dependent bacterial protein acetylation. PLoS One. 2014;9:e94816. doi: 10.1371/journal.pone.0094816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laubacher ME, Ades SE. The Rcs phosphorelay is a cell envelope stress response activated by peptidoglycan stress and contributes to intrinsic antibiotic resistance. J Bacteriol. 2008;190:2065–2074. doi: 10.1128/JB.01740-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D-W, Kim D, Lee Y-J, Kim J-A, Choi JY, Kang S, Pan J-G. Proteomic analysis of acetylation in thermophilic Geobacillus kaustophilus. Proteomics. 2013;13:2278–2282. doi: 10.1002/pmic.201200072. [DOI] [PubMed] [Google Scholar]
- Li R, Gu J, Chen Y-Y, Xiao C-L, Wang L-W, Zhang Z-P, Bi L-J, Wei H-P, Wang X-D, Deng J-Y, et al. CobB regulates Escherichia coli chemotaxis by deacetylating the response regulator CheY. Molecular Microbiology. 2010;76:1162–1174. doi: 10.1111/j.1365-2958.2010.07125.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang W, Malhotra A, Deutscher Murray P. Acetylation Regulates the Stability of a Bacterial Protein: Growth Stage-Dependent Modification of RNase R. Molecular Cell. 2011;44:160–166. doi: 10.1016/j.molcel.2011.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liarzi O, Barak R, Bronner V, Dines M, Sagi Y, Shainskaya A, Eisenbach M. Acetylation represses the binding of CheY to its target proteins. Molecular Microbiology. 2010;76:932–943. doi: 10.1111/j.1365-2958.2010.07148.x. [DOI] [PubMed] [Google Scholar]
- Lima BP, Antelmann H, Gronau K, Chi BK, Becher D, Brinsmade SR, Wolfe AJ. Involvement of Protein Acetylation in Glucose-induced Transcription of a Stress-Responsive Promoter. Molecular Microbiology. 2011;81:1190–1204. doi: 10.1111/j.1365-2958.2011.07742.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima BP, Thanh Huyen TT, Bassell K, Becher D, Antelmann H, Wolfe AJ. Inhibition of Acetyl Phosphate-dependent Transcription by an Acetylatable Lysine on RNA Polymerase. Journal of Biological Chemistry. 2012;287:32147–32160. doi: 10.1074/jbc.M112.365502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard DB, Dash BP, Kumar S. Acetyl-ed question in mitochondrial biology? EMBO J. 2015;34:2597–2600. doi: 10.15252/embj.201592927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Wood TK. Protein acetylation in prokaryotes increases stress resistance. Biochemical and Biophysical Research Communications. 2011;410:846–851. doi: 10.1016/j.bbrc.2011.06.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahon BP, Lomelino CL, Salguero AL, Driscoll JM, Pinard MA, McKenna R. Observed surface lysine acetylation of human carbonic anhydrase II expressed in Escherichia coli. Protein Sci. 2015;24:1800–1807. doi: 10.1002/pro.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majdalani N, Hernandez D, Gottesman S. Regulation and mode of action of the second small RNA activator of RpoS translation, RprA. Mol Microbiol. 2002;46:813–826. doi: 10.1046/j.1365-2958.2002.03203.x. [DOI] [PubMed] [Google Scholar]
- Mizrahi I, Biran D, Ron EZ. Requirement for the acetyl phosphate pathway in Escherichia coli ATP-dependent proteolysis. Molecular Microbiology. 2006;62:201–211. doi: 10.1111/j.1365-2958.2006.05360.x. [DOI] [PubMed] [Google Scholar]
- Mizrahi I, Biran D, Ron EZ. Involvement of the Pta-AckA pathway in protein folding and aggregation. Research in Microbiology. 2009;160:80–84. doi: 10.1016/j.resmic.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Moya G, Gonzalez LJ, Huerta V, Garcia Y, Morera V, Perez D, Brena F, Arana M. Isolation and characterization of modified species of a mutated (Cys125 -Ala) recombinant human interleukin-2. J Chromatogr A. 2002;971:129–142. doi: 10.1016/s0021-9673(02)00845-2. [DOI] [PubMed] [Google Scholar]
- Pflumm MN, Gruber SC, Tsarbopoulos A, Wylie D, Pramanik B, Bausch JN, Patel ST. Isolation and characterization of an acetylated impurity in Escherichia coli-derived recombinant human interleukin-10 (IL-10) drug substance. Pharm Res. 1997;14:833–836. doi: 10.1023/a:1012127228239. [DOI] [PubMed] [Google Scholar]
- Ranjit DK, Young KD. The Rcs stress response and accessory envelope proteins are required for de novo generation of cell shape in Escherichia coli. J Bacteriol. 2013;195:2452–2462. doi: 10.1128/JB.00160-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rardin MJ, Newman JC, Held JM, Cusack MP, Sorensen DJ, Li B, Schilling B, Mooney SD, Kahn CR, Verdin E, et al. Label-free quantitative proteomics of the lysine acetylome in mitochondria identifies substrates of SIRT3 in metabolic pathways. Proc Natl Acad Sci U S A. 2013;110:6601–6606. doi: 10.1073/pnas.1302961110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J, Sang Y, Ni J, Tao J, Lu J, Zhao M, Yao YF. Acetylation regulates survival of Salmonella Typhimurium in acid stress. Appl Environ Microbiol. 2015;81:5675–5682. doi: 10.1128/AEM.01009-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross FE, Zamborelli T, Herman AC, Yeh CH, Tedeschi NI, Luedke ES. Detection of acetylated lysine residues using sequencing by Edman degradation and mass spectrometry. In: Marshak D, editor. Techniques in protein chemistry. New York: Academic; 1996. pp. 201–207. [Google Scholar]
- Schilling B, Christensen D, Davis R, Sahu AK, Hu LI, Walker-Peddakotla A, Sorensen DJ, Zemaitaitis B, Gibson BW, Wolfe AJ. Protein acetylation dynamics in response to carbon overflow in Escherichia coli. Mol Microbiol. 2015 Aug 11; doi: 10.1111/mmi.13161. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling B, Rardin MJ, MacLean BX, Zawadzka AM, Frewen BE, Cusack MP, Sorensen DJ, Bereman MS, Jing E, Wu CC, et al. Platform-independent and Label-free Quantitation of Proteomic Data Using MS1 Extracted Ion Chromatograms in Skyline. Molecular & Cellular Proteomics. 2012;11:202–214. doi: 10.1074/mcp.M112.017707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soppa J. Protein acetylation in archaea, bacteria, and eukaryotes. Archaea. 2010;2010 doi: 10.1155/2010/820681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starai VJ, Celic I, Cole RN, Boeke JD, Escalante-Semerena JC. Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science. 2002;298:2390–2392. doi: 10.1126/science.1077650. [DOI] [PubMed] [Google Scholar]
- Starai VJ, Escalante-Semerena JC. Acetyl-coenzyme A synthetase (AMP forming) Cellular and Molecular Life Sciences. 2004a;61:2020–2030. doi: 10.1007/s00018-004-3448-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starai VJ, Escalante-Semerena JC. Identification of the Protein Acetyltransferase (Pat) Enzyme that Acetylates Acetyl-CoA Synthetase in Salmonella enterica. Journal of Molecular Biology. 2004b;340:1005–1012. doi: 10.1016/j.jmb.2004.05.010. [DOI] [PubMed] [Google Scholar]
- Starai VJ, Takahashi H, Boeke JD, Escalante-Semerena JC. Short-Chain Fatty Acid Activation by Acyl-Coenzyme A Synthetases Requires SIR2 Protein Function in Salmonella enterica and Saccharomyces cerevisiae. Genetics. 2003;163:545–555. doi: 10.1093/genetics/163.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suenaga M, Ohmae H, Tsuji S, Tanaka Y, Koyama N, Nishimura O. epsilon-N-acetylation in the production of recombinant human basic fibroblast growth factor mutein. Prep Biochem Biotechnol. 1996;26:259–270. doi: 10.1080/10826069608000070. [DOI] [PubMed] [Google Scholar]
- Szewczak J, Bierczynska-Krzysik A, Piejko M, Mak P, Stadnik D. Isolation and Characterization of Acetylated Derivative of Recombinant Insulin Lispro Produced in Escherichia coli. Pharm Res. 2015;32:2450–2457. doi: 10.1007/s11095-015-1637-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takao T, Kobayashi M, Nishimura O, Shimonishi Y. Chemical characterization of recombinant human leukocyte interferon A using fast atom bombardment mass spectrometry. J Biol Chem. 1987;262:3541–3547. [PubMed] [Google Scholar]
- Thao S, Escalante-Semerena JC. Biochemical and thermodynamic analyses of Salmonella enterica Pat, a multidomain, multimeric N(epsilon)-lysine acetyltransferase involved in carbon and energy metabolism. MBio. 2011a;2(5):e00216–e00211. doi: 10.1128/mBio.00216-11. pii: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thao S, Escalante-Semerena JC. Control of protein function by reversible N[epsilon]-lysine acetylation in bacteria. Current Opinion in Microbiology. 2011b;14:200–204. doi: 10.1016/j.mib.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdin E, Ott M. 50 years of protein acetylation: from gene regulation to epigenetics, metabolism and beyond. Nat Rev Mol Cell Biol. 2015;16:258–264. doi: 10.1038/nrm3931. [DOI] [PubMed] [Google Scholar]
- Violand BN, Schlittler MR, Lawson CQ, Kane JF, Siegel NR, Smith CE, Kolodziej EW, Duffin KL. Isolation of Escherichia coli synthesized recombinant eukaryotic proteins that contain epsilon-N-acetyllysine. Protein Sci. 1994;3:1089–1097. doi: 10.1002/pro.5560030712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner G, Payne R. Widespread and enzyme-independent N{epsilon}-acetylation and N{epsilon}-succinylation in the chemical conditions of the mitochondrial matrix. J Biol Chem. 2013;288:29036–29045. doi: 10.1074/jbc.M113.486753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Zhang Y, Yang C, Xiong H, Lin Y, Yao J, Li H, Xie L, Zhao W, Yao Y, et al. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science. 2010;327:1004–1007. doi: 10.1126/science.1179687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert Brian T, Iesmantavicius V, Wagner Sebastian A, Scholz C, Gummesson B, Beli P, Nystrom T, Choudhary C. Acetyl-Phosphate Is a Critical Determinant of Lysine Acetylation in E. coli. Molecular Cell. 2013;51:265–272. doi: 10.1016/j.molcel.2013.06.003. [DOI] [PubMed] [Google Scholar]
- Weinert BT, Moustafa T, Iesmantavicius V, Zechner R, Choudhary C. Analysis of acetylation stoichiometry suggests that SIRT3 repairs nonenzymatic acetylation lesions. EMBO J. 2015;34:2620–2632. doi: 10.15252/embj.201591271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe AJ. The Acetate Switch. Microbiology and Molecular Biology Reviews. 2005;69:12–50. doi: 10.1128/MMBR.69.1.12-50.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe AJ. Glycolysis for Microbiome Generation. Microbiol Spectr. 2015;3 doi: 10.1128/microbiolspec.MBP-0014-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Vellaichamy A, Wang D, Zamdborg L, Kelleher NL, Huber SC, Zhao Y. Differential lysine acetylation profiles of Erwinia amylovora strains revealed by proteomics. Journal of Proteomics. 2013;79:60–71. doi: 10.1016/j.jprot.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Barak R, Liarzi O, Shainskaya A, Eisenbach M. In Vivo Acetylation of CheY, a Response Regulator in Chemotaxis of Escherichia coli. Journal of Molecular Biology. 2008;376:1260–1271. doi: 10.1016/j.jmb.2007.12.070. [DOI] [PubMed] [Google Scholar]
- Yang N, Lan L. Pseudomonas aeruginosa Lon and ClpXP proteases: roles in linking carbon catabolite repression system with quorum-sensing system. Curr Genet. 2015 Jun 5; doi: 10.1007/s00294-015-0499-5. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Yang X-J, Seto E. Lysine acetylation: codified crosstalk with other posttranslational modifications. Molecular Cell. 2008a;31:449–461. doi: 10.1016/j.molcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XJ, Seto E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat Rev Mol Cell Biol. 2008b;9:206–218. doi: 10.1038/nrm2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Kim J, Moon J, Ryu S, Pan J. The diversity of lysine-acetylated proteins in Escherichia coli. J Microbiol Biotechnol. 2008;18:1529–1536. [PubMed] [Google Scholar]
- Zhang J, Sprung R, Pei J, Tan X, Kim S, Zhu H, Liu C-F, Grishin NV, Zhao Y. Lysine acetylation is a highly abundant and evolutionarily conserved modification in Escherichia coli. Molecular & Cellular Proteomics. 2009;8:215–225. doi: 10.1074/mcp.M800187-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Zheng S, Yang JS, Chen Y, Cheng Z. Comprehensive Profiling of Protein Lysine Acetylation in Escherichia coli. J Proteome Res. 2013;12:844–851. doi: 10.1021/pr300912q. [DOI] [PubMed] [Google Scholar]
- Zhao K, Chai X, Marmorstein R. Structure and substrate binding properties of cobB, a Sir2 homolog protein deacetylase from Escherichia coli. J Mol Biol. 2004;337:731–741. doi: 10.1016/j.jmb.2004.01.060. [DOI] [PubMed] [Google Scholar]