Abstract
The nuclear pore complex (NPC) constitutes the sole gateway for bidirectional nucleocytoplasmic transport. We present the reconstitution and interdisciplinary analyses of the ~425-kDa inner ring complex (IRC), which forms the central transport channel and diffusion barrier of the NPC, revealing its interaction network and equimolar stoichiometry. The Nsp1•Nup49•Nup57 channel nucleoporin hetero-trimer (CNT) attaches to the IRC solely through the adaptor nucleoporin Nic96. The CNT•Nic96 structure reveals that Nic96 functions as an assembly sensor that recognizes the three dimensional architecture of the CNT, thereby mediating the incorporation of a defined CNT state into the NPC. We propose that the IRC adopts a relatively rigid scaffold that recruits the CNT to primarily form the diffusion barrier of the NPC, rather than enabling channel dilation.
One of the great hallmarks of eukaryotic evolution is the enclosure of genetic information in the nucleus. The spatial segregation of replication and transcription in the nucleus from translation in the cytoplasm imposes the requirement of transporting thousands of macromolecules between these two compartments. Nuclear pore complexes (NPCs) are massive transport channels that allow bidirectional macromolecular exchange across the nuclear envelope (NE) and thus function as key regulators of the flow of genetic information from DNA to RNA to protein (1).
NPCs are formed by multiple copies of ~34 distinct proteins, termed nucleoporins (nups) (1). The docking of the yeast coat nup complex (CNC) crystal structure into a cryo-electron tomographic (ET) reconstruction of the intact human NPC revealed its organization into two sixteen-membered CNC rings on the nuclear and cytoplasmic faces of ~100 nm NE pores (Fig. 1A) (2, 3). This arrangement established that the doughnut-shaped inner ring is composed of adaptor and channel nups (3).
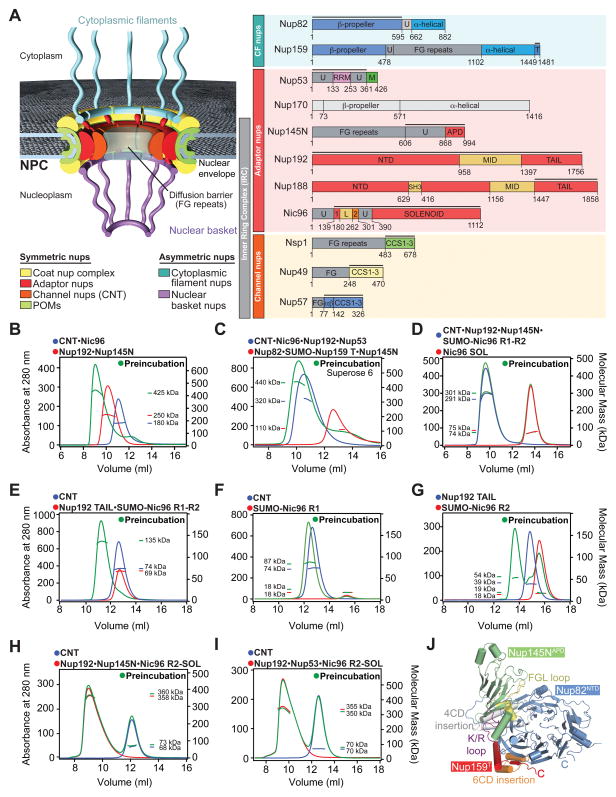
Fig. 1. Reconstitution and Dissection of the IRC.
(A) Cross-sectional schematic representation of the NPC and domain structures of the Chaetomium thermophilum nucleoporins (nups). Black lines indicate regions used for reconstitution. U, unstructured; T, TAIL; NTD, N-terminal domain; MID, middle domain; SH3, Src-homology 3-like domain; L, linker domain; APD, auto-proteolytic domain; RRM, RNA recognition motif; M, membrane-binding motif; FG repeats, phenylalanine-glycine repeats; CCS, coiled-coil segment; α/β, α/β insertion domain; IRC, inner ring complex. (B to I) Pair-wise biochemical interaction analyses of various reconstituted nup complexes. Size-exclusion chromatography coupled to multiangle light scattering (SEC-MALS) profiles of nup or nup complexes are shown individually (red and blue) and after their preincubation (green). Measured molecular masses are indicated for the peak fractions. (J) Structure of the Nup82NTD•Nup159T•Nup145NAPD cytoplasmic filament nup complex (CFC) is shown in a cartoon representation. Nup145NAPD (green), Nup159T (red), and Nup82NTD (blue), the Nup82NTD helical 4CD (gray) and 6CD (orange) insertions and FGL loop (yellow), and the conserved Nup145NAPD K/R loop (purple) are illustrated.
Anchoring of the inner ring in the NE pore is mediated by the membrane recruitment complex, composed of adaptor nups Nup53 and Nup170, and transmembrane nup Ndc1 (1, 4–6). Recently, Nup53 was shown to harbor distinct binding sites for Nic96, Nup170 and Nup192, allowing the Nup53•Nup170 hetero-dimer to interact with either Nic96•Nup192 or Nic96•Nup188 (7, 8).
The inner ring harbors the central transport channel and diffusion barrier of the NPC, preventing macromolecules larger than ~40 kDa to freely diffuse across the NE (1, 9). The channel nups Nsp1, Nup49, and Nup57 constitute part of the central transport channel and form the diffusion barrier with their disordered phenylalanine-glycine (FG) repeats (1, 9–11). Transport factors ferry cargo across the NE by binding to FG repeats in the central transport channel (1, 9). Furthermore, the central transport channel seems to be permanently occupied by transport factors even after biochemical purification of NPCs (10, 12). In addition to their N-terminal FG repeats, the three channel nups are predicted to possess C-terminal coiled-coil regions (13, 14). The knockout of any of the three channel nups is lethal in S. cerevisiae, whereas the deletion of any two N-terminal FG-repeat regions can be tolerated, but reduces transport rates, suggesting an essential function of the coiled-coil regions (13, 15–19).
Arguably, the most important questions about the inner ring architecture pertain to the recruitment and positioning of the channel nups, because of their essential function in forming the diffusion barrier and providing binding sites for transport factors. Native purifications from S. cerevisiae and mammalian cells showed that the channel nups co-purified with Nic96, suggesting the evolutionary conservation of a hetero-tetrameric Nsp1•Nup49•Nup57•Nic96 complex (13, 20, 21). Subsequent biochemical reconstitution attempts yielded channel nup complexes with inconsistent stoichiometries that resisted structural analysis (22–24). Reconstitutions employing channel nup fragments revealed dynamic interactions and generated a series of crystal structures with various homomeric and heteromeric assembly states (22, 25, 26). Biochemical analysis of these structures led to heavily contested models that have resisted physiological validation, including the proposal that the reversible karyopherin-mediated transition between homo- and hetero-oligomers facilitates the constriction and dilation of the central transport channel (25–28).
Despite more than half a century of research, our understanding of the inner ring architecture remains rudimentary. The prevalent assumption is that the inner ring of the NPC is composed of multiple copies of a single NPC subcomplex, but such a complex has remained elusive. Here, we present an in-depth characterization of the NPC’s inner ring. Starting with the reconstitution of a ~425 kDa hetero-hexameric inner ring complex (IRC), we demonstrate its scaffolding function by showing that it interacts with additional peripheral nups. In dissecting the underlying protein interaction network of the IRC, we determined an equimolar stoichiometry for its six components and identified Nic96 as the sole NPC attachment site for the channel nup hetero-trimer (CNT). Nic96 is essential for CNT recruitment and, through its interaction with Nup192, for proper CNT positioning within the inner ring scaffold in vivo. Structural and functional analyses of the intact CNT reveal a defined coiled-coil domain architecture that is specifically recognized by Nic96. Our results differ dramatically from previous characterizations of channel nup fragments and an associated model for a flexible transport channel that is capable of constriction and dilation of up to ~400 Å (25–28). We propose a model for the inner ring architecture in which sixteen copies of the IRC form a relatively rigid scaffold. In the proposed arrangement, the central transport channel would be filled with the channel nup FG repeats to establish the diffusion barrier of the NPC and to provide binding sites for cargo•transport factor complexes.
Reconstitution and dissection of the inner ring complex (IRC) and binding of peripheral nups
To reconstitute and uncover the architectural principles of a putative inner ring complex (IRC), we developed expression and purification protocols for the channel, adaptor, and cytoplasmic filament nups from C. thermophilum on the milligram scale. All purified nups lacked FG-repeat regions to facilitate soluble protein expression (Fig. 1A). We refer to all nups in the remainder of the text according to the C. thermophilum nomenclature and indicate nups from other species with a prefix.
Using recombinant purified nups, we reconstituted a monodisperse hetero-hexameric IRC containing the adaptor nups Nup192, Nic96 and Nup145N, and the Nsp1•Nup49•Nup57 channel nup hetero-trimer (CNT) (Fig. 1, A and B and fig. S1A). The ~425 kDa measured molecular mass of the IRC is consistent with an equimolar stoichiometry (Table S1). No higher-order oligomers formed at concentrations up to ~5 μM. Our reconstitution omitted Nup170 because of its low solubility in standard buffer conditions. However, the interaction of Nup170 with a C-terminal region of Nup53, directly adjacent to the membrane-binding motif, established its proximity to the pore membrane (7).
Further analyses showed that the reconstituted IRC is a bona fide NPC scaffold complex, capable of binding the membrane recruiting Nup53, and the peripheral cytoplasmic filament complex (CFC), forming a stable hetero-nonamer (Fig. 1C and fig. S1B). The measured masses for both complexes, ~100 kDa lower than expected, indicated their dynamic nature at the tested concentrations. The reconstitution and structure determination of a core IRC-CFC attachment complex composed of the CFC nups Nup82NTD and Nup159T and the autoproteolytic domain (APD) of the IRC component Nup145N revealed the molecular details of this inter-subcomplex linkage (Fig. 1J and figs. S2, A and B and Table S2). A structural comparison with its S. cerevisiae homolog showed that the interactions of the core CFC•Nup145NAPD complex are conserved, despite low sequence conservation (29, 30). This analysis supports the hypothesis that nup interactions in the NPC are evolutionary conserved (fig. S3).
Because Nup145N acts as an adaptor that attaches the CFC, we probed its interaction with the other IRC components. Nup145N formed a stoichiometric complex with Nup192, elicited weak CNT association, and barely interacted with the Nic96 solenoid (SOL) (fig. S2, C to E). However, Nup145N was incapable of linking the CNT to Nup192, suggesting mutually exclusive interactions (fig. S2F).
The IRC reconstitution enabled us to identify how the CNT is incorporated into the IRC by defining a minimal CNT attachment complex. We found that Nup145N, Nic96SOL and the N-terminal and middle domains of Nup192 were dispensable for CNT attachment, because a minimal stoichiometric Nup192TAIL•Nic96R1-R2•CNT attachment complex could be reconstituted (Fig. 1D and E and figs. S1, C and D). Nic96R1-R2 is an adaptor linking the CNT with Nup192TAIL (Fig. 1, F and G and figs. S1, E and F, and S4, A and B). Consistently, no interaction between Nup192TAIL and the CNT was observed (fig. S4C). Most importantly, an IRC lacking Nic96R1 failed to incorporate the CNT, demonstrating that Nic96R1 is the sole IRC attachment site for the CNT (Fig. 1H and fig. S1G). We tested whether Nup53 could rescue the deletion of Nic96R1 by providing an additional CNT binding site. However, the CNT did not incorporate into the IRC in the presence of Nup53 (Fig. 1I and fig. S1H).
These data established an equimolar stoichiometry of the six different IRC components and demonstrated that the CNT is solely attached to the adaptor nups through its interaction with Nic96R1.
The Nic96-Nup192 interaction positions the CNT in the central transport channel
Consistent with previous findings that Nup192 and Nup188 form mutually exclusive complexes (7), we were able to reconstitute an architecturally equivalent Nup188TAIL•Nic96R1-R2•CNT hetero-pentamer (fig. S5, A and B). However, salt stability and competition experiments showed that Nup192TAIL interacts more tightly with Nic96R2 than Nup188TAIL does (figs. S5, C and D and S6, A and B).
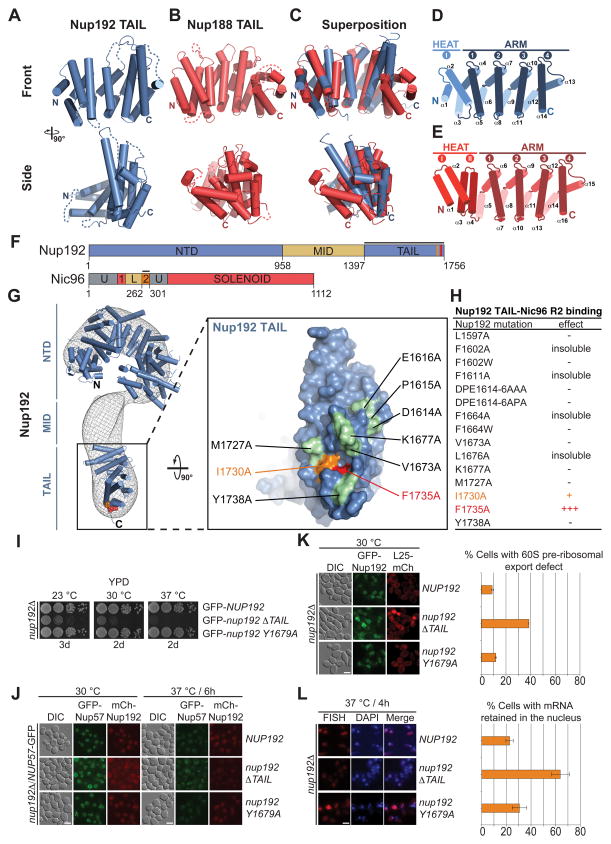
To gain insight into the molecular details of the Nic96R2 interaction with Nup192 and Nup188, we determined the crystal structures of the TAIL domains of Nup192 and Nup188 (Table S2). Nup192TAIL and Nup188TAIL share a similar crescent-shaped architecture, composed of ARM and HEAT repeats with overall similar surface conservation and electrostatic properties. However, Nup188TAIL contains an additional evolutionarily conserved C-terminal ARM repeat (Fig. 2A–E and figs. S7 to S11) (31). When docked into the electron microscopy (EM) reconstruction of S. cerevisiae Nup192, Nup192TAIL was located at the bottom of the question mark-shaped map (Fig. 2G) (32). Based on the structural and biochemical results and previous findings that Nup192, but not Nup188, is essential for viability in yeast, we focused our further analyses on the IRC containing Nup192 (33, 34).
Fig. 2. Structural and functional analyses of the TAIL domains of Nup192 and Nup188.
Cartoon representations of (A) Nup192TAIL (blue), (B) Nup188TAIL (red), and (C) a superposition of the two structures is shown in two different orientations. (D, E) Schematic representation of Nup192TAIL and Nup188TAIL structures. Positions of HEAT and ARM repeats are indicated. (F) Domain structures of Nup192 and Nic96 are shown; black bars indicate fragments used for interaction studies in (H). (G) Docking of Nup192TAIL and the previously determined Nup192NTD crystal structure into the yeast Nup192 EM envelope (8, 32). The Inset illustrates the position of Nup192TAIL and is expanded on the right, rotated by 90º. Surface representation of Nup192TAIL with the location of the analyzed mutations and their effect on Nic96R2 binding indicated; no effect (green), decreased binding (orange), abolished binding (red). (H) Summary of tested Nup192TAIL mutants and their effect on SUMO-Nic96R2 binding; (-) no effect, (+) decreased binding, (+++) abolished binding. (I) Growth analysis of S. cerevisiae strains carrying the indicated GFP-NUP192 variants. Serial dilutions of the respective cells were spotted onto YPD plates and grown for 2–3 days. (J) Subcellular localization of mCherry-Nup192 variants (red) and Nup57-GFP (green) in a nup192Δ̃/NUP57-GFPstrain. (K) Subcellular localization of the 60S ribosomal export reporter Rpl25-mCherry (red) and GFP-tagged Nup192 variants (green) in a nup192Δ strain. Representative images and quantification of nuclear Rpl25-mCherry retention are shown on the right. (L) mRNA export assay in a nup192Δ strain carrying GFP-NUP192 variants. Representative images and quantification of nuclear poly(A)+ RNA retention are shown. Cells were analyzed at the indicated temperatures and incubation times. Error bars represent the standard deviation. Scalebars are 5 μm.
To identify the Nic96R2 interaction surface in Nup192TAIL, we tested alanine mutants of 15 conserved surface residues and identified two adjacent residues, Phe1735 and Ile1730, that abolished binding and decreased the interaction with Nic96R2, respectively (Fig. 2F to H and figs. S12 and S13). Both residues are located in a hydrophobic pocket at the bottom of the Nup192 molecule (Fig. 2G).
To evaluate the physiological relevance of the identified Nup192-Nic96 interaction for NPC function, we analyzed the identified Nup192 mutants in S. cerevisiae. Whereas the F1735A point mutant (Y1679A in yeast) displayed no significant defects in growth and ribosomal or mRNA export, the removal of the entire TAIL domain resulted in a substantial growth defect at all temperatures, and substantial mRNA and ribosomal export defects, at 30 ºC and 37 ºC, respectively (Fig. 2I, K and L). Despite the severity of the ΔTAIL phenotypes, Nup57-GFP yielded strong NE staining (Fig. 2J), consistent with our biochemical analyses that Nup192TAIL is expendable for CNT attachment.
These data established that the Nic96-Nup192 interaction is dispensable for CNT incorporation into the NPC, but required for proper NPC function, suggesting that the Nic96-Nup192 interaction is required for correct CNT positioning, and, in turn, for the proper placement of the FG-repeat meshwork.
Nic96 is the sole NPC attachment site for the CNT in vivo
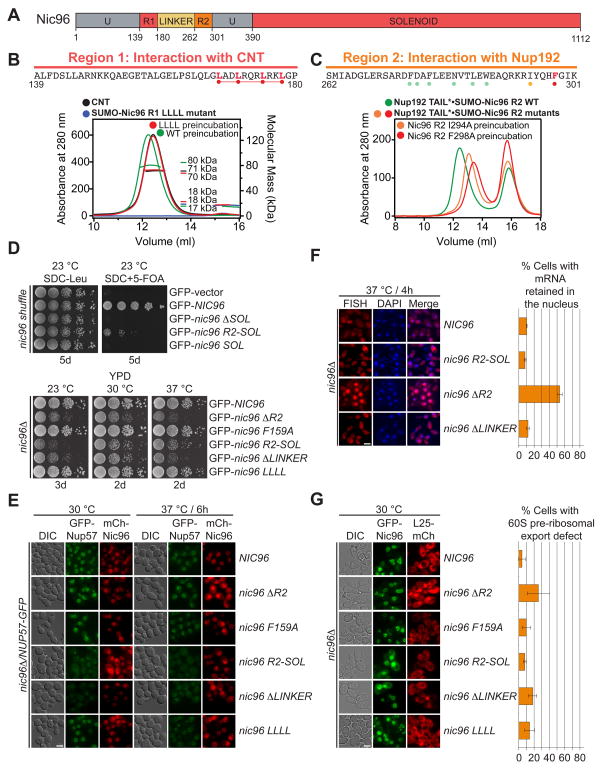
To characterize the adaptor function of Nic96, we carried out a mutational analysis of the conserved R1 and R2 regions of Nic96, which interact with the CNT and Nup192, respectively (Fig. 3, A to C and fig. S13). Because the Nic96R1-CNT interaction is stable at increased salt concentrations but sensitive to C-terminal truncations, we focused our analysis on conserved hydrophobic residues in the C-terminal region of Nic96R1 (figs. S14 and S15A). Indeed, mutating four leucines located at the C-terminal end of R1 to alanines (LLLL mutant) abolished the Nic96R1-CNT interaction (Fig. 3B and figs. S13 and S15B).
Fig. 3. Nic96 is an adaptor protein that attaches the CNT to Nup192.
(A) Domain structure of Nic96 is shown. R1, region 1; R2, region 2. (B) The positions of the mutated leucine residues of the LLLL mutant are indicated below the primary Nic96R1 sequence. SEC-MALS profiles of CNT (black), Nic96R1 LLLL mutant (blue) and after preincubation of the CNT with wild type Nic96R1 (green) or Nic96R1 LLLL (red). Measured molecular masses are indicated for the peak fractions. (C) The positions of the mutated residues are indicated below the primary Nic96R2 sequence and colored according to their effect on Nup192TAIL binding; no effect (green), mild effect (orange), abolished binding (red). SEC profiles of Nup192TAIL preincubated with wild type Nic96R2 (green), Nic96R2 I294A (orange), and Nic96R2 F298A (red). (D) Growth analysis of S. cerevisiae strains carrying the indicated GFP-NIC96 variants. Serial dilutions of the respective cells were spotted onto indicated plates and grown for 3–5 days at the specified temperatures. (E) Subcellular localization of mCherry-Nic96 variants (red) and Nup57-GFP (green) in a nic96Δ̃/NUP57-GFP strain. (F) mRNA export assay in a nic96Δ̃NUP57 -GFP strain carrying GFP-NIC96 variants. Representative images and quantification of nuclear poly(A)+ RNA retention are shown. (G) Subcellular localization of the 60S ribosomal export reporter Rpl25-mCherry (red) and GFP-tagged Nup49 variants (green) in a nic96Δ strain. Quantification of nuclear Rpl25-mCherry retention is shown on the right. Cells were analyzed at the indicated temperatures and incubation times. Error bars represent the standard deviation. Scalebars are 5 μm.
Similarly, because the Nic96R2-Nup192TAIL interaction is also salt stable and was disrupted by mutating a hydrophobic pocket on Nup192TAIL, we focused our mutational analysis on evolutionarily conserved hydrophobic residues of Nic96R2 (figs. S5C and S13). We identified two mutations, I294A and F298A, which reduced and abolished Nup192TAIL binding, respectively (Fig. 3C and fig. S16).
Next, we carried out functional analyses of the identified Nic96 mutants in S. cerevisiae (fig. S17). Nic96 fragments lacking either Nic96SOL (ΔSOL) or Nic96R1-R2 (SOL) were unable to rescue the previously identified lethal nic96Δ phenotype and were not analyzed further (Fig. 3D) (35). All remaining mutants were viable and yielded NE staining, consistent with NPC incorporation (Fig. 3E). A Nic96 mutant lacking the Nup192-interacting region R2 (ΔR2) displayed severe growth and mRNA export defects yet failed to affect Nup57-GFP localization (Fig. 3D to F). Nic96 mutants lacking the CNT-interacting region R1 (R2-SOL) and R1-R2 linker (ΔLINKER) also had severe growth defects accompanied by a marked decrease and no detectable NE staining for Nup57-GFP, respectively (Fig. 3D to F). Additionally, ΔR2 and ΔLINKER displayed a mild ribosome export defect (Fig. 3G). The milder LLLL and F159A mutants failed to yield significant phenotypes (Fig. 3D to G).
Together, these experiments demonstrated that the channel nup Nup57 is solely anchored to the NPC through its interaction with Nic96 in vivo, in agreement with our biochemical studies.
Channel nup fragments do not capture the solution behavior of the intact CNT
A secondary structure analysis revealed that the channel nups Nsp1, Nup49, and Nup57 possessed evolutionarily conserved domain architectures with an N-terminal FG-repeat region of varying length, followed by three predicted α-helical coiled-coil segments, CCS1-3 (fig. S18A). Nup57 contained an additional evolutionarily conserved α/β region, which preceded the three coiled-coil regions.
Lacking a fully assembled CNT structure, we attempted to obtain structural insight from short channel nup fragments. Using fragments of Nic96 and the three channel nups from three different species, we carried out a systematic interaction and crystallization analysis, which led to the structure determination of multiple channel nup fragments (figs. S19 to S21). This approach yielded six different crystal structures of homomeric and heteromeric channel nup assemblies with different stoichiometries (figs. S18B, S22 and S23). Two structures revealed novel assembly states (figs. S18B and S24). The remaining four were almost identical to previously determined R. norvegicus structures (22, 25, 26). These assembly states constituted the basis for the proposal of a dilating transport channel, whose diameter is modulated by karyopherin-mediated transitioning between these homomeric and heteromeric assembly states (25–28). Despite the remarkable degree of sequence conservation between human and rat fragments with identical domain boundaries, we observed different assembly states that behaved inconsistently when mutated or further truncated (figs. S25 and S26).
In contrast, only two states were detected for the intact CNT in solution, corresponding to monodisperse equimolar monomeric and dimeric species (fig. S18C). At concentrations up to ~30 mg/ml, the predominant species was the CNT monomer. Both the CNT monomer and dimer were capable of forming monodisperse hetero-tetrameric complexes with Nic96R1 in solution (fig. S18D).
To identify which of the three channel nups form a specific interaction with Nic96R1, we carried out a GST-pull down interaction assay. We found that GST-Nic96R1 did not interact with separately purified Nsp1 or Nup49•Nup57 in isolation, but formed a complex in the presence of both (fig. S18E).
These data show that the coiled-coil architecture of the intact CNT cannot be elucidated by a reductionist approach that involves channel nup coiled-coil fragments, because of their inconsistent behavior in solution and their inability to capture the correct stoichiometry of the intact CNT.
Crystal structure of CNT•Nic96R1
Our biochemical and in vivo data established that the CNT architecture can only be elucidated by a structure of the intact CNT and that the physiologically relevant state is the CNT•Nic96R1 attachment complex. However, extensive crystallization attempts for the CNT•Nic96R1 yielded crystals that diffracted to ~10 Å resolution, at best. To improve the diffraction quality, we generated a series of conformation-specific, high-affinity monobodies (MBs) and synthetic antibody Fab fragments (sABs) by phage display selection methods as crystallization chaperones. Up to two different MBs or sABs could bind to the intact IRC and CNT•Nic96R1, forming monodisperse stoichiometric complexes (figs. S27 and S28). These data established that access to the CNT was sterically unrestricted by its incorporation into the IRC, suggesting that the CNT protrudes from the IRC.
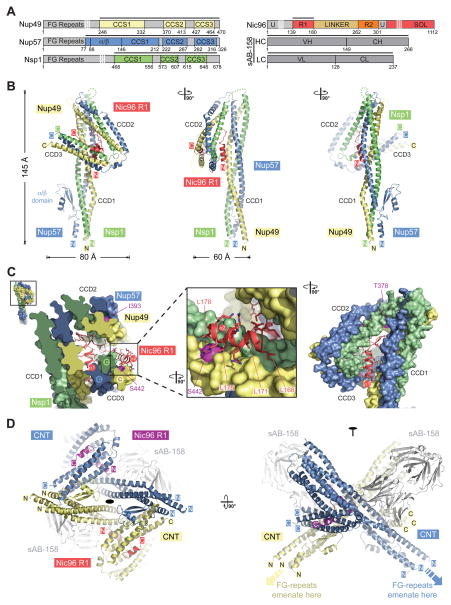
Crystals of CNT•Nic96R1•sAB-158 diffracted to a 3.77 Å resolution, facilitating structure determination by single anomalous dispersion (SAD) (fig. S29 and Table S4). The crystal contacts were primarily mediated by sAB-158, demonstrating the effectiveness of chaperoning reagents in aiding the crystallization of difficult structural targets (fig. S30). The three channel nups formed a parallel, three-stranded, hetero-trimeric coiled-coil structure with two sharp kinks that divided the CNT into three left-handed coiled-coil domains (CCD1-3), with an overall resemblance to the numeral “4” and maximum dimensions of ~145 Å × ~80 Å × ~60 Å (Fig. 4, A and B and movies S1 and S2). CCD1 formed the ~145 Å long stalk and was composed of the N-terminal coiled-coil segments (CCS1) of Nsp1, Nup49, and Nup57, each containing twelve heptad repeats. Analogously, CCS2 and CCS3 were each composed of five heptad repeats that were ~70 Å in length. The three CCDs were connected by unstructured linker regions, of varying length, between the three coiled-coil segments of each channel nup. CCS1 of Nup57 was interrupted close to the base of the stalk by a 56-residue insertion forming a novel fold with two parallel β-sheets and one α-helix, termed the α/β insertion domain (Fig. 4B).
Fig. 4. Crystal structure of the intact Nsp1•Nup49•Nup57 CNT bound to Nic96R1 and sAB-158.
(A) Domain structures of the C. thermophilum channel nups Nsp1, Nup49, Nup57, the adaptor nup Nic96, and sAB-158. Black lines indicate the construct boundaries of the crystallized CNT•Nic96R1•sAB-158 complex. The constructs of the three channel nups comprise all regions with predicted secondary structure elements and only lack their unstructured N-terminal FG repeat regions. VH, heavy chain variable region; CH, heavy chain constant region; VL, light chain variable region; CL, light chain constant region. (B) Cartoon representation of the Nic96R1•CNT crystal structure viewed from three sides. For clarity, sAb-158 has been omitted. (C) Details of the Nic96R1-CNT interaction, illustrating the three channel nups and Nic96R1 in surface and cartoon representation, respectively. The Inset marks the region enlarged and 90º rotated in the middle panel. A further 90º rotated view is shown on the right. Nic96R1 (red) and Nup49 (magenta) residues that abolish CNT-Nic96R1 complex formation upon mutation (Figures 3 and 5) are indicated. (D) Structure of the CNT•Nic96R1•sAB-158 dimer shown in cartoon representation. The two copies of the CNT, Nic96R1, and sAB-158 are shown in blue/yellow, magenta/red, and gray, respectively. A 90º rotated view is shown on the right. The two CNT•Nic96R1•sAB-158 complexes in the asymmetric unit are related by two-fold rotational symmetry (black oval). Notably, the N-termini of all channel nups in the dimer point in the same direction, which would permit the FG repeats to project towards the central transport channel of the NPC.
A salient feature of the structure was the extensive interaction of Nic96R1 with all three CCDs of the CNT, located in the triangular opening at the apex of the “4” (Fig. 4B and movie S3). Nic96R1 formed two 11-residue α-helices, α1 and α2, which were linked by a sharply kinked 16-residue connector (Fig. 4C). One face of helix α1 recognized CCD1 by binding the Nup57-Nup49 surface, while the opposite face of helix α1 interacted with the Nsp1-Nup57 surface of CCD3. The kinked loop inserted as a wedge between the top of the long stalk-forming CCD1 and CCD2. Residues at the N- and C-termini of the loop formed additional electrostatic contacts with the Nup57-Nup49 surface of CCD1 and extensive hydrophobic contacts with the Nup49-Nsp1 surface of CCD2, respectively. Finally, helix α2, which harbors our LLLL mutant, bound exclusively to the hydrophobic Nsp1-Nup49 surface of CCD3. Not only did Nic96R1 recognize the proper assembly of all three CCDs by forming specific interactions with composite CCD surfaces that are each formed by two different channel nups, but it also locked the CNT into a specific conformation. Overall, ~4,150 Å2 of surface area are buried in the Nic96R1-CNT interface, involving 38 and 64 residues of Nic96R1 and the CNT, respectively. The evolutionary conservation of these interface residues further corroborates the evolutionary conservation of the CNT•Nic96R1 architecture (fig. S13, S31 to S34).
CNT•Nic96R1•sAB-158 forms a cross-handshake dimer in the asymmetric unit of the crystal that is generated by two identical dimerization interfaces, involving the Nup57 α/β insertion domain and CCD3 (Fig. 4D and movie S4). The CNT•Nic96R1 dimer buried ~2,600 Å2 of surface area. The N-termini of all channel nups in the dimer point in the same direction, which would permit the FG repeats to be projected towards the central transport channel. Notably, sAB-158 did not participate in CNT•Nic96R1 dimer formation, as it recognized a composite Nup57-Nup49 surface in the middle of the CCD1 stalk.
The N-terminal regions of Nsp1, Nup49, and Nup57 in the three-stranded CCD1 stalk appeared to contain seven-residue signature motifs akin to coiled-coil trigger sequences that were previously shown to facilitate proper three-stranded coiled-coil assembly (fig. S35A) (36). An N-terminal truncation of Nup57, Δα/β, which lacked four helical turns of CCS1 and the α/β insertion domain, transformed the equimolar and monodisperse CNT into a polydisperse mixture (fig. S35, B to E). These data provided a molecular explanation for the observed heterogeneous stoichiometries and dynamic nature of previous mammalian CNT reconstitutions, which also lacked this region (22, 23, 26, 28). In addition to removing the trigger sequence, Nup57 Δα/β exposed a hydrophobic surface of approximately four unpaired heptad repeats (~50 Å in length). Thus, misfolding and aggregation would be expected.
We conclude that the CNT adopts a robust coiled-coil domain architecture with a single defined assembly state that is specifically recognized by Nic96R1 to ensure NPC incorporation of only properly assembled CNT. None of the interactions from previously determined channel nup fragment crystal structures occured in the intact CNT. Thus, these data strongly disagree with the model in which the CNT undergoes dynamic rearrangements to facilitate NPC constriction and dilation, as previously proposed based on the dynamic nature of various channel nup fragment interactions (25, 26, 28).
Nic96 is an assembly sensor for the properly assembled CNT
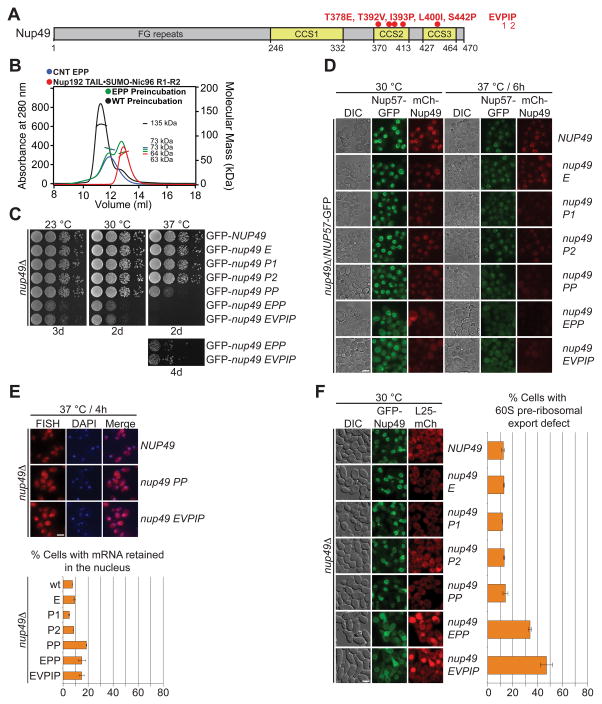
Next, we identified channel nup mutants that disrupted the Nic96R1-CNT interaction. As we deemed alanine scanning mutagenesis unlikely to disrupt the extensive CNT-Nic96R1 interface, we characterized a previously identified quintuple S. cerevisiae Nup49 mutant (EVPIP; K376E, I390V, I391P, V398I, and L449P) (37). The five Nup49 EVPIP mutations mapped to two Nic96R1 interfaces located in CCD2 and CCD3 (Figs. 4C and 5A). The corresponding C. thermophilum CNTEVPIP mutant failed to interact with the minimal IRC in SEC-MALS experiments (Fig. 5A and fig. S36A). Further analysis showed that CNTEPP was sufficient to abolish binding and CNTPP severely reduced binding, but no single mutant alone affected complex formation (Fig. 5B and fig. S36, B to F). The severity of the required mutations demonstrated the robustness of the CNT-Nic96R1 interaction.
Fig. 5. The intact CNT is recruited to the NPC by its interaction with Nic96.
(A) Domain structure of Nup49, indicating the corresponding EVPIP mutations (red) of the S. cerevisiae nup49-313 allele (37). (B) SEC-MALS profiles of the CNTEPP mutant (blue), Nic96R1-R2•Nup192TAIL (red) and after their preincubation (green). As reference, wild type CNT•Nic96R1-R2•Nup192TAIL is shown (black). Measured molecular masses are indicated for the peak fractions. (C) Growth analysis of S. cerevisiae strains carrying the indicated GFP-NUP49 variants. Serial dilutions of the respective cells were spotted onto YPD plates and grown for 2–4 days at the specified temperatures. (D) Subcellular localization of mCherry-Nup49 variants (red) and Nup57-GFP (green) in a nup49Δ̃/NUP57-GFPs strain. (E) mRNA export assay in a nup49Δ strain carrying GFP-NUP49 variants. Representative images and quantification of nuclear poly(A)+ RNA retention are shown. (F) Subcellular localization of the 60S ribosomal export reporter Rpl25-mCherry (red) and GFP-tagged Nup49 variants (green) in a nup49Δ strain. Quantification of nuclear Rpl25-mCherry retention is shown on the right. Cells were analyzed at the indicated temperatures and incubation times. Error bars represent the standard deviation. Scalebars are 5 μm.
Next, we tested the effects of these Nup49 mutations in S. cerevisiae. Whereas, the single mutants displayed no phenotypes, PP displayed a temperature-sensitive growth defect, which was intensified in EPP and EVPIP, consistent with their decreased thermostability (Fig. 5C and fig. S37). In line with the growth defects, PP showed wild type levels of mCherry-Nup49 and Nup57-GFP NE staining at 30 °C, but barely detectable levels at 37 °C (Fig. 5D). The temperature-sensitive localization defect for both nups was escalated in EPP and EVPIP, which showed severe reduction and complete loss of NE staining at 30 °C and 37 °C, respectively (Fig. 5D). Interestingly, despite an almost complete loss of Nup49 and Nup57 from the NE, we only observed a mild effect on mRNA export (Fig. 5E). Regardless, the EPP and EVPIP mutants resulted in a ribosomal export defect at 30 °C. (Fig. 5F).
These data established that NE recruitment of Nup57 and Nup49 are co-dependent, consistent with our biochemical and structural analyses that Nic96R1 only interacts with the intact CNT, validating that the CNT•Nic96R1 structure represents the physiologically relevant state in the assembled NPC.
Conclusions
Through reconstitution and systematic structural and functional dissection of the IRC, we established the equimolar stoichiometry of its six components and uncovered the physiologically relevant underlying interaction network. We showed that Nup53 and the CFC could be attached to the IRC, facilitating membrane attachment and functionalization at the cytoplasmic face, respectively. Surprisingly, none of the previously determined dynamic channel nup fragment assemblies occur in the intact CNT•Nic96R1 structure. In fact, the CNT adopts a robust parallel three-stranded coiled-coil domain architecture resembling the numeral 4. This organization guarantees that the N-terminal FG repeats of the three channel nups emanate from a single site on the CNT surface. Because Nic96 harbors an assembly sensor for this specific CNT state, the NPC incorporation of the three channel nups is co-dependent and thereby enables the concomitant generation of the diffusion barrier and cargo•transport factor complex docking sites. Additionally, we demonstrated that efficient nucleocytoplasmic transport also requires proper CNT positioning, which is achieved by the Nic96-Nup192 interaction. The conclusion that the inner ring of the NPC adopts a relatively rigid architecture with a transport channel filled with channel nup FG repeats, is also supported by a channel nup FG repeat-coated artificial nanopore, which mimicked the NPC’s transport selectivity (38).
NPC recruitment of a single robust CNT assembly state rules out the possibility that channel nups dynamically rearrange into different assembly states with various stoichiometries to facilitate karyopherin-assisted dilation and constriction of the central transport channel, as previously proposed (25–28). Indeed, dilation is unnecessary as the central transport channel of the human NPC can easily accommodate even one of the largest cargoes, the pre-60S ribosomal particle (fig. S38) (2, 39).
Apart from its role in the CNT, Nsp1 forms a mutually exclusive complex with the C-terminal coiled-coil relions coilded of Nup82 and Nup159 at the cytoplasmic face of the NPC (40). The CNT•Nic96R1 structure suggests that Nsp1•Nup82•Nup159 might adopt a similar three-stranded coiled-coil domain architecture. More generally, our biochemical and structural analyses of the three channel nups instruct that utmost caution is required when analyzing heteromeric coiled-coil interactions.
The finding that the IRC and the IRC•CFC•Nup53 hetero-nonamer are monomeric in solution is not unexpected. The CNC is also monomeric in solution, but organizes into densely packed, sixteen-membered peripheral rings in the intact NPC (2, 3). The cryo-ET reconstruction of the human NPC suggests that the inner ring is large enough to accommodate sixteen copies of the IRC, and we propose that they are recruited to the nuclear pore membrane by their association with the membrane anchored Nup53•Nup170•Ndc1 complex (Fig. 6) (5, 41). Stabilization of the inner ring scaffold could occur through multiple weak interactions between adjacent IRCs, including the dimerization of neighboring CNTs, the formation of a FG-repeat hydrogel in the central transport channel (9), and/or transport factor•cargo complex binding to such a FG-repeat meshwork. A sixteen-membered inner ring scaffold accounts for ~10-MDa of the NPC mass. Our results represent a major step forward towards the in vitro reconstitution of the entire NPC, which is essential for the development of in vitro assays to quantitatively characterize nucleocytoplasmic transport.
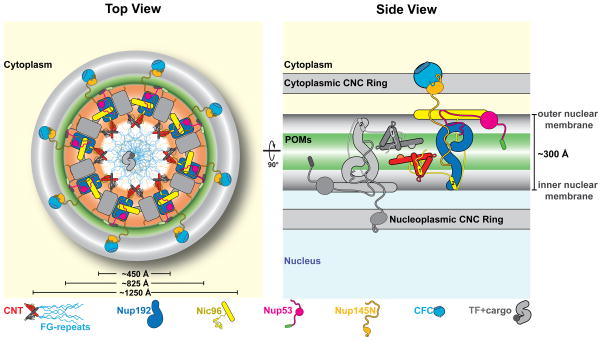
Fig. 6. Proposed architecture of the NPC inner ring scaffold.
Sixteen copies of the IRC are anchored to the nuclear pore membrane and arranged in a ring-shaped scaffold in an anti-parallel fashion to satisfy the established 8-fold and 2-fold symmetry axes of the NPC. Each IRC is composed of the channel nucleoporins Nsp1, Nup49, and Nup57 (CNT, red), the adaptor nucleoporins Nup192 (blue), Nic96 (yellow), Nup53 (magenta), and Nup145N (orange). The cytoplasmic and nuclear CNC rings (gray), the putative location of the POMs (green), and the inner ring (red) are shown. On the cytoplasmic side, Nup145N attaches the cytoplasmic filament nucleoporins Nup82 and Nup159 (CFC, cyan). The primarily unstructured adaptor nucleoporins Nup53 and Nup145N mediate the association of various structured IRC components and thus are critical for the IRC scaffold integrity. Nic96 functions as an assembly sensor that recognizes the conformation of the overall 4-shaped three-stranded coiled-coil domain architecture of the CNT, thereby mediating the selective incorporation of a defined CNT state into the NPC. The CNTs are positioned in the equatorial plane of the inner ring with the FG repeats of all three channel nups projecting towards the central transport channel. The anti-parallel orientation of adjacent IRCs would generate two CNT rings, one at the top and another at the bottom of the central transport channel. This arrangement would allow for the formation of a transport factor mediated FG repeat meshwork or hydrogel, which would further reinforce the inner ring scaffold. For size reference, a β-karyopherin•cargo complex is shown in gray.
Supplementary Material
Acknowledgments
We thank W. M. Clemons, A. Correia, G. Mobbs, A. Patke, D. C. Rees, S. O. Shan, and E. Stuwe for critical reading of the manuscript, M. Budd for help with yeast experiments, J. Herrmann, R. Kunze, and L. Zhang for technical support, and J. Kaiser and the scientific staff of the Stanford Synchrotron Radiation Laboratory (SSRL) Beamline 12-2, the National Institute of General Medical Sciences and National Cancer Institute Structural Biology Facility (GM/CA) at the Advanced Photon Source (APS), and the Advanced Light Source (ALS) beamline 8.2.1 for their support with x-ray diffraction measurements. We acknowledge the Gordon and Betty Moore Foundation, the Beckman Institute, and the Sanofi-Aventis Bioengineering Research Program for their support of the Molecular Observatory at the California Institute of Technology (Caltech). The operations at the SSRL, ALS, and APS are supported by the U.S. Department of Energy and the National Institutes of Health (NIH). GM/CA has been funded in whole or in part with federal funds from the National Cancer Institute (ACB-12002) and the National Institute of General Medical Sciences (AGM-12006). T.S. was supported by a Postdoctoral Fellowship of the Deutsche Forschungsgemeinschaft. S.P. and D.H.L are Amgen Graduate Fellows, supported through the Caltech-Amgen Research Collaboration. F.M.H. was supported by a PhD student fellowship of the Boehringer Ingelheim Fonds. S.K. was supported by NIH Awards R01-GM090324 and U54-GM087519 and by the University of Chicago Comprehensive Cancer Center (P30-CA014599). A.A.K. was supported by NIH awards U01-GM094588 and U54-GM087519 and by Searle Funds at The Chicago Community Trust. A.H. was supported by Caltech startup funds, the Albert Wyrick V Scholar Award of the V Foundation for Cancer Research, the 54th Mallinckrodt Scholar Award of the Edward Mallinckrodt Jr. Foundation, a Kimmel Scholar Award of the Sidney Kimmel Foundation for Cancer Research, a Camille-Dreyfus Teacher Scholar Award of The Camille & Henry Dreyfus Foundation, and NIH grant R01-GM111461. The coordinates and structure factors have been deposited with the Protein Data Bank with accession codes 5CWV (Nup192TAIL), 5CWU (Nup188TAIL), 4JQ5 (hsNup49CCS2+3*), 4JNV and 4JNU (hsNup57CCS3*), 5CWT (Nup57CCS3*), 4JO7 (hsNup49CCS2+3*•hsNup57CCS3*, 2:2 stoichiometry), 4JO9 (hsNup49CCS2+3*•hsNup57CCS3*, 1:2 stoichiometry), 5CWW (Nup82NTD•Nup159T•Nup145NAPD) and 5CWS (CNT•Nic96R1•sAB-158).
Footnotes
The authors declare no financial conflicts of interest.
Materials and Methods
References (42–77)
References and Notes
- 1.Hoelz A, Debler EW, Blobel G. The structure of the nuclear pore complex. Annu Rev Biochem. 2011;80:613–643. doi: 10.1146/annurev-biochem-060109-151030. [DOI] [PubMed] [Google Scholar]
- 2.Bui KH, von Appen A, et al. Integrated structural analysis of the human nuclear pore complex scaffold. Cell. 2013;155:1233–1243. doi: 10.1016/j.cell.2013.10.055. [DOI] [PubMed] [Google Scholar]
- 3.Stuwe T, Correia AR, et al. Nuclear pores. Architecture of the nuclear pore complex coat. Science. 2015;347:1148–1152. doi: 10.1126/science.aaa4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eisenhardt N, Redolfi J, Antonin W. Interaction of Nup53 with Ndc1 and Nup155 is required for nuclear pore complex assembly. J Cell Sci. 2014;127:908–921. doi: 10.1242/jcs.141739. [DOI] [PubMed] [Google Scholar]
- 5.Onischenko E, Stanton LH, Madrid AS, Kieselbach T, Weis K. Role of the Ndc1 interaction network in yeast nuclear pore complex assembly and maintenance. J Cell Biol. 2009;185:475–491. doi: 10.1083/jcb.200810030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitchell JM, Mansfeld J, Capitanio J, Kutay U, Wozniak RW. Pom121 links two essential subcomplexes of the nuclear pore complex core to the membrane. J Cell Biol. 2010;191:505–521. doi: 10.1083/jcb.201007098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amlacher S, Sarges P, Flemming D, van Noort V, et al. Insight into structure and assembly of the nuclear pore complex by utilizing the genome of a eukaryotic thermophile. Cell. 2011;146:277–289. doi: 10.1016/j.cell.2011.06.039. [DOI] [PubMed] [Google Scholar]
- 8.Stuwe T, Lin DH, Collins LN, Hurt E, Hoelz A. Evidence for an evolutionary relationship between the large adaptor nucleoporin Nup192 and karyopherins. Proc Natl Acad Sci USA. 2014;111:2530–2535. doi: 10.1073/pnas.1311081111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cook A, Bono F, Jinek M, Conti E. Structural biology of nucleocytoplasmic transport. Annu Rev Biochem. 2007;76:647–671. doi: 10.1146/annurev.biochem.76.052705.161529. [DOI] [PubMed] [Google Scholar]
- 10.Rout MP, et al. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frey S, Richter RP, Gorlich D. FG-rich repeats of nuclear pore proteins form a three-dimensional meshwork with hydrogel-like properties. Science. 2006;314:815–817. doi: 10.1126/science.1132516. [DOI] [PubMed] [Google Scholar]
- 12.Yang Q, Rout MP, Akey CW. Three-dimensional architecture of the isolated yeast nuclear pore complex: functional and evolutionary implications. Mol Cell. 1998;1:223–234. doi: 10.1016/s1097-2765(00)80023-4. [DOI] [PubMed] [Google Scholar]
- 13.Grandi P, Schlaich N, Tekotte H, Hurt EC. Functional interaction of Nic96p with a core nucleoporin complex consisting of Nsp1p, Nup49p and a novel protein Nup57p. EMBO J. 1995;14:76–87. doi: 10.1002/j.1460-2075.1995.tb06977.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu T, Guan T, Gerace L. Molecular and functional characterization of the p62 complex, an assembly of nuclear pore complex glycoproteins. J Cell Biol. 1996;134:589–601. doi: 10.1083/jcb.134.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wimmer C, Doye V, Grandi P, Nehrbass U, Hurt EC. A new subclass of nucleoporins that functionally interact with nuclear pore protein NSP1. EMBO J. 1992;11:5051–5061. doi: 10.1002/j.1460-2075.1992.tb05612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wente SR, Rout MP, Blobel G. A new family of yeast nuclear pore complex proteins. J Cell Biol. 1992;119:705–723. doi: 10.1083/jcb.119.4.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nehrbass U, et al. NSP1: a yeast nuclear envelope protein localized at the nuclear pores exerts its essential function by its carboxy-terminal domain. Cell. 1990;61:979–989. doi: 10.1016/0092-8674(90)90063-k. [DOI] [PubMed] [Google Scholar]
- 18.Strawn LA, Shen T, Shulga N, Goldfarb DS, Wente SR. Minimal nuclear pore complexes define FG repeat domains essential for transport. Nat Cell Biol. 2004;6:197–206. doi: 10.1038/ncb1097. [DOI] [PubMed] [Google Scholar]
- 19.Hurt EC. A novel nucleoskeletal-like protein located at the nuclear periphery is required for the life cycle of Saccharomyces cerevisiae. EMBO J. 1988;7:4323–4334. doi: 10.1002/j.1460-2075.1988.tb03331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guan T, et al. Structural analysis of the p62 complex, an assembly of O-linked glycoproteins that localizes near the central gated channel of the nuclear pore complex. Mol Biol Cell. 1995;6:1591–1603. doi: 10.1091/mbc.6.11.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grandi P, et al. Nup93, a vertebrate homologue of yeast Nic96p, forms a complex with a novel 205-kDa protein and is required for correct nuclear pore assembly. Mol Biol Cell. 1997;8:2017–2038. doi: 10.1091/mbc.8.10.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Melcak I, Hoelz A, Blobel G. Structure of Nup58/45 suggests flexible nuclear pore diameter by intermolecular sliding. Science. 2007;315:1729–1732. doi: 10.1126/science.1135730. [DOI] [PubMed] [Google Scholar]
- 23.Ulrich A, Partridge JR, Schwartz TU. The stoichiometry of the nucleoporin 62 subcomplex of the nuclear pore in solution. Mol Biol Cell. 2014;25:1484–1492. doi: 10.1091/mbc.E13-12-0745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schlaich NL, Haner M, Lustig A, Aebi U, Hurt EC. In vitro reconstitution of a heterotrimeric nucleoporin complex consisting of recombinant Nsp1p, Nup49p, and Nup57p. Mol Biol Cell. 1997;8:33–46. doi: 10.1091/mbc.8.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Solmaz SR, Blobel G, Melcak I. Ring cycle for dilating and constricting the nuclear pore. Proc Natl Acad Sci USA. 2013;110:5858–5863. doi: 10.1073/pnas.1302655110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Solmaz SR, Chauhan R, Blobel G, Melcak I. Molecular architecture of the transport channel of the nuclear pore complex. Cell. 2011;147:590–602. doi: 10.1016/j.cell.2011.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koh J, Blobel G. Allosteric Regulation in Gating the Central Channel of the Nuclear Pore Complex. Cell. 2015;161:1361–1373. doi: 10.1016/j.cell.2015.05.013. [DOI] [PubMed] [Google Scholar]
- 28.Sharma A, Solmaz SR, Blobel G, Melcak I. Ordered Regions of Channel Nucleoporins Nup62, Nup54 and Nup58 Form Dynamic Complexes in Solution. J Biol Chem. 2015 doi: 10.1074/jbc.M115.663500. jbc.M115.663500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stuwe T, von Borzyskowski LS, Davenport AM, Hoelz A. Molecular basis for the anchoring of proto-oncoprotein Nup98 to the cytoplasmic face of the nuclear pore complex. J Mol Biol. 2012;419:330–346. doi: 10.1016/j.jmb.2012.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoshida K, Seo HS, Debler EW, Blobel G, Hoelz A. Structural and functional analysis of an essential nucleoporin heterotrimer on the cytoplasmic face of the nuclear pore complex. Proc Natl Acad Sci USA. 2011;108:16571–16576. doi: 10.1073/pnas.1112846108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andersen KR, et al. Scaffold nucleoporins Nup188 and Nup192 share structural and functional properties with nuclear transport receptors. Elife. 2013;2:e00745. doi: 10.7554/eLife.00745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sampathkumar P, et al. Structure, dynamics, evolution, and function of a major scaffold component in the nuclear pore complex. Structure. 2013;21:560–571. doi: 10.1016/j.str.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kosova B, Pante N, Rollenhagen C, Hurt E. Nup192p is a conserved nucleoporin with a preferential location at the inner site of the nuclear membrane. J Biol Chem. 1999;274:22646–22651. doi: 10.1074/jbc.274.32.22646. [DOI] [PubMed] [Google Scholar]
- 34.Aitchison JD, Rout MP, Marelli M, Blobel G, Wozniak RW. Two novel related yeast nucleoporins Nup170p and Nup157p: complementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore-membrane protein Pom152p. J Cell Biol. 1995;131:1133–1148. doi: 10.1083/jcb.131.5.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grandi P, Doye V, Hurt EC. Purification of NSP1 reveals complex formation with 'GLFG' nucleoporins and a novel nuclear pore protein NIC96. EMBO J. 1993;12:3061–3071. doi: 10.1002/j.1460-2075.1993.tb05975.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frank S, Lustig A, Schulthess T, Engel J, Kammerer RA. A distinct seven-residue trigger sequence is indispensable for proper coiled-coil formation of the human macrophage scavenger receptor oligomerization domain. J Biol Chem. 2000;275:11672–11677. doi: 10.1074/jbc.275.16.11672. [DOI] [PubMed] [Google Scholar]
- 37.Doye V, Wepf R, Hurt EC. A novel nuclear pore protein Nup133p with distinct roles in poly(A)+ RNA transport and nuclear pore distribution. EMBO J. 1994;13:6062–6075. doi: 10.1002/j.1460-2075.1994.tb06953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jovanovic-Talisman T, et al. Artificial nanopores that mimic the transport selectivity of the nuclear pore complex. Nature. 2009;457:1023–1027. doi: 10.1038/nature07600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bradatsch B, et al. Structure of the pre-60S ribosomal subunit with nuclear export factor Arx1 bound at the exit tunnel. Nat Struct Mol Biol. 2012;19:1234–1241. doi: 10.1038/nsmb.2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bailer SM, Balduf C, Hurt E. The Nsp1p carboxy-terminal domain is organized into functionally distinct coiled-coil regions required for assembly of nucleoporin subcomplexes and nucleocytoplasmic transport. Mol Cell Biol. 2001;21:7944–7955. doi: 10.1128/MCB.21.23.7944-7955.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hawryluk-Gara LA, Shibuya EK, Wozniak RW. Vertebrate Nup53 interacts with the nuclear lamina and is required for the assembly of a Nup93-containing complex. Mol Biol Cell. 2005;16:2382–2394. doi: 10.1091/mbc.E04-10-0857. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.