Summary
Histone deacetylase 3 (HDAC3), a chromatin modifying enzyme, requires association with the deacetylase containing domain (DAD) of the nuclear receptor co-repressors NCOR1 and SMRT for its stability and activity. Here we show that aldose reductase (AR), the rate-limiting enzyme of the polyol pathway, competes with HDAC3 to bind the NCOR1/SMRT DAD. Increased AR expression leads to HDAC3 degradation followed by increased PPARγ signaling resulting in lipid accumulation in the heart. AR also downregulates expression of nuclear corepressor complex cofactors including Gps2 and Tblr1, thus affecting activity of the nuclear corepressor complex itself. Though AR reduces HDAC3-corepressor complex formation, it specifically de-represses the retinoic acid receptor (RAR), but not other nuclear receptors such as the thyroid receptor (TR) and liver X receptor (LXR). In summary, this work defines a distinct role for AR in lipid and retinoid metabolism through HDAC3 regulation and consequent de-repression of PPARγ and RAR.
Introduction
Acetylation orchestrates genome wide transcriptional changes through chromatin decondensation and is the net result of the opposing actions of histone acetyl transferases (HATs) and histone deacetylases (HDACs). HATs and HDACs form large activator and repressor complexes resulting in transcriptional gene regulation (Kouzarides, 2000; Sterner and Berger, 2000). HDAC3, belonging to Class I HDAC family, is specifically recruited to the SMRT (Silencing mediator of retinoid and thyroid receptor) complex, where it interacts with the DAD (deacetylase activation domain) domain of either NCOR1 (Nuclear CoRepressor1) or SMRT (Nuclear CoRepressor 2) (Guenther et al., 2001). This domain in NCOR1/SMRT activates HDAC3 and is essential for the maintenance of normal circadian metabolic physiology. Reductions in HDAC3 expression/activity influence lipogenesis in mouse models (Liu et al., 2013; Sun et al., 2012; Sun et al., 2011). Cardiac-specific deletion of HDAC3 leads to severe cardiac hypertrophy due to myocardial lipid accumulation and elevated triglyceride levels (Montgomery et al., 2008). De novo HDAC3 synthesis is an ongoing process and this protein is degraded if unbound to the corepressors. The possibility of co-occurrence of corepressors (NCOR1 and SMRT) in the same complex has been refuted (Zhang et al., 2002). Hence specific HDAC3- nuclear repressor complex is achieved by dissociation of the existing corepressor complex followed by de novo synthesis of both the corepressor and HDAC3 protein in the cytoplasm. (Guenther et al., 2000; Guo et al., 2012).
The HDAC3- corepressor complex represses several nuclear receptors including thyroid receptor (TR), retinoic acid receptor (RAR), vitamin D receptor (VDR), androgen receptor (AR), glucocorticoid receptor (GR),and others (Dwivedi et al., 1998; Shang et al., 2000; Wagner et al., 2003; Wong et al., 2014). Upon ligand binding, the HDAC3-NCOR1/SMRT complex dissociates, paving the way for activator complexes to bind the receptors and activate gene transcription (Atsumi et al., 2006; Ishizuka and Lazar, 2003; Li et al., 2011). Despite their critical roles in controlling gene expression, factors involved in regulation of repressor complex dissociation remain unidentified.
Aldose reductase (AR), the rate limiting enzyme of the polyol pathway, plays a critical role in mediating cardiovascular complications in diabetes, aging, and I/R injury (Ananthakrishnan et al., 2011; Hwang et al., 2004; Ramasamy and Goldberg, 2010; Ramasamy et al., 1997). Since mice display low AR expression and activity, transgenic expression of human AR (hAR) in mice is essential to recapitulate the activity and expression levels observed in humans, including the diabetes-associated cardiac complications (Ramasamy and Goldberg, 2010). In mice, cardiomyocyte-specific overexpression of human AR (Myosin Heavy Chain (MHC)-hAR) causes increased ceramide levels and accompanying heart failure in ischemic and aging hearts (Son et al., 2012). Furthermore, our recent studies demonstrated decreased HDAC activity, increased acetylation of hypoxia-responsive Egr-1 and induction of inflammatory genes in hAR-overexpressing diabetic apoE null mice (Vedantham et al., 2014). These studies led us to investigate if hAR driven lipid changes in cardiac cells are mediated, in part, via HDACs.
Our studies revealed that hAR expression in mice hearts leads to a decrease in HDAC3 levels accompanied by an increase in PPARγ activity thereby contributing to lipid-enriched environment. Human AR expression leads to reduced HDAC3 corepressor complexes with consequent increases in RARB in the nucleus and derepression of its target gene expression. The observed degradation of HDAC3 was due, in part, to competition between hAR and HDAC3 for interaction with the DAD domain of SMRT/NCOR1. Our findings reveal a role for hAR as a dissociation factor acting upon the HDAC3-corepressor complex, thereby specifically derepressing RAR and expression of its target genes.
Results
hAR overexpression leads to lipid accumulation
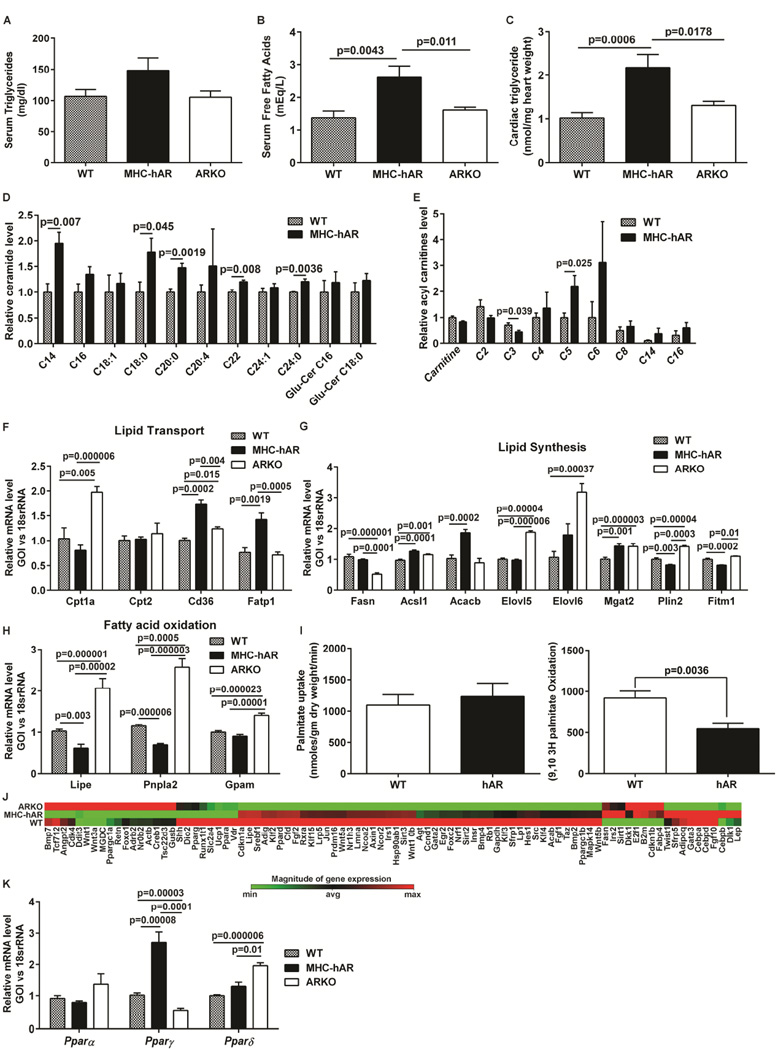
To elucidate mechanisms by which hAR drives lipid accumulation in the heart, we performed comprehensive measurements of triglycerides, fatty acids, ceramides and carnitines in hearts and plasma of cardiomyocyte-specific human AR (MHC-hAR) expressing mice, global AR knockout (ARKO), and non-transgenic littermates (WT). Left ascending coronary artery ligation followed by 48 hrs of reperfusion (LADp48h) was used as an ischemia-reperfusion (I/R) model and the tissues were harvested at 48hrs post-surgery (LADp48h). We observed a two-fold increase in serum and myocardial triglycerides in MHC-hAR mice compared to WT, whereas in ARKO mice, triglyceride levels were lower or comparable to WT subjected to I/R (Figure S1A–B). Increased serum free fatty acids levels were observed in MHC-hAR after I/R, though the increase was observed in ARKO I/R mice as well (Figure S1C). We confirmed triglyceride accumulation in hAR expressing H9c2 cells (rat myoblast cell line) (Figure S1–D). Under basal conditions (in the absence of I/R), we observed no changes triglyceride, while free fatty acids were increased in serum of MHC-hAR mice vs. WT and the levels were comparable in WT and ARKO mice (Figure 1A–B). Interestingly, the cardiac triglyceride levels were doubled in MHC-hAR mice compared to WT and ARKO mice (Figure 1C). Comprehensive measurements of the lipid species using mass spectrometry revealed significant increases in deleterious C14, C18:0, C20:0, C22 and C24:0 ceramide species (Figure 1D), while medium and long chain acyl carnitines showed an increasing trend in MHC-hAR vs. WT hearts (Figure 1E). Transcript levels of fatty acid transporters Cd36, Fatp1 were increased in MHC-hAR hearts (Fig 1 F). While the transcript levels of genes responsible for lipid synthesis were not different (Fig 1G), the genes essential for fatty acid oxidation (Lipe and Pnpla2) were 40% lower in MHC-hAR hearts vs. WT hearts (Fig 1 H). Transcripts of fatty acid oxidation genes were increased by 2 fold in ARKO mice compared to WT hearts (Fig 1H). Measurements of H-palmitate uptake and oxidation revealed marginal increase in fatty acid uptake (Fig 1 I) and reduction in fatty acid oxidation (Fig 1 I) in MHC-AR expressing hearts. We observed 1.5 fold increase in triglyceride content in MHC-hAR hearts compared to WT hearts (Fig S1–E). Same trend of reduced H-fatty acid oxidation was observed in adenovirally transduced hAR-expressing H9c2 (Ad-hAR) cells compared to the GFP-transduced (control) cells ( H-fatty acid oxidation was 1.39±0.1 µmoles/10*6 cells in control vs. 0.72±0.1 µmoles/10*6 cells in Ad hAR, p<0.05). Taken together, these results suggested the lipid accumulation observed in MHC-hAR mice is mainly due to reduced oxidation in MHC-hAR hearts. RT-PCR array studies revealed an upregulation of several of the PPARγ target genes, including AdipoQ, Cfd, Leptin, and Fabp4 in MHC-hAR hearts compared to WT, and a downregulation of these PPARγ target genes in ARKO mice (Figure 1J). Real time PCR analysis of PPAR isoforms (a, P and 5) revealed a 2.5 fold increase in Pparγ mRNA in MHC-hAR mice and a concordant reduction in ARKO mice, (Figure 1K), whereas Pparα and δ were unaffected. Increase in Pparγ mRNA was also observed in Ad-hAR cells compared to the GFP-transduced (control) cells (Fig S1–F).
Figure 1. AR overexpression leads to lipid accumulation.
Hearts from normally fed young mice were used. (A). serum triglyceride (n=7 mice/group), (B). serum free fatty acids (n=7–10 mice/group), and (C) cardiac tissue triglyceride measurements were measured (n=6 mice/group) using biochemical assays. (D & E). Cardiac ceramide and acyl carnitines levels were measured by Mass spectrometry. (n=5 mice/group). (F–H). qRT-PCR analysis of genes involved in fatty acid transport, synthesis and oxidation from the cardiac tissue of mice. (n=6 mice/group). (I). Fatty acid uptake and oxidation in hearts were measured using [9,10 (N)-3H] palmitate (n=5/group). (J) Heat map of adipogenesis PCR array from the heart tissues (n=4 mice/group). (K). qRT-PCR analysis of Ppar genes from heart tissue. (n=6 mice/group). See also Figure S1F.
Increased hAR mediates HDAC3 downregulation and increased PPARγ
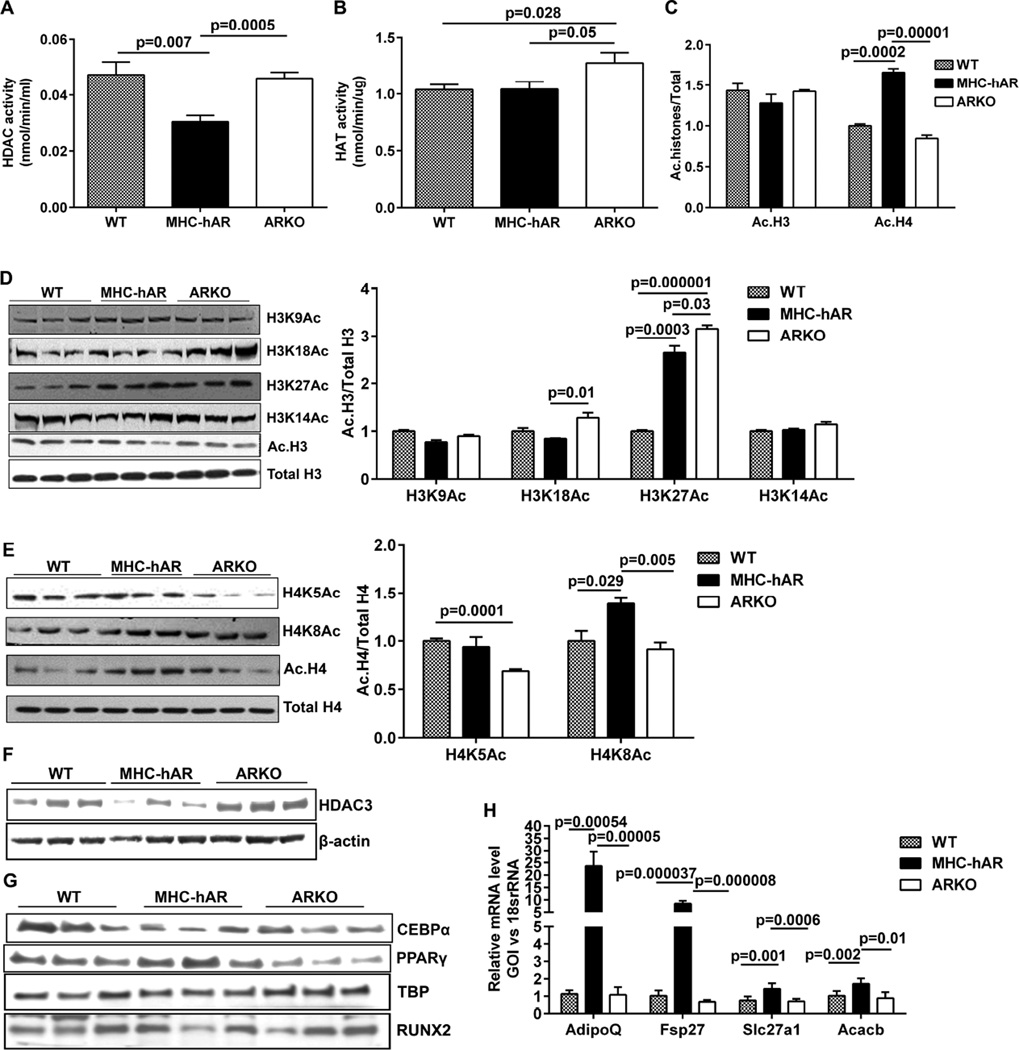
Since our previous findings linked hAR expression to increased acetylation of Egr-1 and induction of inflammatory genes, we investigated if hAR-driven lipid changes are mediated via acetylation (Vedantham et al., 2014). Measurements of total HDAC activity from the nuclear lysates revealed a 35% reduction in HDAC activity but no significant change in total HAT activity in MHC-hAR vs WT hearts (Figure 2A–B). Western blot analysis of WT, MHC-hAR, and ARKO heart lysates revealed increased histone H4 acetylation in MHC-hAR mice and reduced histone H4 acetylation in ARKO mice. No significant changes were observed in histone H3 acetylation (Figure 2C). We assessed individual histone modifications and observed significant increases in H4K8Ac and H3K27Ac in MHC-hAR hearts, whereas the other modifications like H3K9Ac, H3K14Ac, H3K18Ac, H4K5Ac was not significantly changed or was decreased (Figure 2D–E). In Ad-hAR expressing H9c2 cells, decreased HDAC activity, and increased histone acetylation at the H4K5 and H4K8 sites was observed (Figure S2A–C). Taken together these data suggest that increased acetylation observed in cells and tissue due to hAR expression was the result of decreased HDAC activity, mainly at the histone H4K8 site. To elucidate the histone deacetylases responsible for increased acetylation, we performed HDAC profiling in Ad-hAR H9c2 cells. We found no changes in HDAC 1, 2 and 10, increases in HDAC 6, 8 and 11, and reduced HDAC 3 and 4 (Figure S2D). We observed 40% reduction in HDAC3 protein level in cells expressing hAR vs. controls (Figure S2E). These results in hAR cells were recapitulated in hearts from MHC-hAR mice (Figure 2F).
Figure 2. AR overexpression leads to reduced HDAC3.
Nuclear fractions were isolated from 2 months old WT, MHC-hAR and ARKO mice hearts. (A). HDAC activity (n=6 mice/group). (B). HAT activity (n=6 mice/group). (C). Quantification of acetylated H3 and H4 histones normalized to total H3 and H4.Western blot analysis and quantification of (D). Acetyl H3 modifications normalized to total H3. (E). Acetyl H4 modifications normalized to total H4. (F). Western blot analysis of cardiac total lysates using HDAC3 antibody, normalized to β-actin. (G). Western blot analysis of cardiac nuclear lysates using CEBPα and PPARγ antibody, normalized to TBP. (n=3/ group). (H). qRT-PCR analysis of PPARγ target genes using cardiac tissues. (n=6 mice/group). See also Figure S2 and S3.
hAR activates PPARγ target genes
HDAC3 serves as a repressor of many transcription factors such as MEF2, KLF6, PPARγ, Runx2, CEBP, TFII-1, and NF-kB, and a reduction in HDAC3 can lead to activation of these factors (Gao et al., 2006; Gregoire et al., 2007; Li et al., 2005; Paz-Priel et al., 2011; Ren et al., 2013; Schroeder et al., 2004; Tussie-Luna et al., 2002). We observed increased PPARγ and reduced CEBPα expression in the nuclear fractions of MHC-hAR hearts compared to WT and ARKO, indicating increased PPARγ to be the driving force for increased lipid accumulation by hAR (Figure 2G). No significant changes were observed in RUNX2 levels (Figure 2G). Purity of cytoplasmic and nuclear fraction was verified using specific markers by western blot analysis (Figure S2G). Real time PCR analysis of PPARγ target genes revealed increased AdipoQ, Fsp27, and Acacb expression in MHC-hAR hearts, thus confirming the activation of PPARγ in vivo (Figure 2H). Consistently, in hAR-expressing H9c2 cells, we observed increases in PPARγ protein (Figure2E). Out of 7 PPARγ target genes examined, 5 of the genes involved in fatty acid metabolism including Fasn, Cd36, Fatp1, AdipoQ, and Sirt3 were upregulated, indicative of enhanced PPARγ transcriptional activation, although there were no significant changes in Slc2a4 and Srebf1 (Figure S2H). A 2.5-fold increase in protein levels of CD36, a prototype target gene of PPARγ, in hAR vs. control cells was observed (Figure S3A). Chromatin immunoprecipitation (ChIP) studies using the PPARγ antibody revealed increased binding to the promoter of the Cd36 gene in cells expressing hAR vs. control (Figure S3B). Thus these results confirmed the decreased HDAC3 protein level and enhanced PPARγ activation in hAR expressing cells and mice.
hAR-mediated lipid changes in aging and diabetes
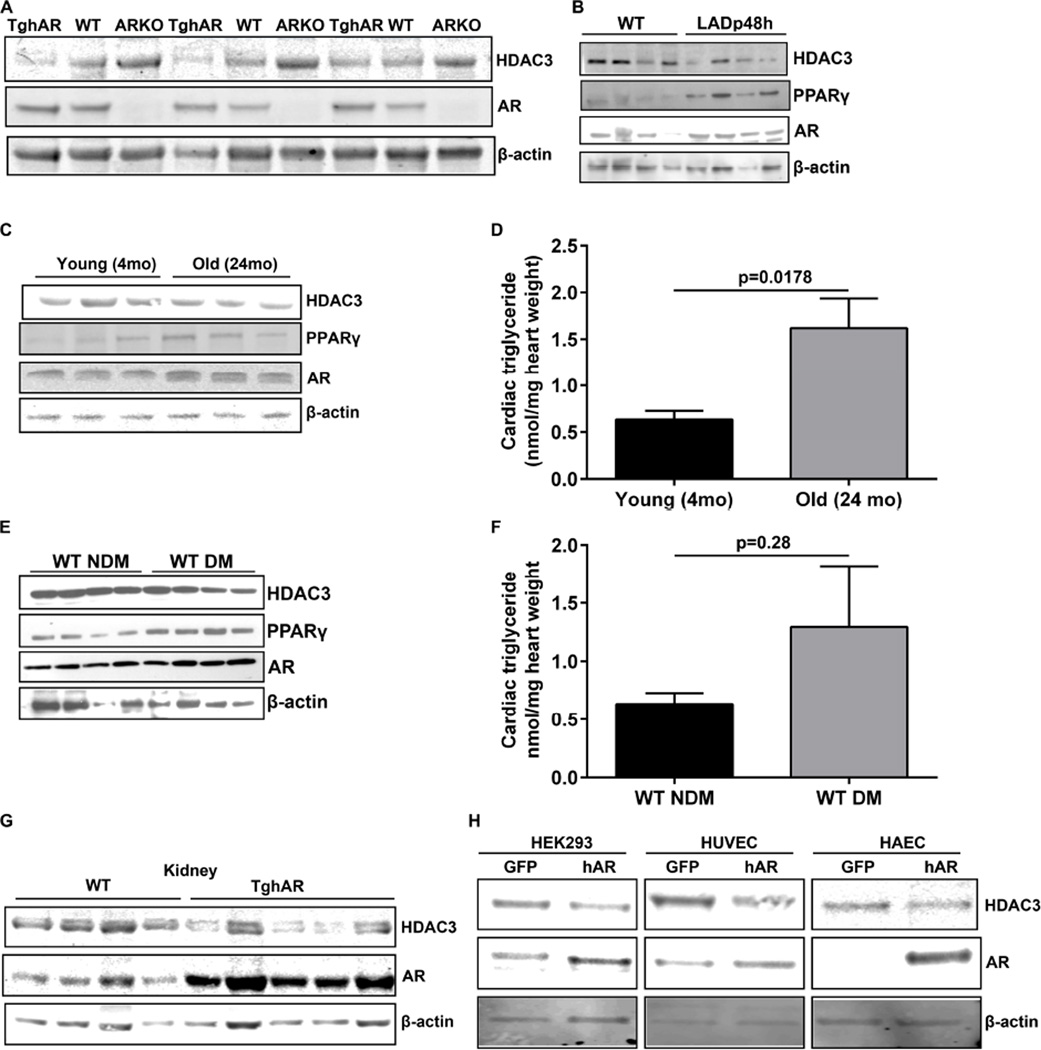
We then assessed the effect of HDAC3 in globally expressing transgenic hAR (TghAR) mice and observed reduced HDAC3 in TghAR mice and elevated HDAC3 protein levels in ARKO mice (Figure 3A). To demonstrate that pathological conditions, which lead to increased AR activity, can subsequently reduce HDAC3 and activate PPARγ, we examined hearts from I/R, aging mice and diabetic mice. We observed AR-mediated changes both in HDAC3 and PPARγ in I/R (Figure 3B) and aging hearts (Figure 3 C). We observed increases in cardiac triglyceride levels in aging hearts compared to control (Figure 3C–D). Similar changes in HDAC3 and PPARγ were observed in diabetic mouse hearts (DM) compared to the non-diabetic hearts (NDM) (Figure 3E). DM hearts showed a non-significant trend for increased triglyceride accumulation (Figure 3F). Kidney lysates from TghAR mice showed reduced HDAC3 expression (Figure 3G). Reduced HDAC3 expression was noted in hAR-transfected human umbilical cord endothelial cells (HUVEC), human aortic endothelial cells (HAEC), and HEK293T cells (Figure 3H). These data indicated that AR-mediated changes in HDAC3, and PPARγ, are not only induced by exogenous overexpression, but also seen in pathological (diabetes, I/R), physiological (aging) conditions and in other tissue and cell types as well.
Figure 3. AR mediated HDAC3 downregulation observed in various pathological settings.
Western blot analysis of cardiac lysates using specific antibodies to HDAC3, PPARγ, AR and β-actin. (A). Hearts from 4 months old mice obtained from WT, TghAR and ARKO. (n=5 group) (B). from wild type and mice subjected to left occlusion ascending coronary artery ligation surgery post 48 hrs (LADp48h) were used for western analysis (n=6/ group). (C). Young and old hearts from wild type mice. (n=5/ group) were analyzed by western blots. (D). Cardiac tissue triglyceride measurements from young and old mice. (n=4 mice/group). (E). 6 months old diabetic and non-diabetic mice lysates were used for western analysis and (F). their corresponding cardiac triglycerides (n=4 mice/group). (G). Western blots on kidney lysates from wild type and TghAR mice. (n=5 mice/group) and in (H). HEK293, HAEC and HUVEC cells transfected with GFP or hAR encoding plasmids in cells (n=3).
hAR competes with HDAC3 for DAD
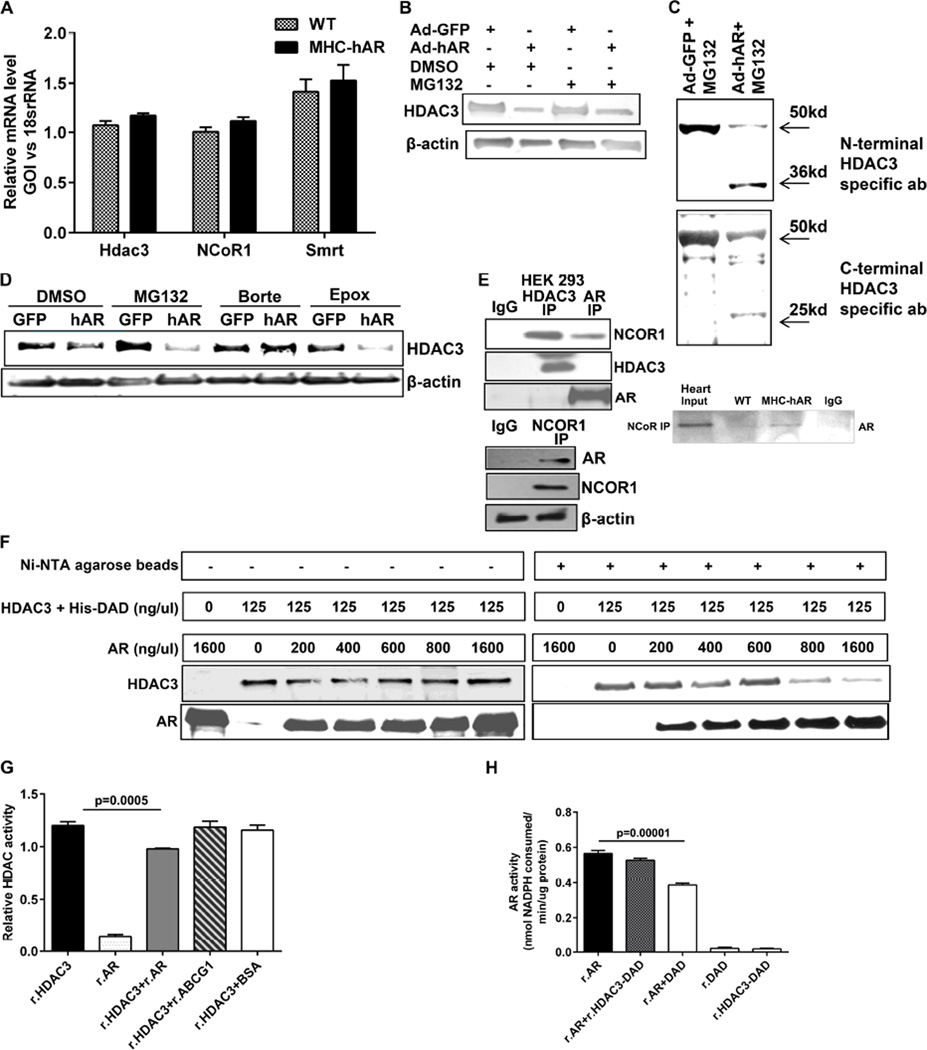
We then assessed the possibility that the mechanism underlying HDAC3 downregulation by hAR was transcriptionally regulated. Real time PCR analysis of Hdac3 and its corepressors Ncor1 and Smrt did not reveal any differences, indicating that it was not at the transcriptional or corepressor level in tissue or cells (Figure 4A, Figure S3C). To determine the role of ubiquitin-mediated proteasomal degradation, we treated Ad-hAR cells with MG132, a potent proteasome inhibitor and found no recovery of HDAC3 expression (Figure 4B). Interestingly, MG132 treatment in Ad-hAR cells revealed an additional lower molecular weight band when probed with the HDAC3 antibody. To confirm that the additional bands in Ad-hAR cells were specific to HDAC3, we employed two different (N- and C-terminal specific) HDAC3 antibodies. In both cases, we detected the presence of the additional lower molecular weight HDAC3 band, indicating HDAC3 cleavage in the presence of hAR (Figure 4C). Since HDAC3 protein degradation appeared to be insensitive to MG132, testing with other proteasome inhibitors, revealed that bortezomib prevented HDAC3 degradation, confirming that HDAC3 undergoes cleavage through the ubiquitin-mediated proteasome pathway (Figure 4D). To identify the underlying mechanisms of enhanced proteasomal degradation, we probed for a protein-protein interaction between AR and the HDAC3 corepressor complex. Co-immunoprecipitation using HDAC3 antibody and probed for AR failed to show positive interaction, whereas immunoprecipitation using NCoR1 or AR antibody revealed a positive interaction (Figure 4E). Since corepressor-free HDAC3 has been shown to be prone to proteasomal degradation (Guo et al., 2012), we hypothesized that AR competes with HDAC3 to bind to the corepressor. To address our hypothesis, we performed competition binding assays by incubating equal amounts of the HDAC3-DAD-His complex with increasing amounts of recombinant AR (r.AR) and precipitated using Ni-NTA beads. As depicted, increasing AR displaces the HDAC3 from the complex (Figure 4F). We then questioned whether the interaction between AR and DAD abrogates their activity. We performed in vitro HDAC activity assays with the active HDAC3 fraction (HDAC3 and GST-DAD (SMRT) (r.HDAC3) and human AR protein (r.AR). r.AR did not exhibit HDAC activity. When r.AR was incubated with HDAC3, there was a 10% reduction in HDAC activity (Figure 4G). To confirm the reduction in HDAC activity was due to AR presence, we incubated r.HDAC3 with either purified BSA or recombinant ABCG1 protein and observed no reduction in HDAC activity, suggesting that the reduction in activity is specific to AR. Incubation of AR with HDAC3-DAD did not lead to significant reductions in AR activity (Figure 4H), while DAD in the absence of HDAC3, reduced AR activity by 30% (Figure 4H). Thus these results confirm the interaction between AR and the DAD domain of nuclear corepressors.
Figure 4. AR interacts with the DAD of nuclear corepressors.
(A). qRT-PCR analysed for Hdac3, Ncor1 and Smrt transcripts from the cardiac tissues of MHC-hAR and wild type littermates. (n=6 mice/group). (B). Ad-GFP or Ad-hAR transduced cells were treated with 5uM MG132 for overnight, 24hrs post transfection. Expression of HDAC3 was evaluated using western blot. (C). Western blot analysis of MG132 treated cells with both N-terminal and C-terminal HDAC3 specific antibodies. (D). Adenovirally transduced H9c2 cells were treated with one of the inhibitors- MG132 (5uM), bortezomib (1uM) and epoxomicin (1uM) overnight and lysates were analyzed for HDAC3 expression by western blot. €. Expression analysis of HEK293 lysates after immunoprecipitation with either HDAC3 or AR antibody and probed for NCOR1 (top). Immunoprecipitation with NCOR1 antibody and probed for AR (bottom). Expression analysis of WT and MHC-hAR heart lysates after immunoprecipitation with either NCOR1 antibody and probed for AR (side). (F). Competition binding assay Ni-NTA agarose beads. Input control without beads (left) and pull down fraction subjected to western blot (right) showing reduced HDAC3 with increasing AR. (G). HDAC activity performed with the purified recombinant proteins obtained as indicated. Relative activity changes have been shown. (H). AR activity performed with the purified recombinant proteins. (n=6). Specific activity of AR was measured as nmoles of NADPH consumed per min per ug protein. (n=9).
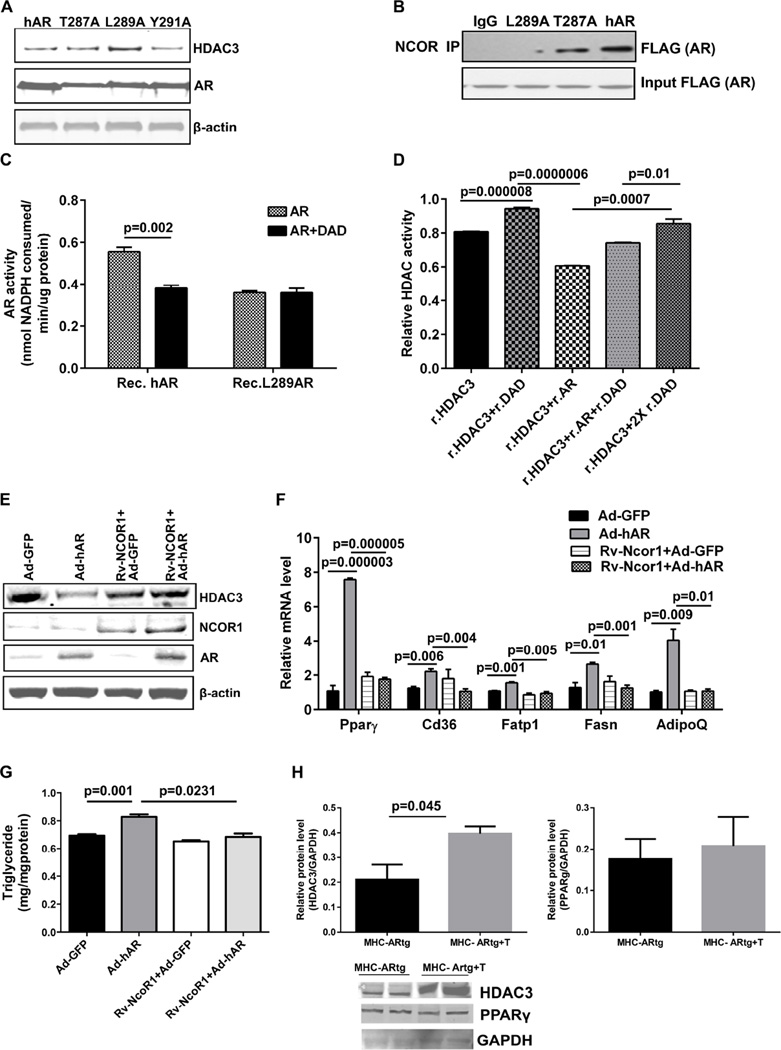
L289-hAR is essential for DAD interaction
Crystal structures of AR and the HDAC3-DAD complex have previously been elucidated (Watson et al., 2012; Wilson et al., 1993). Overlapping the structures of AR and HDAC3-DAD using Pymol program revealed a particular stretch of amino acids (30–39aa) in HDAC3 resembling the residue (286–293aa) of AR. Analysis of this sequence indicated that the key sites were 287, 289, and 291 of AR (Figure S3D). Three site directed mutants of hAR in the pTCN-Sport6 vector with Flag tag were generated (T287A, L289A, Y291A). Transfection of wild type hAR along with these mutants in HEK293 cells showed that L289A maximally rescued the HDAC3 protein whereas the wild type and other two mutants failed to do so (Figure 5A). Co-immunoprecipitation using the Flag antibody followed by probing with the NCOR1 antibody revealed that L289A mutant fails to interact with NCOR1 (Figure 5B). Thus, Leu 289 in hAR is essential for its interaction with the DAD domain. We further confirmed the lack of interaction between the L289A mutant and DAD by comparing AR activity in L289A mutant and the wild type hAR protein (Figure 5C). Furthermore, L289A-hAR expressing H9c2 cells failed to show increases in Pparγ target genes and triglycerides when compared to hAR expressing cells (Figure S3E–F).
Figure 5. DAD supplementation rescues HDAC3 degradation.
(A). Western blot analysis of site directed mutants of AR-Flag in HEK293 cells using Flag antibody, normalized to actin. (B). NCOR immunoprecipitated fraction on HEK293 lysates transfected with AR mutants were probed with Flag antibody. (C). AR activity performed with purified recombinant proteins. (n=6). (D). HDAC activity performed with purified recombinant proteins and relative HDAC activity changes has been shown. (n=9). H9c2 cells were retrovirally transduced with mock or Ncor1 expressing viral particles and were selected for 15 days with puromycin. Stably expressing cells were then transduced with Ad-GFP or Ad-hAR and (E). Representative western blot analysis of cell lysates. (F). qRT-PCR analysis on Pparγ and its target genes (n=6–9). (G). Triglyceride measurements were made in H9c2 cells (n=5). (H). Representative western blot analysis on the theophylline (10uM) treated and untreated MHC-hAR heart lysates. (n=4)
We next determined that DAD supplementation rescued the reduction in HDAC3 activity in the presence of r.AR (Figure 5D). To confirm the rescue of HDAC3 by modulation of NCOR1 expression, we performed retroviral-mediated transduction of NCOR1 in H9c2 cells followed by adenoviral transduction with GFP (control) or hAR. HDAC3 levels were rescued to that of Ad-GFP cells, confirming that NCOR1 availability is the determining factor in HDAC3 degradation (Figure 5E). The transcript levels of Pparγ, and its downstream target genes AdiopoQ, Cd36,and Fatp1 were not increased and HDAC3 levels were not lowered upon hAR expression in NCOR1 expressing cells (Figure 5F), reaffirming the DAD availability as the criteria to rescue HDAC3 degradation. Triglyceride measurement in NCOR1 stably expressing H9c2 cells failed to show increased accumulation despite hAR overexpression (Figure 5G). Treatment of MHC-hAR hearts with theophylline enhanced HDAC3 expression without attenuating PPARγ (Figure 5H). Taken together, these data indicate that increasing HDAC3 along with equivalent increases in NCOR1 may be required for repression of PPARγ (Figure 5H).
hAR derepress RAR pathway
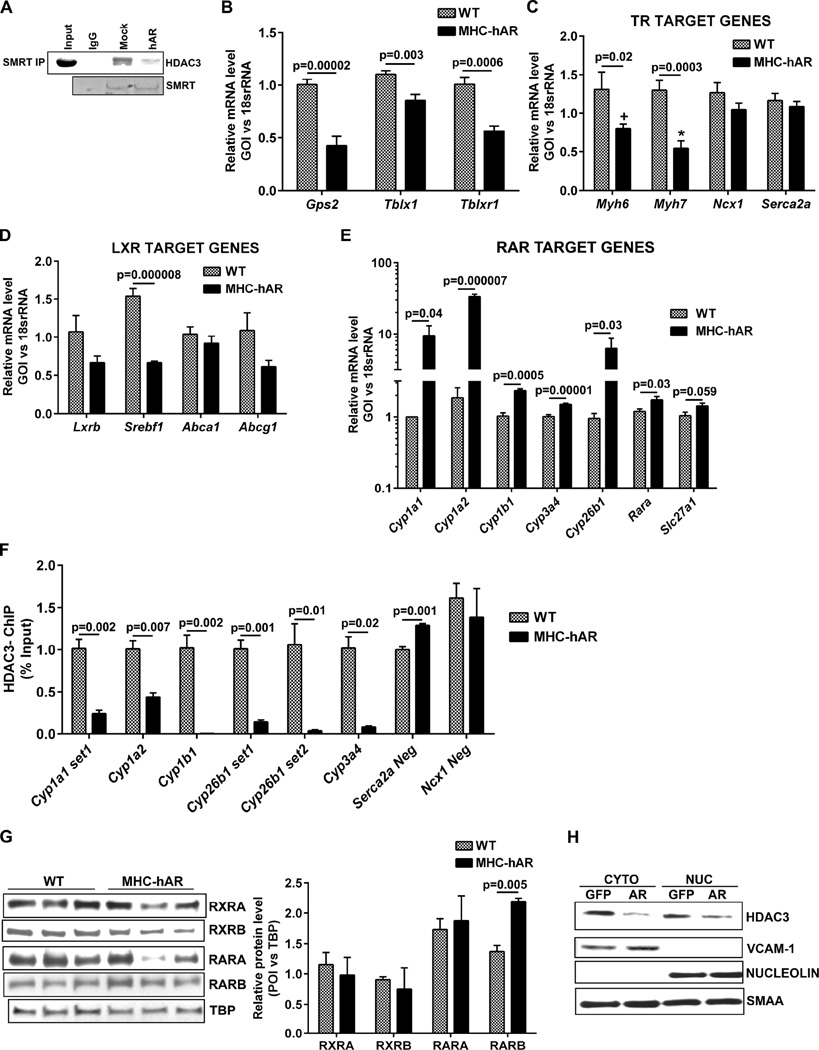
Immunoprecipitation of the nuclear corepressor complex using the SMRT antibody followed by probing with HDAC3 revealed reduced HDAC3-corepressor complex formation in Ad-hAR expressing cells compared to control cells (Figure 6A). In addition to HDAC3 and its corepressors, the HDAC3-corepressor complex consists of cofactors including GPS2, TBL1, TBLX1 and TBLXR1. GPS2 has been reported to interact with various transcriptional factors including p53, LXR, MSH4, and MSH5. Similar to GPS2, both TBL1 and TBLR1 are involved in transcriptional regulation (Li and Wang, 2008; Oberoi et al., 2011; Zhang et al., 2006). We observed that the expression of these accessory cofactors essential for nuclear corepressor complex formation including Gps2, Tbl1, and Tbl1xr were downregulated in MHC-hAR mice (Figure 6B). Taken together, these data indicate that that reduced HDAC3 corepressor complex in MHC-hAR mice is likely due to both HDAC3 degradation and cofactor downregulation.
Figure 6. AR derepress RAR target genes.
(A). Immunoprecipitation of Ad-mock or Ad-hAR transduced cell lysates using SMRT antibody and western blot analysis performed with HDAC3 antibody. AR expression and SMRT pull down fractions were shown below. qRT-PCR analysis on cardiac tissues (B) for the expression of cofactors of nuclear corepressor complex, (C) TR target genes (D) LXR target genes (E) RAR target genes. (n=6 mice/group). (F). ChiP assay performed using HDAC3 antibody in heart tissues of wild type and MHC-hAR mice. For negative control, Serca2A and Ncx.1 promoter regions were used. (n=4 mice/group). (G). Representative western blot using nuclear lysates of wild type and MHC-hAR mice for the expression of nuclear receptors like RXRA, RXRB, RARA and RARB. (n=6–9 mice/group). Results were normalized to TBP. H. Western blots analysis on cytoplasmic and nuclear fractions for the expression of HDAC3. VCAM1 and Nucleolin were used as localization specific controls. Smooth muscle alpha actin (SMAA) was used as loading control. See also Figure S4.
Nuclear receptors transduce ligand-mediated signals into changes in expression of gene networks (Evans, 1988; Glass and Rosenfeld, 2000; Mangelsdorf et al., 1995; Xu et al., 1999; Yen et al., 2003). Nuclear receptors including TR, RAR, LXR, and PXR form large molecular complexes with the HDAC3-nuclear corepressor complexes, binding DNA and repressing their target genes. Since hAR expression reduces HDAC3-nuclear corepressor complexes, we hypothesized that the nuclear receptors which rely on HDAC3-nuclear corepressor for their repression will be derepressed. To test this, we probed for TR target genes, both positively (Myh6 and Serca2a) and negatively (Myh7 and Ncx1) modulated by thyroid hormone (Reed et al., 2000; Rohrer et al., 1991; Yen et al., 2006). To our surprise, we found no increases of TR target genes in MHC-hAR hearts compared to WT (Figure 6C). To determine if the lack of increment in TR target genes were due to dysregulation of thyroid hormones, we measured serum T3 and T4 levels and observed no significant changes (Figure S4A). Next, we probed for the expression of Lxrb and its target genes, Srebf1, Abca1, and Abcg1 (Calkin and Tontonoz, 2012; Torra et al., 2008) and failed to observe any increases (Figure 6D). Probing of the RAR target genes, the CypP450 enzymes (Bray et al., 2001; Cai et al., 2003; Cai et al., 2002), revealed an upregulation of these genes in MHC-hAR hearts compared to WT (Figure 6E). To confirm that increased transcription of these RAR target Cyp genes was due to decreased HDAC3 binding in their promoter region, we performed chromatin immunoprecipitation studies and confirmed decreased HDAC3 binding in MHC-hAR hearts (Figure 6F). Western blot analysis of nuclear fractions from the heart revealed a significant increase in RARB in the MHC-hAR mice whereas RARA, RXRA, and RXRB were unchanged (Figure 6G). Since these mice express hAR specifically in the heart, there were no differences in RARB target genes in the livers of MHC-hAR mice compared to WT (Figure S4B). This led us to conclude that hAR expression leads to reduced HDAC3 corepressor complexes with consequent increases in RARB in the nucleus and derepression of its target gene expression. Since AR is a cytoplasmic enzyme and HDAC3 resides both in the nucleus and cytoplasm, we performed western blotting and determined that the cytoplasmic HDAC3 was more prominently downregulated by AR than the nuclear fraction (Figure 6H).This led us to conclude that the cytoplasmic HDAC3-corepressor complex formation is inhibited by AR (Rajendran et al., 2011).
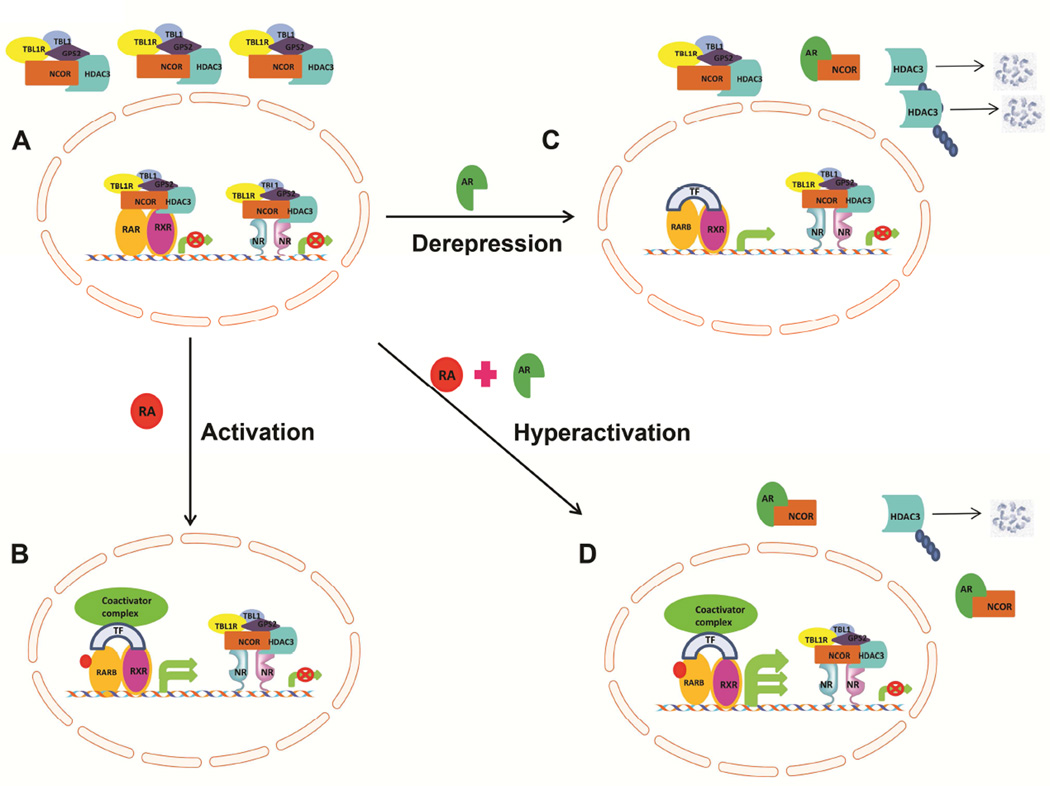
AR-mediated derepression of RARB targets in the absence of the RARB ligand, all trans retinoic acid (RA), led us to hypothesize that AR in the presence of the ligand would lead to hyper activation of RARB targets as proposed in Figure 7. To test the hypothesis at a preliminary level, we treated Ad-hAR H9c2 cells with RA. Indeed, we observed further increases in RARB target genes with RA treatment as shown in Figure S4C. Reduced HDAC3-corepressor complex leads to the activation of PPARγ as well as RARB, leading to the activation of target genes. Taken together, these data support our hypothesis that AR plays a key role in regulating nuclear receptor pathways.
Figure 7. Schematic model of AR mediated derepression of RAR target genes.
In unliganded condition, nuclear receptor RARB, bound to HDAC3 nuclear corepressor complex and remains inactive and repress the target genes. During activation, i.e, in the presence of ligand, all trans retinoic acid (atRA), RARB is relieved from the corepressor complex. Liganded RARB binds to coactivator complexes along with transcription factors (TF) and activates the target gene expression. In the presence of AR, nuclear corepressor binds to AR, free HDAC3 is subjected to proteolytic cleavage. Though the coactivator fails to bind RARB, transcriptional machinery binds and activates the basal transcription, termed as derepression. When both ligand and AR is present, hyperactivation of RAR target genes is observed. This is achieved because of enhanced degradation of nuclear corepressor complexes (AR effect) combined with RARB activation (ligand effect), to reach the hyperactive state.
Discussion
HDAC3 plays an important role in the maintenance of cardiac function through the regulation of substrate metabolism (Montgomery et al., 2008). Here we show that AR interacts with the DAD domain of nuclear corepressors NCOR1/SMRT, thereby specifically attenuating their ability to regulate HDAC3 levels. Chief metabolic consequences of this interaction include induction of PPARγ and a host of genes that mediate accumulation of pathological amounts of lipid. Increasing the concentrations of NCOR1 restored HDAC3 levels and its downstream effects. Concomitantly, AR-mediated reductions in the HDAC3-corepressor complex led to derepression of RARB target genes. We demonstrate a link between the glucose metabolizing enzyme, AR, and transcriptional regulation via nuclear receptors, through the HDAC3-corepressor complex.
Ectopic lipid accumulation in the heart is one of the major mechanisms underlying cardiac failure in diabetic conditions (Kankaanpaa et al., 2006; Montani et al., 2004; Ruberg et al., 2010). Accumulation of long chain acyl CoAs (substrate for ceramide synthesis) and ceramides have been implicated in lipotoxic cardiomyopathy (Gao et al., 2015; Park et al., 2008). In this study, we observed a 2-fold increase in cardiac triglyceride levels along with increased ceramide species and acyl carnitines, suggesting a lipotoxic like environment in human AR expressing cardiac cells and tissues.
In both cell and mouse models, we show reduced HDAC activity, HDAC3 levels, PPARγ activation, and increased H4K8Ac modification by hAR. The increased acetylation of histone H4 at both H4K5 and H4K8 along with reduced HDAC3 levels are consistent with the earlier observations that the HDAC3/NCOR complex favors histone H4 rather than H3 (Bhaskara et al., 2008; Hartman et al., 2005; Knutson et al., 2008). Though HDAC3 belongs to the Class I HDAC family, it is essential for the enzymatic activity for Class II HDAC family members, specifically HDAC 4, 5 and 7 (Fischle et al., 2001; Fischle et al., 2002). Hence, the decrease in HDAC3 protein mediated by hAR, that we observed, contributes to the reduced activity of both Class I and Class II HDACs. Taken together, these observations strengthen our hypothesis that AR-mediated increases in acetyl H4 are likely to be secondary to decreased HDAC3/NCOR complex.
It is well established that PPARγ is a master transcriptional regulator of lipid metabolism and that HDAC3 functions as a repressor of PPARγ (Gao et al., 2006; Jiang et al., 2014; Tian et al., 2014). In the heart, modulation of PPARγ has been shown to drive lipid accumulation (Duan et al., 2005; Fajas et al., 2002; Fu et al., 2005; Lai et al., 2008; Son et al., 2007; Son et al., 2010). (Marfella et al., 2009a; Marfella et al., 2009b). In our study, we show that both PPARγ expression and activity are upregulated in hAR-expressing hearts, whereas in mice lacking AR, PPARγ is downregulated. Though increased PPARγ activity is well characterized by the degradation of its repressor, HDAC3, the potential mechanism by which Pparγ mRNA expression increases in hAR expressing cells is likely to be complex. We speculate that the change in PPARγ activity increases its target gene expression AdipoQ and consequently, Tnf-α level. Tnf-α increases may mediate the increase in Pparγ mRNA expression(Figure S4D) as demonstrated earlier in other studies (Guo et al., 2013; Jin et al., 2014; Liu et al., 2012; Qiao et al., 2011; Yamaguchi et al., 2008).
Consistent with generalized effects of the hAR pathway, our data revealed that hAR mediates HDAC3 reduction and increased PPARγ activation in other tissues and cell lines distinct from the heart. Further, aging has been shown to increase lipid accumulation in various tissues, including the heart (Petersen et al., 2003; Unger, 2005). Our earlier studies showed that AR expression and activity are upregulated in aging hearts (Ananthakrishnan et al., 2011). Here we show increased cardiac triglycerides along with reduced HDAC3 activity in aging hearts, thus providing a potential AR driven mechanism of HDAC3 reduction and lipid accumulation in this setting.
Our study shows that AR interacts with the DAD of NCOR1/SMRT complex, leaving free, unstable HDAC3 to be degraded as reported earlier (Guo et al., 2012). HDAC3 undergoes ubiquitin mediated proteasomal degradation and this degradation is essential for the transcriptional activation of nuclear hormones as well as transcription factors like TR, RAR, PPARγ, NF-kB (Perissi et al., 2004; Zhao et al., 2010). Among the proteasome inhibitors tested, only bortezomib rescued HDAC3 from cleavage. This could be due to their sensitivity towards Cyp450 enzymes. AR induced many of the RAR target genes, such as the Cyp450 enzymes including Cyp3a4. It has previously been shown that bortezomib is insensitive to Cyp3a4, while MG132 and epoxomicin are degraded by the action of Cyp3a4 enzymes (Lee et al., 2010). Rescue of HDAC3 from degradation by bortezomib, confirms that the degradation is via the ubiquitin-mediated proteasome pathway. Though our study strongly supports that AR-mediated HDAC3 downregulation due to degradation via proteosomal pathways, the role of post transcriptional or translational mechanisms as contributors to this HDAC3 downregulation cannot be ruled out.
AR interaction with corepressors not only restricts complex formation, but also reduces the activity in vitro. The presence of AR reduced HDAC3 activity by 10%; however, the effect of AR on HDAC3 activity is very low. This indicates that the affinity of HDAC3 for DAD is greater compared to the affinity of AR for DAD. It is important to note that the HDAC3-DAD complex was already formed and used for the assay, which might not be the actual scenario in vivo in the cellular milieu. HDAC3 is synthesized and subjected to degradation continuously in the cytoplasm and the degradation is inhibited by its interaction with either of its corepressors NCOR1 or SMRT. Apart from HDAC3 and corepressors, the HDAC3-corepressor complex consists of proteins including GPS2, TBL1, and TBL1XR (Li and Wang, 2008; Oberoi et al., 2011; Zhang et al., 2006). GPS2 has been reported to interact with various transcriptional factors including p53, LXR, MSH4, and MSH5. The role of GPS2 in both activation and repression complexes is poorly understood (Jakobsson et al., 2009; Lee et al., 2006; Peng et al., 2001). Both TBL1 and TBLR1 possess F-box like motifs which mediate interaction with ubiquitin ligases. Like GPS2, both TBL1 and TBLR1 are also involved in transcriptional activation (Adikesavan et al., 2014; Li and Wang, 2008; Perissi et al., 2004). AR downregulates the expression of all three cofactors of the corepressor complex.AR expression leads to reduced HDAC3-corepressor complex formation in two ways: (i) AR competes with HDAC3 for its interaction with the corepressor, leading to free HDAC3 for degradation; (ii) AR leads to downregulation of all accessory factors required for corepressor complex formation including Gps2, Tbl1, and Tbl1xr.
Our result that downregulation of HDAC3 mediated by AR is more pronounced in the cytoplasm is consistent with earlier findings which showed that the association and dissociation of the HDAC3-NCOR complexes take place mostly in the cytoplasm (Rajendran et al., 2011). The HDAC3-corepressor complex represses several nuclear receptors like TR, RXR, RAR, VDR (Vitamin D Receptor), AR (Androgen receptor), GR (Glucocorticoid receptor), PXR (Pregnane X Receptor), and LXR (Dwivedi et al., 1998; Shang et al., 2000; Wagner et al., 2003; Zhang et al., 1998). One of the targets of HDAC3 corepressor complex is circadian rhythmic genes and RT-PCR array on these genes in hAR expressing heart tissues failed to show any changes (Figure S4E). In our study, though we have shown low HDAC3-corepressor complex formation, derepression is specific to PPARγ and RARB, altering lipid and retinoid metabolism. The relationship between lipid and retinoid metabolism in the heart is unclear, though recent studies suggest that both pathways are interlinked (Kim et al., 2015; Lee et al., 2014).
In conclusion, our study reveals a key role for AR as a dissociation factor of the HDAC3-corepressor complex via interaction with the DAD domain, thereby derepressing target gene expression, mimicking the liganded PPARγ and RAR state. Our findings define mechanism of AR-mediated lipid accumulation and indicate that AR, via its ability to dissociate the HDAC3-corepressor complexes, may function as a regulator of retinoic acid pathways.
Experimental Procedures
Animals used: All animal experimentations were approved by the Institutional Animal Care and Use Committee at New York University School of Medicine. MHC-hAR, TghAR and ARKO and wild type littermates mice were used as described earlier (Gabbay et al., 2010; Hwang et al., 2004; Son et al., 2012).
Cell culture: H9c2, HEK293, HUVEC and HAEC cells were purchased from ATCC. For adenoviral transduction, adenoviral particles encoding GFP or hAR (Vector Biolabs) was used at a concentration of 0.5 × 109 pfu/ml.
Adipogenesis PCR array: PCR array was performed and the data analyzed as per manufacturer’s protocol (SABiosciences Mouse adipogenesis PCR array).
Serum and tissue triglyceride and free fatty acid measurements were made using commercially available kits from Wako chemicals and Biovision, as per manufacturer instructions.
Recombinant proteins: For experiments mentioned in Figure 4F, recombinant AR, purchased from Wako was used. HDAC3 (transomic) and SMRT-DAD cDNA (a kind gift from Dr. Schwabe JW) were subcloned in pET-Duet1 vector (Novagen). SMRT-DAD cDNA had His tag and hence HDAC3-SMRT-DAD complex was purified using Ni-NTA agarose beads (Invitrogen). The expression of both proteins was verified through western blot. For the reagents mentioned in Figure 4H and Figure 5C, hAR and L289A AR cDNA (transomic) was subcloned in pET28b vector and purified using Ni-NTA agarose beads. For experiments mentioned in Figure 4G and Figure 5D, r.HDAC3 (Active Motif), SMRT-DAD (Active Motif), r.AR (Wako), r.ABCG1 (Active Motif), BSA (Sigma) were used. hAR cDNA (transomic) was subcloned in pcDNA3.1Myc HisA vector in fusion with Flag tag at the C-terminal end. The plasmid was used to create site directed mutagenesis T287A, L289A and L291A clones using site directed mutagenesis kit (Invitrogen). Flag antibody used for immunoprecipitation was obtained from Sigma.
HDAC activity in tissue and cell lysates was measured using fluorometric activity assay kit (Caymen Chemicals). HAT activity was performed using HAT activity assay kit (Biovision). Cytoplasmic and nuclear fractionations were prepared using kit (Thermo scientific) as per manufacturer’s protocol. Proteins were quantified using BCA reagent (BioRad). Competitive binding assay: Equal amount of HDAC3-DAD-His complex was incubated with increasing amounts of AR using binding buffer (40mM HEPES, 5% glycerol, 100mM KCl, 1mM DTT, 10mM MgCl2 and 1mM ATP) at room temperature for 30min. Ni-NTA agarose beads were blocked using BSA (1mg/ml) for 1 hr and then washed 3X with the binding buffer. DAD was pulled down using Ni-NTA beads for 2hrs and then subjected to western blot for detecting AR.
Fatty acid uptake and oxidation in hearts: Freshly isolated hearts were subjected to a retrograde Langendorff perfusion in KH buffer containing 5 mM glucose and, 0.4 mmol/L palmitate and [9,10- 3H] palmitate (80,000 dpm/ml) prebound to 0.4 mmol/L fatty acid–free bovine serum albumin as described in (Hwang et al., 2004). [9, 10-3H] palmitate uptake (Kienesberger et al., 2012) and oxidation rates were measured as published in the literature. (Kuang et al., 2004; Larsen et al., 1999). Pymol program: Snapshots were taken with molecular visualization software PyMOL initiated from 2DUX (AR) and 4A69 (HDAC3) PDB structures. Using the display option of secondary structure cartoon residues 9–49aa from 4A69 and 299–312aa (2DUX) were overlapped (Watson et al., 2012; Wilson et al., 1993).
Mass spectrometry: Acyl carnitines and ceramides were quantified by liquid chromatography-electron spray ionization-tandem mass spectrometry (LC/ESI-MS/MS) in the multiple reaction monitoring (MRM) mode as previously described (Kasumov et al., 2010; Umemoto et al., 2012).
Statistical methods: Data was analyzed using INSTAT (GraphPad, San Diego, CA) software. Differences between groups were assessed using ANOVA for repeated measures, with subsequent student-Newman-Keuls multiple comparisons post-test if the p value for ANOVA was significant. All data are expressed as mean+standard error of the mean (SEM).
Detailed methods sections are available in Supplementary information.
Supplementary Material
Highlights.
AR, the first enzyme in the polyol pathway, influences cardiac lipid accumulation.
AR competes with HDAC3 for corepressor complex binding, leading to HDAC3 degradation.
AR reduces HDAC3-corepressor complex formation.
AR specifically derepresses the PPARγ and RAR pathways.
Acknowledgments
We thank Ms. Latoya Woods for assistance with figure preparation and Dr. Leenus Martin for sharing plasmid reagents. This work was supported by: NIH (HL60901, AG026467, HL102022 and RO1 HL61783).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
D.T designed research, conducted research, analyzed data and wrote the paper. R.A, X.J, S.K conducted research. K.O.S, N.Q, Q.L, J.Z and R.Ra prepared critical reagents. R.Rb, S.P and A.M.S. designed and directed research, analyzed data and wrote the paper.
The authors declare no competing financial interests.
References
- Adikesavan AK, Karmakar S, Pardo P, Wang L, Liu S, Li W, Smith CL. Activation of p53 transcriptional activity by SMRT: a histone deacetylase 3-independent function of a transcriptional corepressor. Mol Cell Biol. 2014;34:1246–1261. doi: 10.1128/MCB.01216-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananthakrishnan R, Li Q, Gomes T, Schmidt AM, Ramasamy R. Aldose reductase pathway contributes to vulnerability of aging myocardium to ischemic injury. Exp Gerontol. 2011;46:762–767. doi: 10.1016/j.exger.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atsumi A, Tomita A, Kiyoi H, Naoe T. Histone deacetylase 3 (HDAC3) is recruited to target promoters by PML-RARalpha as a component of the N-CoR co-repressor complex to repress transcription in vivo. Biochem Biophys Res Commun. 2006;345:1471–1480. doi: 10.1016/j.bbrc.2006.05.047. [DOI] [PubMed] [Google Scholar]
- Bhaskara S, Chandrasekharan MB, Ganguly R. Caffeine induction of Cyp6a2 and Cyp6a8 genes of Drosophila melanogaster is modulated by cAMP and D-JUN protein levels. Gene. 2008;415:49–59. doi: 10.1016/j.gene.2008.02.017. [DOI] [PubMed] [Google Scholar]
- Bray BJ, Goodin MG, Inder RE, Rosengren RJ. The effect of retinol on hepatic and renal drug-metabolising enzymes. Food Chem Toxicol. 2001;39:1–9. doi: 10.1016/s0278-6915(00)00110-1. [DOI] [PubMed] [Google Scholar]
- Cai Y, Dai T, Ao Y, Konishi T, Chuang KH, Lue Y, Chang C, Wan YJ. Cytochrome P450 genes are differentially expressed in female and male hepatocyte retinoid X receptor alpha-deficient mice. Endocrinology. 2003;144:2311–2318. doi: 10.1210/en.2002-0129. [DOI] [PubMed] [Google Scholar]
- Cai Y, Konishi T, Han G, Campwala KH, French SW, Wan YJ. The role of hepatocyte RXR alpha in xenobiotic-sensing nuclear receptor-mediated pathways. Eur J Pharm Sci. 2002;15:89–96. doi: 10.1016/s0928-0987(01)00211-1. [DOI] [PubMed] [Google Scholar]
- Calkin AC, Tontonoz P. Transcriptional integration of metabolism by the nuclear sterol-activated receptors LXR and FXR. Nat Rev Mol Cell Biol. 2012;13:213–224. doi: 10.1038/nrm3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan SZ, Ivashchenko CY, Russell MW, Milstone DS, Mortensen RM. Cardiomyocyte-specific knockout and agonist of peroxisome proliferator-activated receptor-gamma both induce cardiac hypertrophy in mice. Circ Res. 2005;97:372–379. doi: 10.1161/01.RES.0000179226.34112.6d. [DOI] [PubMed] [Google Scholar]
- Dwivedi PP, Muscat GE, Bailey PJ, Omdahl JL, May BK. Repression of basal transcription by vitamin D receptor: evidence for interaction of unliganded vitamin D receptor with two receptor interaction domains in RIP13delta1. J Mol Endocrinol. 1998;20:327–335. doi: 10.1677/jme.0.0200327. [DOI] [PubMed] [Google Scholar]
- Evans RM. The steroid and thyroid hormone receptor superfamily. Science. 1988;240:889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajas L, Egler V, Reiter R, Hansen J, Kristiansen K, Debril MB, Miard S, Auwerx J. The retinoblastoma-histone deacetylase 3 complex inhibits PPARgamma and adipocyte differentiation. Dev Cell. 2002;3:903–910. doi: 10.1016/s1534-5807(02)00360-x. [DOI] [PubMed] [Google Scholar]
- Fischle W, Dequiedt F, Fillion M, Hendzel MJ, Voelter W, Verdin E. Human HDAC7 histone deacetylase activity is associated with HDAC3 in vivo. J Biol Chem. 2001;276:35826–35835. doi: 10.1074/jbc.M104935200. [DOI] [PubMed] [Google Scholar]
- Fischle W, Dequiedt F, Hendzel MJ, Guenther MG, Lazar MA, Voelter W, Verdin E. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol Cell. 2002;9:45–57. doi: 10.1016/s1097-2765(01)00429-4. [DOI] [PubMed] [Google Scholar]
- Fu M, Rao M, Bouras T, Wang C, Wu K, Zhang X, Li Z, Yao TP, Pestell RG. Cyclin D1 inhibits peroxisome proliferator-activated receptor gamma-mediated adipogenesis through histone deacetylase recruitment. J Biol Chem. 2005;280:16934–16941. doi: 10.1074/jbc.M500403200. [DOI] [PubMed] [Google Scholar]
- Gabbay KH, Bohren KM, Morello R, Bertin T, Liu J, Vogel P. Ascorbate synthesis pathway: dual role of ascorbate in bone homeostasis. J Biol Chem. 2010;285:19510–19520. doi: 10.1074/jbc.M110.110247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Feng XJ, Li ZM, Li M, Gao S, He YH, Wang JJ, Zeng SY, Liu XP, Huang XY, et al. Downregulation of adipose triglyceride lipase promotes cardiomyocyte hypertrophy by triggering the accumulation of ceramides. Arch Biochem Biophys. 2015;565:76–88. doi: 10.1016/j.abb.2014.11.009. [DOI] [PubMed] [Google Scholar]
- Gao Z, He Q, Peng B, Chiao PJ, Ye J. Regulation of nuclear translocation of HDAC3 by IkappaBalpha is required for tumor necrosis factor inhibition of peroxisome proliferator-activated receptor gamma function. J Biol Chem. 2006;281:4540–4547. doi: 10.1074/jbc.M507784200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- Gregoire S, Xiao L, Nie J, Zhang X, Xu M, Li J, Wong J, Seto E, Yang XJ. Histone deacetylase 3 interacts with and deacetylates myocyte enhancer factor 2. Mol Cell Biol. 2007;27:1280–1295. doi: 10.1128/MCB.00882-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Barak O, Lazar MA. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol Cell Biol. 2001;21:6091–6101. doi: 10.1128/MCB.21.18.6091-6101.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Lane WS, Fischle W, Verdin E, Lazar MA, Shiekhattar R. A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes Dev. 2000;14:1048–1057. [PMC free article] [PubMed] [Google Scholar]
- Guo C, Gow CH, Li Y, Gardner A, Khan S, Zhang J. Regulated clearance of histone deacetylase 3 protects independent formation of nuclear receptor corepressor complexes. J Biol Chem. 2012;287:12111–12120. doi: 10.1074/jbc.M111.327023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo R, Zhang Y, Turdi S, Ren J. Adiponectin knockout accentuates high fat diet-induced obesity and cardiac dysfunction: role of autophagy. Biochim Biophys Acta. 2013;1832:1136–1148. doi: 10.1016/j.bbadis.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman HB, Yu J, Alenghat T, Ishizuka T, Lazar MA. The histone-binding code of nuclear receptor co-repressors matches the substrate specificity of histone deacetylase 3. EMBO Rep. 2005;6:445–451. doi: 10.1038/sj.embor.7400391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang YC, Kaneko M, Bakr S, Liao H, Lu Y, Lewis ER, Yan S, Ii S, Itakura M, Rui L, et al. Central role for aldose reductase pathway in myocardial ischemic injury. FASEB J. 2004;18:1192–1199. doi: 10.1096/fj.03-1400com. [DOI] [PubMed] [Google Scholar]
- Ishizuka T, Lazar MA. The N-CoR/histone deacetylase 3 complex is required for repression by thyroid hormone receptor. Mol Cell Biol. 2003;23:5122–5131. doi: 10.1128/MCB.23.15.5122-5131.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson T, Venteclef N, Toresson G, Damdimopoulos AE, Ehrlund A, Lou X, Sanyal S, Steffensen KR, Gustafsson JA, Treuter E. GPS2 is required for cholesterol efflux by triggering histone demethylation, LXR recruitment, and coregulator assembly at the ABCG1 locus. Mol Cell. 2009;34:510–518. doi: 10.1016/j.molcel.2009.05.006. [DOI] [PubMed] [Google Scholar]
- Jiang X, Ye X, Guo W, Lu H, Gao Z. Inhibition of HDAC3 promotes ligand-independent PPARgamma activation by protein acetylation. J Mol Endocrinol. 2014;53:191–200. doi: 10.1530/JME-14-0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin D, Sun J, Huang J, He Y, Yu A, Yu X, Yang Z. TNF-alpha reduces g0s2 expression and stimulates lipolysis through PPAR-gamma inhibition in 3T3-L1 adipocytes. Cytokine. 2014;69:196–205. doi: 10.1016/j.cyto.2014.06.005. [DOI] [PubMed] [Google Scholar]
- Kankaanpaa M, Lehto HR, Parkka JP, Komu M, Viljanen A, Ferrannini E, Knuuti J, Nuutila P, Parkkola R, Iozzo P. Myocardial triglyceride content and epicardial fat mass in human obesity: relationship to left ventricular function and serum free fatty acid levels. J Clin Endocrinol Metab. 2006;91:4689–4695. doi: 10.1210/jc.2006-0584. [DOI] [PubMed] [Google Scholar]
- Kasumov T, Huang H, Chung YM, Zhang R, McCullough AJ, Kirwan JP. Quantification of ceramide species in biological samples by liquid chromatography electrospray ionization tandem mass spectrometry. Anal Biochem. 2010;401:154–161. doi: 10.1016/j.ab.2010.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kienesberger PC, Pulinilkunnil T, Sung MM, Nagendran J, Haemmerle G, Kershaw EE, Young ME, Light PE, Oudit GY, Zechner R, et al. Myocardial ATGL overexpression decreases the reliance on fatty acid oxidation and protects against pressure overload-induced cardiac dysfunction. Mol Cell Biol. 2012;32:740–750. doi: 10.1128/MCB.06470-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YK, Zuccaro MV, Costabile BK, Rodas R, Quadro L. Tissue- and sex-specific effects of beta-carotene 15,15’ oxygenase (BCO1) on retinoid and lipid metabolism in adult and developing mice. Arch Biochem Biophys. 2015 doi: 10.1016/j.abb.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutson SK, Chyla BJ, Amann JM, Bhaskara S, Huppert SS, Hiebert SW. Liver-specific deletion of histone deacetylase 3 disrupts metabolic transcriptional networks. EMBO J. 2008;27:1017–1028. doi: 10.1038/emboj.2008.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Acetylation: a regulatory modification to rival phosphorylation? EMBO J. 2000;19:1176–1179. doi: 10.1093/emboj/19.6.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang M, Febbraio M, Wagg C, Lopaschuk GD, Dyck JR. Fatty acid translocase/CD36 deficiency does not energetically or functionally compromise hearts before or after ischemia. Circulation. 2004;109:1550–1557. doi: 10.1161/01.CIR.0000121730.41801.12. [DOI] [PubMed] [Google Scholar]
- Lai PH, Wang WL, Ko CY, Lee YC, Yang WM, Shen TW, Chang WC, Wang JM. HDAC1/HDAC3 modulates PPARG2 transcription through the sumoylated CEBPD in hepatic lipogenesis. Biochim Biophys Acta. 2008;1783:1803–1814. doi: 10.1016/j.bbamcr.2008.06.008. [DOI] [PubMed] [Google Scholar]
- Larsen TS, Belke DD, Sas R, Giles WR, Severson DL, Lopaschuk GD, Tyberg JV. The isolated working mouse heart: methodological considerations. Pflugers Arch. 1999;437:979–985. doi: 10.1007/s004240050870. [DOI] [PubMed] [Google Scholar]
- Lee CM, Kumar V, Riley RI, Morgan ET. Metabolism and action of proteasome inhibitors in primary human hepatocytes. Drug Metab Dispos. 2010;38:2166–2172. doi: 10.1124/dmd.110.035501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SA, Jiang H, Trent CM, Yuen JJ, Narayanasamy S, Curley RW, Jr, Harrison EH, Goldberg IJ, Maurer MS, Blaner WS. Cardiac dysfunction in beta-carotene-15,15’-dioxygenase-deficient mice is associated with altered retinoid and lipid metabolism. Am J Physiol Heart Circ Physiol. 2014;307:H1675–H1684. doi: 10.1152/ajpheart.00548.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TH, Yi W, Griswold MD, Zhu F, Her C. Formation of hMSH4-hMSH5 heterocomplex is a prerequisite for subsequent GPS2 recruitment. DNA Repair (Amst) 2006;5:32–42. doi: 10.1016/j.dnarep.2005.07.004. [DOI] [PubMed] [Google Scholar]
- Li D, Wang X, Ren W, Ren J, Lan X, Wang F, Li H, Zhang F, Han Y, Song T, et al. High expression of liver histone deacetylase 3 contributes to high-fat-diet-induced metabolic syndrome by suppressing the PPAR-gamma and LXR-alpha-pathways in E3 rats. Mol Cell Endocrinol. 2011;344:69–80. doi: 10.1016/j.mce.2011.06.028. [DOI] [PubMed] [Google Scholar]
- Li D, Yea S, Li S, Chen Z, Narla G, Banck M, Laborda J, Tan S, Friedman JM, Friedman SL, et al. Kruppel-like factor-6 promotes preadipocyte differentiation through histone deacetylase 3-dependent repression of DLK1. J Biol Chem. 2005;280:26941–26952. doi: 10.1074/jbc.M500463200. [DOI] [PubMed] [Google Scholar]
- Li J, Wang CY. TBL1-TBLR1 and beta-catenin recruit each other to Wnt target-gene promoter for transcription activation and oncogenesis. Nat Cell Biol. 2008;10:160–169. doi: 10.1038/ncb1684. [DOI] [PubMed] [Google Scholar]
- Liu Q, Yuan B, Lo KA, Patterson HC, Sun Y, Lodish HF. Adiponectin regulates expression of hepatic genes critical for glucose and lipid metabolism. Proc Natl Acad Sci U S A. 2012;109:14568–14573. doi: 10.1073/pnas.1211611109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Brown JD, Stanya KJ, Homan E, Leidl M, Inouye K, Bhargava P, Gangl MR, Dai L, Hatano B, et al. A diurnal serum lipid integrates hepatic lipogenesis and peripheral fatty acid use. Nature. 2013;502:550–554. doi: 10.1038/nature12710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marfella R, Di Filippo C, Portoghese M, Barbieri M, Ferraraccio F, Siniscalchi M, Cacciapuoti F, Rossi F, D’Amico M, Paolisso G. Myocardial lipid accumulation in patients with pressure-overloaded heart and metabolic syndrome. J Lipid Res. 2009a;50:2314–2323. doi: 10.1194/jlr.P900032-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marfella R, Portoghese M, Ferraraccio F, Siniscalchi M, Babieri M, Di Filippo C, D’Amico M, Rossi F, Paolisso G. Thiazolidinediones may contribute to the intramyocardial lipid accumulation in diabetic myocardium: effects on cardiac function. Heart. 2009b;95:1020–1022. doi: 10.1136/hrt.2009.165969. [DOI] [PubMed] [Google Scholar]
- Montani JP, Carroll JF, Dwyer TM, Antic V, Yang Z, Dulloo AG. Ectopic fat storage in heart, blood vessels and kidneys in the pathogenesis of cardiovascular diseases. Int J Obes Relat Metab Disord. 2004;(28 Suppl 4):S58–S65. doi: 10.1038/sj.ijo.0802858. [DOI] [PubMed] [Google Scholar]
- Montgomery RL, Potthoff MJ, Haberland M, Qi X, Matsuzaki S, Humphries KM, Richardson JA, Bassel-Duby R, Olson EN. Maintenance of cardiac energy metabolism by histone deacetylase 3 in mice. J Clin Invest. 2008;118:3588–3597. doi: 10.1172/JCI35847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberoi J, Fairall L, Watson PJ, Yang JC, Czimmerer Z, Kampmann T, Goult BT, Greenwood JA, Gooch JT, Kallenberger BC, et al. Structural basis for the assembly of the SMRT/NCoR core transcriptional repression machinery. Nat Struct Mol Biol. 2011;18:177–184. doi: 10.1038/nsmb.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park TS, Hu Y, Noh HL, Drosatos K, Okajima K, Buchanan J, Tuinei J, Homma S, Jiang XC, Abel ED, et al. Ceramide is a cardiotoxin in lipotoxic cardiomyopathy. J Lipid Res. 2008;49:2101–2112. doi: 10.1194/jlr.M800147-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz-Priel I, Houng S, Dooher J, Friedman AD. C/EBPalpha and C/EBPalpha oncoproteins regulate nfkb1 and displace histone deacetylases from NF-kappaB p50 homodimers to induce NF-kappaB target genes. Blood. 2011;117:4085–4094. doi: 10.1182/blood-2010-07-294470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng YC, Kuo F, Breiding DE, Wang YF, Mansur CP, Androphy EJ. AMF1 (GPS2) modulates p53 transactivation. Mol Cell Biol. 2001;21:5913–5924. doi: 10.1128/MCB.21.17.5913-5924.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perissi V, Aggarwal A, Glass CK, Rose DW, Rosenfeld MG. A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell. 2004;116:511–526. doi: 10.1016/s0092-8674(04)00133-3. [DOI] [PubMed] [Google Scholar]
- Petersen KF, Befroy D, Dufour S, Dziura J, Ariyan C, Rothman DL, DiPietro L, Cline GW, Shulman GI. Mitochondrial dysfunction in the elderly: possible role in insulin resistance. Science. 2003;300:1140–1142. doi: 10.1126/science.1082889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao L, Kinney B, Schaack J, Shao J. Adiponectin inhibits lipolysis in mouse adipocytes. Diabetes. 2011;60:1519–1527. doi: 10.2337/db10-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendran P, Delage B, Dashwood WM, Yu TW, Wuth B, Williams DE, Ho E, Dashwood RH. Histone deacetylase turnover and recovery in sulforaphane-treated colon cancer cells: competing actions of 14-3-3 and Pin1 in HDAC3/SMRT corepressor complex dissociation/reassembly. Mol Cancer. 2011;10:68. doi: 10.1186/1476-4598-10-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramasamy R, Goldberg IJ. Aldose reductase and cardiovascular diseases, creating human-like diabetic complications in an experimental model. Circ Res. 2010;106:1449–1458. doi: 10.1161/CIRCRESAHA.109.213447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramasamy R, Oates PJ, Schaefer S. Aldose reductase inhibition protects diabetic and nondiabetic rat hearts from ischemic injury. Diabetes. 1997;46:292–300. doi: 10.2337/diab.46.2.292. [DOI] [PubMed] [Google Scholar]
- Reed TD, Babu GJ, Ji Y, Zilberman A, Ver Heyen M, Wuytack F, Periasamy M. The expression of SR calcium transport ATPase and the Na(+)/Ca(2+)Exchanger are antithetically regulated during mouse cardiac development and in Hypo/hyperthyroidism. J Mol Cell Cardiol. 2000;32:453–464. doi: 10.1006/jmcc.1999.1095. [DOI] [PubMed] [Google Scholar]
- Ren J, Li D, Li Y, Lan X, Zheng J, Wang X, Ma J, Lu S. HDAC3 interacts with sumoylated C/EBPalpha to negatively regulate the LXRalpha expression in rat hepatocytes. Mol Cell Endocrinol. 2013;374:35–45. doi: 10.1016/j.mce.2013.04.013. [DOI] [PubMed] [Google Scholar]
- Rohrer DK, Hartong R, Dillmann WH. Influence of thyroid hormone and retinoic acid on slow sarcoplasmic reticulum Ca2+ ATPase and myosin heavy chain alpha gene expression in cardiac myocytes. Delineation of cis-active DNA elements that confer responsiveness to thyroid hormone but not to retinoic acid. J Biol Chem. 1991;266:8638–8646. [PubMed] [Google Scholar]
- Ruberg FL, Chen Z, Hua N, Bigornia S, Guo Z, Hallock K, Jara H, LaValley M, Phinikaridou A, Qiao Y, et al. The relationship of ectopic lipid accumulation to cardiac and vascular function in obesity and metabolic syndrome. Obesity (Silver Spring) 2010;18:1116–1121. doi: 10.1038/oby.2009.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder TM, Kahler RA, Li X, Westendorf JJ. Histone deacetylase 3 interacts with runx2 to repress the osteocalcin promoter and regulate osteoblast differentiation. J Biol Chem. 2004;279:41998–42007. doi: 10.1074/jbc.M403702200. [DOI] [PubMed] [Google Scholar]
- Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103:843–852. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- Son NH, Ananthakrishnan R, Yu S, Khan RS, Jiang H, Ji R, Akashi H, Li Q, O’Shea K, Homma S, et al. Cardiomyocyte aldose reductase causes heart failure and impairs recovery from ischemia. PLoS One. 2012;7:e46549. doi: 10.1371/journal.pone.0046549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son NH, Park TS, Yamashita H, Yokoyama M, Huggins LA, Okajima K, Homma S, Szabolcs MJ, Huang LS, Goldberg IJ. Cardiomyocyte expression of PPARgamma leads to cardiac dysfunction in mice. J Clin Invest. 2007;117:2791–2801. doi: 10.1172/JCI30335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son NH, Yu S, Tuinei J, Arai K, Hamai H, Homma S, Shulman GI, Abel ED, Goldberg IJ. PPARgamma-induced cardiolipotoxicity in mice is ameliorated by PPARalpha deficiency despite increases in fatty acid oxidation. J Clin Invest. 2010;120:3443–3454. doi: 10.1172/JCI40905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev. 2000;64:435–459. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Miller RA, Patel RT, Chen J, Dhir R, Wang H, Zhang D, Graham MJ, Unterman TG, Shulman GI, et al. Hepatic Hdac3 promotes gluconeogenesis by repressing lipid synthesis and sequestration. Nat Med. 2012;18:934–942. doi: 10.1038/nm.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Singh N, Mullican SE, Everett LJ, Li L, Yuan L, Liu X, Epstein JA, Lazar MA. Diet-induced lethality due to deletion of the Hdac3 gene in heart and skeletal muscle. J Biol Chem. 2011;286:33301–33309. doi: 10.1074/jbc.M111.277707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Wang C, Hagen FK, Gormley M, Addya S, Soccio R, Casimiro MC, Zhou J, Powell MJ, Xu P, et al. Acetylation-defective mutant of Ppargamma is associated with decreased lipid synthesis in breast cancer cells. Oncotarget. 2014;5:7303–7315. doi: 10.18632/oncotarget.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torra IP, Ismaili N, Feig JE, Xu CF, Cavasotto C, Pancratov R, Rogatsky I, Neubert TA, Fisher EA, Garabedian MJ. Phosphorylation of liver X receptor alpha selectively regulates target gene expression in macrophages. Mol Cell Biol. 2008;28:2626–2636. doi: 10.1128/MCB.01575-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tussie-Luna MI, Bayarsaihan D, Seto E, Ruddle FH, Roy AL. Physical and functional interactions of histone deacetylase 3 with TFII-I family proteins and PIASxbeta. Proc Natl Acad Sci U S A. 2002;99:12807–12812. doi: 10.1073/pnas.192464499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umemoto T, Subramanian S, Ding Y, Goodspeed L, Wang S, Han CY, Teresa AS, Kim J, O’Brien KD, Chait A. Inhibition of intestinal cholesterol absorption decreases atherosclerosis but not adipose tissue inflammation. J Lipid Res. 2012;53:2380–2389. doi: 10.1194/jlr.M029264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unger RH. Longevity, lipotoxicity and leptin: the adipocyte defense against feasting and famine. Biochimie. 2005;87:57–64. doi: 10.1016/j.biochi.2004.11.014. [DOI] [PubMed] [Google Scholar]
- Vedantham S, Thiagarajan D, Ananthakrishnan R, Wang L, Rosario R, Zou YS, Goldberg I, Yan SF, Schmidt AM, Ramasamy R. Aldose reductase drives hyperacetylation of Egr-1 in hyperglycemia and consequent upregulation of proinflammatory and prothrombotic signals. Diabetes. 2014;63:761–774. doi: 10.2337/db13-0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner BL, Valledor AF, Shao G, Daige CL, Bischoff ED, Petrowski M, Jepsen K, Baek SH, Heyman RA, Rosenfeld MG, et al. Promoter-specific roles for liver X receptor/corepressor complexes in the regulation of ABCA1 and SREBP1 gene expression. Mol Cell Biol. 2003;23:5780–5789. doi: 10.1128/MCB.23.16.5780-5789.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson PJ, Fairall L, Santos GM, Schwabe JW. Structure of HDAC3 bound to co-repressor and inositol tetraphosphate. Nature. 2012;481:335–340. doi: 10.1038/nature10728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DK, Tarle I, Petrash JM, Quiocho FA. Refined 1.8 A structure of human aldose reductase complexed with the potent inhibitor zopolrestat. Proc Natl Acad Sci U S A. 1993;90:9847–9851. doi: 10.1073/pnas.90.21.9847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong MM, Guo C, Zhang J. Nuclear receptor corepressor complexes in cancer: mechanism, function and regulation. Am J Clin Exp Urol. 2014;2:169–187. [PMC free article] [PubMed] [Google Scholar]
- Xu L, Glass CK, Rosenfeld MG. Coactivator and corepressor complexes in nuclear receptor function. Curr Opin Genet Dev. 1999;9:140–147. doi: 10.1016/S0959-437X(99)80021-5. [DOI] [PubMed] [Google Scholar]
- Yamaguchi N, Kukita T, Li YJ, Kamio N, Fukumoto S, Nonaka K, Ninomiya Y, Hanazawa S, Yamashita Y. Adiponectin inhibits induction of TNF-alpha/RANKL-stimulated NFATc1 via the AMPK signaling. FEBS Lett. 2008;582:451–456. doi: 10.1016/j.febslet.2007.12.037. [DOI] [PubMed] [Google Scholar]
- Yen PM, Ando S, Feng X, Liu Y, Maruvada P, Xia X. Thyroid hormone action at the cellular, genomic and target gene levels. Mol Cell Endocrinol. 2006;246:121–127. doi: 10.1016/j.mce.2005.11.030. [DOI] [PubMed] [Google Scholar]
- Yen PM, Feng X, Flamant F, Chen Y, Walker RL, Weiss RE, Chassande O, Samarut J, Refetoff S, Meltzer PS. Effects of ligand and thyroid hormone receptor isoforms on hepatic gene expression profiles of thyroid hormone receptor knockout mice. EMBO Rep. 2003;4:581–587. doi: 10.1038/sj.embor.embor862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Kalkum M, Chait BT, Roeder RG. The N-CoR-HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. Mol Cell. 2002;9:611–623. doi: 10.1016/s1097-2765(02)00468-9. [DOI] [PubMed] [Google Scholar]
- Zhang X, Jeyakumar M, Petukhov S, Bagchi MK. A nuclear receptor corepressor modulates transcriptional activity of antagonist-occupied steroid hormone receptor. Mol Endocrinol. 1998;12:513–524. doi: 10.1210/mend.12.4.0089. [DOI] [PubMed] [Google Scholar]
- Zhang XM, Chang Q, Zeng L, Gu J, Brown S, Basch RS. TBLR1 regulates the expression of nuclear hormone receptor co-repressors. BMC Cell Biol. 2006;7:31. doi: 10.1186/1471-2121-7-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao HL, Ueki N, Hayman MJ. The Ski protein negatively regulates Siah2-mediated HDAC3 degradation. Biochem Biophys Res Commun. 2010;399:623–628. doi: 10.1016/j.bbrc.2010.07.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.