SUMMARY
The presence of chromatin containing the histone H3 variant CENP-A dictates the location of the centromere in a DNA sequence–independent manner. But the mechanism by which centromere inheritance occurs is largely unknown. We previously reported that CENP-A K124 ubiquitylation, mediated by CUL4A-RBX1-COPS8 E3 ligase activity, is required for CENP-A deposition at the centromere. Here we show that pre-existing ubiquitylated CENP-A is necessary for recruitment of newly synthesized CENP-A to the centromere and that CENP-A ubiquitylation is inherited between cell divisions. In vivo and in vitro analyses using dimerization mutants and dimerization domain fusion mutants revealed that the inheritance of CENP-A ubiquitylation requires CENP-A dimerization. Therefore, we propose models in which CENP-A ubiquitylation is inherited and, through dimerization, determines centromere location. Consistent with this model is our finding that overexpression of a monoubiquitin-fused CENP-A mutant induces neocentromeres at noncentromeric regions of chromosomes.
INTRODUCTION
CENP-A is a centromere-specific histone H3 variant that is required to ensure kinetochore assembly for proper chromosome segregation; defects in CENP-A function lead to aneuploidy and thereby cancer (Amato et al., 2009; Au et al., 2008; Heun et al., 2006; Howman et al., 2000; Li et al., 2011; Tomonaga et al., 2003). In most species except the budding yeast, centromere identity relies not on the DNA sequence but on the presence of a special nucleosome that contains CENP-A (Fukagawa and Earnshaw, 2014; Karpen and Allshire, 1997). CENP-A– containing nucleosomes are formed with canonical histones H2A, H2B, and H4 at the active centromeres, but the nucleosome structure remains controversial (Fukagawa and Earnshaw, 2014; Hasson et al., 2013; Miell et al., 2013; Padeganeh et al., 2013). CENP-A nucleosomes localize to the inner plate of mammalian kinetochores (Warburton et al., 1997) and bind to the 171-bp alpha-satellite DNA in humans (Vissel and Choo, 1987). Active centromeres require CENP-A nucleosomes to direct the recruitment of a constitutive centromere-associated network (CCAN) and the kinetochore proteins in a DNA sequence– independent manner, and together this CCAN and kinetochore proteins orchestrate the kinetochore-microtubule attachment and regulate cycle progression through the spindle checkpoint (Fukagawa and Earnshaw, 2014). Therefore, CENP-A is proposed to be the epigenetic mark of the centromere (Karpen and Allshire, 1997), and recently, through the use of gene targeting in human cells and fission yeast, this mark was demonstrated to act through a two-step mechanism to identify, maintain, and propagate centromere function indefinitely (Fachinetti et al., 2013).
Evidence regarding the mechanism by which the epigenetic mark of the centromere is generated has been divergent and somewhat contradictory. This variation appears to be derived from the variety of species and cell types studied. In gametes of the holocentric nematode Caenorhabditis elegans and possibly in plants, centromere marking is independent of CENP-A/CenH3 (Gassmann et al., 2012; Ingouff et al., 2010; Monen et al., 2005). A recent study suggested that in C. elegans, pre-existing CENP-A/HCP-3 nucleosomes are not necessary to guide the recruitment of new CENP-A nucleosomes (Gassmann et al., 2012). In contrast, in Drosophila melanogaster, CENP-A/CID is present in mature sperm, and the amount of CID that is loaded during each cell cycle appears to be determined primarily by the pre-existing centromeric CID, a finding that is consistent with a “template-governed” mechanism (Raychaudhuri et al., 2012). However, it is unclear how CENP-A works as the epigenetic mark at the molecular level in humans.
Numerous studies have found that CENP-A can be experimentally mistargeted to noncentromeric regions of chromatin and that this mistargeting leads to the formation of ectopic centromeres in model organisms (Fukagawa and Earnshaw, 2014). Chromosome engineering has allowed the efficient isolation of neocentromeres on a wide range of both transcriptionally active and inactive sequences in chicken DT40 cells (Shang et al., 2013). More than 100 neocentromeres in human clinical samples have been described (Marshall et al., 2008). They form on diverse DNA sequences and are associated with CENP-A localization but not with alpha-satellite arrays; thus, these findings provide strong evidence that human centromeres result from sequence-independent epigenetic mechanisms. However, neocentromeres have not yet been created experimentally in humans; overexpression of CENP-A induces mislocalization of CENP-A but not the formation of functional neocentromeres (Van Hooser et al., 2001). Lacoste et al. reported that mislocalization of CENP-A in human cells depends on the chaperone DAXX (Lacoste et al., 2014). Identifying and analyzing factors essential to the generation of human neocentromeres are important in clarifying the mechanism of epigenetic inheritance of centromeres.
In our previous study, we showed that CENP-A K124 ubiquitylation serves as a signal for the deposition of CENP-A at centromeres (Niikura et al., 2015). Here we report that CENP-A K124 ubiquitylation is epigenetically inherited through dimerization. Based on this molecular mechanism, models in which the location of the centromere is inherited are proposed.
RESULTS
CENP-A Monoubiquitylation or Diubiquitylation Is Epigenetically Inherited between Cell Divisions
It has been suggested that the epigenetic centromere mark is generated through a “template-governed” mechanism (Mendiburo et al., 2011; Raychaudhuri et al., 2012): the pre-assembled “old” CENP-A nucleosomes may act as a template, allowing the local stoichiometric loading of new CENP-A nucleosomes during each cell cycle. We have previously shown that CENP-A K124 ubiquitylation serves as a signal for the deposition of CENP-A at centromeres (Niikura et al., 2015). Therefore, we hypothesized a model in which CENP-A K124 ubiquitylation is epigenetically inherited (Figure 1A). This model predicts that CENP-A K124 ubiquitylation depends on pre-existing K124-ubiquitylated CENP-A.
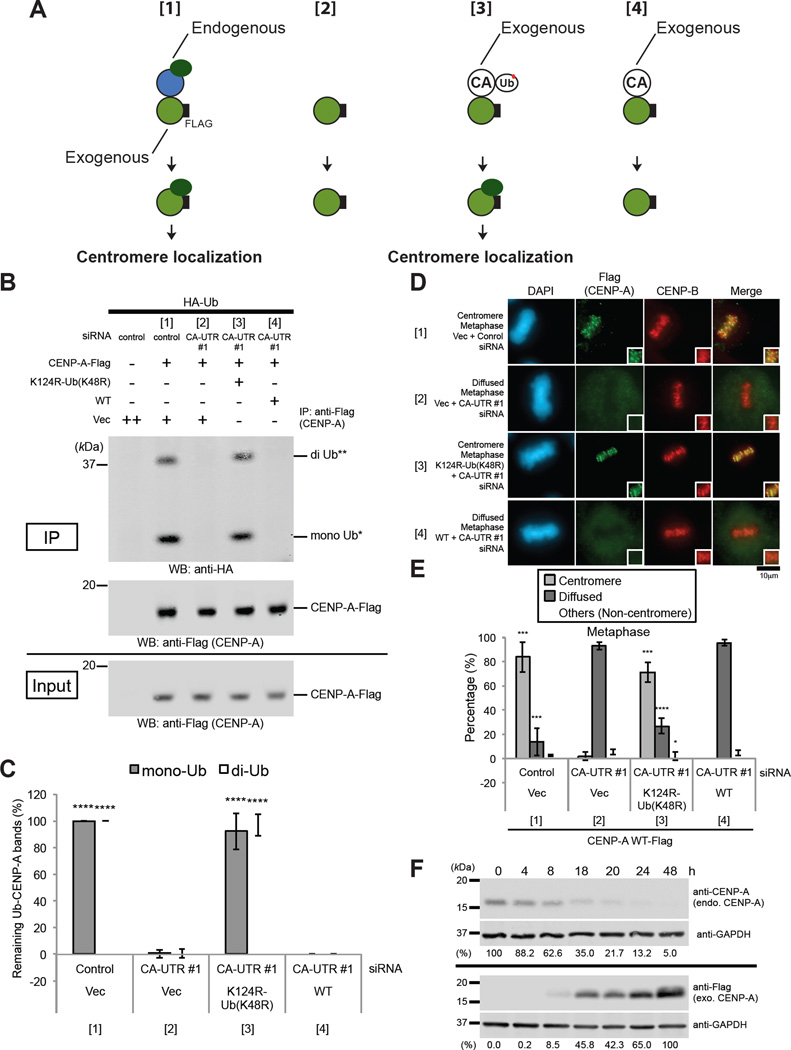
Figure 1. CENP-A Monoubiquitylation or Diubiquitylation Is Epigenetically Inherited between Cell Divisions.
(A) Schematic of the experimental protocol for Figures 1 and S1. When endogenous CENP-A (blue) was reduced to less than 10% of the normal level, exogenously expressed Flag-tagged CENP-A (green) was not ubiquitylated and did not localize to the centromere (panel [2]). However, coexpression of the monoubiquitin-fused CENP-A K124R mutant (white) restored monoubiquitylation and diubiquitylation of Flag-tagged CENP-A and centromeric localization of Flag-tagged CENP-A (panel [3]). The C-terminal Flag tag on CENP-A (white “F” in black rectangle) is indicated.
(B) In vivo ubiquitylation assay (see CENP-A In Vivo Ubiquitylation Assay in Experimental Procedures). HeLa Tet-Off cells were cotransfected with the indicated constructs (Table S3) plus siRNAs (Table S2, CA-UTR #1 or #2 [control]). Proteins in 5% of the total cell lysates (Input) and immunoprecipitates (IP) were detected by Western blot analysis using the indicated antibodies. K124R-Ub (K48R): pTRM4-CENP-A K124R-Ub (K48R), WT: pTRM4-CENP-A WT transfectant (Table S3).
(C) Histogram of quantified ubiquitylated CENP-A bands shown in (B). (Ratio of Flag band signal normalized with the sample in the left column). Experiments were repeated (n ≥ 3 experiments), and mean percentages (±SD) are shown. **** P < 0.0001 compared with the sample [2] (Student’s t test).
(D) Immunostaining of HeLa Tet-Off cells cotransfected with indicated constructs (Table S3) plus siRNAs (Table S2, CA-UTR #1 or #2 [control]). Representative images of other cell-cycle stages are shown in Figure S1D (see also Immunofluorescence in Supplemental Experimental Procedures). DAPI (blue), Flag (green), and endogenous CENP-B (red), which served as a centromere location control, were visualized. Scale bar, 10 µm.
(E) Histograms summarizing the localization patterns shown in (D). Representative data of other cell-cycle stages are shown in Figure S1E. More than 50 metaphase cells were counted per experiment (n ≥ 3 experiments), and the mean percentages (±SD) are shown. “Others (Non-centromere)” indicates mostly cells that were damaged or died, presumably as a result of transfection or other treatments. ****P < 0.0001, ***P < 0.001, and *P < 0.05 compared with the sample [2] (Student’s t test).
(F) Western blot analysis of lysates of HeLa Tet-Off cells transfected at 0 h with (top) CENP-A siRNAs targeting 5’ UTR and 3’ UTR sequences (Table S2, CA-UTR #1) or (bottom) pTRM4-CENP-A-Flag. Endogenous CENP-A and exogenous CENP-A-Flag protein levels were evaluated with the indicated antibodies (Table S1; see also Immunoblotting in Supplemental Experimental Procedures). GAPDH protein was used as a loading control. Percentages (%) of signals compared with that of the 0 h sample (top) or 48 h sample (bottom) are indicated (normalized to anti-GAPDH signals).
In human cells, 10% of the normal level of CENP-A is sufficient to drive kinetochore assembly (Liu et al., 2006). We performed CENP-A siRNA knockdown, targeting the 5’ and 3’ untranslated regions of CENP-A mRNA to reduce the quantity of endogenous CENP-A to 7.2% of its normal level in HeLa cells (Figure S1A and Table S2, CA-UTR #1). This severe loss of endogenous CENP-A prevented endogenous CENP-C localization at centromeres (Figures S1B and S1C); this effect confirmed the previous result (Liu et al., 2006). The severe loss of endogenous CENP-A also substantially abrogated ubiquitylation (Figure 1A, panel [2]; Figures 1B and 1C, sample [2] in the histogram) and centromere localization of exogenously coexpressed CENP-A WT (wild type)-Flag (Figure 1A, panel [2]; Figures 1D and 1E, sample [2] in the histogram; representative images and histograms of other cell cycle stages are shown in Figures S1D and S1E). We further confirmed the localization of Flag-tagged CENP-A proteins by chromosome spreading (Figure S1F, sample [2]). These results suggest that the presence of endogenous CENP-A is required for ubiquitylation of newly synthesized CENP-A and for centromere localization. In this experiment, we confirmed that the maximum expression level of CENP-A WT-Flag was achieved when a substantial amount of endogenous CENP-A was already depleted from cells (reduced to 5.0% of the normal level) 48 h after cotransfection (Figure 1F).
We utilized the constitutively monoubiquitylated CENP-A “mutant” to test the requirement of the presence of ubiquitylated CENP-A (Figure 1A, panel [3]). Interestingly, coexpression of untagged, monoubiquitin-fused CENP-A K124R (CENP-A K124R-Ub [K48R]) (Figure S1G) restored the monoubiquitylation and diubiquitylation of CENP-A WT-Flag (Figure 1A, panel [3]; Figures 1B and 1C, sample [3] in histogram) and the localization of CENP-A WT-Flag at centromeres (Figure 1A, panel [3]; Figures 1D, 1E, S1D, and S1E, sample [3] in histograms). However, coexpression of untagged CENP-A WT (Figure S1H) did not restore monoubiquitylation and diubiquitylation of CENP-A WT-Flag (Figure 1A, panel [4]; Figures 1B and 1C, sample [4] in histogram), nor did this coexpression restore the localization of CENP-A WT-Flag at centromeres (Figure 1A, panel [4]; Figures 1D, 1E, S1D, and S1E, sample [4] in histograms). We further confirmed the localization of Flag-tagged CENP-A proteins by chromosome spreading (Figure S1F, samples [3] and [4]). These results indicated that the presence of ubiquitylated CENP-A is required for ubiquitylation of newly synthesized CENP-A and for centromere localization. We confirmed that exogenous “untagged” CENP-A WT (Figure S1H) did not localize to the centromeres when endogenous CENP-A was decreased to less than 10% of the normal level (Figure S1I and S1J, sample [3]); thus, our result eliminated the possibility that “Flag-tagged” CENP-A WT did not localize to the centromere because it was not functional.
To confirm the result obtained in our experiment in which CENP-A was depleted, we used CENP-A KO (knock-out) cells (CENP-A−/F RPE1 cells) (Fachinetti et al., 2013) instead of siRNA. Fachinetti et al. reported that only 1% of the initial CENP-A level is detectable in 7 days (as expected for the 1/27 dilution, which results in a concentration of approximately 0.8%) after Ad-Cre infection and that no centromere-bound CENP-A was detected 9 days following the excision of CENP-A alleles (Fachinetti et al., 2013). Consistent with these findings, the endogenous CENP-A level was reduced to approximately 2% of the initial level in 6 days (Figure S2A, endo. CENP-A; as expected for the 1/26 dilution, which results in a concentration of approximately 1.6%) and to less than 1% of the initial level in 7 days (data not shown) after transient expression of Cre recombinase (using retro-Cre, a retrovirus vector that induces excision of exons 3 and 4 of the floxed allele). In our experiment, the retrovirus expressed exogenous Flag-CENP-A WT 4 days after retro-Cre infection (Figure S2A, exo. Flag-CENP-A). Two days after infection by the Flag-CENP-A–expressing retrovirus, the severe loss of endogenous centromeric CENP-A substantially abrogated ubiquitylation (Figure 1A, panel [2]; Figure S2B, second lane from left) and centromere localization of exogenously expressed Flag-CENP-A WT (Figure 1A, panel [2]; Figures S2D and S2E, sample [2]). Again, coexpression of untagged, monoubiquitin-fused CENP-A K124R (CENP-A K124R-Ub [K48R]) (Figure 1A, panel [3]; Figure S2C, third lane from left) restored monoubiquitylation and diubiquitylation of Flag-CENP-A WT (Figure 1A, panel [3]; Figure S2B, third lane from left) as well as the localization of Flag-CENP-A WT at centromeres (Figure 1A, panel [3]; Figures S2D and S2E, sample [3]). We confirmed that exogenous “untagged” CENP-A WT (Figure S2C, second lane from left) did not localize to the centromere when endogenous CENP-A was reduced to approximately to 2% of the initial level after 6 days of retro-Cre infection (Figures S2F and S2G, sample [2]); this result eliminated the possibility that “Flag-tagged” CENP-A WT did not localize to the centromere because it is dysfunctional. These results confirmed the finding that the presence of ubiquitylated CENP-A is required for ubiquitylation and centromere localization of newly synthesized CENP-A.
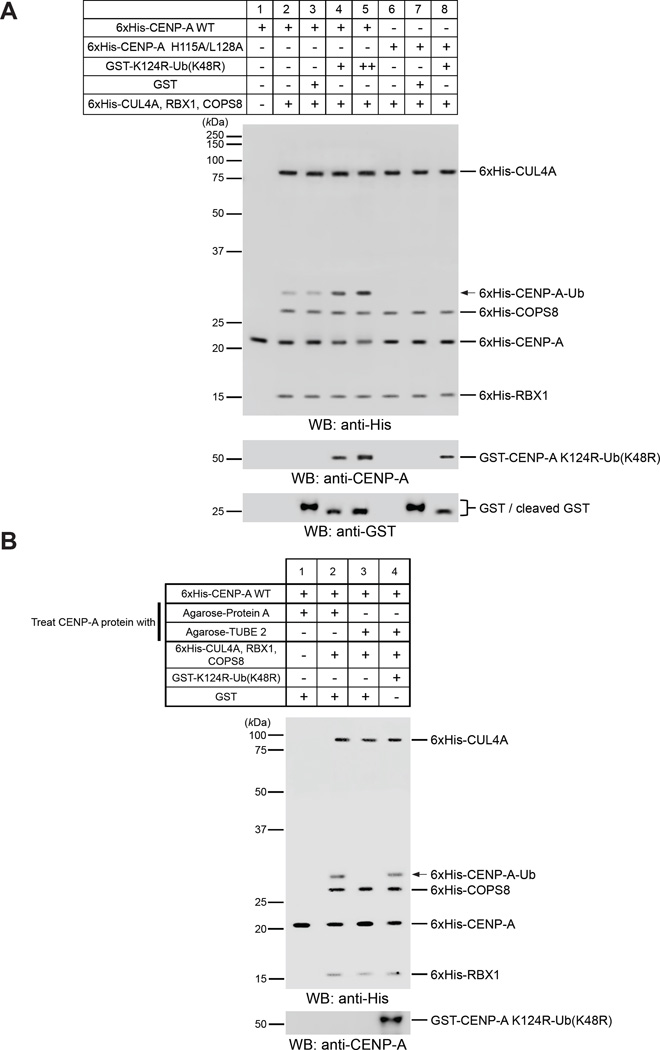
Previously, we showed that 6xHis-CENP-A WT is monoubiquitylated by the purified Cul4A-Rbx1-COPS8 complex in vitro (Niikura et al., 2015). Therefore, we examined whether the in vivo results are true in vitro. Addition of purified monoubiquitin-fused GST-CENP-A K124R (GST-CENP-A K124R-Ub (K48R]) into the reactions enhanced ubiquitylation of 6xHis-CENP-A WT (Figure 2A, compare lanes 3–5; see Figure S3A for verification of purified recombinant proteins). Such enhancement supports the in vivo results.
Figure 2. Pre-existing CENP-A Ubiquitylation Is Required for K124 Ubiquitylation of New CENP-A by the CUL4A Complex in Vitro.
(A) Adding monoubiquitin-fused CENP-A K124R-Ub (K48R) enhanced CENP-A ubiquitylation in vitro. Putative 6xHis-CENP-A-Ub is indicated by the arrow. See CENP-A In Vitro Ubiquitylation Assay in Supplemental Experimental Procedures and Figure S3A for verification of purified recombinant proteins.
(B) Adding monoubiquitin-fused CENP-A K124R-Ub (K48R) after removal of ubiquitylated CENP-A from Sf9 lysates restored CENP-A ubiquitylation in vitro. Putative 6xHis-CENP-A-Ub is indicated by the arrow. See CENP-A In Vitro Ubiquitylation Assay in Supplemental Experimental Procedures and Figure S3A for verification of purified recombinant proteins.
If the presence of ubiquitylated CENP-A is required for ubiquitylation of newly synthesized CENP-A, why can purified 6xHis-CENP-A WT be ubiquitylated in vitro? We assumed that 6xHis-CENP-A WT expressed in insect cells contained some ubiquitylated 6xHis-CENP-A, but the levels were undetectable. Therefore, we depleted presumably existing ubiquitylated CENP-A from purified 6xHis-CENP-A WT by using Agarose-TUBE 2 (LifeSensors). In vitro ubiquitylation was then performed with the remaining nonubiquitylated 6xHis-CENP-A WT. Indeed, depletion of ubiquitylated CENP-A abolished ubiquitylation of 6xHis-CENP-A WT (Figuress 2B and S3B, lane 3). Addition of GST-CENP-A K124R-Ub (K48R) restored ubiquitylation of 6xHis-CENP-A WT (Figures 2B and S3B, compare lanes 3 and 4), a result that is consistent with those of the in vivo experiments described in the preceding text.
We also tested bacterially expressed and purified CENP-A, which is not ubiquitylated (Figure S3C), as substrate for in vitro ubiquitylation assay, and consistent results were obtained (Figure S3D, compare lanes 2 and 3).
Taken together, our results indicate that pre-existing ubiquitylated CENP-A is required for ubiquitylation of newly synthesized CENP-A and for centromere localization. Thus, CENP-A ubiquitylation appears to be inherited epigenetically between cell divisions.
CENP-A Dimerization through H115/L128 Is Required for CENP-A K124 Ubiquitylation
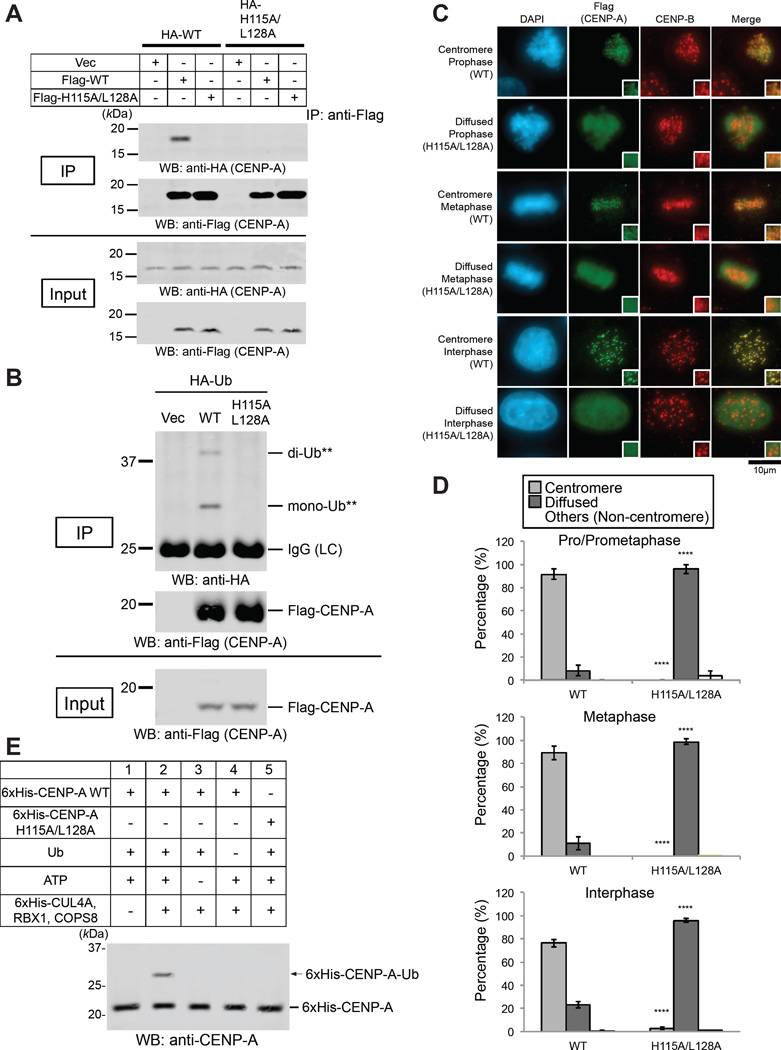
If our “epigenetic” model is correct (Figure 1A), then the CUL4A E3 complex should recognize a heterodimer of nonubiquitylated CENP-A and K124-ubiquitylated CENP-A (hemiubiquitylation) but not a homodimer of nonubiquitylated CENP-A (Figure 1A). In this case, heterodimerization would be required for K124 ubiquitylation of nonubiquitylated CENP-A. Thus, CENP-A K124 ubiquitylation would depend on pre-existing K124-ubiquitylated CENP-A. In Drosophila melanogaster, disruption of the dimerization interface of CENP-A/CID reduces its centromere localization in vivo (Zhang et al., 2012). In human cells, SDS-resistant CENP-A dimers have been reported (Niikura et al., 2015; Shelby et al., 1997; Yoda et al., 2000). Bassett et al. reported that a human CENP-A dimerization mutant (CENP-AH115A/L128A) cannot stably assemble into chromatin (Bassett et al., 2012). Consistent with this finding, the CENP-A H115A/L128A mutation reduced dimerization of CENP-A in cell lysates (Figure S3F) and abrogated dimerization in immunoprecipitation analysis (Figure 3A). Moreover, the CENP-A H115A/L128A mutation abrogated ubiquitylation of CENP-A in vivo (Figure 3B) and localization of CENP-A to the centromeres (Figures 3C and 3D; chromosome spreads are shown in Figures S3G). It should be noted that monoubiqutin-fused CENP-AH115A/L128A was not able to interact with CENP-A WT properly (Figure S3H, lanes 4 and 5). These results suggested that dimerization is required for CENP-A localization to the centromere.
Figure 3. The CENP-A H115A/L128A Mutation Abrogated Dimerization and Ubiquitylation of CENP-A and Localization of CENP-A to the Centromere.
(A) The CENP-A H115A/L128A mutation abrogated dimerization of CENP-A. HeLa Tet-Off cells were cultured and transfected with pQCXIP-HA-CENP-A (WT or H115A/L128A) plus pTRM4-Flag-CENP-A (WT or H115A/L128A) or pTRM4 vector (Table S3) (see Immunoprecipitation Assay in Supplemental Experimental Procedures). Proteins in 3% of the total cell lysates (Input) and immunoprecipitates (IP) were detected.
(B) The CENP-A H115A/L128A mutation abrogated ubiquitylation of CENP-A in vivo (see CENP-A In Vivo Ubiquitylation Assay in Experimental Procedures).
(C) The CENP-A H115A/L128A mutant delocalized from centromeres. HeLa Tet-Off cells were cultured, cotransfected with indicated constructs plus CA-UTR #2 siRNAs (Table S2), and immunostained (see Immunofluorescence in Supplemental Experimental Procedures). DAPI (blue), Flag (green), and endogenous CENP-B (red) as a centromere location control were visualized. Diffuse signals appeared in the exogenous CENP-A-Flag overexpression, presumably because its expression level was approximately 1.0 to 1.4 orders of magnitude (10- to 25-fold) higher than endogenous CENP-A (data not shown). WT: pTRM4-Flag-CENP-A WT, H115A/L128A: pTRM4-Flag-CENP-A H115A/L128A transfectant (Table S3). Asterisks show a non-transfected, Flag-negative cell. Scale bar, 10 µm.
(D) Histograms summarizing the localization patterns shown in (C). More than 50 pro/prometaphase, 50 metaphase cells, and 200 interphase cells were counted per experiment (n ≥ 3 experiments), and the mean percentages (±SD) are shown. “Others (Non-centromere)” indicates mostly damaged cells, dead cells, or cells with nucleolar localization in interphase, presumably because of transfection or other treatments. ****P < 0.0001 compared with WT (Student’s t test).
(E) The CENP-A H115A/L128A mutation abrogated ubiquitylation of CENP-A in vitro (see CENP-A In Vitro Ubiquitylation Assay in Supplemental Experimental Procedures). Purified 6xHis-tagged components (CLU4A, RBX1, and COPS8) are shown in the upper table. Putative 6xHis-CENP-A-Ub is indicated by the arrow. See Figure S3A for verification of purified recombinant proteins.
We could not conclude that dimerization is directly required for CENP-A K124 ubiquitylation, because the dimerization mutant may disturb proper nucleosome formation. Therefore, we examined ubiquitylation of CENP-AH115A/L128A in vitro. Indeed, CENP-AH115A/L128A protein was not ubiquitylated in vitro (Figure 2A, lanes 6–8; Figure 3E, lane 5; see Figure S3A for verification of purified recombinant proteins), and this absence of ubiquitylation strongly suggested that dimerization is required for CENP-A K124 ubiquitylation.
Heterodimerization with Ubiquitylated CENP-A Is Required to Ubiquitylate New CENP-A
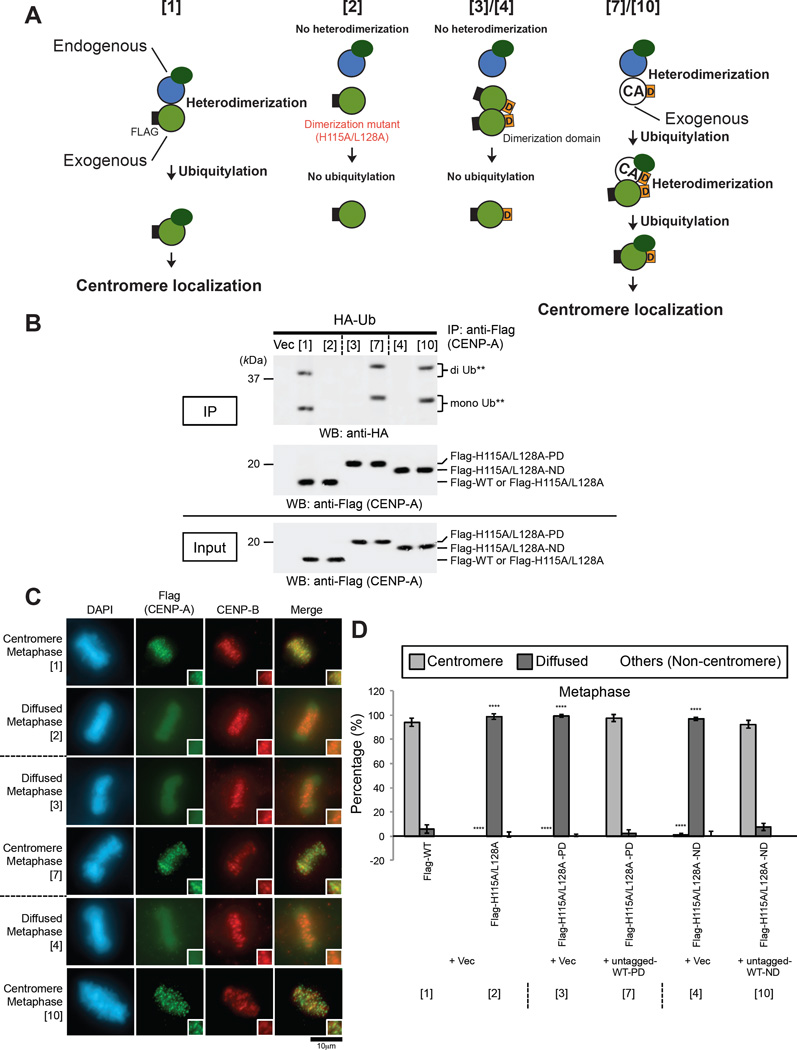
To evaluate the importance of heterodimerization (i.e., a dimerization of old CENP-A –new CENP-A) for CENP-A localization at the centromere, we hypothesized that the loss of CENP-A dimerization and centromere localization by the H115A/L128A mutation could be rescued by the addition of a dimerization domain that has previously been used to force the dimerization of specific proteins (Wang et al., 2002). To test this hypothesis, we fused 28 or 24 amino acids derived from the dimerization domain of the Saccharomyces cerevisiae PUT3 protein (ScPUT3 dimerization domain; Figure S4A, “PD”) or the Drosophila melanogaster Ncd (DmNcd dimerization domain; Figure S4A, “ND”) protein to the C-terminal end of the CENP-A H115A/L128A protein. In our hypothetical scheme (Figures 4A and S4A), in the presence of endogenous CENP-A proteins, overexpression of Flag-H115A/L128A or Flag-H115A/L128A-D (“D” refers to the dimerization domain) does not lead to heterodimer formation with endogenous CENP-A. Thus, neither subsequent K124 ubiquitylation nor centromere localization of these exogenous proteins occurs (Figures 4A and S4A, [2] and [3]/[4] compared with [1] single Flag-WT overexpression). If untagged WT-D is coexpressed, Flag-H115A/L128A-D is hypothesized to form heterodimers with it through the dimerization domain and to localize to the centromere (Figures 4A and S4A, [7]/[10] compared with [6]/[9] negative control and [5]/[8] positive control). We confirmed the coexpression of each exogenous protein in cell lysates (Figure S4B), and overexpression of Flag-H115A/L128A, Flag-H115A/L128A-PD, or Flag-H115A/L128A-ND alone did not result in significant ubiquitylation (Figures 4B and S4C, [2]-[4] compared with [1] positive control) nor centromere localization (Figures 4C, 4D, and S5, [2]-[4] compared with [1] positive control), whereas the loss of Flag-H115A/L128A homodimer formation (Figure S4B, [2]) was rescued by the addition of PD or ND (Figure S4B, [3] and [4]). Indeed, Flag-H115A/L128A-PD or Flag-H115A/L128A-ND localized to the centromere when untagged WT-PD or untagged WT-ND, respectively, were coexpressed; Flag-H115A/L128A that lacked PD or ND did not colocalize to the centromere (Figures 4C, 4D, and S5, [7]/[10] compared with [6]/[9] negative control). In addition, we confirmed the localization of Flag-tagged CENP-A proteins by chromosome spreading (Figure S6). In summary, the status of centromere localization of each Flag-tagged CENP-A protein matches that of ubiquitylation (Figure S4A, right two columns). These results indicate that CENP-A heterodimerization with pre-existing ubiquitylated CENP-A is required for ubiquitylation and centromere localization of new CENP-A.
Figure 4. Heterodimarization of New CENP-A with Pre-existing Ubiquitylated CENP-A Is Required for Ubiquitylation of the New CENP-A and Localization of the New CENP-A to the Centromere.
(A) Schematic of the experimental protocol for Figures 4 and S4–S6. Heterodimerization between endogenous CENP-A and Flag-CENP-A WT led to K124 ubiquitylation of Flag-CENP-A WT and localization of Flag-CENP-A WT to the centromere (sample [1]). Single overexpression of Flag-CENP-A H115A/L128A or Flag-CENP-A H115A/L128A-D (“D” refers to the dimerization domain) did not lead to heterodimerization or K124 ubiquitylation of Flag-tagged exogenous CENP-A (sample [2] or [3]/[4]). Overexpression of both Flag-CENP-A H115A/L128A-D and untagged CENP-A WT-D led to heterodimerization and the subsequent K124 ubiquitylation of Flag-tagged exogenous CENP-A (sample [7]/[10]). The N-terminal Flag tag on CENP-A (white “F” in black rectangle) is indicated. See Figure S4A for full set of samples.
(B) In vivo ubiquitylation assay (See CENP-A In Vivo Ubiquitylation Assay in Experimental Procedures). HeLa Tet-Off cells were cotransfected with the indicated constructs as numbered in Figures 4 and S4A, plus CA-UTR #2 siRNA (Table S2). Proteins in 5% of the total cell lysates (Input) and immunoprecipitates (IP) were detected by Western blot analysis using the indicated antibodies. See Figures 4A and S4C for full set of samples.
(C) Immunostaining of HeLa Tet-Off cells cotransfected with the indicated constructs as numbered in Figure 4A and S4C, plus CA-UTR #2 siRNA (Table S2). Representative images of the full set of samples in the same and other cell-cycle stages are shown in Figures S5A, S5C, and S5E. Images of samples [1]-[4], [7], and [10] are the same as those in Figure S5A (see Immunofluorescence in Supplemental Experimental Procedures). DAPI (blue), Flag (green), and endogenous CENP-B (red), which served as a centromere localization control during metaphase, were visualized. Scale bar, 10 µm.
(D) Histograms summarizing the localization patterns shown in (C). More than 50 metaphase cells were counted per experiment (n ≥ 3 experiments), and the mean percentages (±SD) are shown. “Others (Non-centromere)” indicates mostly cells that were damaged or died presumably because of transfection or other treatments. ****P < 0.0001 compared with the sample [1] (Student’s t test). See also Figures S5B, S5D, and S5F for full set of samples in the same and other cell-cycle stages; results for samples [1]-[4], [7], and [10] are the same as those in Figure S5B.
Taken together, our results suggest that the CUL4A E3 complex recognizes a heterodimer of nonubiquitylated CENP-A and K124-ubiquitylated CENP-A (hemiubiquitylation) to ubiquitylate nonubiquitylated CENP-A. Thus, CENP-A K124 ubiquitylation is epigenetically inherited (see Discussion and Figure 7).
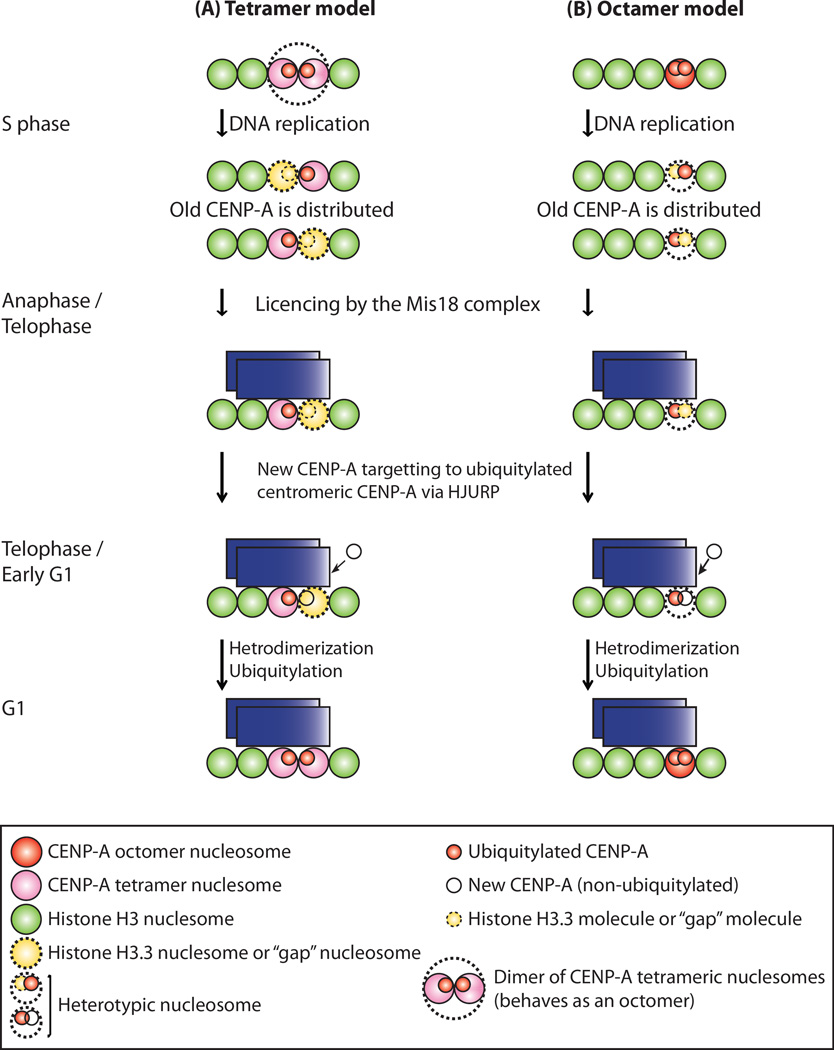
Figure 7. Models of Epigenetic Inheritance of CENP-A Ubiquitylation.
HJURP preferentially binds to ubiquitylated, preassembled “old” CENP-A, which resides predominantly in nucleosomes. A new CENP-A monomer targets ubiquitylated centromeric CENP-A via preassembled HJURP. New CENP-A is ubiquitylated in a heterodimerization-dependent manner (i.e., dimers of old CENP-A with new CENP-A). In this way, the ubiquitylation and the location of the centromere are inherited epigenetically (see Discussion).
(A) Tetramer model. Because of the dimerization of CENP-A tetrameric nucleosomes (larger dots circle in top), octamer formation can be achieved only during S phase as Bui et al. suggested (Bui et al., 2012). Heterodimerization could be internucleosomal.
(B) Octamer model. Two CENP-A dimers in one nucleosome are split/diluted between the two daughter centromere-DNA sequences, and one CENP-A molecule either is replaced with one H3 molecule or leaves a molecule-free gap during replication/S phase. Histone H4 is omitted from both models for simplicity.
Overexpression of Monoubiquitin-Fused CENP-A K124R Induces Formation of Neocentromeres in Human Cells
In Drosophila cell lines, CENP-A overexpression causes mislocalization of CENP-A into noncentromeric regions (Heun et al., 2006). These ectopic centromeres are able to attract downstream kinetochore proteins and cause chromosome segregation defects, presumably as a result of dicentric activity (Heun et al., 2006). In humans, overexpression of CENP-A induces misloading of CENP-A at noncentromeric regions and assembly of a subset of kinetochore components, including CENP-C, hSMC1, and HZwint-1 (Van Hooser et al., 2001). However, the microtubule-associated proteins CENP-E and HZW10 were not recruited, and neocentromeric activity was not detected (Van Hooser et al., 2001). Recently, Lacoste et al. reported that ectopic mislocalization of CENP-A in human cells depends on the H3.3 chaperone DAXX rather than the specific centromeric CENP-A chaperone HJURP (Lacoste et al., 2014). We previously reported that addition of monoubiquitin to the CENP-A K124R mutant restores centromere targeting and interaction with HJURP; this finding demonstrated that monoubiquitylation is an essential signaling modification required for efficient interaction with HJURP and subsequent recruitment of CENP-A to centromeres (Niikura et al., 2015). Therefore, we hypothesized that if CENP-A K124 ubiquitylation is required to determine the location of the centromere, overexpression of constitutively ubiquitylated CENP-A K124R-Ub (K48R) could induce the formation of neocentromeres at noncentromeric locations.
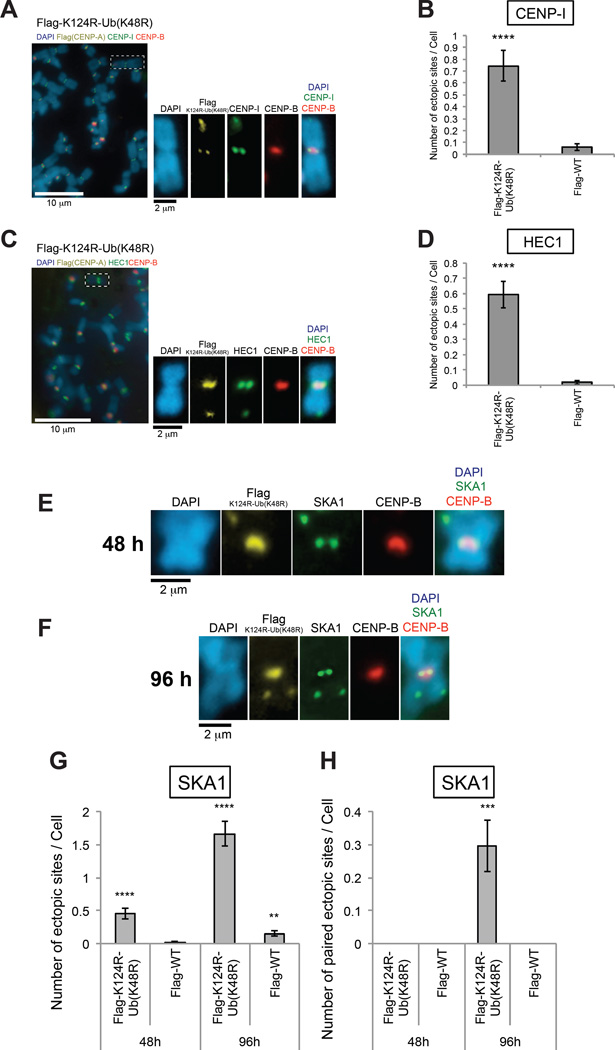
We induced transient overexpression of Flag-CENP-A K124R-Ub (K48R) or Flag-CENP-A WT in HeLa Tet-Off cells for 48 h and performed 4-color immunostaining of chromosome spreads (staining for DAPI; Flag; endogenous CENP-B; and endogenous central-outer kinetochore proteins CENP-I, HEC1, or SKA1). Endogenous CENP-B was stained as preassembled native centromeres, and endogenous central-outer kinetochore proteins (CENP-I, HEC1, or SKA1) or chaperone proteins (HJURP or DAXX) were stained as both native and “putative” ectopic (neo-) centromeres. Structures with Flag and central-outer kinetochore protein–positive but CENP-B–negative signals were counted as “putative” neocentromeres at ectopic sites in HeLa Tet-Off cells (the average number of neocentromeres per cell of Flag-K124R-Ub [K48R] was 0.75 as determined by CENP-I immunostaining, 0.59 as determined by HEC1 immunostaining, and 0.46 as determined by SKA1 immunostaining) (Figures 5A–5E, 5G, and 5H).
Figure 5. Neocentromeres Were Ectopically Generated by Overexpression of Monoubiquitin-fused CENP-A K124R.
(A–D) Overexpression of monoubiquitin-fused CENP-A K124R ectopically generates neocentromeres, including central-outer kinetochore proteins (CENP-I and HEC1). (Left array) HeLa Tet-Off cells were cultured without tetracycline/doxycycline and harvested 48 h after transfection with pTRM4-Flag-CENP-A K124R-Ub (K48R) or pTRM4-Flag-CENP-A WT, plus CA-UTR #2 siRNAs (Table S2; see also Chromosome Spreading in Supplemental Experimental Procedures). Forty-six hours after transfection, cells were arrested at prometaphase by treatment with nocodazole for 2 h, and their chromosomes were examined after staining with antibodies against Flag (CENP-A), endogenous central-outer kinetochore protein (A) CENP-I or (C) HEC1, and CENP-B. DNA was costained with DAPI. Representative images including magnified paired sister chromatids are shown (insets). K124R-Ub (K48R): pTRM4-Flag-CENP-A K124R-Ub (K48R), WT: pTRM4-Flag-CENP-A WT transfectant (Table S3). Scale bar, 10 µm or 2 µm.
(Right array) The average number (±SEM) of neocentromeres per cell (n ≥ 100 cells) was obtained: (B) 0.75 (with CENP-I) and (D) 0.59 (with HEC1) from the experiment performed in (left array). ****P < 0.0001 compared with Flag-CENP-A WT transfectant (Student’s t test).
(E) Chromosome spreads 48 h after transfection A) but with anti-SKA1. Scale bar, 10 µm or 2 µm.
(F) Chromosome spreads 96 h after transfection as in (E). Scale bar, 10 µm or 2 µm.
(G) The average number (±SEM) of ectopic sites per cell (n ≥ 100 cells) was obtained with SKA1: 0.46 at 48 h and 1.67 at 96 h from the experiment performed in (E) and (F), respectively. ****P < 0.0001 and **P < 0.01 compared with Flag-CENP-A WT transfectant at 48 h (Student’s t test).
(H) The average number (±SEM) of paired ectopic sites per cell (n ≥ 100 cells) was obtained with SKA1: 0 at 48 h and 0.30 at 96 h from the experiment performed in (E) and (F), respectively. ***P < 0.001 compared with Flag-CENP-A WT transfectant at 48 h (Student’s t test).
Because SKA1 requires microtubules to localize to centromeres (Hanisch et al., 2006), we interpreted the result regarding SKA1 immunostaining as indicating that overexpression of constitutively monoubiquitylated CENP-A induces functional neocentromeres. This finding suggests that CENP-A ubiquitylation plays a role to determine the location of the centromere. The number of “paired” putative neocentromeres with SKA1-positive sister chromatids significantly increased from 48 h to 96 h after the induction of Flag-CENP-A K124R-Ub (K48R) expression (Figure 5F–5H; from 0 per cell [48 h] to 0.30 per cell [96 h]). This result indicates that newly created neocentromeres are duplicated and inherited epigenetically between cell divisions.
CENP-A Ubiquitylation Contributes to Kinetochore Assembly in Addition to CENP-A Deposition at the Centromere
New centromeres can be generated artificially at ectopic sites through the assembly of CENP-A nucleosomes at Lac operator (LacO)-containing arrays (Barnhart et al., 2011; Logsdon et al., 2015; Mendiburo et al., 2011). We applied this LacO/LacI ectopic centromeric chromatin assembly system to address the ability of CENP-A (WT or mutants) fused to the Lac repressor (LacI) to recruit CENP-A chaperons and/or kinetochore components at arrays of Lac operator (LacO) sequences on chromosome 1 of U2OS cells (Janicki et al., 2004; Logsdon et al., 2015).
First, we confirmed that expression levels of HA-LacI-CENP-A (WT and mutants: K124R and K124R-Ub [K48R]) are comparable in the cells (Figure S7A). In addition to the loci of LacO arrays, HA-LacI-CENP-A WT localized at endogenous centromeres, whereas the K124R mutation substantially abrogated the centromere localization of HA-LacI-CENP-A (Figures S7C and S7E, ‘HA’; Figure S7B: Note that HEC1 and SKA1 localize at the centromere in mitosis) (Hanisch et al., 2006; Lin et al., 2006). Compared with the expression of the Vec (LacI) control, the expression of HA-LacI-CENP-A WT also increased signals of “punctuated” localization of CENP-A chaperons (HJURP and DAXX) and outer kinetochore proteins (HEC1 and SKA1) at endogenous centromeres (Figure S7C and S7E, ‘red’ and ‘Merge’). The K124R mutation diminished the localization of CENP-A chaperons (HJURP and DAXX) and that of outer kinetochore proteins (HEC1 and SKA1) at endogenous centromeres (Figure S7C and S7E, ‘red’ and ‘Merge’), presumably because of dominant negative effects of the K124R mutation. The HA-Lac-I-CENP-A K124R-Ub (K48R) mutant, which mimics monoubiquitylated CENP-A, localized to endogenous centromeres (Figures S7C and S7E, ‘HA’; Figure S7B) as previously observed when Flag- tagged CENP-A proteins were used (Niikura et al., 2015). Also, the HA-LacI-CENP-A K124R-Ub (K48R) mutant, compared with the K124R mutant, increased signals of “punctuated” centromere localization of CENP-A chaperons (HJURP and DAXX) and outer kinetochore proteins (HEC1 and SKA1) (Figures S7C and S7E, ‘red’ and ‘Merge’).
The K124R mutation significantly reduced the recruitment of CENP-A chaperons (HJURP and DAXX) and outer kinetochore proteins (HEC1 and SKA1) at LacO arrays, after ectopic loci forcibly were determined through LacO-LacI interaction (Figures S7C–S7F). Addition of monoubiquitin to the CENP-A K124R mutant, when compared with CENP-A WT, significantly restored and enhanced the recruitment of CENP-A chaperons and outer kinetochore proteins at LacO arrays (Figures S7C–S7F).
Taken together, these results suggest that CENP-A ubiquitylation contributes not only to determining the position of centromeric loci, but also to the assembly of kinetochores after ectopic loci were forcibly determined (see Discussion).
DISCUSSION
Our results of in vivo and in vitro ubiquitylation assays using the constitutively ubiquitylated CENP-A mutant clearly showed that ubiquitylated CENP-A is required for ubiquitylation of nonubiquitylated CENP-A. Therefore, the heterodimer (i.e., a dimer of old CENP-A and new CENP-A) is presumably recognized by the CUL4A complex, and the new CENP-A is ubiquitylated and maintained at the centromeres (Niikura et al., 2015). Our previous studies showed that CENP-A K124 is ubiquitylated in the M and G1 phases (i.e., at the time of CENP-A deposition at centromeres) (Jansen et al., 2007; Niikura et al., 2015). Based on these results, we provide two models of epigenetic inheritance of CENP-A ubiquitylation for the control of CENP-A deposition and maintenance at centromeres (Figure 7 and see further discussion below).
Models of the Inheritance of the Centromere Location
Bui et al. reported that native CENP-A nucleosomes are tetrameric during the early G1 phase, are converted to octamers at the transition from the G1 phase to the S phase, and revert to tetramers after DNA replication (Bui et al., 2012). CENP-A binds to HJURP during the G1 and G2 phases but not during the S phase (Bui et al., 2012). However, the current model of interconversion between tetrameric and octameric CENP-A nucleosomes in the cell cycle remains controversial (Fukagawa and Earnshaw, 2014; Hasson et al., 2013; Miell et al., 2013; Padeganeh et al., 2013), although the structures of the homotypic and heterotypic CENP-A particles have been solved (Arimura et al., 2014; Tachiwana et al., 2011). Therefore, we provide two models of epigenetic inheritance of CENP-A ubiquitylation: a tetramer model (Figure 7A) and an octamer model (Figure 7B).
Dunleavy et al. reported that histone H3.3 is deposited at centromeres in the S phase as a placeholder for CENP-A that is newly assembled in the G1 phase (Dunleavy et al., 2011) (Figure 7, S phase). Thus, in the tetramer model, formation of the CENP-A octamer nucleosome can be established only during the S phase as Bui et al. suggested (Bui et al., 2012), because of the dimerization of the CENP-A tetrameric nucleosome (Figure 7A, larger dots circle in top). CENP-A nucleosomes, where each nucleosome has one single CENP-A molecule, are divided/diluted between the two daughter centromere-DNA sequences, and either is replaced with an H3 nucleosome or leaves a nucleosome-free gap during replication/S phase (Figure 7A). In the octamer model, two CENP-A dimers in one nucleosome are split/diluted between the two daughter centromere-DNA sequences, and one CENP-A molecule either is replaced with one H3 molecule or leaves a molecule-free gap during replication/S phase (Figure 7B).
The following lines of evidence were collected from our studies and others. Zasadzinska et al. demonstrated that HJURP itself dimerizes through a C-terminal repeat region, which is essential for centromeric assembly of nascent CENP-A (Zasadzinska et al., 2013). Dunleavy et al. showed that HJURP localizes at centromeres during late telophase, which is when newly synthesized CENP-A is incorporated at centromeres in humans (Dunleavy et al., 2009). Phosphorylation and DNA binding of HJURP were suggested to determine its centromeric requirement and function in CENP-A loading (Muller et al., 2014). Our previous UbFC analysis suggested that K124-ubiquitylated CENP-A exists at centromeres and the nuclear region (Niikura et al., 2015). In addition, we previously showed that K124-ubiquitylated CENP-A is found in the insoluble chromatin fraction (Niikura et al., 2015). Our previous study indicated that CENP-A K124 ubiquitylation is required for efficient interaction with HJURP (Niikura et al., 2015), and in the present study, our in vitro and in vivo ubiquitylation assays revealed that HJURP itself contributes to CENP-A K124 ubiquitylation (Figures S3B, S3D, and S3E). Addition of purified HJURP itself did not induce ubiquitylation of CENP-A in vitro (in the absence of ubiquitylated CENP-A) but enhanced CENP-A ubiquitylation about 2-fold in the presence of CENP-A K124R-Ub (K48R) (Figure S3B, compare lanes 4–6; Figure S3D, compare lanes 3–5). Heterodimerization of new CENP-A with pre-existing ubiquitylated CENP-A is required for ubiquitylation of new CENP-A and localization of new CENP-A to the centromere (Figures 4 and S4–S6). K124R-Ub (K48R) mutant increased signals of “punctuated” centromere localization of HJURP, whereas K124R mutation abrogated the centromere localization of HJURP (Figure S7E, ‘HJURP’ and ‘Merge’). The evidence summarized in this paragraph supports our proposed models (Figures 7A and 7B) in which HJURP preferentially binds to ubiquitylated, preassembled “old” CENP-A, which resides predominantly in nucleosomes, especially at the initial step of the ubiquitylation of nascent (“new”) CENP-A (Figure 7, anaphase/telophase). During this process, newly synthesized, free CENP-A targets ubiquitylated centromeric CENP-A through its attraction to HJURP, which is preassembled with “old” ubiquitylated, centromeric CENP-A (Figure 7, telophase/early G1). Subsequently, new CENP-A is ubiquitylated in the proximity of the nucleosome and/or inside the nucleosomes in a heterodimerization-dependent manner (old CENP-A – new CENP-A) during the M and G1 phases (Figure 7, telophase/early G1), and HJURP partly contributes to ubiquitylation (Figures S3B, S3D, and S3E). In the tetramer model, heterodimerization could be internucleosomal (Figure 7A, telophase/early G1). Thus, in these models, ubiquitylation and the location of the centromere are inherited epigenetically.
Neocentromere Formation
To date, “functional” neocentromeres have not resulted from the experimental mistargeting of overexpressed CENP-A in humans (Van Hooser et al., 2001). In our study, overexpression of the monoubiquitin fusion protein Flag-CENP-A K124R-Ub (K48R) led to sufficient recruitment of HJURP and central-outer kinetochore components (i.e., CENP-I, HEC1, and SKA1) to noncentromeric chromatin regions. In particular, SKA1 recruitment on the ectopic centromere verified that the neocentromeres are functional, because SKA1 centromere localization requires the formation of kinetochore–microtubule interactions (Hanisch et al., 2006; Schmidt et al., 2012). In our experiment, SKA1-positive, putative neocentromeres replicated and were inherited epigenetically between cell divisions.
Functions of CENP-A Chaperons, HJURP, and DAXX
Our assay using the LacO/LacI ectopic centromeric chromatin assembly system clearly revealed that CENP-A ubiquitylation contributes to the recruitment of CENP-A chaperons (HJURP and DAXX) and outer kinetochore components (HEC1 and SKA1) at LacO arrays (Figure S7). It is possible that ubiquitylation of CENP-A contributes to maintain and stabilize ectopic neocentromeres in humans; in Drosophila melanogaster the E3 Ligase CUL3/RDX controls centromere maintenance by ubiquitylating and stabilizing CENP-A in a CAL1-dependent manner (Bade et al., 2014).
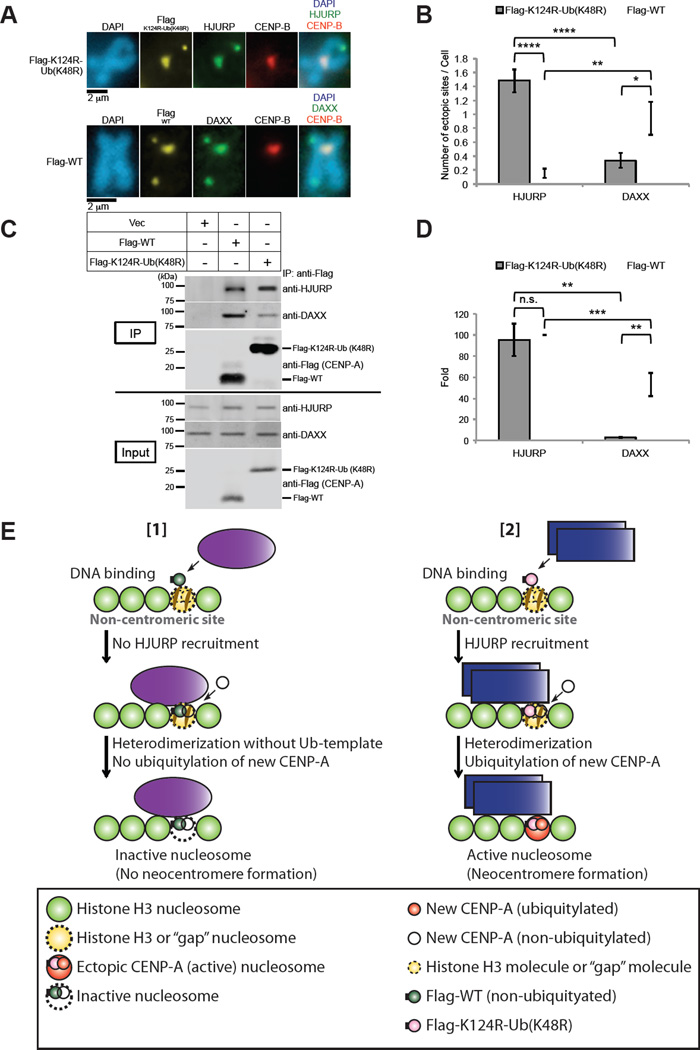
Overexpression of Flag-CENP-A-K124R-Ub (K48R) induced the colocalization of more endogenous HJURP at putative neocentromeres (noncentromere regions where Flag-CENP-A localized) than Flag-CENP-A WT did (Figures 6A and 6B, ‘HJURP’), whereas the colocalization of endogenous DAXX at putative neocentromeres was greater with Flag-CENP-A-WT than with Flag-K124R-Ub (K48R) (Figures 6A and 6B, ‘DAXX’). We previously found that the affinity of the K124R-Ub (K48R) mutant is greater for HJURP than for CENP-A WT in vitro (Niikura et al., 2015). Therefore, we hypothesized that overexpression of Flag-CENP-A-K124R-Ub (K48R) can induce neocentromere formation as Flag-CENP-A-K124R-Ub (K48R) recruits HJURP efficiently to noncentromere sites where Flag-CENP-A localizes, whereas overexpression of Flag-CENP-A WT recruits DAXX to noncentromere sites where Flag-CENP-A localizes, which is consistent with the results from the study by Lacoste et al. (Lacoste et al., 2014) (Figure 6E).
Figure 6. Functions of CENP-A Chaperons, HJURP and DAXX.
(A) Exogenously expressed Flag-CENP-A WT and Flag-CENP-A K124R-Ub (K48R) are incorporated predominantly by DAXX and HJURP, respectively, at ectopic noncentromeric sites. HeLa Tet-Off cells were cultured, transfected, and treated with nocodazole as described for Figure 5. Their chromosomes were examined after staining with antibodies against Flag (CENP-A), endogenous chaperone protein HJURP or DAXX, and CENP-B. DNA was costained with DAPI. Representative images including those of magnified paired sister chromatids are shown. Flag-K124R-Ub (K48R): pTRM4-Flag-CENP-A K124R-Ub (K48R), Flag-WT: pTRM4-Flag-CENP-A WT transfectant (Table S3). Scale bar, 2 µm.
(B) The average number (±SEM) of ectopic sites per cell (n ≥ 50 cells) was obtained: 1.48 for Flag-K124-Ub (K48R), 0.15 for Flag-WT (with HJURP); 0.34 for Flag-K124-Ub (K48R), 0.94 for Flag-WT (with DAXX) from the experiment performed in (A). ****P < 0.0001, **P < 0.01, and *P < 0.05 (Student’s t test).
(C) The in vivo immunoprecipitation assay was performed with constructs shown in the upper table. HeLa Tet-Off cells were cultured and transfected with pTRM4-Flag-CENP-A WT or pTRM4-Flag-CENP-A K124R-Ub (K48R), or pTRM4 vector (Table S3; see Immunoprecipitation Assay in Supplemental Experimental Procedures). Proteins in 3% of the total extracts (Input) and immunoprecipitates (IP) obtained by using ANTI-FLAG M2 Affinity Gel (SIGMA-ALDRICH) were detected by immunoblotting with the indicated antibodies. The level of endogenous HJURP protein was enhanced by Flag-CENP-A overexpression (input, anti-HJURP), as observed with CENP-A-Flag overexpression in Figure S3E.
(D) Histogram summarizing quantified bands of anti-HJURP and anti-DAXX shown in (C) (ratio of the intensity of each band signal to Flag band signal was normalized with the intensity of HJURP band signal of Flag-WT [2nd column from left]). Experiments were repeated (n ≥ 4 experiments), and the mean percentages (±SEM) are shown. ***P < 0.001, **P < 0.01, and n.s. (no significant) comparing the samples of Flag-K124R-Ub (K48R) and Flag-WT (Student’s t test).
(E) Models of neocentromere formation for Figures 5, 6A–6D, and S7. The model with the octameric nucleosome is adapted for simplicity (see Figure 7A for the tetramer model in which a similar concept is applied to a tetrameric nucleosome). (Panel [1]) Exogenously expressed Flag-CENP-A WT (nonubiquitylated) does not recruit HJURP but DAXX, and forms a heterodimer with new, endogenously expressed CENP-A (nonubiquitylated) at the proximity of ectopic nucleosome, and this heterodimer is incorporated by DAXX at the ectopic site; nevertheless, both monomers are not ubiquitylated as a result of the loss of the ubiquitylated CENP-A template. Therefore, the inactive nucleosome remains. (Panel [2]) Exogenously expressed Flag-CENP-A K124R-Ub (K48R) recruits HJURP and forms heterodimers, and this formation leads to ubiquitylation of new CENP-A by virtue of the ubiquitylated template: Flag-CENP-A K124R-Ub (K48R). This heterodimer is incorporated by HJURP; thus, a neocentromere is formed by the active CENP-A nucleosome at the ectopic site. Histone H4 is omitted from both models for simplicity.
Results of our immunoprecipitation assays using cell lysates showed that the affinity of Flag-CENP-A-K124R-Ub (K48R) with endogenous DAXX is lower than that of Flag-CENP-A WT (Figures 6C and 6D); this difference in affinity supports this model (Figure 6E). However, we found no significant difference between CENP-A WT and K124R-Ub (K48R) mutant regarding their affinity to endogenous HJURP (Figures 6C and 6D). We speculate that the CENP-A binding to HJURP may have been saturated in the immunoprecipitation experiments using the cell lysates. Otherwise, this might be because the binding of CENP-A with HJURP is not stable through the cell cycle (Bui et al., 2012; Foltz et al., 2009)
We also found that the fusion of monoubiquitin to the CENP-A K124R mutant significantly restored and enhanced the recruitment of CENP-A chaperons (HJURP and DAXX) and outer kinetochore proteins (HEC1 and SKA1) at LacO arrays (Figures S7C–S7F); this result seemed to be inconsistent with the results regarding neocentromeres in Figures 6A and 6B. However, the results of the LacO/LacI ectopic centromeric chromatin assembly assay should represent the status “after” CENP-A assembly into chromatin, because in this system HA-LacI-CENP-A WT and mutants are forcibly incorporated into chromatin containing LacO arrays. Therefore, we speculate that HJURP and DAXX function in two different ways before and after CENP-A deposition at the centromere. Indeed, a dual chaperone function of HJURP in coordinating CENP- A and CENP-C recruitment was recently proposed (Tachiwana et al., 2015). DAXX also may have a dual role, because heat shock increases the accumulation of DAXX at CEN/periCEN (Morozov et al., 2012) and DAXX interacts with CENP-C (Pluta et al., 1998).
The Epigenetic Mark of Centromere Identity
CENP-A has been proposed to be the epigenetic mark of the centromere identity (Karpen and Allshire, 1997) on the basis of the following findings: CENP-A is localized only at active centromeres on dicentric chromosomes (Earnshaw et al., 1989; Sullivan and Schwartz, 1995; Sullivan and Willard, 1998; Warburton et al., 1997), CENP-A can be experimentally mistargeted to noncentromeric regions of chromatin, and this mistargeting leads to the formation of ectopic centromeres in model organisms (Fukagawa and Earnshaw, 2014). However, in humans, we have shown that overexpression of CENP-A itself is not sufficient for the creation of a neocentromere at a noncentromeric region. Ubiquitylation of CENP-A is necessary for the formation of neocentromeres and for the epigenetic inheritance of the centromere location. Considering that histone posttranslational modifications are defined as “epigenetic marks” traditionally, we propose that CENP-A ubiquitylation is a candidate for the epigenetic mark of centromere location, i.e., the centromere identity.
CENP-A Overexpression and Cancer
Ectopic incorporation of overexpressed CENP-A can lead to genomic instability (Amato et al., 2009; Au et al., 2008; Heun et al., 2006; Van Hooser et al., 2001), which occurs in particularly aggressive cancer cells and tissues (Biermann et al., 2007; McGovern et al., 2012; Tomonaga et al., 2003; Vardabasso et al., 2014; Wu et al., 2012). In humans, CENP-A overexpression can lead to its ectopic localization to chromosome regions with active histone turnover, as seen in cancer cell lines (Lacoste et al., 2014; Tomonaga et al., 2003). At these ectopic loci, CENP-A forms heterotypic nucleosomes (CENP-A/H3.3) occluding CTCF binding, and their presence may increase DNA damage tolerance in cancer cells (Lacoste et al., 2014). Arimura et al. revealed a “hybrid” structure of the heterotypic CENP-A/H3.3 and suggested that the stable existence of the CENP-A/H3.3 nucleosome may cause ectopic kinetochore assembly (Arimura et al., 2014), which could lead to neocentromere formation and chromosome instability in cancer cells (Biermann et al., 2007; Lacoste et al., 2014; McGovern et al., 2012; Tomonaga et al., 2003; Wu et al., 2012). Our models of epigenetic inheritance of CENP-A ubiquitylation suggest that errors in CENP-A targeting, heterodimerization, and/or ubiquitylation induce abnormal accumulation of heterotypic nucleosomes (Figure 7B, bottom). Hence, our findings may provide a basis for potential insights into understanding the mechanisms of cancer development.
EXPERIMENTAL PROCEDURES
Antibodies and Reagents
Please see Table S1 for antibodies, Table S2 for siRNA sequences, Table S3 for plasmid vectors, and Table S4 for baculovirus extracts used in this study.
Cell Culture and Transfection
HeLa or HeLa Tet-Off cells (Clontech, Mountain View, CA) were cultured in high-glucose Dulbecco’s modified Eagle’s medium (BioWhittaker, Walkersville, MD) with 10% fetal bovine serum (FBS) (Invitrogen, Grand Island, NY). Cells were grown at 37°C in 5% CO2 in a humidified incubator. Cells were transfected with annealed double-stranded siRNA(s) or mammalian expression plasmids by using Lipofectamine 2000 (Invitrogen), Lipofectamine 3000 (Invitrogen), Lipofectamine LTX (Invitrogen), Lipofectamine RNAiMAX (Invitrogen), or linear polyethylenimine (PEI) (Reed et al., 2006). HeLa Tet-Off cells were cultured without tetracycline/doxycycline and transiently transfected with the pTRM4 overexpression vector whose transcription was regulated by the TRE promoter. In Figures 1B–1E (“control”), 3C, 3D, 4B–4D, 5, 6A, 6B, S1D (“control”), S1E (“control”), S1F (“control”), S3G, S4B, S4C, S5, and S6, cells were also cotransfected with CA-UTR #2 siRNAs (Table S2) for partial depletion of endogenous CENP-A, but this partial depletion did not disrupt endogenous CENP-C localization at centromeres (ca. 80% of endogenous CENP-A remained at 48 h after transfection; data not shown).
CENP-A−/F hTERT RPE1 cells were generously given Dr. Don W. Cleveland, Ludwig Institute for Cancer Research, Department of Cell and Molecular Medicine, University of California at San Diego, and cultured as described previously (Fachinetti et al., 2013). Retro-Cre was added to CENP-A−/F RPE1 cells infected with retrovirus produced by transient co-transfection of 293T cells with pBabe-puro-Cre, psPAX2, and pMD2.G (Table S3). Four days after retro-Cre infection (Figure S2A), cells were further infected with retrovirus produced by stable Plat-GP cells (Cell Biolabs, Inc) with pQCXIP-Flag-CENP-A or transient cotransfection of Plat-GP cells with the indicated pQCXIP constructs and pCMV-VSG-G (Table S3). For in vivo ubiquitylation assay of CENP-A−/F RPE1 cells, pCGN-HA-Ubiquitin was also transfected by using Fugene HD (Promega) 4 days after retro-Cre infection (Figure S2A; see also CENP-A In Vivo Ubquitylation Assay). Cells were collected or fixed 6 days after retro-Cre infection and used in each analysis.
Taxol or TN16 treatment was performed as described previously (Jansen et al., 2007; Niikura et al., 2015; Perpelescu et al., 2009). For indirect immunofluorescent staining of mitotic CENP-A−/F RPE1 cells, Taxol (10 nM) or TN16 (0.5 µM) was added 24 h or 2.5 h before cell fixation, respectively.
CENP-A In Vivo Ubiquitylation Assay
The in vivo CENP-A ubiquitylation assay was performed as described previously with the following minor modifications (Niikura et al., 2015). HeLa Tet-Off cells were transfected with the indicated expression vectors (see Cell Culture and Transfection) and incubated with 5 µM MG132 (CALBIOCHEM) for 24 h. Cells were then collected and lysed in denaturing buffer A1 (20 mM Tris-HCl pH 7.4, 50 mM NaCl, 0.5% Nonidet P-40, 0.5% deoxycholate, 0.5% SDS, 1 mM EDTA, 50 µM MG132, and complete EDTA-free protease inhibitor cocktail [Roche]) by a sonication and freeze-thaw process. Proteins were immunoprecipitated, and the immunoprecipitates underwent Western blot analysis with the indicated antibodies.
For CENP-A−/F RPE1 cells, the experiment was performed according to the time-course scheme in Figure S2A. Cells were infected with retroviruses harboring the indicated vector constructs, cultured (see Cell Culture and Transfection in Experimental Procedures), collected 6 days after retro-Cre infection, lysed, and analyzed as described for the HeLa Tet-Off cells.
Supplementary Material
Acknowledgments
The authors thank Yue Xiong, Beezly Groh, Vivien Measday, Iain Cheeseman, Lars E.T. Jansen, Paul S. Maddox, Dorota Skowyra, Yoshinori Watanabe, Tatsuo Fukugawa, and past and current researchers at The Research Institute at Nationwide Children’s Hospital and St. Jude Children’s Research Hospital for their helpful discussion, experimental guidance, and reagents; and Don W. Cleveland, Daniele Fachinetti, Daniel Foltz, Gustavo W. Leone, John Thompson, Lawrence S. Kirschner, Amruta Ashtekar, Hitoshi Kurumizaka, Ben E. Black, and Glennis A. Logston for their generous gifts of reagents. This study was supported by NIH grants GM68418 and CancerFree KIDS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental information is linked to the online version of the paper at http://www.cell.com/cell-reports/home
Supplemental Experimental Procedures
Supplemental Tables S1 to S4
Supplemental References
Supplemental Figure Legends
Supplemental Figures S1 to S7
Author Contributions
Conceptualization and Methodology, Y.N. and K.K.; Investigation, Y.N. and R.K.; Writing – Original Draft, Y.N. and K.K.; Writing – Review & Editing, Y.N. and K.K.; Funding Acquisition and Supervision, K.K.
References
- Amato A, Schillaci T, Lentini L, Di Leonardo A. CENPA overexpression promotes genome instability in pRb-depleted human cells. Mol Cancer. 2009;8:119. doi: 10.1186/1476-4598-8-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimura Y, Shirayama K, Horikoshi N, Fujita R, Taguchi H, Kagawa W, Fukagawa T, Almouzni G, Kurumizaka H. Crystal structure and stable property of the cancer-associated heterotypic nucleosome containing CENP-A and H3.3. Sci Rep. 2014;4:7115. doi: 10.1038/srep07115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au WC, Crisp MJ, DeLuca SZ, Rando OJ, Basrai MA. Altered dosage and mislocalization of histone H3 and Cse4p lead to chromosome loss in Saccharomyces cerevisiae. Genetics. 2008;179:263–275. doi: 10.1534/genetics.108.088518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bade D, Pauleau AL, Wendler A, Erhardt S. The E3 ligase CUL3/RDX controls centromere maintenance by ubiquitylating and stabilizing CENP-A in a CAL1-dependent manner. Dev Cell. 2014;28:508–519. doi: 10.1016/j.devcel.2014.01.031. [DOI] [PubMed] [Google Scholar]
- Barnhart MC, Kuich PH, Stellfox ME, Ward JA, Bassett EA, Black BE, Foltz DR. HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. J Cell Biol. 2011;194:229–243. doi: 10.1083/jcb.201012017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett EA, DeNizio J, Barnhart-Dailey MC, Panchenko T, Sekulic N, Rogers DJ, Foltz DR, Black BE. HJURP uses distinct CENP-A surfaces to recognize and to stabilize CENP-A/histone H4 for centromere assembly. Dev Cell. 2012;22:749–762. doi: 10.1016/j.devcel.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biermann K, Heukamp LC, Steger K, Zhou H, Franke FE, Guetgemann I, Sonnack V, Brehm R, Berg J, Bastian PJ, et al. Gene expression profiling identifies new biological markers of neoplastic germ cells. Anticancer Res. 2007;27:3091–3100. [PubMed] [Google Scholar]
- Bui M, Dimitriadis EK, Hoischen C, An E, Quenet D, Giebe S, Nita-Lazar A, Diekmann S, Dalal Y. Cell-Cycle-Dependent Structural Transitions in the Human CENP-A Nucleosome In Vivo. Cell. 2012;150:317–326. doi: 10.1016/j.cell.2012.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunleavy EM, Almouzni G, Karpen GH. H3.3 is deposited at centromeres in S phase as a placeholder for newly assembled CENP-A in G(1) phase. Nucleus. 2011;2:146–157. doi: 10.4161/nucl.2.2.15211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, Daigo Y, Nakatani Y, Almouzni-Pettinotti G. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 2009;137:485–497. doi: 10.1016/j.cell.2009.02.040. [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, Ratrie H, 3rd, Stetten G. Visualization of centromere proteins CENP-B and CENP-C on a stable dicentric chromosome in cytological spreads. Chromosoma. 1989;98:1–12. doi: 10.1007/BF00293329. [DOI] [PubMed] [Google Scholar]
- Fachinetti D, Folco HD, Nechemia-Arbely Y, Valente LP, Nguyen K, Wong AJ, Zhu Q, Holland AJ, Desai A, Jansen LE, et al. A two-step mechanism for epigenetic specification of centromere identity and function. Nat Cell Biol. 2013;15:1056–1066. doi: 10.1038/ncb2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz DR, Jansen LE, Bailey AO, Yates JR, 3rd, Bassett EA, Wood S, Black BE, Cleveland DW. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell. 2009;137:472–484. doi: 10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukagawa T, Earnshaw WC. The centromere: chromatin foundation for the kinetochore machinery. Dev Cell. 2014;30:496–508. doi: 10.1016/j.devcel.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann R, Rechtsteiner A, Yuen KW, Muroyama A, Egelhofer T, Gaydos L, Barron F, Maddox P, Essex A, Monen J, et al. An inverse relationship to germline transcription defines centromeric chromatin in C. elegans. Nature. 2012;484:534–537. doi: 10.1038/nature10973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanisch A, Sillje HH, Nigg EA. Timely anaphase onset requires a novel spindle and kinetochore complex comprising Ska1 and Ska2. EMBO J. 2006;25:5504–5515. doi: 10.1038/sj.emboj.7601426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasson D, Panchenko T, Salimian KJ, Salman MU, Sekulic N, Alonso A, Warburton PE, Black BE. The octamer is the major form of CENP-A nucleosomes at human centromeres. Nat Struct Mol Biol. 2013;20:687–695. doi: 10.1038/nsmb.2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heun P, Erhardt S, Blower MD, Weiss S, Skora AD, Karpen GH. Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Dev Cell. 2006;10:303–315. doi: 10.1016/j.devcel.2006.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howman EV, Fowler KJ, Newson AJ, Redward S, MacDonald AC, Kalitsis P, Choo KH. Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. Proc Natl Acad Sci U S A. 2000;97:1148–1153. doi: 10.1073/pnas.97.3.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingouff M, Rademacher S, Holec S, Soljic L, Xin N, Readshaw A, Foo SH, Lahouze B, Sprunck S, Berger F. Zygotic resetting of the HISTONE 3 variant repertoire participates in epigenetic reprogramming in Arabidopsis. Curr Biol. 2010;20:2137–2143. doi: 10.1016/j.cub.2010.11.012. [DOI] [PubMed] [Google Scholar]
- Janicki SM, Tsukamoto T, Salghetti SE, Tansey WP, Sachidanandam R, Prasanth KV, Ried T, Shav-Tal Y, Bertrand E, Singer RH, et al. From silencing to gene expression: real-time analysis in single cells. Cell. 2004;116:683–698. doi: 10.1016/s0092-8674(04)00171-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen LE, Black BE, Foltz DR, Cleveland DW. Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol. 2007;176:795–805. doi: 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpen GH, Allshire RC. The case for epigenetic effects on centromere identity and function. Trends Genet. 1997;13:489–496. doi: 10.1016/s0168-9525(97)01298-5. [DOI] [PubMed] [Google Scholar]
- Lacoste N, Woolfe A, Tachiwana H, Garea AV, Barth T, Cantaloube S, Kurumizaka H, Imhof A, Almouzni G. Mislocalization of the centromeric histone variant CenH3/CENP-A in human cells depends on the chaperone DAXX. Mol Cell. 2014;53:631–644. doi: 10.1016/j.molcel.2014.01.018. [DOI] [PubMed] [Google Scholar]
- Li Y, Zhu Z, Zhang S, Yu D, Yu H, Liu L, Cao X, Wang L, Gao H, Zhu M. ShRNA-targeted centromere protein A inhibits hepatocellular carcinoma growth. PLoS One. 2011;6:e17794. doi: 10.1371/journal.pone.0017794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YT, Chen Y, Wu G, Lee WH. Hec1 sequentially recruits Zwint-1 and ZW10 to kinetochores for faithful chromosome segregation and spindle checkpoint control. Oncogene. 2006;25:6901–6914. doi: 10.1038/sj.onc.1209687. [DOI] [PubMed] [Google Scholar]
- Liu ST, Rattner JB, Jablonski SA, Yen TJ. Mapping the assembly pathways that specify formation of the trilaminar kinetochore plates in human cells. J Cell Biol. 2006;175:41–53. doi: 10.1083/jcb.200606020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logsdon GA, Barrey EJ, Bassett EA, DeNizio JE, Guo LY, Panchenko T, Dawicki-McKenna JM, Heun P, Black BE. Both tails and the centromere targeting domain of CENP-A are required for centromere establishment. J Cell Biol. 2015;208:521–531. doi: 10.1083/jcb.201412011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall OJ, Chueh AC, Wong LH, Choo KH. Neocentromeres: new insights into centromere structure, disease development, and karyotype evolution. Am J Hum Genet. 2008;82:261–282. doi: 10.1016/j.ajhg.2007.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGovern SL, Qi Y, Pusztai L, Symmans WF, Buchholz TA. Centromere protein-A, an essential centromere protein, is a prognostic marker for relapse in estrogen receptor-positive breast cancer. Breast Cancer Res. 2012;14:R72. doi: 10.1186/bcr3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendiburo MJ, Padeken J, Fulop S, Schepers A, Heun P. Drosophila CENH3 is sufficient for centromere formation. Science. 2011;334:686–690. doi: 10.1126/science.1206880. [DOI] [PubMed] [Google Scholar]
- Miell MD, Fuller CJ, Guse A, Barysz HM, Downes A, Owen-Hughes T, Rappsilber J, Straight AF, Allshire RC. CENP-A confers a reduction in height on octameric nucleosomes. Nat Struct Mol Biol. 2013;20:763–765. doi: 10.1038/nsmb.2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monen J, Maddox PS, Hyndman F, Oegema K, Desai A. Differential role of CENP-A in the segregation of holocentric C. elegans chromosomes during meiosis and mitosis. Nat Cell Biol. 2005;7:1248–1255. doi: 10.1038/ncb1331. [DOI] [PubMed] [Google Scholar]
- Morozov VM, Gavrilova EV, Ogryzko VV, Ishov AM. Dualistic function of Daxx at centromeric and pericentromeric heterochromatin in normal and stress conditions. Nucleus. 2012;3:276–285. doi: 10.4161/nucl.20180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S, Montes de Oca R, Lacoste N, Dingli F, Loew D, Almouzni G. Phosphorylation and DNA binding of HJURP determine its centromeric recruitment and function in CenH3(CENP-A) loading. Cell Rep. 2014;8:190–203. doi: 10.1016/j.celrep.2014.06.002. [DOI] [PubMed] [Google Scholar]
- Niikura Y, Kitagawa R, Ogi H, Abdulle R, Pagala V, Kitagawa K. CENP-A K124 Ubiquitylation Is Required for CENP-A Deposition at the Centromere. Dev Cell. 2015 doi: 10.1016/j.devcel.2015.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padeganeh A, Ryan J, Boisvert J, Ladouceur AM, Dorn JF, Maddox PS. Octameric CENP-A nucleosomes are present at human centromeres throughout the cell cycle. Curr Biol. 2013;23:764–769. doi: 10.1016/j.cub.2013.03.037. [DOI] [PubMed] [Google Scholar]
- Perpelescu M, Nozaki N, Obuse C, Yang H, Yoda K. Active establishment of centromeric CENP-A chromatin by RSF complex. The Journal of cell biology. 2009;185:397–407. doi: 10.1083/jcb.200903088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluta AF, Earnshaw WC, Goldberg IG. Interphase-specific association of intrinsic centromere protein CENP-C with HDaxx, a death domain-binding protein implicated in Fas-mediated cell death. J Cell Sci. 1998;111(Pt 14):2029–2041. doi: 10.1242/jcs.111.14.2029. [DOI] [PubMed] [Google Scholar]
- Raychaudhuri N, Dubruille R, Orsi GA, Bagheri HC, Loppin B, Lehner CF. Transgenerational propagation and quantitative maintenance of paternal centromeres depends on cid/cenp-a presence in Drosophila sperm. PLoS Biol. 2012;10:e1001434. doi: 10.1371/journal.pbio.1001434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed SE, Staley EM, Mayginnes JP, Pintel DJ, Tullis GE. Transfection of mammalian cells using linear polyethylenimine is a simple and effective means of producing recombinant adeno-associated virus vectors. J Virol Methods. 2006;138:85–98. doi: 10.1016/j.jviromet.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Schmidt JC, Arthanari H, Boeszoermenyi A, Dashkevich NM, Wilson-Kubalek EM, Monnier N, Markus M, Oberer M, Milligan RA, Bathe M, et al. The kinetochore-bound Ska1 complex tracks depolymerizing microtubules and binds to curved protofilaments. Dev Cell. 2012;23:968–980. doi: 10.1016/j.devcel.2012.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang WH, Hori T, Martins NM, Toyoda A, Misu S, Monma N, Hiratani I, Maeshima K, Ikeo K, Fujiyama A, et al. Chromosome engineering allows the efficient isolation of vertebrate neocentromeres. Dev Cell. 2013;24:635–648. doi: 10.1016/j.devcel.2013.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelby RD, Vafa O, Sullivan KF. Assembly of CENP-A into centromeric chromatin requires a cooperative array of nucleosomal DNA contact sites. J Cell Biol. 1997;136:501–513. doi: 10.1083/jcb.136.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan BA, Schwartz S. Identification of centromeric antigens in dicentric Robertsonian translocations: CENP-C and CENP-E are necessary components of functional centromeres. Hum Mol Genet. 1995;4:2189–2197. doi: 10.1093/hmg/4.12.2189. [DOI] [PubMed] [Google Scholar]
- Sullivan BA, Willard HF. Stable dicentric X chromosomes with two functional centromeres. Nat Genet. 1998;20:227–228. doi: 10.1038/3024. [DOI] [PubMed] [Google Scholar]
- Tachiwana H, Kagawa W, Shiga T, Osakabe A, Miya Y, Saito K, Hayashi-Takanaka Y, Oda T, Sato M, Park SY, et al. Crystal structure of the human centromeric nucleosome containing CENP-A. Nature. 2011;476:232–235. doi: 10.1038/nature10258. [DOI] [PubMed] [Google Scholar]
- Tachiwana H, Muller S, Blumer J, Klare K, Musacchio A, Almouzni G. HJURP involvement in de novo CenH3(CENP-A) and CENP-C recruitment. Cell Rep. 2015;11:22–32. doi: 10.1016/j.celrep.2015.03.013. [DOI] [PubMed] [Google Scholar]
- Tomonaga T, Matsushita K, Yamaguchi S, Oohashi T, Shimada H, Ochiai T, Yoda K, Nomura F. Overexpression and mistargeting of centromere protein-A in human primary colorectal cancer. Cancer Res. 2003;63:3511–3516. [PubMed] [Google Scholar]
- Van Hooser AA, Ouspenski II, Gregson HC, Starr DA, Yen TJ, Goldberg ML, Yokomori K, Earnshaw WC, Sullivan KF, Brinkley BR. Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J Cell Sci. 2001;114:3529–3542. doi: 10.1242/jcs.114.19.3529. [DOI] [PubMed] [Google Scholar]
- Vardabasso C, Hasson D, Ratnakumar K, Chung CY, Duarte LF, Bernstein E. Histone variants: emerging players in cancer biology. Cell Mol Life Sci. 2014;71:379–404. doi: 10.1007/s00018-013-1343-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissel B, Choo KH. Human alpha satellite DNA--consensus sequence and conserved regions. Nucleic Acids Res. 1987;15:6751–6752. doi: 10.1093/nar/15.16.6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Basler CF, Williams BR, Silverman RH, Palese P, Garcia-Sastre A. Functional replacement of the carboxy-terminal two-thirds of the influenza A virus NS1 protein with short heterologous dimerization domains. J Virol. 2002;76:12951–12962. doi: 10.1128/JVI.76.24.12951-12962.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton PE, Cooke CA, Bourassa S, Vafa O, Sullivan BA, Stetten G, Gimelli G, Warburton D, Tyler-Smith C, Sullivan KF, et al. Immunolocalization of CENP-A suggests a distinct nucleosome structure at the inner kinetochore plate of active centromeres. Curr Biol. 1997;7:901–904. doi: 10.1016/s0960-9822(06)00382-4. [DOI] [PubMed] [Google Scholar]
- Wu Q, Qian YM, Zhao XL, Wang SM, Feng XJ, Chen XF, Zhang SH. Expression and prognostic significance of centromere protein A in human lung adenocarcinoma. Lung Cancer. 2012;77:407–414. doi: 10.1016/j.lungcan.2012.04.007. [DOI] [PubMed] [Google Scholar]
- Yoda K, Ando S, Morishita S, Houmura K, Hashimoto K, Takeyasu K, Okazaki T. Human centromere protein A (CENP-A) can replace histone H3 in nucleosome reconstitution in vitro. Proc Natl Acad Sci U S A. 2000;97:7266–7271. doi: 10.1073/pnas.130189697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zasadzinska E, Barnhart-Dailey MC, Kuich PH, Foltz DR. Dimerization of the CENP-A assembly factor HJURP is required for centromeric nucleosome deposition. EMBO J. 2013;32:2113–2124. doi: 10.1038/emboj.2013.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Colmenares SU, Karpen GH. Assembly of Drosophila centromeric nucleosomes requires CID dimerization. Mol Cell. 2012;45:263–269. doi: 10.1016/j.molcel.2011.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.