Abstract
Abiotic stresses, including high soil salinity, significantly reduce crop production worldwide. Salt tolerance in plants is a complex trait and is regulated by multiple mechanisms. Understanding the mechanisms and dissecting the components on their regulatory pathways will provide new insights, leading to novel strategies for the improvement of salt tolerance in agricultural and economic crops of importance. Here we report that soybean salt tolerance 1, named GmST1, exhibited strong tolerance to salt stress in the Arabidopsis transgenic lines. The GmST1-overexpressed Arabidopsis also increased sensitivity to ABA and decreased production of reactive oxygen species under salt stress. In addition, GmST1 significantly improved drought tolerance in Arabidopsis transgenic lines. GmST1 belongs to a 3-prime part of Glyma.03g171600 gene in the current version of soybean genome sequence annotation. However, comparative reverse transcription-polymerase chain reaction analysis around Glyma.03g171600 genomic region confirmed that GmST1 might serve as an intact gene in soybean leaf tissues. Unlike Glyma.03g171600 which was not expressed in leaves, GmST1 was strongly induced by salt treatment in the leaf tissues. By promoter analysis, a TATA box was detected to be positioned close to GmST1 start codon and a putative ABRE and a DRE cis-acting elements were identified at about 1 kb upstream of GmST1 gene. The data also indicated that GmST1-transgenic lines survived under drought stress and showed a significantly lower water loss than non-transgenic lines. In summary, our results suggest that overexpression of GmST1 significantly improves Arabidopsis tolerance to both salt and drought stresses and the gene may be a potential candidate for genetic engineering of salt- and drought-tolerant crops.
Keywords: soybean, GmST1 overexpression, salt tolerance, drought tolerance, ROS production, ABA sensitivity
Introduction
Soil salinity is one of the major environmental factors that significantly affected crop productivity and quality (Allakhverdiev et al., 2000). Large number of arable lands are being removed from crop production due to increased soil salinity (Epstein et al., 1980), and over 25% of the world’s potential arable land is currently contaminated by salt in different ways including irrigation, overuse of fertilizers and/or seawater intrusion (Pathan et al., 2007). Decreasing of the acreage of arable land for crop production has become a severe threat to global food security as more food will be needed to feed the growing population.
Soybean is considered a salt-sensitive glycophyte, with all developmental stages adversely affected by salinity stress (Phang et al., 2008). High salt levels generate a two-component stress on plants: an osmotic stress caused by reduced water availability in soil and an ionic stress due to imbalance of solutes in the cytosol (Blumwald et al., 2000; Conde et al., 2011). During soybean development, salt stress significantly reduces plant height and leaf size (Wang et al., 2001; Essa, 2002), decreases the number of internodes and pods (Phang et al., 2008), decreases protein content and seed quality, and causes a reduction in chlorophyll content (Lu et al., 2009). Salt stress also significantly affects seed germination, seedling growth, biomass, and seed yield (Abel and MacKenzie, 1964; Katerji et al., 1998; Wang and Shannon, 1999; Essa, 2002). The increase in salt content in soil could cause a concomitant decrease of up to 40% in soybean yield (Chang et al., 1994). Development of more precise salt tolerant cultivars will help reduce the detrimental loss of yield in soybean production in the areas with elevated salt levels.
Mechanisms of plant salt tolerance have been extensively studied and well characterized in many species (Lauchli, 1984; Zhu, 2002; Lenis et al., 2011). Both ABA-dependent and –independent pathways have been characterized, including identification of many key genes (Zhu, 2002). In addition, the role of calcium as a key secondary messenger in plant salt tolerance is well established in Arabidopsis (Mahajan et al., 2008). Soybean plants have developed several mechanisms to cope with salt stress conferring a wide spectrum of salt tolerance among genotypes (Phang et al., 2008). Mechanisms of salt tolerance include maintaining ion homeostasis by withholding toxic ions from sensitive aerial parts, adjusting osmotic potential in cells by accumulating metabolites, and restoring oxidative balance to prevent further damage due to excess accumulation of reactive oxygen species (ROS) (Phang et al., 2008). Understanding the mechanisms and identifying the genes involved in salt stress tolerance of soybeans will enable breeders to develop new strategies to enhance salt tolerance.
Due to the complexity of the soybean genome, mechanisms conferring salt tolerance are often overlooked. Nevertheless, with advances in high throughput sequencing technology, the full soybean genome has recently been sequenced (Schmuta et al., 2010). The availability of the entire soybean genome sequence provides ample opportunities for basic research to reveal the underlying mechanisms of salt tolerance. Many putative components of the salt tolerance signaling network homologous to those identified in Arabidopsis have been elucidated in soybean using the reverse genetic approach. Of these, GmSCA1, GmCaMs, GmSTL, GmAAPK, GmSTY1, and GmCIPK1 are involved in calcium signaling and eventually regulated downstream transcription factors and the effector genes for salt stress responses (Chung et al., 2000; Park et al., 2004; Luo et al., 2005; Li et al., 2006; Xu et al., 2006). Several soybean DREB homologs identified as regulators on the ABA-independent pathway were also shown to play roles in the regulation of salt tolerance (Chen et al., 2007). In addition, the ABA-dependent pathway also plays important functions in regulating soybean salt tolerance. Several transcriptional factors induced by both ABA and salt stress were successfully identified in soybean, including GmTDF-5, GmbZIPs, WRKY transcriptional factor family members, GmAP2/ERFs, and GmNACs (NAC transcriptional regulators) (Aoki et al., 2005; Meng et al., 2007; Liao et al., 2008, 2010; Zhang et al., 2008; Zhou et al., 2008; Zhai et al., 2013). These genes have a similar function in Arabidopsis.
Ion transporter genes are also believed to play essential roles in salt tolerance. Many putative ion transporters identified in soybean have also demonstrated a relationship between gene expression and salt tolerance (Luo et al., 2005; Li et al., 2006; Sun et al., 2006). More recently, through integration of whole genome sequencing/transcriptome sequencing and map-based cloning approaches, Guan et al. (2014b) and Qi et al. (2014) independently identified Glyma03g32900 as a potential candidate gene of salt tolerance in both wild and cultivated soybeans. Glyma03g32900 encodes a putative cation proton antiporter, GmCHX1, and is homologous to Arabidopsis cation proton antiporter AtCHX20 (Qi et al., 2014). However, neither report provided solid in-planta evidence demonstrating a strong salt tolerant phenotype as a result of overexpressing the gene. Moreover, the gene structure described by Qi et al. (2014) is different from what it was deposited in NCBI (http://www.ncbi.nlm.nih.gov/nuccore/kf879912), and the expression patterns observed by Guan et al. (2014b) are inconsistent with the role of this gene played in salt tolerance.
Previously, we characterized soybean salt tolerance in WF-7 and mapped a major QTL in the same region on linkage group N that was closely linked with a microsatellite marker, JD33–432 (Ren et al., 2009, 2012). Using the first draft of the soybean genome sequence, and through bioinformatics analysis, we identified two putative cation/proton antiporters on scaffold 63 nearby JD33–432. These two genes were, at the time, named Gm0063×00340 and Gm0063×00341. According to the update soybean sequence database available currently, these two genes are overlapped with Glyma03g32900 (Guan et al., 2014b; Qi et al., 2014) and are predicted as one gene, Glyma.03g171600 (Qi et al., 2014), in the current Wm82.a2 assembly. Here we present the evidence that overexpression of Gm0063×00340, renamed GmST1, produced strong tolerance to salt stress in Arabidopsis seedling and adult stages. In addition, the GmST1-transgenic Arabidopsis lines also increased sensitivity to ABA and decreased production of ROS under salt stress. Furthermore, the Arabidopsis GmST1-transgenic lines also improved tolerance to drought stress. Based on the current version of soybean genome sequence annotation, GmST1 was located within Glyma03g171600, and Glyma03g171600 overlaps with Glyma03g32900. Thus we propose that at least two different models of gene prediction exist for this region. By comparative RT-PCR analysis, GmST1 has been demonstrated as an intact gene functioning in soybean leaf tissues, not a part of Glyma03g171600.
Materials and Methods
Soybean Materials
Two soybean varieties, WF-7, salt tolerant (Ren et al., 2009, 2012), and Union, sensitive to salt stress, were used in this study. WF-7 was kindly provided by Dr. Lijuan Qiu, Chinese Academy of Agricultural Science, Beijing, China. Union was originally received from the USDA Soybean Germplasm Collection (PI548622) and was maintained at the Virginia State University Soybean Breeding Program. Both varieties were grown in a greenhouse with ambient temperature and natural light.
Arabidopsis Transgenic Lines
To produce Arabidopsis transgenic lines, Arabidopsis ecotype Col-0 was used. The DNA fragment covering start and stop codons of GmST1 was amplified from WF-7 genomic DNA using primers GmST1-F (P4) and GmST1-R (P6), cloned into TOPO vector PCR2.1 and sequence-confirmed. The confirmed DNA fragment was then inserted into binary vector pCBK05 between XbaI (5′ end) and SacI (3′ end) sites, driven by the CaMV 35S promoter. The plasmid containing 35S::GmST1 construct was transformed first into Agrobacterium tumefaciens GV3101 and then into Arabidopsis ecotype Col-0 by the floral dipping method (Clough and Bent, 1998). Two independent single-copy homozygous transgenic lines were generated through herbicide resistance selection. Briefly, primary transformants were identified and determined based on the presence of herbicide resistance that is completely linked with the target gene. Individual transgenic lines were further genetically analyzed using herbicide resistance as the indicator in the following T1 and T2 generations to identify single copy T-DNA insertion lines. Two independent homozygous transgenic lines were then bred and the seeds from T3 and/or T4 generations were subjected to various stress tests.
Arabidopsis wild type Col-0 and homozygous GmST1-transgenic lines were grown in soil in a growth chamber with 14/10 light-dark cycles at a light intensity of approximately 130 μmol m-2s-1, 21°C temperature and 60% relative humidity. For germination and seedling assays, Arabidopsis seeds were first surface-sterilized with 50% (v/v) bleach and 0.1% (v/v) Triton X-100 for 7.5 min, and were cold-treated at 4°C for 24 h. The cold-treated seeds were then grown on Murashige and Skoog (MS) solid media with 1% sucrose under continuous light of approximately 50 μmol m-2s-1. The seedlings/plants were maintained in the growth chamber under the same conditions until sampling for experiments as describe below.
GmST1 Overexpressing in Arabidopsis and Transgenic Line Confirmation
To determine the expression of GmST1 in the transgenic Arabidopsis lines, total RNA was isolated from leaves of 3-week-old seedlings of wild type and two independent transgenic lines using TRI reagent (Sigma; St. Louis, MO, USA), following manufacture’s instruction. RT-PCR was conducted to examine the expression of GmST1 in Arabidopsis transgenic lines using primer pair of P4 and P6. After reverse transcription step (Invitrogen Reverse Transcriptase III was used), 30 cycles of PCR amplification (95°C, 30 s-60°C, 30 s-72°C, 1 min) was performed. Arabidopsis actin 1 gene was served as a loading control.
Seed Germination under ABA and NaCl Stress and Seedling Salt Tolerance Assay
To investigate the role and mechanism(s) of GmST1 in abiotic stress tolerance, the wild type and transgenic lines were first subjected to seed germination tests in response to ABA and NaCl stresses. Surface-sterilized and cold-treated seeds were planted on MS medium supplemented with (1) 1, 2.5, and 5 μm ABA; and (2) 50 and 100 mM NaCl, with no ABA- and/or NaCl-supplemented treatment used as the control. All seeds were germinated under continuous light, and germination rate was recorded daily for 4 days. To test salt tolerance ability at seedling stage, 4-day-old germinated seeds of wild type and transgenic lines were transferred to fresh MS medium with or without 100 mM NaCl. All seedlings were grown vertically for additional 6 days under the same growing conditions. Photos were taken to visualize the phenotypes, and root lengths were measured. All tests were repeated twice, and Student’s t-test was used for statistical analysis.
Detection of H2O2 Production
Salt stress-induced H2O2 production was detected using 3,3′-diaminobenzidine (DAB) staining as described by Xia et al. (2009). Four-week-old homozygous transgenic lines and wild type Col-0 were watered with 200 mM NaCl (treated) or water (untreated). After 24 h of treatment, two leaves from each individual plant were detached from salt treated and untreated plants, placed in 1 mg/ml DAB, and incubated at room temperature for 5 h. Samples were de-stained in boiling absolute ethanol, and then pictures were captured for record. Five plants from each transgenic line and wild type were used for H2O2 detection.
Salt Tolerance Test at Adult Stage
Four-week-old wild type and transgenic lines grown in the growth chamber were tested for their salt tolerance ability. All pots containing tested lines were saturated with 200 mM NaCl for 6 days. Control pots were watered with the same amount of water. After the treatment, survival rates were recorded, and pictures were captured to show visible phenotypes. The experiment was repeated twice for statistical analysis.
Water Loss Assay and Drought Tolerance Analysis
To examine the possible role of GmST1 in drought stress tolerance, we first conducted water loss experiments using detached leaves. Three leaves were excised from each 4-week-old plant grown in the growth chamber, and the fresh weight was immediately determined. All leaves were placed on a laboratory bench at room temperature for 210 min. Every 30 min, leaf weight was recorded in the same order that leaves were detached from their corresponding plants. Water loss was calculated as the percentage of the initial fresh weight at each time point. For each genotype (Col-0, GmST1–1, and GmST1–2), five plants were tested for water loss.
To test drought tolerance at adult stage, wild type and a transgenic line were grown under normal watering conditions for 4 weeks and then subjected to drought stress by withdrawing irrigation. After 11 days of drought stress, drought tolerance phenotypes were examined, and pictures were captured to show the difference between the wild type and the transgenic line. A total of 15 plants for each of Col-0 and GmST1–1 were evaluated for drought tolerance.
RNA Extraction and RT-PCR Analysis
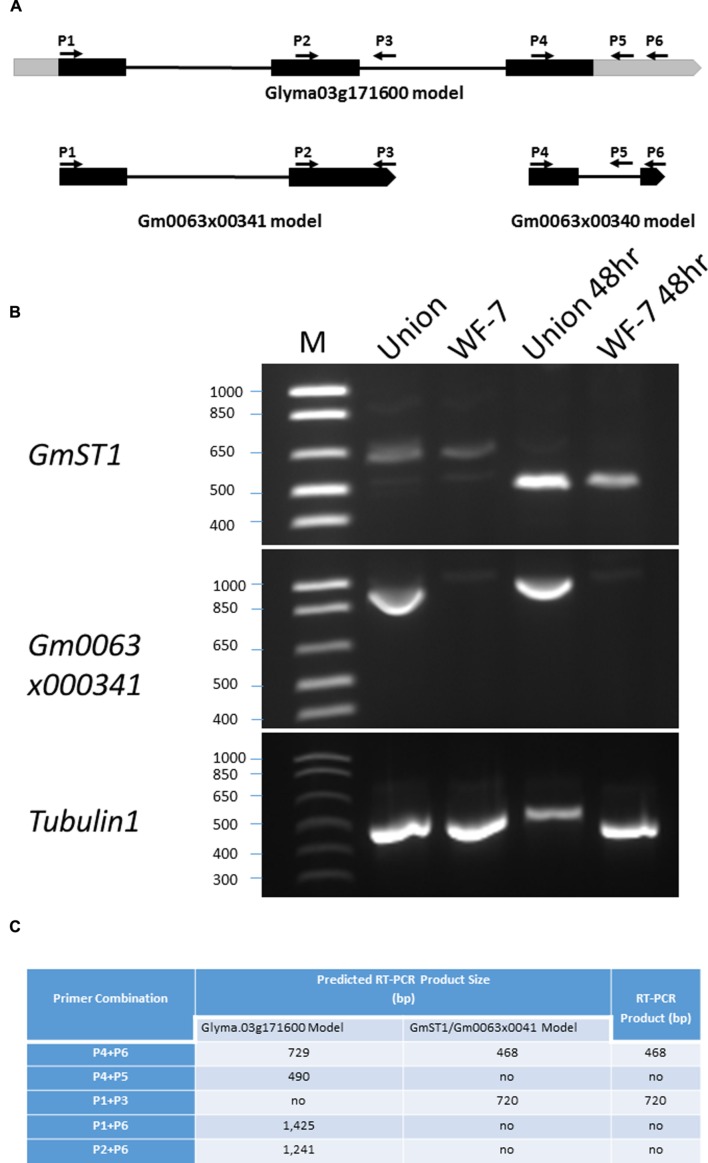
Given that different gene models might exist in the soybean genomic region where GmST1 is located (Ren et al., 2009, 2012), we designed serial primers (Table 1) according to the different gene prediction models and examined GmST1 structure in relation to other predicted models using RT-PCR analysis. WF-7 and Union seedlings at V3 stage were treated with 200 mM NaCl or H2O (control) for 48 h prior to RNA isolation. Total RNAs were extracted from the leaves using TRI® reagent (Sigma; St. Louis, MO, USA) as per the manufacturer’s instructions. RNA was quantified using NanoDrop 2000 (Thermo Scientific; Wilmington, DE, USA), and its quality was monitored on 1.2% agarose gel. Five μg of total RNA for each sample was used for cDNA synthesis in a 20 μl reaction using SuperScript® III (Invitrogen; Carlsbad, CA, USA) according to the manufacturer’s instructions, followed by PCR of 1 μl of the RT products using Taq DNA polymerase (New England BioLabs; Ipswich, MA, USA) with 35 cycles of amplification. The relative position of each designed primer in relation to the gene prediction models was illustrated in Figure 7A, and primers’ names and sequences were listed in Table 1.
Table 1.
List of primers used in reverse transcription-polymerase chain reaction (RT-PCR) analysis of soybean and Arabidopsis lines.
| Primer Name | Sequences |
|---|---|
| GmST1 F (P4) | 5′-TCTAGAATGGCGTTTGTTGCAGCCATG -3′ |
| GmST1 R(P6) | 5′-GAGCTCTCATAAGGTTCGGGGATCCTTTC -3′ |
| GmST1 Intron (P5) | 5′-TTATTTAATTCAAGACTGTGCATC -3′ |
| Gm0063×00341 F (P1) | 5′-GGATCCATGACGTTCAACGCGAGCAC -3′ |
| Gm0063×00341 R (P3) | 5′-ACTAGTTTAAGTTATAACTATAGTAGGTCC -3′ |
| Gm0063×00341 Exon2 (P2) | 5′-TGGAATTTTGTTGGGGCCTTC-3′ |
| GmTubulin1 F | 5′-ATGAGAGAAATCTTGCACATCCAG-3′ |
| GmTubulin1 R | 5′-TAAGGCTCCACAACGGTATC3′-3′ |
| AtActin1 F | 5′-ATGCTGGTATCCATGAAACCACCT-3′ |
| AtActin1 R | 5′-CCTGTGAACAATCGATGGACCTGA-3′ |
FIGURE 7.
Characterization of gene structure around GmST1 genomic region. (A) Schematic summary of different gene prediction models around GmST1 genomic region. Black bars represent exons, gray bars are untranslated regions (5′-UTR and 3′-UTR), and lines represent introns in their expected gene models. Primer positions (P1 through P6) are indicated in response to different gene prediction models. (B) GmST1 and Gm0063×00341 expression pattern analysis in soybean leaf tissues. RT-PCR was conducted using primer pairs corresponding to GmST1 and Gm0063×00341, respectively. Total RNA was isolated from the salt treated and untreated leaves of soybean varieties WF-7 and Union. (C) Prediction and experimental RT-PCR products around GmST1 genomic region.
Analysis of Regulatory Elements on Putative Promoter of GmST1
To further define GmST1 gene structure, the putative promoter, a 1,269 bp DNA fragment from the start codon of GmST1 to the stop codon of the upstream gene (Gm0063×00341), was analyzed for its possible regulatory elements. TATA box was predicted using TSSP (Solovyev, 2001) and other regulatory elements were analyzed in NSite-PL software (Solovyev et al., 2010).
Results
Overexpressing Soybean Gene GmST1 in Arabidopsis
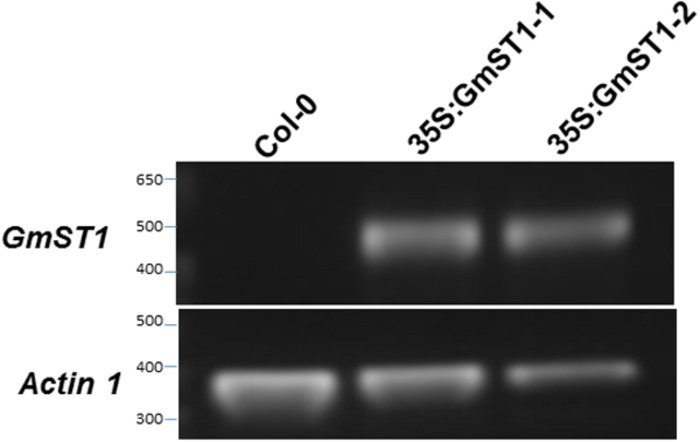
To examine the role of GmST1 in salt tolerance in plants, we cloned a genomic DNA fragment covering the full length of cDNA of GmST1 from soybean variety WF-7. Driven by CAMV 35S promoter, we transferred the construct into Arabidopsis ecotype Col-0. Transformants were selected based on herbicide resistance as the maker, and two independent homozygous transgenic lines carrying a single copy of T-DNA insertion were developed for further characterization of the roles of GmST1 in abiotic stress tolerance. RT-PCR analysis showed high expression of GmST1 in both Arabidopsis transgenic lines (Figure 1).
FIGURE 1.
Reverse transcription polymerase chain reaction (RT-PCR) confirmation of GmST1 overexpression in Arabidopsis thaliana. Total RNA was isolated from leaves of wild type and two independent CAMV 35S:GmST1 Arabidopsis transgenic lines. RT-PCR was conducted using soybean primers P4 and P6. Arabidopsis actin 1 was used for loading control.
GmST1 Enhances Seed Germination and Root Elongation under Salt Stress
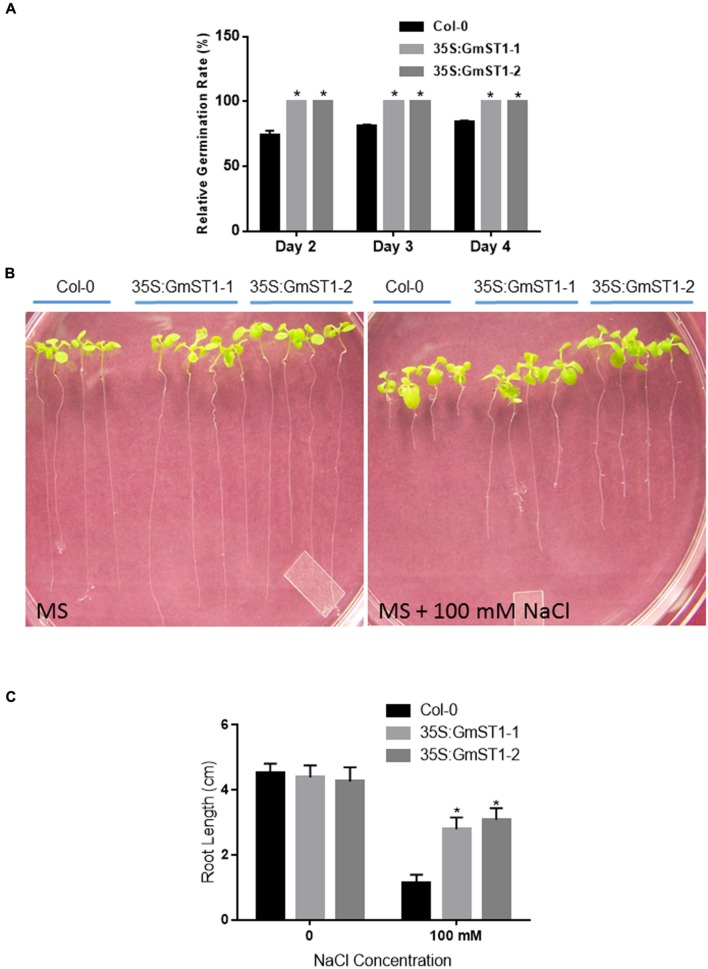
The transgenic lines were first investigated for their responses to salt stress during the germination and the seedling stages. Both transgenic lines, together with wild type Col-0, were germinated on MS medium supplemented with 50 mM or 100 mM NaCl. Comparing to MS control, there were no significant differences in seed germination between Col-0 and GmST1 transgenic lines under 50 mM NaCl stress. All seeds were fast-germinated on day 2 (data not shown). However, under the stress treated with 100 mM NaCl, the seed germination of Col-0 was significantly inhibited, with only about 70% of seeds germinated on day 2 and 83% germinated by the end of experiment day 4 (Figure 2A). On the contrary, 100% of seeds were geminated at day 2 in both transgenic lines. These results demonstrated that GmST1 expression improved seed germination under high salt stress.
FIGURE 2.
Effect of GmST1 on Arabidopsis seed germination and seedling development under salt stress. (A) Seed germination rate of Arabidopsis transgenic lines was enhanced by GmST1 under 100 mM NaCl condition. (B) Effect of GmST1 overexpression on Arabidopsis root development under control and 100 mM NaCl conditions. (C) Quantitative measurements of GmST1 effects on root growth. Experiments were biologically duplicated and bars represent means ± SD. ∗P < 0.05, as determined by Student’s t-test.
Furthermore, we examined the effect of GmST1 expression on root elongation under salt stress. Synchronized Arabidopsis seedlings with similar initial root length were vertically grown on MS medium with or without 100 mM NaCl. As shown in Figures 2B,C, on MS only medium, wild type and both GmST1 transgenic lines grew normally with similar root elongation. However, under 100 mM NaCl stress, root elongation of Col-0 was significantly inhibited, with about 75% inhibition observed compared with the control without stress. On the other hand, both transgenic lines showed significant greater root elongation relative to that of Col-0 under the same salt condition. The root length of transgenic lines grew two times more than that of Col-0, and only about 25% inhibition was produced by salt stress compared with no salt stress applied. These results confirmed the role of GmST1 in salt tolerance in responses both to seed germination and root elongation at seedling stage.
GmST1 Expression Confers Strong Salt Tolerance in Arabidopsis at Adult Stage
Salt tolerance phenotype caused by GmST1 at the seedling development stage prompted us to further test whether or not GmST1 could improve salt tolerance at later stages during plants’ growth. As shown in the duplicated experiments with 4-week-old wild type Col-0 and transgenic lines (Figures 3A,B), all lines grew normally without NaCl treatment. The treatment with 200 mM NaCl caused severe damage on wild type Col-0, while no or less damage by NaCl was observed on either of the GmST1 overexpression lines (Figure 3A). After 6 days of treatment, only 10% of wild type plants survived; however, 95 and 78% of survival rates were recorded for the two transgenic lines. These data strongly suggest that soybean GmST1 expression would improve salt tolerance in Arabidopsis.
FIGURE 3.
Effect of GmST1 overexpression on Arabidopsis salt tolerance at adult stage. (A) Overexpression of GmST1 improves salt tolerance ability in Arabidopsis. 200 mM NaCl were used to treat 3-week-old wild type Col-0, and two overexpressing lines (GmST1–1 and GmST1–2). Control plot was watered with tap water. (B) Survival rate of NaCl treated lines of Col-0, GmST1–1, and GmST1–2. Two independent experiments (with 25 plants each) were conducted.
GmST1 Expression Reduces ROS Production during Salt Stress
Abiotic stresses cause damage to plants and lead to over production of ROS, such as H2O2 (Mustilli et al., 2002; Apel and Hirt, 2004). Regulation of ROS production is one of the mechanisms that crops use to defend against different biotic and abiotic stresses (Mustilli et al., 2002; Apel and Hirt, 2004; Jaspers and Kangasjarvi, 2010; Torres, 2010). To investigate if GmST1’s salt tolerance response involves ROS-related mechanism(s), we examined H2O2 production during salt stress. Leaves from both NaCl-treated and untreated control plants (4-week-old) of Col-0 and transgenic lines (five individual plants from each line/treatment) were excised and stained by DAB to detect the production of H2O2. As shown in Figure 4, there was no difference between Col-0 and transgenic lines in H2O2 production under the control condition (Figure 4, upper). However, when plants treated with 200 mM NaCl for 48 h, wild type Col-0 produced more H2O2 than that of either transgenic lines (Figure 4, lower), indicating a possible involvement of ROS signaling in GmST1-mediated salt tolerance.
FIGURE 4.
Effect of GmST1 overexpression on reactive oxygen species (ROS) production under control and salt stress. Hydrogen peroxide was detected by incubating leaves excised from Arabidopsis lines treated or untreated with 200 mM NaCl and stained by 1 mg/ml DAB solution for 5 h followed by de-staining. Upper panel represents Arabidopsis lines under control condition, and lower panel represents Arabidopsis lines treated with 200 mM NaCl.
GmST1 Enhances ABA Sensitivity in Arabidopsis
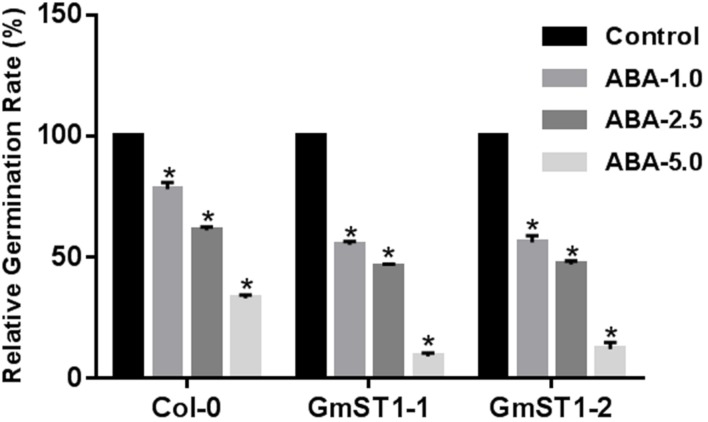
To further examine the role of GmST1 in abiotic stress tolerance, we tested whether or not GmST1 overexpression could alter ABA sensitivity during the seed germination stage. Seeds of Col-0 and two transgenic lines were surface-sterilized and germinated on MS media containing none or 1 μM, 2.5 μM, or 5 μM ABA. Shown in Figure 5, all seeds of either the transgenic lines or Col-0 were germinated on MS medium without ABA treatment. On the MS medium containing ABA, seed germination was inhibited by ABA in a dosage-dependent manner. Relatively, seed germination of the two transgenic lines were significantly more inhibited than that of Col-0 at all ABA concentration levels. At 5 μM ABA level, over 30% of Col-0 seeds germinated, while only about 10% seeds of transgenic lines were able to germinate. These results clearly demonstrate that GmST1 regulates salt tolerance, at least in part, through an ABA-dependent pathway.
FIGURE 5.
Overexpression of GmST1 enhances ABA sensitivity. Seeds of Arabidopsis lines (Col-0, GmST1–1, and GmST1–2) were germinated on MS medium supplemented with 0, 1.0 μM, 2.5 μM, and 5.0 μM ABA. Relative germination rate was calculated. Experiments were biologically duplicated. Bars represent means ± SD. ∗P < 0.05, as determined by Student’s t-test.
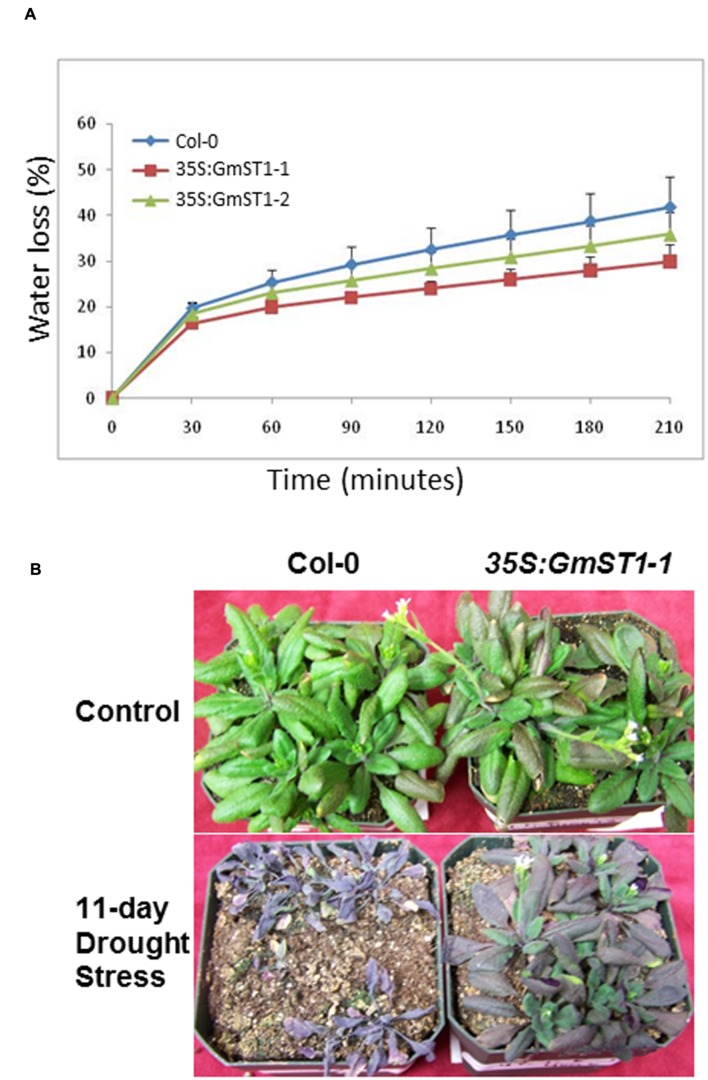
GmST1 Reduces Leaf Water Loss and Improves Drought Tolerance in Arabidopsis
Since both ABA and ROS are regarded as classical signal pathways that plants evolved for defending against abiotic stresses, the fact that GmST1 both increased ABA sensitivity and decreased ROS production under salt stress prompted us to further examine if it could also strengthen drought tolerance in Arabidopsis. We first conducted water loss experiments using detached leaves. As shown in Figure 6A, during a 210-min period, wild type Col-0 lost more than 40% of water due to evaporation, while two GmST1 transgenic lines evaporated only less than 30% of water. The significant difference in water loss between Col-0 and the transgenic lines indicated a possible role of GmST1 in plants’ drought tolerance. We further compared the drought tolerance between Col-0 and a transgenic line for an extended period of drought stress. After 11 days of water withholding, Col-0 plants died of stress (Figure 6B), while almost all the plants from the transgenic line survived. It suggests that GmST1 may play an important role in drought stress tolerance in plants.
FIGURE 6.
Effect of GmST1 overexpression on Arabidopsis drought tolerance. (A) Overexpression of GmST1 reduces water loss through transpiration. Leaves were excised from 4-week-old Arabidopsis lines (Col-0, GmST1–1, and GmST1–1) and monitored their water losses under ambient temperature for 210 min. Water loss was calculated as the percentage of initial fresh weight. (B) Overexpression of GmST1 confers drought tolerance in Arabidopsis. 4-week-old Arabidopsis plants (Col-0 and GmST1–1) were subjected to a water stress by withholding water. Photos were taken after 11 days of drought stress.
GmST1 Serves as an Intact Gene, Not Part of Glyma.03g171600, in Soybean Leaf Tissue
In the first draft of the soybean genome sequence, GmST1 and Gm0063×00341 were initially annotated as two independent genes, both encoding putative cation/proton antiporters. In soybean genome sequence versions Glyma 1.0 and Glyma 1.1, this region was predicted to be a part of Glyma03g32900 (Schmuta et al., 2010). Later, in the newly released version Glyma 2.0, these two genes (GmST1 and Gm0063×00341) were predicted to be a single gene, named Glyma.03g171600 (Qi et al., 2014), but with different intron-exon predictions. Given that different gene models were proposed in this region, we further characterized the GmST1 gene structure in relation to Gm0063×00341 and Glyma.03g171600. The designated primers are listed in Table 1 and their relative positions in the region are illustrated in Figure 7A.
In the GmST1 model, the GmST1 gene covers 729 bp genomic sequence with two exons and one intron. Its full length cDNA is 468 bp. In the Glyma.03g171600 model, the GmST1 DNA fragment lies in its 4th exon with a 2 bp shift to the left and a 121 bp extra sequence at 3 prime end. However, no intron was predicted in this region in the Glyma.03g171600 model. Therefore, we first conducted RT-PCR analysis using Primer pair P4 and P6, which are GmST1 forward and reverse primers covering the full length cDNA. As shown in Figure 7B, Primer combination P4 and P6 successfully amplified a strong RT-PCR product at about 500 bp in both salt-treated WF-7 and Union leaves, indicating that GmST1 was induced by salt and that there was a predicted intron in GmST1. We then designed a reverse primer (P5) within the predicted intron region of GmST1 but annotated as 3′ UTR in the Glyma.03g171600 model, and we performed RT-PCR using the P4 and P5 combination. If the Glyma.03g171600 model was correct for the case in our study, this combination should produce a 490 bp band. However, the RT-PCR analysis using P4 and P5 failed to amplify any PCR product (Figure 7C), indicating that P5 was located within the intron region.
In the Glyma.03g171600 model, Gm0063×00341 belongs to the 5′ part of the Glyma.03g171600 gene, and the primer P3 (Figure 7A), located at the 3′ end of Gm0063×00341, is predicted to be in the intron of the Glyma.03g171600 gene. We conducted RT-PCR using P1 and P3. If Gm0063×00341 was an intact gene, then the P1 and P3 combination should yield 720 bp RT-PCR product; and if the Glyma.03g171600 prediction model was correct, then it should not amplify any RT-PCR product. Shown in Figure 7B, we successfully amplified an RT-PCR product at about 720 bp in both salt-treated and untreated plants of the variety Union. These results clearly demonstrate that both GmST1 and Gm0063×00341 are not part of the Glyma.03g171600 gene in soybean leaf tissue.
Furthermore, we discovered that the expression patterns of GmST1 and Gm0063×00341 were significantly different. GmST1 was induced by salt and strongly expressed in both WF-7 and Union upon salt treatment. On the other hand, Gm0063×00341 was highly expressed only in Union (both control and salt-treated samples) and its expression was not responsive to salt stress. To further examine whether or not GmST1 and Gm0063×00341 belong to the same gene, we conducted RT-PCR analyses using primer pairs of P1 and P6, and P2 and P6 (where P2 is located on the second exon of the Gm0063×00341 gene). As predicted, neither primer combination yielded RT-PCR products at the corresponding sizes (Figure 7C). Taken together, our results strongly suggest that GmST1 and Gm0063×00341 serve as independent genes in soybean leaf tissues.
GmST1 Promoter Analysis Reveals a Putative ABA Responsive Element and a Dehydration Responsive Element
To further confirm that GmST1 served as an intact gene, we analyzed a 1,269 bp DNA fragment covering from the stop codon of the upstream gene and the start codon of GmST1. This fragment covers the whole sequence of the 2nd intron of Glyma.03g171600. Sequence analysis revealed 78% AT composition and a TATA box (TATAAA) at position -122. Furthermore, through NSite-PL prediction, we identified an ABRE element (GTCAAGTGTC) (Yamamoto et al., 2009) at position -985, and a DRE element (TTAGTCGGTT) (Chen H. et al., 2010) at position -1,049. The discovery of a putative functional promoter and possible stress-related cis-acting elements further indicate that GmST1 may serve as an intact gene and play roles in soybean abiotic stress tolerance.
Discussion
Abiotic stresses, including salinity, are a great challenge and major restricting factor for crop production worldwide. It was predicted that about 50% of the world arable land would be affected by salinity by 2050 (Blumwald and Grover, 2006). One of the best solutions to such agricultural challenge is to efficiently use genetic resources to develop salt tolerant cultivars. We previously characterized WF-7 as a salt-tolerant soybean variety (Ren et al., 2012) and mapped a major QTL associated with salt tolerance to the linkage group N near a microsatellite marker JD33–432 in the mapping population of WF-7 × Union (Ren et al., 2009). By initial bioinformatics search in the rough draft of the soybean genome sequence, a putative cation/proton antiporter was identified nearby JD33–432. In this study, we cloned this putative cation/proton antiporter, named soybean salt tolerance 1 (GmST1), and investigated its roles in abiotic stress tolerance when overexpressed in Arabidopsis thaliana. The results showed that overexpression of GmST1 in Arabidopsis produced strong tolerance to salt stress at both seedling and adult stages (Figures 2 and 3). Moreover, given that GmST1 caused transgenic Arabidopsis lines to exhibit more sensitivity to ABA (Figure 5), we speculate that GmST1 may regulate salt tolerance through an ABA-dependent pathway. The GmST1 encodes a putative cation/proton antiporter. Many cation/proton antiporters have been cloned from different plant species, and their potential roles in salt tolerance also have been clearly demonstrated (Shi et al., 2000; Kato et al., 2001; Brini et al., 2007). However, mechanisms in regulating salt tolerance by cation/proton antiporters still remain uncovered. For example, Arabidopsis SOS1 and AtHKT1 both conferred salt tolerance (Shi et al., 2000; Kato et al., 2001; Rus et al., 2001). Even though ABA-responsive elements were discovered in their promoter regions (Osakabe et al., 2014), SOS1 seemed to be regulated by an ABA-independent pathway (Yamaguchi-Shinozaki and Shinozaki, 2006), while AtHKT1 was regulated partially by ABA-dependent signaling pathway (Shkolnik-Inbar et al., 2013). We previously found that the salt tolerance in WF-7 was partially regulated by ABA (Ren et al., 2012). Although we did not directly test if GmST1 was induced by ABA in soybean, an ABRE cis-acting element (Bonetta and McCourt, 1998; Yamamoto et al., 2009) and a DRE cis-acting element (Chen H. et al., 2010) were identified in its putative promoter region. Additionally, overexpression of GmST1 in Arabidopsis enhanced ABA sensitivity. Taken together, these results assured at least a partial involvement of ABA in GmST1-mediated salt tolerance.
ABA is considered an abiotic stress hormone, and in most cases, an ABA-dependent signaling pathway regulates plant responses to multiple stresses (Larkindale and Knight, 2002; Zhu, 2002; Fujita et al., 2005; Chen L.T. et al., 2010). Thus, we further tested GmST1’s ability to defend against drought stress, the major cause of crop losses worldwide. The results indicated that GmST1 expression prevented water loss from leaves and improved drought tolerance during the adult stage (Figure 6). This is not surprising, though, as many other cation/proton antiporters from different species also enhanced both salt and drought tolerance when being overexpressed in Arabidopsis (Brini et al., 2007; Wei et al., 2011). Such dual functions in salt and drought tolerance would allow scientists to efficiently engineer the abiotic stress tolerance of crops, thus leading to improved agricultural production on lands that are marginal due to salt and/or drought problems. Both drought and salt tolerance in soybean were shown to be related to the accumulation of soluble saccharides and proline, and less damage of chlorophyll when stresses occurred (Abd El-Samad and Shaddad, 1997). Such physiological traits and Na+, K+, and Cl- accumulations in shoots, together with possible alternations of the expressions for genes on ABA signaling pathways, would be excellent targets for the future investigation on the precise physiological and molecular mechanism(s) that drive GmST1’s abiotic stress tolerance.
Abiotic stresses usually cause physiological damage to crops and lead to more ROS production that, in turn, damages cell and DNA structures, eventually leading to apoptosis. ROS signaling is known to be involved in regulating plants’ responses to both biotic and abiotic stresses (Mustilli et al., 2002; Jaspers and Kangasjarvi, 2010; Torres, 2010). One way to improve abiotic stress tolerance is to alleviate ROS production during stress periods. In the present study, we discovered that, after a 48-h salt treatment, GmST1 transgenic lines produced less H2O2 than that of the wild type control (Figure 4). In one way, this result can be interpreted to indicate that salt stress caused less damage to GmST1 transgenic lines than that to the wild type control. On the other hand, it also implies a possible mechanism that GmST1 uses in regulating plant abiotic stress tolerance.
In recent studies, Guan et al. (2014b) and Qi et al. (2014) independently identified Glyma03g32900 as a potential candidate gene governing soybean salt tolerance. Our present study demonstrated that GmST1 conferred strong salt tolerance in Arabidopsis transgenic lines. GmST1 is located on linkage group N and is overlapped with Glyma03g32900. In consistent with these findings, multiple QTL mapping studies for soybean salt tolerance, independently conducted involving different salt-tolerant cultivars and/or different species (wild and cultivated soybeans), also identified a major QTL in this region on linkage group N and suggested that it plays a critical role in improving soybean salt tolerance under different genetic backgrounds (Lee et al., 2004; Hamwieh and Xu, 2008; Tuyen et al., 2010; Hamwieh et al., 2011; Ha et al., 2013; Guan et al., 2014a,b; Qi et al., 2014). The importance of this genomic region in control of soybean salt tolerance warrants a further analysis of genome/gene structures, and molecular and physiological mechanisms of this locus that controls salt tolerance.
In search of salt tolerance candidate genes in soybean, previously a cation/proton antiporter (GmCAX1) was identified (Luo et al., 2005). Transgenic Arabidopsis lines overexpressing GmCAX1 accumulated less Na+ and were more resistant to salt stress during germination stage (Luo et al., 2005). Additionally, novel sodium/proton antiporters, GmNHX1, and GmNHX2, were also cloned from soybean (Sun et al., 2006; Zhou et al., 2009). Overexpression of GmNHX1 in Lotus corniculatus also exhibited strong salt tolerance and reduced accumulation of Na+ in shoots (Sun et al., 2006), while overexpression of GmNHX2 in Arabidopsis also resulted in salt tolerance at germination and seedling stages (Zhou et al., 2009). GmST1 was annotated as a cation/proton antiporter. However, the size of GmST1 is relatively small (Accession KU871394), in comparison with other homologous genes isolated in other plant species (Shi et al., 2000; Brini et al., 2007; Wei et al., 2011). Since cation/proton antiporters belong to transmembrane proteins, the size of GmST1 does not fit into this group of genes. According to the newly annotated soybean genome database, GmST1 overlaps with the Glyma.03g171600 gene and belongs to the three prime part of the Glyma.03g171600. In addition, Gm0063x0041, an adjacent (upstream) gene to GmST1, was annotated as the five prime part of Glyma.03g171600. However, intron-exon splicing sites predicted in the Glyma.03g171600 model were completely different from those based on the model which interpreted GmST1 and Gm0063x0041 as individual genes.
Recently Glyma.03g171600 was identified as GmCHX1 and contributed to soybean salt tolerance (Qi et al., 2014). The intact GmCHX1 gene was highly expressed in soybean roots, but was barely detected in soybean shoots (Guan et al., 2014b; Qi et al., 2014). Through RT-PCR analysis, we were able to examine if GmST1 and Gm0063x0041 belong to parts of GmCHX1 or if they act as individual genes. Using primer combinations of Gm0063x0041 forward (P1 and P2, respectively) and GmST1 reverse (P6), we failed to amplify any RT-PCR products from either salt-treated or untreated soybean leaves. Though, we could not rule out the GmCHX1 model given that GmCHX1 was not expressed in leaf tissues (Guan et al., 2014b; Qi et al., 2014). By examining the expressions of GmST1 and Gm0063x0041 as individual genes, however, we found that both genes were expressed in soybean leaves. GmST1 was highly induced by salt treatment in both salt-sensitive and salt-tolerant soybean varieties, but less expressed in the conditions with no salt treatment. On the other hand, Gm0063x0041 was highly expressed in the salt-sensitive soybean Union under both control and salt-treated conditions but only a very low level of expression was detected in the salt-tolerant soybean WF-7. Interestingly, according to the GmCHX1 model, Gm0063x0041 reverse primer (P3) was located in the intron of GmCHX1 and should not be able to amplify the RT-PCR product if the GmCHX1 model was correct, but it did amplify the RT-PCR product. Furthermore, we designed another reverse primer, P5, to further verify the model. In the GmST1 model, P5 was located in the intron region, but in the GmCHX1 model, it was located at 3′ UTR. Compared with the P4+P6 combination, the P4+P5 combination did not yield any RT-PCR product at the predicted size. It is clearly indicated that both GmST1 and Gm0063×00341 are not part of the Glyma.03g171600 gene but two individual genes in soybean leaf tissues, though they are overlapped by the latter. It also implies that the current Glyma.03g171600 gene annotation might need further revision. Based on our data and those reported by Guan et al. (2014b) and Qi et al. (2014), we hypothesize that different intron-exon splicing sites exist in this genomic region. In soybean root tissues, a splicing pattern forms one gene, GmCHX1, while in soybean leaf tissues, different splicing patterns create two distinct genes in the same genomic region. This phenomena is an exciting observation and warrants further investigation into the details of intron-exon site reorganization in different organs and genome structures in this genomic region.
Conclusion
We have cloned GmST1 from the salt-tolerant soybean variety WF-7, and we have demonstrated that GmST1 enhances both salt and drought tolerance through Arabidopsis in planta study. Furthermore, GmST1 also increases ABA sensitivity and reduces ROS production upon salt treatment. Additionally, GmST1 is strongly induced by salt stress in soybean leaf tissues and functions as an intact gene, rather than as a part of the Glyma.03g171600 gene. Further comparative studies between GmST1 and Glyma.03g171600, as well as on the possible different intron-exon splicing patterns in different organs, will be required to fully describe the real gene structure and gene models within this genomic region. Regardless, the fact that GmST1 overexpression improved both salt and drought tolerance in Arabidopsis suggests that it may be worthwhile to elucidate the contribution of this gene to multi-abiotic tolerance in crops. The overexpression of GmST1 in crops, including soybean, may provide a new approach to genetically engineering agricultural and economical plants of importance with tolerance to multiple abiotic stresses simultaneously.
Accession Number
cDNA sequence of GmST1 gene is submitted to GeneBank with accession number KU871394.
Author Contributions
SR designed experiments, SR, CL, and AP conducted experiments, SR and G-LJ analyzed data and wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank USDA for its Evans Allen formula funds to support the research programs at the Virginia State University (to SR and G-LJ). Dr. Sarah Weeda, who passed away in a car accident in summer of 2015, contributed significantly in conduction of the designed experiments. We would also like to thank Ms. Roz Stein for her editing and proof reading of the manuscript. The authors would also like to thank reviewers for their valuable comments. This article is a contribution of the Virginia State University, Agricultural Research Station (Journal series No. 329).
References
- Abd El-Samad M., Shaddad M. (1997). Salt tolerance of soybean cultivars. Biol. Plant. 39 263–269. 10.1023/A:1000309407275 [DOI] [Google Scholar]
- Abel G., MacKenzie A. (1964). Salt tolerance of soybean varieties (Glycine max L. Merrill) during germination and later growth. Crop Sci. 4 157–161. 10.2135/cropsci1964.0011183X000400020010x [DOI] [Google Scholar]
- Allakhverdiev S., Sakamoto A., Nishiyama Y., Inaba M., Murata N. (2000). Ionic and osmotic effects of NaCl-induced inactivation of photosystems I and II in Synechococcus sp. Plant Physiol. 123 1047–1056. 10.1104/pp.123.3.1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki A., Kanegami A., Mihara M., Kojima T., Shiraiwa M., Takahara H. (2005). Molecular cloning and characterization of a novel soybean gene encoding a leucine-zipper-like protein induced to salt stress. Gene 356 135–145. 10.1016/j.gene.2005.04.014 [DOI] [PubMed] [Google Scholar]
- Apel K., Hirt H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55 373–399. 10.1146/annurev.arplant.55.031903.141701 [DOI] [PubMed] [Google Scholar]
- Blumwald E., Aharon G. S., Apse M. P. (2000). Sodium transport in plant cells. Biochim. Biophys. Acta 1465 140–151. 10.1016/S0005-2736(00)00135-8 [DOI] [PubMed] [Google Scholar]
- Blumwald E., Grover A. (2006). “Salt tolerance,” in Plant Biotechnology: Current and Future Uses of Genetically Modified Crops, ed. Halford N. G. (Chichester: John Wiley and Sons Ltd; ), 206–224. [Google Scholar]
- Bonetta D., McCourt P. (1998). Genetic analysis of ABA signal transduction pathways. Trends Plant Sci. 3 231–235. 10.1016/S1360-1385(98)01241-2 [DOI] [Google Scholar]
- Brini F., Hanin M., Mezghani I., Berkowitz G., Masmoudi K. (2007). Overexpression of wheat Na+/H+ antiporter TNHX1 and H+-pyrophosphatase TVP1 improve salt- and drought-stress tolerance in Arabidopsis thaliana plants. J. Exp. Bot. 58 301–308. 10.1093/jxb/erl251 [DOI] [PubMed] [Google Scholar]
- Chang R., Chen Y., Shao G., Wan C. (1994). Effect of salt stress on agronomic characters and chemical quality of seed in soybean. Soybean Sci. 13 101–105. [Google Scholar]
- Chen H., Hwang J. E., Lim C. J., Kim D. Y., Lee S. Y., Lim C. (2010). Arabidopsis DREB2C functions as a transcriptional activator of HsfA3 during the heat stress response. Biochem. Biophys. Res. Commun. 401 238–244. 10.1016/j.bbrc.2010.09.038 [DOI] [PubMed] [Google Scholar]
- Chen L. T., Luo M., Wang Y. Y., Wu K. (2010). Involvement of Arabidopsis histone deacetylase HDA6 in ABA and salt stress response. J. Exp. Bot. 61 3345–3353. 10.1093/jxb/erq154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Wang Q., Chen X., Xu Z., Li L., Ye X. (2007). GmDREB2 a soybean DRE-binding transcription factor, conferred drought and high-salt tolerance in transgenic plants. Biochem. Biophys. Res. Commun. 353 299–305. 10.1016/j.bbrc.2006.12.027 [DOI] [PubMed] [Google Scholar]
- Chung W., Lee S., Kim J., Heo W., Kim M., Park C. (2000). Identification of a calmodulin-regulated soybean Ca2+ATPase (SCA1) that is located in the plasma membrane. Plant Cell 12 1393–1407. 10.2307/3871138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Conde A., Chaves M. M., Gero’s H. (2011). Membrane transport, sensing and signaling in plant adaptation to environmental stress. Plant Cell Physiol. 52 1583–1602. 10.1093/pcp/pcr107 [DOI] [PubMed] [Google Scholar]
- Epstein E., Norlyn J. D., Rush D. W., Kings R. W., Kelly D. B. (1980). Saline culture of crops. A general approach. Science 210 399–404. 10.1126/science.210.4468.399 [DOI] [PubMed] [Google Scholar]
- Essa T. (2002). Effect of salinity stress on growth and nutrient composition of three soybean (Glycine max) cultivars. J. Agron. Crop Sci. 188 86–93. 10.1046/j.1439-037X.2002.00537.x [DOI] [Google Scholar]
- Fujita Y., Fujita M., Satoh R., Maruyama K., Parvez M., Seki M., et al. (2005). AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17 3470–3488. 10.1105/tpc.105.035659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan R., Chen J., Jiang J., Liu G., Liu Y., Tian L., et al. (2014a). Mapping and validation of a dominant salt tolerance gene in the cultivated soybean variety Tiefeng8. Crop J. 2 358–365. 10.1016/j.cj.2014.09.001 [DOI] [Google Scholar]
- Guan R., Qu Y., Guo Y., Yu L., Jiang J., Chen J., et al. (2014b). Salinity tolerance in soybean is modulated by natural variation in GmSALT3. Plant J. 80 937–950. 10.1111/tpj.12695 [DOI] [PubMed] [Google Scholar]
- Ha B., Vuong T., Velusamy V., Nguyen H., Shannon J., Lee J. (2013). Genetic mapping of quantitative trait loci conditioning salt tolerance in wild soybean (Glycine soja) PI 483463. Euphytica 193 79–88. 10.1007/s10681-013-0944-9 [DOI] [Google Scholar]
- Hamwieh A., Tuyen D., Cong H., Benitez E., Takahashi R., Xu D. (2011). Identification and validation of a major QTL for salt tolerance in soybean. Euphytica 179 451–459. 10.1007/s10681-011-0347-8 [DOI] [Google Scholar]
- Hamwieh A., Xu D. (2008). Conserved salt tolerance quantitative trait locus (QTL) in wild and cultivated soybean. Breed. Sci. 58 355–359. 10.1270/jsbbs.58.355 [DOI] [Google Scholar]
- Jaspers P., Kangasjarvi J. (2010). Reactive oxygen species in abiotic stress signaling. Physiol. Plant. 138 405–413. 10.1111/j.1399-3054.2009.01321.x [DOI] [PubMed] [Google Scholar]
- Katerji N., van Hoorn J., Hamdy A., Mastrorilli M., Karam F. (1998). Salinity and drought, a comparison of their effects on the relationship between yield and evapotranspiration. Agric. Water Manag. 36 45–54. 10.1016/S0378-3774(97)00049-8 [DOI] [Google Scholar]
- Kato Y., Sakaguchi M., Mori Y., Satio K., Nakamura T., Baker E. P., et al. (2001). Evidence in support of a four transmembrance-pore-transmember topology model for the Arabidopsis thaliana Na+/K+ translocating AtHKT1 protein, a member of the superfamily of K+ transporters. Proc. Natl. Acad. Sci. U.S.A. 98 6488–6493. 10.1073/pnas.101556598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkindale J., Knight M. R. (2002). Protection against heat stress-induced oxidative damage in Arabidopsis involves calcium, Abscisic Acid, Ethylene, and Salicylic Acid. Plant Physiol. 128 682–695. 10.1104/pp.010320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauchli A. (1984). “Salt exclusion: an adaptation of legume crops and pastures under saline conditions,” in Salinity Tolerance in Plants: Strategies for Crop Improvement, eds Staples R. C., Toeniessen G. H. (New York, NY: John Wiley and Sons; ), 171–187. [Google Scholar]
- Lee G., Boerma H., Villagarcia M., Zhou X., Carter T., Li Z., et al. (2004). A major QTL conditioning salt tolerance in S-100 soybean and descendent cultivars. Theor. Appl. Genet. 109 1610–1619. 10.1007/s00122-004-1783-9 [DOI] [PubMed] [Google Scholar]
- Lenis J., Ellersieck M., Blevins D., Sleper D., Nugyen H., Dunn D., et al. (2011). Differences in ion accumulation and salt tolerance among Glycine accessions. J. Agron. Crop Sci. 197 302–310. 10.1111/j.1439-037X.2011.00466.x [DOI] [Google Scholar]
- Li F., Zhang L., Wang G., Cao Y., Wang J., Tang K. (2006). Cloning and characterization of a salt tolerance related gene from Glycine max. Mol. Plant Breed. 4 464–468. [Google Scholar]
- Liao Y., Zhang J., Chen S., Zhang W. (2008). Role of soybean GmZIP132 under abscisic acid and salt stresses. J. Integr. Plant Biol. 50 221–230. 10.1111/j.1744-7909.2007.00593.x [DOI] [PubMed] [Google Scholar]
- Liao Y., Zou H., Wei W., Hao Y., Tian A., Huang J., et al. (2010). Soybean GmbZIP44 GmbZIP62 and GmbZIP78 genes function as negative regulator of ABA signaling and confer salt and freezing tolerance in transgenic Arabidopsis. Theor. Appl. Genet. 121 229–236. 10.1007/s00425-008-0731-3 [DOI] [PubMed] [Google Scholar]
- Lu K. X., Cao B. H., Feng X. P., He Y., Jiang D. A. (2009). Photosynthetic response of salt tolerant and sensitive soybean varieties. Photosynthesis 47 381–387. 10.1007/s11099-009-0059-7 [DOI] [Google Scholar]
- Luo G., Wang H., Huang J., Tian A., Wang Y., Zhang J. (2005). A putative plasma membrane cation/proton antiporter from soybean confers salt tolerance in Arabidopsis. Plant Mol. Biol. 59 809–820. 10.1007/s11103-005-1386-0 [DOI] [PubMed] [Google Scholar]
- Mahajan S., Pandey G., Tuteja N. (2008). Calcium- and salt-stress signaling in plants: shedding light on SOS pathway. Arch. Biochem. Biophys. 471 146–158. 10.1016/j.abb.2008.01.010 [DOI] [PubMed] [Google Scholar]
- Meng Q., Zhang C., Gai J., Yu D. (2007). Molecular cloning, sequence characterization and tissue-specific expression of six NAC-like genes in soybean (Glycine max). J. Plant Physiol. 164 1002–1012. 10.1016/j.jplph.2006.05.019 [DOI] [PubMed] [Google Scholar]
- Mustilli A., Merlot S., Vavasseur A., Fenzi F., Giraudat J. (2002). Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 14 3089–3099. 10.1105/tpc.007906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakabe Y., Yamaguchi-Shinozaki K., Shinozaki K., Tran L. P. (2014). ABA control of plant macroelement membrance transport systems in response to water deficit and high salinity. New Phytol. 202 35–49. 10.1111/nph.12613 [DOI] [PubMed] [Google Scholar]
- Park H., Kim M., Kang Y., Jeon J., Yoo J., Kim M. (2004). Pathogen- and NaCl-induced expression of the SCaM-4 promoter is mediated in part by a GT-1 box that interacts with a GT-1-like transcription factor. Plant Physiol. 135 2150–2161. 10.1104/pp.104.041442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathan M., Lee J., Shannon J., Nguyen H. (2007). “Recent advances in breeding for drought and salt stress tolerance in soybean,” in Advances in Molecular Breeding Toward Drought and Salt Tolerant Crops, eds Jenks M., Hasegawa P., Jain S. (Dordrecht: Springer; ), 739–773. [Google Scholar]
- Phang T., Shao G., Lam H. (2008). Salt tolerance in soybean. J. Integr. Plant Biol. 50 1196–1212. 10.1111/j.1744-7909.2008.00760.x [DOI] [PubMed] [Google Scholar]
- Qi X., Li M., Xie M., Liu X., Ni M., Shao G., et al. (2014). Identification of a novel salt tolerance gene in wild soybean by whole genome sequencing. Nat. Commun. 5:4340 10.1038/ncomms5340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren S., Omololu A., Brodie W., Tadesse M., Guan R., Qiu L., et al. (2009). Physiological and molecular characterization of salt tolerance in soybean cultivar WF-7. World Soybean Res. Conf. 8:218. [Google Scholar]
- Ren S., Weeda S., Li H., Whitehead B., Guo Y., Atalay A., et al. (2012). Salt tolerance in soybean WF-7 is partially regulated by ABA and ROS signaling and involves withholding toxic Cl- ions from aerial tissues. Plant Cell Rep. 31 1527–1533. 10.1007/s00299-012-1268-2 [DOI] [PubMed] [Google Scholar]
- Rus A., Yokoi S., Sharkhuu A., Reddy M., Lee B. H., Matsumoto T. K., et al. (2001). AtHKT1 is asalt tolerance determinant that controls Na+ entry into plant roots. Proc. Natl. Acad. Sci. U.S.A. 98 14150–14155. 10.1073/pnas.241501798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmuta J., Cannon S., Schlueter J., Ma J., Jackson S. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463 178–183. 10.1038/nature08670 [DOI] [PubMed] [Google Scholar]
- Shi H., Ishitani M., Kim C., Zhu J. K. (2000). The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. U.S.A. 97 6896–6901. 10.1073/pnas.120170197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shkolnik-Inbar D., Adler G., Bar-Zvi D. (2013). ABI4 downregulates expression of the sodium transporter HKT1;1 in Arabidopsis roots and affects salt tolerance. Plant J. 73 993–1005. 10.1111/tpj.12091 [DOI] [PubMed] [Google Scholar]
- Solovyev V. V. (2001). “Statistical approaches in Eukaryotic gene prediction,” in Handbook of Statistical Genetics, eds Balding D., Bishop M., Wiley C. C. (Chichester: John Wiley & Sons, Ltd.), 83–127. [Google Scholar]
- Solovyev V. V., Shahmuradov I. A., Salamov A. A. (2010). Identification of promoter regions and regulatory sites. Methods Mol. Biol. 674 57–83. 10.1007/978-1-60761-854-6_5 [DOI] [PubMed] [Google Scholar]
- Sun Y., Wang D., Bai Y. L., Wang N. N., Wang Y. (2006). Studies on the overexpression of the soybean GmNHX1in Lotus corniculatus: the reduced Na+ level is the basis of the increased salt tolerance. Chin. Sci. Bull. 51 1306–1315. 10.1007/s11434-006-1306-y [DOI] [Google Scholar]
- Torres M. (2010). ROS in biotic interactions. Physiol. Plant. 138 414–429. 10.1111/j.1399-3054.2009.01326.x [DOI] [PubMed] [Google Scholar]
- Tuyen D., Lal S., Xu D. (2010). Identification of a major QTL allele from wild soybean (Glycine soja Sieb. & Zucc.) for increasing alkaline salt tolerance in soybean Theor. Appl. Genetics 121 229–236. 10.1007/s00122-010-1304-y [DOI] [PubMed] [Google Scholar]
- Wang D., Shannon M. C. (1999). Emergence and seedling growth of soybean cultivars and maturity groups under salinity. Plant Soil 214 117–124. 10.1023/A:1004719420806 [DOI] [Google Scholar]
- Wang D., Shannon M. C., Grieve C. M. (2001). Salinity reduces radiation absorption and use efficiency in soybean. Field Crop Res. 69 267–277. 10.1016/S0378-4290(00)00154-4 [DOI] [Google Scholar]
- Wei Q., Guo Y. J., Cao H. M., Kuai B. K. (2011). Cloning and characterizationof an AtNHX2-like Na+/H+ antiporter gene from Ammopiptanthus mongolicus and its ectopic expression enhanced drought and salt tolerance in Arabidopsis thaliana. Plant Cell Tiss Organ Cult. 105 309–316. 10.1007/s11240-010-9869-3 [DOI] [Google Scholar]
- Xia X. J., Wang Y. J., Zhou Y. H., Tao Y., Mao W. H., Shi K., et al. (2009). Reactive oxygen species are involved in Brassinosteroid-induced stress tolerance in cucumber. Plant Physiol. 150 801–814. 10.1104/pp.109.138230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Ma Y., Cheng X., Cao L., Li L., Chen M. (2006). Isolation and characterization of GmSTY1 a novel gene encoding a dual-specificity protein kinase in soybean (Glycine max L.). J. Integr. Plant Biol. 48 857–866. 10.1111/j.1744-7909.2006.00277.x [DOI] [Google Scholar]
- Yamaguchi-Shinozaki K., Shinozaki K. (2006). Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 57 781–803. 10.1146/annurev.arplant.57.032905.105444 [DOI] [PubMed] [Google Scholar]
- Yamamoto A., Kagaya Y., Toyoshima R., Kagaya M., Takeda S., Hattori T. (2009). Arabidopsis NF-YB subunits LEC1 and LEC1-LIKE activate transcription by interacting with seed-specific ABRE-binding factors. Plant J. 58 843–856. 10.1111/j.1365-313X.2009.03817.x [DOI] [PubMed] [Google Scholar]
- Zhai Y., Wang Y., Li Y., Lei T., Yan F., Su L., et al. (2013). Isolation and molecular characterization of GmERF7, a soybean ethylene-response factor that increases salt stress tolerance in tobacco. Gene 513 174–183. 10.1016/j.gene.2012.10.018 [DOI] [PubMed] [Google Scholar]
- Zhang G., Chen M., Li L., Xu Z., Chen X., Guo J., et al. (2008). Overexpression of the soybean GmERF3 gene, an AP2/ERF type transcription factor for increased tolerance to salt, drought, and diseases in transgenic tobacco. Aust. J. Agric. Res. 59 1086–1091. 10.1093/jxb/erp214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G., Guan R., Li Y., Chang R., Qiu L. (2009). Molecular characterization of GmNHX2. A Na+/H+ antiporter gene homolog from soybean, and its heterologous expression to improve salt tolerance in Arabidopsis. Chinese Sci. Bull. 54 3536–3545. [Google Scholar]
- Zhou Q., Tian A., Zou H., Xie Z., Lei G., Huang J., et al. (2008). Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol. J. 6 486–503. 10.1111/j.1467-7652.2008.00336.x [DOI] [PubMed] [Google Scholar]
- Zhu J. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53 247–273. 10.1146/annurev.arplant.53.091401.143329 [DOI] [PMC free article] [PubMed] [Google Scholar]