Abstract
Background
The purpose of this study was to test the hypothesis that the genetic backgrounds of lung cancers could affect the spatial distribution of brain metastases.
Methods
CT or MR images of 200 patients with a total of 1033 treatment-naive brain metastases from lung cancer were retrospectively reviewed (23 by CT and 177 by MRI). All images were standardized to the human brain MRI atlas provided by the Montreal Neurological Institute 152 database. Locations, depths from the brain surface, and sizes of the lesions after image standardization were analyzed.
Results
The posterior fossa, the anatomic “watershed areas,” and the gray-white matter junction were confirmed to be more commonly affected by lung cancer brain metastases, and brain metastases with epidermal growth factor receptor (EGFR) L858R mutation occurred more often in the caudate, cerebellum, and temporal lobe than those with exon 19 deletion of EGFR. Median depths of the lesions from the brain surface were 13.7 mm (range, 8.6–21.9) for exon 19 deleted EGFR, 11.5 mm (6.6–16.8) for L858R mutated, and 15.0 mm (10.0–20.7) for wild-type EGFR. Lesions with L858R mutated EGFR were located significantly closer to the brain surface than lesions with exon 19 deleted or wild-type EGFR (P = .0032 and P < .0001, respectively). Furthermore, brain metastases of adenocarcinoma lung cancer patients with a history of chemotherapy but not molecular targeted therapy were located significantly deeper from the brain surface (P = .0002).
Conclusion
This analysis is the first to reveal the relationship between EGFR mutation status and the spatial distribution of brain metastases of lung cancer.
Keywords: EGFR, lung cancer brain metastasis, location analysis, MRI
Brain metastases (BMs) are diagnosed in more than 200 000 patients in the United States each year and are the most prevalent malignant tumors of the central nervous system, outnumbering primary CNS tumors by more than 10-fold.1 Over 20% of all cancer patients develop BMs and have a 9-month median survival with maximal treatment,2–4 making BMs among the major causes of systemic cancer mortality.2 Among many types of cancers, lung and breast cancers are by far the most common tumors to present with BMs,5,6 and recent years' improvements in imaging and treatments for primary cancer lesions seem to have increased the incidence of BMs.4,7–9
The heterogeneity of BMs has been reported since the 1950s. In early years, such studies were performed postmortem, whereas image analysis with CT or MRI has been the main approach in recent years, demonstrating preferential involvement of the anatomic “watershed areas,” the gray-white matter junction, and cerebellum; and a relationship between the spatial distribution of BMs and primary cancer types has been shown.10–16 In most image analyses, researchers used standard anatomic templates for anatomic reference, and they documented the anatomic locations of tumors by mere observation of those templates. Today, with the advent of image registration and 3D structure deformation algorithms, it is now possible to deform and register individual brain images onto an averaged standard brain image, enabling detection of even the slightest difference in spatial distribution.17 Using this technique for cross-patient lesion location analysis, Bender and Tomé18 and Quattrocchi et al19 analyzed the spatial distribution of BMs more objectively than before, finding that BMs preferentially involve the parieto-occipital lobes and cerebellum in lung cancer patients and the cerebellum in breast cancer patients.
On the other hand, molecular and genetic information about tumors, such as epidermal growth factor receptor (EGFR) mutations in non–small cell lung cancer (NSCLC), has recently become increasingly important. EGFR mutations have been reported to be highly associated with sensitivity to EGFR tyrosine kinase inhibitors,20,21 and nearly 90% of all known mutations are located in exon 19 (in-frame deletion; exon 19 deletion of EGFR) and exon 21 (L858R point mutation; EGFR L858R mutation).22 There have been a few reports regarding the relationship between BMs and EGFR mutation status. Patients with EGFR-mutated NSCLC have been reported to develop more BMs, in number23 and frequency,24 than those with wild-type EGFR NSCLC. Moreover, patients with NSCLC harboring exon 19 deletion of EGFR have been reported to have BMs more often,25 especially miliary BMs,26 than those with EGFR L858R mutated NSCLC. To the best of our knowledge, however, no report has discussed the relationship between the spatial distribution of BMs and EGFR mutation status. We hypothesized that the molecular and genetic background of tumors could affect the spatial distribution of BMs. Comprehensive understanding of the molecular mechanisms that affect BMs could lead to development of novel agents that prevent this disease. Understanding different spatial distributions of BMs according to their genetic background might also make possible more precise prophylactic irradiation adjustments in lung cancer. To test this hypothesis, the spatial distribution of BMs was analyzed using cross-patient lesion location analysis focusing on lung cancer BMs. More specifically, the spatial distribution of BMs was compared by EGFR mutation status.
Materials and Methods
Patients and Data Collection
CT or MR images of 254 lung cancer patients with radiation- and operation-naive BMs from 2006 to 2014 were retrospectively reviewed. Patients who underwent systemic chemotherapy, molecular targeted therapy with agents such as gefitinib and erlotinib, or radiation therapy were included, while patients who underwent previous neurosurgery or brain radiation therapy were excluded. In addition, 6 patients were excluded because of other malignant diseases within 5 years prior to BM identification, and 48 patients were excluded because enhanced CT or MRI of the brain was unavailable. Thus, iodine-enhanced CT or gadolinium-enhanced MRI T1-weighted images (1.5 or 3.0T) of 200 patients in whom initial BMs were found were included in the analysis (23 by CT, 95 by 1.5T MRI, 82 by 3.0T MRI; 79 by thin slice with <1.5 mm thickness, 121 with 5 mm thickness). All patients had histopathological diagnoses of lung cancer by bronchoscopic, percutaneous needle-guided, or surgical biopsies, and some of the tumor samples were analyzed for EGFR mutation status by polymerase chain reaction (PCR). The study was conducted in accordance with the Declaration of Helsinki, and the internal review board of the institution approved clinical data to be used for this research.
Image Registration and Frequency Map Reconstruction
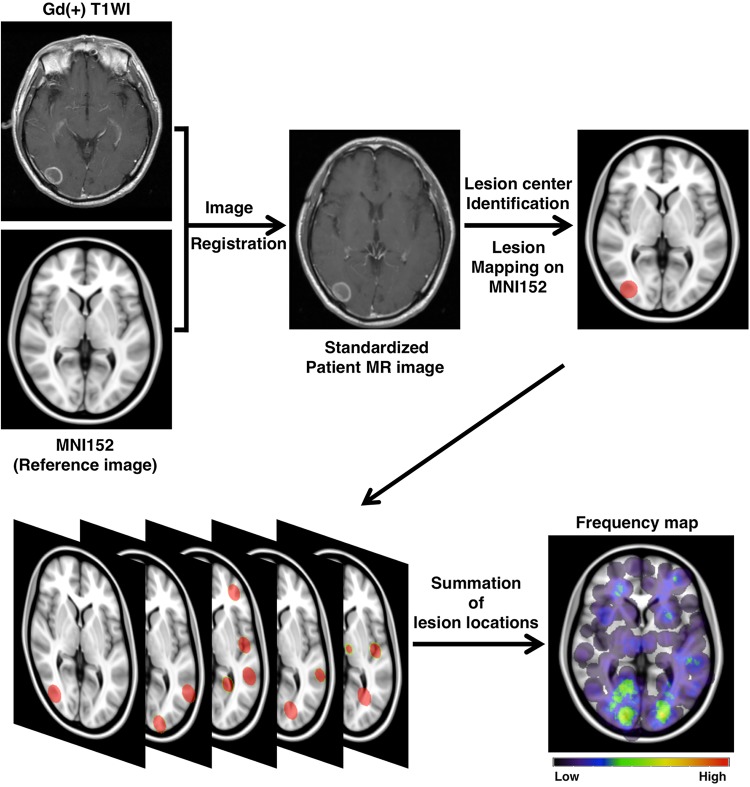
All DICOM (Digital Imaging and Communications in Medicine) images were converted to the NIfTI (Neuroimaging Informatics Technology Initiative) format using the dcm2nii software (http://www.cabiatl.com/mricro/mricron/dcm2nii.html). These NIfTI data were registered to a 1.0-mm isotropic, high-resolution T1-weighted brain atlas provided by the Montreal Neurological Institute (MNI152 database) using a mutual information algorithm with a 12-degree of freedom transformation using the Functional Magnetic Resonance Imaging of the Brain (FMRIB) Software Library/FMRIB Linear Image Registration Tool (FSL-FLIRT; http://fsl.fmrib.ox.ac.uk/fsl/fslwiki/FSL) (Fig. 1). Image registrations were visually confirmed in all cases, and some required rough manual adjustment before using FSL-FLIRT.17,27
Fig. 1.
Schematic illustration of the flow of image analysis. Each patient's brain MR or CT image was deformed and registered to standard brain MRI (MNI152) using FSL-FLIRT. The center of the tumor was manually located, and a 20-mm-diameter sphere was created to visualize the selected location. Finally, spheres from each patient were overlaid to create a frequency map. Gd(+) T1WI, gadolinium T1-weighted image.
All datasets were exported to in-house software written in MatLab 7.14 (MathWorks) for further analysis. The center voxel of the contrast-enhanced lesion was manually identified for each lesion, and that voxel was defined as the primary site of occurrence (Supplementary Fig. S1). For frequency map reconstruction, all lesions were reconstructed to a site-centered spherical shape with a diameter of 20 mm (Fig. 1). All spherical lesions were summed and averaged by the number of patients. A heat-map for the lesion occurrence frequency was reconstructed and superimposed on the reference MNI152 (Fig. 1).
Analysis of Location in the Brain
To reveal the heterogeneity of BMs in the brain, the location of each lesion was determined. The center voxel of the contrast-enhanced lesion, defined as the tumor location, was registered to the MNI structural atlas28 and automatically assigned to 10 segments: deep white matter and brainstem, caudate, cerebellum, frontal lobe, insula, occipital lobe, parietal lobe, putamen, temporal lobe, and thalamus (Supplementary Fig. S2). The metastatic frequency of occurrence per unit volume of each segment was compared among primary lung cancer subgroups: adenocarcinoma (Ad), small cell carcinoma (SCC), squamous cell carcinoma (Sq), and large cell carcinoma (LCC). In the Ad group, it was also compared according to EGFR mutation status: wild-type, exon 19 deletion, and L858R point mutation. Moreover, the metastatic frequency in the hippocampus was analyzed using the Harvard-Oxford cortical and subcortical atlases provided with FSL (Supplementary Fig. S3).
Analysis of Distance from the Brain Surface to the Lesion
To demonstrate preferential involvement of BMs at the gray-white matter junction, the distance from the brain surface to the center of each lesion (DEPTH) was calculated automatically using in-house software. The surface of the brain was extracted from MNI152, and the shortest distance from the brain surface to the center of the lesion was calculated automatically. Subsequently, DEPTH was compared among primary lung cancer subgroups and according to EGFR mutation status in the Ad group. In addition, the maximal diameter of the lesion after MNI152 registration was manually measured in 3 dimensions for adenocarcinomas.
Statistical Analysis
Statistical analysis was performed by JMP version 10 and SAS version 9.3 (SAS Institute). A threshold level of 0.05 was established for significance. The chi-square test and Poisson regression with the volume of each segment being handled as an offset were used for location analysis. The Kruskal–Wallis test and the Steel–Dwass test (the nonparametric analog comparable to Tukey's range test) were used for group comparisons. For multivariate analysis, a linear regression model was used to identify significant variables. Data are shown as medians with interquartile ranges.
Results
Frequency Map of Brain Metastatic Lesions from Lung Cancer
In total, 1033 lesions in 200 patients were analyzed: 712 lesions in 129 patients for Ad, 186 lesions in 28 patients for SCC, 105 lesions in 27 patients for Sq, and 11 lesions in 5 patients for LCC. Diagnoses of 19 lesions in 11 patients could not be reached in detail, being ascribed as “non-small cell lung cancer (NSCLC)” or “Ad or Sq.” Among the 129 patients with Ad, 115 were examined for EGFR mutation status, resulting in 62 patients with EGFR mutation and 53 patients with wild-type EGFR. In patients with EGFR mutation, 28 patients had exon 19 deletion, and 31 patients had L858R point mutation, while 3 patients had other mutations. No patient had both exon 19 deletion and L858R point mutation. The number of BMs in each patient was not significantly different among primary lung cancer subgroups (2 [1–6] for Ad, 2 [1–5] for SCC, 2 [1–4] for Sq, 3 [1–3] for LCC; P = .92), while it showed a borderline difference by EGFR mutation status (4 [1.25–6.75] for exon 19 deletion, 2 [1–4] for L858R point mutation, 2 [1–6] for wild-type; P = .080). Detailed characteristics of the patients are shown in Table 1.
Table 1.
Patient characteristics
| All cancers | Adenocarcinoma |
Small Cell Carcinoma | Squamous Cell Carcinoma | Large Cell Carcinoma | Unable to Classify | ||
|---|---|---|---|---|---|---|---|
| Patients, n | 200 | 129 | 28 | 27 | 5 | 11 | |
| M/F, n | 134/66 | 72/57 | 25/3 | 24/3 | 4/1 | 9/2 | |
| Age, y, at diagnosis of | 62 ± 9 | 62 ± 9 | 65 ± 10 | 62 ± 10 | 65 ± 7 | 65 ± 7 | |
| primary tumor | |||||||
| (mean ± SD) | |||||||
| Number of | 1033 | 712 | 186 | 105 | 11 | 19 | |
| metastases, n | |||||||
| EGFR mutation status | 62/53 | ||||||
| (mutation/wild-type), n | exon 19 deletion | 28 | |||||
| L858R point mutation | 31 | ||||||
| others | 3 | ||||||
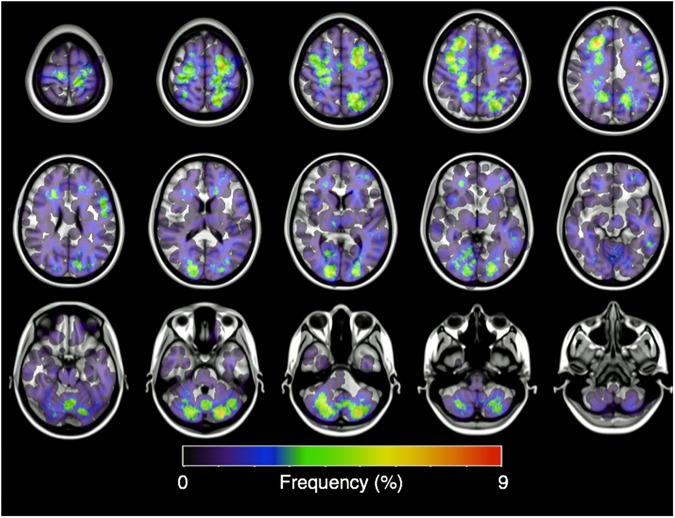
One hundred and seventy-seven MR images and 23 CT images were successfully transformed, and the coordinates corresponding to the center of each lesion were registered onto MNI152. All of the lesions were successfully reconstructed to a spherical shape with a 20-mm diameter on MNI152 (Fig. 2 and Supplementary Figs. S4–S7). The posterior fossa,11–13,18,19 the anatomic “watershed areas,”10,15,16,19 and the gray-white matter junction16 were visually confirmed to be the most commonly involved areas of lung cancer BMs, consistent with past studies.
Fig. 2.
Axial image of the frequency map of all brain metastases from lung cancer. Note that the lesions are mostly concentrated at the gray-white matter junction and the cerebellum.
Distribution of BMs Is Not Uniform in the Brain
The number of BMs in each segment is shown in Table 2. In gross BMs, the frequency of occurrence in each segment did not correlate with the volume of the segment (P < .0001), indicating the heterogeneity of BMs in the brain. There were significant biases for cerebellar, occipital, and temporal metastases compared with what would be predicted based solely on relative brain volume (chi-square test: P = .012, P = .002, P < .0001, respectively). The heterogeneity of BMs was also confirmed in Ad and SCC, but not in Sq (P < .0001, P < .0001, P = .075, respectively), and the data were not applicable in LCC because of the small number of BMs. On the other hand, BMs within the hippocampus were detected in only 3 patients (one with exon 19 deleted EGFR, one with L858R mutated EGFR, and another with SCC).
Table 2.
Distribution of brain metastases
| Volume, mm3 | All Tumors | Adenocarcinoma | Adenocarcinoma |
Large Cell Carcinoma | Small Cell Carcinoma | Squamous Cell Carcinoma | |||
|---|---|---|---|---|---|---|---|---|---|
| With Wild-Type EGFR | With Exon 19 Deletion of EGFR | With L858R Mutated EGFR | |||||||
| Deep white matter and brainstem | 161 082 | 82 | 41 | 20 | 11 | 7 | 0 | 26 | 14 |
| Caudate | 32 353 | 14 | 6 | 3 | 1 | 2 | 0 | 3 | 1 |
| Cerebellum | 294 552 | 159 | 113 | 49 | 30 | 29 | 2 | 38 | 7 |
| Frontal lobe | 708 087 | 310 | 229 | 96 | 72 | 48 | 3 | 39 | 34 |
| Insula | 33 041 | 13 | 8 | 5 | 3 | 0 | 0 | 2 | 2 |
| Occipital lobe | 236 594 | 136 | 89 | 35 | 36 | 12 | 2 | 25 | 14 |
| Parietal lobe | 421 103 | 206 | 142 | 52 | 58 | 23 | 3 | 41 | 21 |
| Putamen | 26 171 | 8 | 6 | 4 | 2 | 0 | 0 | 0 | 2 |
| Temporal lobe | 356 227 | 87 | 64 | 29 | 15 | 13 | 1 | 12 | 8 |
| Thalamus | 34 802 | 18 | 14 | 4 | 8 | 1 | 0 | 0 | 2 |
| Total | 2 304 012 | 1033 | 712 | 297 | 236 | 135 | 11 | 186 | 105 |
Distribution of BMs Is Different by EGFR Mutation Status as well as Primary Lung Cancer Subgroup
There was a significant difference in the distribution of BMs (P = .0074) among primary lung cancer subgroups (Table 2). In greater detail, there was a difference between Ad and SCC (P = .0002) and between SCC and Sq (P = .0097). There was also a significant difference in the distribution of BMs between patients with exon 19 deleted EGFR and those with L858R mutated EGFR (P = .020), specifically in the caudate, cerebellum, and temporal lobe. BMs with L858R mutated EGFR occurred more often in these segments than in those with exon 19 deleted EGFR.
Brain Metastases with L858R Mutated EGFR Prefer to Nest Closer to the Brain Surface
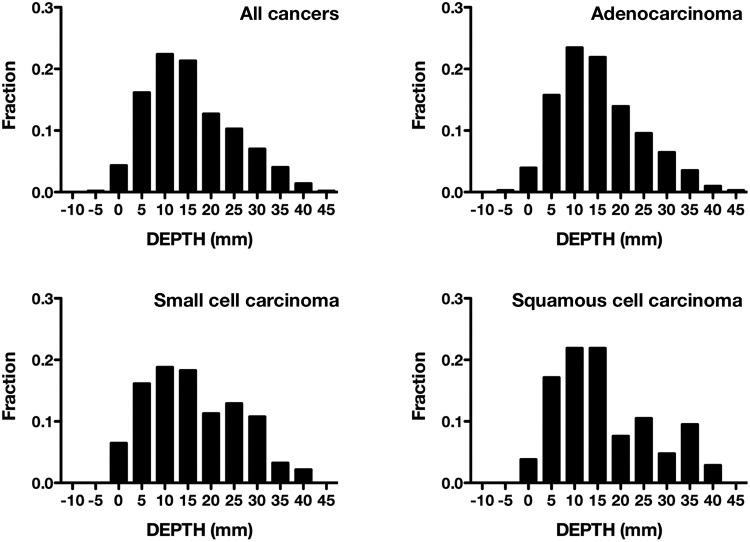
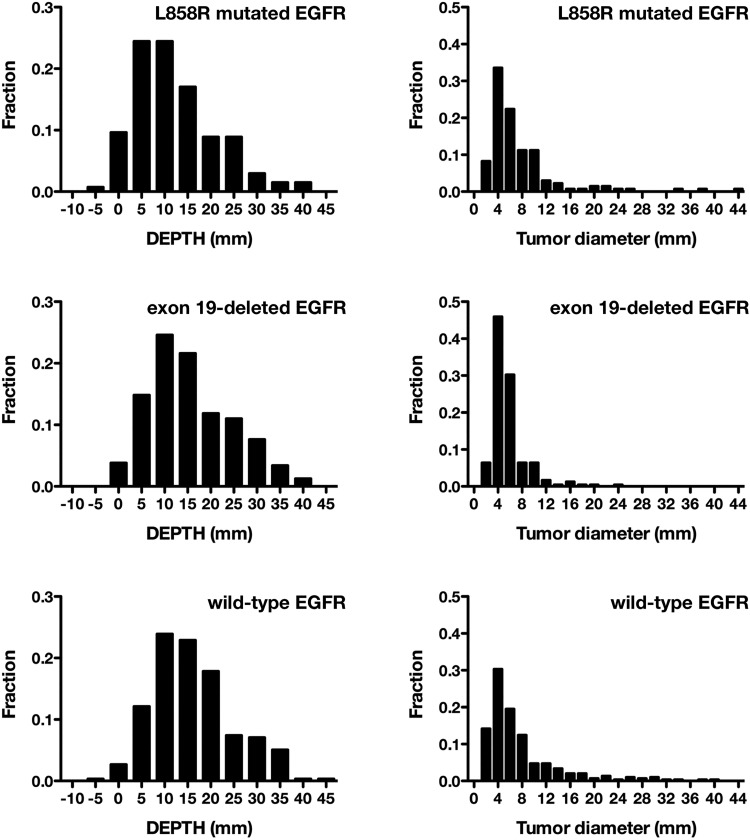
Mean DEPTH was 13.9 mm (8.5–21.9) for all patients; 13.9 mm (8.7–20.6) for Ad, 14.1 mm (8.5–24.5) for SCC, 14.2 mm (8.8–24.2) for Sq, and 9.8 mm (5.9–14.3) for LCC. There was no significant difference among lung cancer subgroups (P = .43), although it was noticed that SCC and Sq showed a bimodal distribution, while Ad showed a single peak of distribution (Fig. 3). Further analyzing patients with Ad, mean DEPTH was 13.7 mm (8.6–21.9) for exon 19 deletion, 1.5 mm (6.6–16.8) for L858R mutation, and 15.0 mm (10.0–20.7) for wild-type EGFR. Lesions with L858R mutated EGFR were located significantly closer to the brain surface than lesions with exon 19 deleted or wild-type EGFR (P = .0032 and P < .0001, respectively) (Fig. 4). On the other hand, the mean diameters of the lesions after image transformation were 4.8 mm (3.9–6.4) for exon 19 deleted, 5.2 mm (3.9–8.8) for L858R mutated, and 5.6 mm (3.6–8.7) for wild-type EGFR. There was a borderline difference by EGFR mutation status (P = .086) (Fig. 4).
Fig. 3.
Histogram of the distance of brain metastases from the surface of the brain (DEPTH). There is no significant difference among lung cancer subgroups (P = .43).
Fig. 4.
Histogram of the distance of brain metastases from the surface of the brain (DEPTH) and the maximal diameters of brain metastases after image transformation. The histogram compares adenocarcinoma patients grouped by EGFR mutation status. The lesions with L858R mutated EGFR are located significantly closer to the brain surface than the lesions with exon 19 deleted or wild-type EGFR (P = .0032 and P < .0001, respectively). As to the diameters of brain metastases after image transformation, there is a borderline difference by EGFR mutation status (P = .086).
Both L858R Point Mutation and Chemotherapy Affect the Spatial Distribution of BMs from Lung Adenocarcinoma
To analyze the effects of systemic chemotherapy and molecular targeted therapy on the distribution of BMs, DEPTH was compared on the basis of treatment history for primary and metastatic lesions except for the brain. Any chemotherapy and molecular targeted therapy longer or shorter than 3 months was included in the analysis.
Mean DEPTH was 13.0 mm (7.5–20.4) for patients with Ad who were treated with any tyrosine kinase inhibitors for more than 3 months, while it was 14.3 mm (9.0–20.6) for patients treated with tyrosine kinase inhibitors for less than 3 months (P = .13). Mean DEPTH was, however, 14.7 mm (9.3–22.2) for patients with Ad who were treated with any chemotherapies for more than 3 months, while it was 11.7 mm (7.8–18.0) for patients treated for less than 3 months. Thus, lesions of patients treated with any chemotherapy for more than 3 months were located significantly deeper from the brain surface (P = .0002).
A multiple regression model was constructed including both EGFR mutation status and length of chemotherapy as contributors to BM location. Both L858R point mutation and any chemotherapy longer than 3 months were significant factors for model construction, with L858R point mutation weighing more (P = .0030 and P = .0107, respectively).
Discussion
One of the purposes of the present study was to evaluate the spatial distribution of BMs from lung cancer more objectively than past reports. Individual images were registered to a standard brain atlas for spatial standardization. Taking the different sizes of the lesions into account, the center of the lesion was selected for analysis on the assumption that this site would reflect the origin more accurately than the whole lesion. This procedure could be considered as “rewinding” the chronological changes of the lesions to the initial stage of lesion development. By combining spatial and chronological standardization, the frequency map shown in Fig. 2 and Supplementary Figs. S4–S7 could be called the 4-dimensional standardized frequency map of BMs from lung cancer. Preferential involvement by lung cancer BMs of the posterior fossa, the anatomic watershed areas, and the gray-white matter junction was visually reconstructed, confirming findings from past reports.10,12,13,15,16,18,19 Furthermore, the heterogeneity of BMs in the brain was statistically verified, confirming past reports with more objectivity.10–16.18,19 With a more detailed frequency map of BMs, prophylactic irradiation for patients with malignant tumor could be optimized to enhance preventive effects and reduce side effects.18 Intensity-modulated radiotherapy with hippocampal avoidance, which was performed in the Radiation Therapy Oncology Group trial 0933, could be an effective radiotherapy for lung cancer patients, as BM occurrence within the hippocampus seems to be infrequent, as revealed by the current study.29
In the present study, we hypothesized that BMs may distribute differently within the brain according to their molecular biological characteristics. First, the spatial distribution of BMs was compared among lung cancer subtypes, revealing a significant difference between SCC and Ad or Sq. This observation confirms that histological subtypes of lung cancer have significant influence over the location of preference for brain metastasis (Table 2). When BMs were further compared according to EGFR mutation status among Ad lung cancers, there were no significant differences in the number or size of BMs, which was inconsistent with the past report that showed that patients with exon 19 deleted EGFR have more and smaller BMs than patients with wild-type EGFR.26 This contradiction may be because of the number of analyzed patients. Further analysis with a larger cohort is required to reach definite conclusions. There were, however, significant differences in the spatial distribution of BMs between EGFR mutation statuses with 2 different analytical methods. BMs with L858R mutated EGFR showed preferential involvement of the caudate, cerebellum, and temporal lobe and a tendency to nest nearer to the brain surface. It was also confirmed that this tendency was independent of tumor size or history of prior systemic chemotherapy.
Organ-specific metastasis of cancer has been proposed for more than a century, known as “Paget's theory” or “seed and soil theory.” Paget suggested that the site of metastasis depends on the affinity of the tumor (the “seed”) to the microenvironment (the “soil”),30 and the molecular mechanism of this has been partially revealed. In breast cancer, chemokine receptors such as CXCR4 and CCR7 have been reported to have a critical role in determining the metastatic destination of tumor cells, as well as leukocytes.31 For metastases to brain, some molecules have been reported to participate in crossing over the blood–brain barrier,32 and some have been reported to be necessary to defend tumor cells from highly expressed tissue plasminogen activator in the brain.33 On the other hand, the molecular mechanism of preferential involvement of specific areas within the brain remains largely unknown. Although a definite explanation of our findings is challenging, the affinity of the tumor to microenvironment of specific areas within the brain is thought to affect the spatial distribution of BMs, as well as organ-specific metastasis, and mutated EGFR or correlative molecules with EGFR mutation status may be among the key molecules.
Systemic chemotherapy was also a factor affecting DEPTH in the present study. To the best of our knowledge, no previous analysis focused on the effects of systemic chemotherapy on the spatial distribution of BMs. Although it has not been confirmed, it is possible that the distance from the cerebrospinal fluid could be a key factor related to this result, since the tissue concentration of anticancer drugs is thought to be higher at the brain surface directly contacting the CSF.
Limitations of our study should also be noted. Firstly, although statistical spatial distribution analysis with 3D structure deformation has already been adopted in many studies,17–19,27 “4-dimensional standardization” has not yet been widely accepted. Since BMs are usually round in shape and develop concentrically, unlike primary CNS tumors such as glioblastoma, the center of the tumor could be approximated as the occurrence point, especially when the BMs are small. Secondarily, we included CT and MRI with variable resolutions together in this analysis, which could affect different sensitivities for BMs. In order to answer this concern, we further analyzed DEPTH, size, and number of BMs within MRI only (n = 177) or 3.0T MRI with 0.8 mm thickness only (n = 76), obtaining nearly identical results compared with those done using all the data available (Supplementary Table S1).
Finally, it should be emphasized that the analytical method used for this study is useful to detect significant factors affecting the spatial distribution of metastases within the brain, and the current study is the first to uncover the relationship between a tumor's molecular biological characteristics and the spatial distribution of BMs. It seems safe to speculate that this kind of analysis enables the identification of more molecules affecting the spatial distribution of BM, which would lead to the development of new molecular targeted drugs to prevent brain metastases and novel radiological diagnostic methods that reveal the biological characteristics of the tumor.
Supplementary Material
Funding
This investigation was supported by the Aichi Cancer Research Foundation, the SENSIN Medical Research Foundation, the Life Science Foundation of Japan, the Japanese Foundation for Multidisciplinary Treatment of Cancer, and JSPS KAKENHI (25462256 and 26670642).
Conflict of interest statement. The authors made no disclosures.
Supplementary Material
References
- 1.Maher EA, Mietz J, Arteaga CL et al. Brain metastasis: opportunities in basic and translational research. Cancer Res. 2009;69:6015–6020. [DOI] [PubMed] [Google Scholar]
- 2.Nathoo N, Chahlavi A, Barnett GH et al. Pathobiology of brain metastases. J Clin Pathol. 2005;58(3):237–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sperduto PW, Berkey B, Gaspar LE et al. A new prognostic index and comparison to three other indices for patients with brain metastases: an analysis of 1,960 patients in the RTOG database. Int J Radiat Oncol Biol Phys. 2008;70(2):510–514. [DOI] [PubMed] [Google Scholar]
- 4.Weil RJ, Palmieri DC, Bronder JL et al. Breast cancer metastasis to the central nervous system. Am J Pathol. 2005;167(4):913–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang EL, Lo S. Diagnosis and management of central nervous system metastases from breast cancer. Oncologist. 2003;8(5):398–410. [DOI] [PubMed] [Google Scholar]
- 6.Lassman AB, Deangelis LM. Brain metastases. Neurol Clin. 2003;21(1):1–23. [DOI] [PubMed] [Google Scholar]
- 7.Nagao E, Yoshiura T, Hiwatashi A et al. 3D turbo spin-echo sequence with motion-sensitized driven-equilibrium preparation for detection of brain metastases on 3 T MR imaging. Am J Neuroradiol. 2011;32(4):664–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tham Y-L, Sexton K, Kramer R et al. Primary breast cancer phenotypes associated with propensity for central nervous system metastases. Cancer. 2006;107(4):696–704. [DOI] [PubMed] [Google Scholar]
- 9.Qian Y-F, Yu C-L, Zhang C et al. MR T1-weighted inversion recovery imaging in detecting brain metastases: could it replace T1-weighted spin-echo imaging? Am J Neuroradiol. 2008;29(4):701–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kindt GW. The pattern of location of cerebral metastatic tumors. J Neurosurg. 1964;21(1):54–57. [DOI] [PubMed] [Google Scholar]
- 11.Chason JL, Walker FB, Landers JW. Metastatic carcinoma in the central nervous system and dorsal root ganglia. A prospective autopsy study. Cancer. 1963;16:781–787. [DOI] [PubMed] [Google Scholar]
- 12.Meyer PC, Reah TG. Secondary neoplasms of the central nervous system and meninges. Br J Cancer. 1953;7(4):438–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lesse S, Netsky MG. Metastasis of neoplasms to the central nervous system and meninges. AMA Arch Neurol Psychiatry. 1954;72(2):133–153. [DOI] [PubMed] [Google Scholar]
- 14.Ask-Upmark E. Metastatic tumours of the brain and their localization. Acta Med Scand. 1956;154(1):1–9. [DOI] [PubMed] [Google Scholar]
- 15.Delattre JY, Krol G, Thaler HT et al. Distribution of brain metastases. Arch Neurol. 1988;45(7):741–744. [DOI] [PubMed] [Google Scholar]
- 16.Hwang TL, Close TP, Grego JM et al. Predilection of brain metastasis in gray and white matter junction and vascular border zones. Cancer. 1996;77(8):1551–1555. [DOI] [PubMed] [Google Scholar]
- 17.Kinoshita M, Sasayama T, Narita Y et al. Different spatial distribution between germinal center B and non-germinal center B primary central nervous system lymphoma revealed by magnetic resonance group analysis. Neuro Oncol. 2014;16(5):728–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bender ET, Tomé WA. Distribution of brain metastases: implications for non-uniform dose prescriptions. Br J Radiol. 2011;84(1003):649–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quattrocchi CC, Errante Y, Gaudino C et al. Spatial brain distribution of intra-axial metastatic lesions in breast and lung cancer patients. J Neurooncol. 2012;110(1):79–87. [DOI] [PubMed] [Google Scholar]
- 20.Lynch TJ, Bell DW, Sordella R et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350(21):2129–2139. [DOI] [PubMed] [Google Scholar]
- 21.Mok TS, Wu Y-L, Thongprasert S et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361(10):947–957. [DOI] [PubMed] [Google Scholar]
- 22.Sharma SV, Bell DW, Settleman J et al. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7(3):169–181. [DOI] [PubMed] [Google Scholar]
- 23.Eichler AF, Kahle KT, Wang DL et al. EGFR mutation status and survival after diagnosis of brain metastasis in nonsmall cell lung cancer. Neuro Oncol. 2010;12(11):1193–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iuchi T, Shingyoji M, Itakura M et al. Frequency of brain metastases in non-small-cell lung cancer, and their association with epidermal growth factor receptor mutations. Int J Clin Oncol. 2015;20(4):674–679. [DOI] [PubMed] [Google Scholar]
- 25.Heon S, Yeap BY, Britt GJ et al. Development of central nervous system metastases in patients with advanced non-small cell lung cancer and somatic EGFR mutations treated with gefitinib or erlotinib. Clin Cancer Res. 2010;16(23):5873–5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sekine A, Kato T, Hagiwara E et al. Metastatic brain tumors from non-small cell lung cancer with EGFR mutations: distinguishing influence of exon 19 deletion on radiographic features. Lung Cancer. 2012;77(1):64–69. [DOI] [PubMed] [Google Scholar]
- 27.Ellingson BM, Cloughesy TF, Pope WB et al. Anatomic localization of O6-methylguanine DNA methyltransferase (MGMT) promoter methylated and unmethylated tumors: a radiographic study in 358 de novo human glioblastomas. NeuroImage. 2012;59(2):908–916. [DOI] [PubMed] [Google Scholar]
- 28.Mazziotta J, Toga A, Evans A et al. A probabilistic atlas and reference system for the human brain: International Consortium for Brain Mapping (ICBM). Philos Trans R Soc Lond B Biol Sci. 2001;356(1412):1293–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gondi V, Pugh SL, Tome WA et al. Preservation of memory with conformal avoidance of the hippocampal neural stem-cell compartment during whole-brain radiotherapy for brain metastases (RTOG 0933): a phase II multi-institutional trial. J Clin Oncol. 2014;32(34):3810–3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ribatti D, Mangialardi G, Vacca A. Stephen Paget and the “seed and soil” theory of metastatic dissemination. Clin Exp Med 2006;6(4):145–149. [DOI] [PubMed] [Google Scholar]
- 31.Müller A, Homey B, Soto H et al. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410(6824):50–56. [DOI] [PubMed] [Google Scholar]
- 32.Bos PD, Zhang XH-F, Nadal C et al. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459(7249):1005–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Valiente M, Obenauf AC, Jin X et al. Serpins promote cancer cell survival and vascular co-option in brain metastasis. Cell. 2014;156(5):1002–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.