Abstract
The SNP-based high-resolution QTL mapping mapped eight major genomic regions harbouring robust QTLs governing seed-Fe and Zn concentrations (39.4% combined phenotypic variation explained/PVE) on six chromosomes of an intra-specific high-density genetic linkage map (1.56 cM map-density). 24620 SNPs discovered from genome-wide GBS (genotyping-by-sequencing) and 13 known cloned Fe and Zn contents-related chickpea gene-orthologs were genotyped in a structured population of 92 sequenced desi and kabuli accessions. The large-scale 16591 SNP genotyping- and phenotyping-based GWAS (genome-wide association study) identified 16 genomic loci/genes associated (29% combined PVE) with seed-Fe and Zn concentrations. Of these, 11 trait-associated SNPs in the genes linked tightly with eight QTLs were validated by QTL mapping. The seed-specific expression, including pronounced differential-regulation of 16 trait-associated genes particularly in accessions/mapping individuals with contrasting level of seed-Fe and Zn contents was apparent. Collectively, the aforementioned rapid integrated genomic strategy led to delineate novel functional non-synonymous and regulatory SNP allelic-variants from 16 known/candidate genes, including three strong trait-associated genes (encoding late embryogenesis abundant and yellow stripe-like 1 protein, and vacuolar protein sorting-associated protein) and eight major QTLs regulating seed-Fe and Zn concentrations in chickpea. These essential inputs thus have potential to be deployed in marker-assisted genetic enhancement for developing nutritionally-rich iron/zinc-biofortified chickpea cultivars.
The global population strength is fast rising in an alarming rate and anticipated to cross the mark of nine billion by 20501,2. Ensuring food and nutritional security of this fast growing population will pose a huge challenge especially in the era of climatic variability and resource scarcity. Adequate intake of nutritious food enriched with essential micronutrients is prerequisite for humans to meet their metabolic need and maintain good health. However, over three billion people, including one third of the children in developing countries are suffering from micronutrient malnutrition or hidden hunger world-wide3,4,5. Henceforth, micronutrient malnutrition/hidden hunger is a major global health concern and require immediate attention6. Iron (Fe) and Zinc (Zn) are the major components of micronutrient and play a crucial role in growth and development of human by acting as co-factors for several proteins, including haemoglobin, cytochrome and transcription factors. The Fe and Zn deficiencies are reportedly the most widespread and commonly observed micronutrient deficiencies in human beings7,8,9. Fe and Zn deficiency leads to several physiological disorders, including anaemia, tissue hypoxia, impaired physical growth, hypogonadism, dwarfism, orificial and acral dermatitis. About half of the world population is at risk for inadequate intake of Fe and Zn in their diet3,10. Consequently, several strategies, including mineral supplementation, dietary diversification and food fortification have been implemented to mitigate the micronutrient deficiency/hidden hunger4,11. Nonetheless, these attempts achieved a limited success due to lack of proper cultural awareness and inappropriate socio-economic infrastructure11.
With the advancement of cutting edge next generation sequencing (NGS)- and array-based genotyping strategies, the genomics-assisted breeding, which primarily involves identification and introgression of useful genes/QTLs (quantitative trait loci) regulating Fe and Zn contents, can be deployed as a powerful intervention tool for biofortification of food crops4,11,12. The essential inputs obtained from the above-mentioned strategy will be useful to increase the bioavailable micronutrients and thus enhance the quality component traits in food crops for nutritional security. However, the accumulation of mineral nutrients, including Fe and Zn in the edible parts (mostly seeds/grains) of crops is tightly regulated through complex cellular and genetic mechanism. The proportionate quantity of Fe and Zn present in seeds is a complex quantitative trait and governed by many major genes/QTLs13,14. Therefore, it is highly imperative to dissect the genetic basis of variability governing Fe and Zn concentrations in seeds of food crops. Tremendous efforts have been made to delve deeper into the complex genetic architecture of this trait in several crop species, including cereals and legumes2,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19. Among legumes with much emphasis on common bean, attempts have been made to decipher the underlying genetic factors regulating Fe and Zn concentrations in seeds of Medicago, lentil, field pea and chickpea8,9,13,14,20,21,22,23,24.
Chickpea is a self-pollinated, annual diploid and highly nutritious legume with a genome size of ~740 Mbp that is considered as a staple source of protein25,26. This legume crop ranks second in consumption and third in production among pulses grown world-wide25,26. The genomes of two chickpea cultivars (desi and kabuli) with contrasting agronomic traits representing diverse genepools, have been sequenced27,28,29. Chickpea seeds constitute several nutritional composition, including protein, carbohydrate and minerals like Fe and Zn5,30. Considering the importance of this crop as a dietary element across a major part of global population, especially in developing countries, it is desirable to understand the complex genetic inheritance pattern and gene regulatory function of seed-Fe and Zn concentrations in chickpea leading to their bio-fortification through marker-assisted genetic enhancement. To date, only one 1536 SNP markers-based trait association mapping study has been reported to identify the major genomic loci/genes regulating seed-Fe and Zn concentrations in chickpea14. Unfortunately, no informative markers tightly linked to the major QTLs/genes governing seed-Fe and Zn contents are available that could be deployed in marker-assisted genetic improvement of chickpea. An integrated genomic approach involving traditional QTL mapping, association analysis and differential gene expression profiling is currently the most efficient strategy for rapid dissection of complex yield and quality component quantitative traits in diverse crop plants, including chickpea31,32,33,34,35,36,37,38,39,40,41,42,43.
In this aspect, the current study has made an effort to integrate QTL mapping with genome-wide association study (GWAS), candidate gene-based association mapping and expression profiling using large-scale natural germplasm lines and mapping populations to delineate functionally relevant molecular tags (markers, genes, QTLs and alleles) regulating seed-Fe and Zn concentrations, with an ultimate objective of genetic enhancement in chickpea.
Results and Discussion
Genome-wide distribution of SNPs
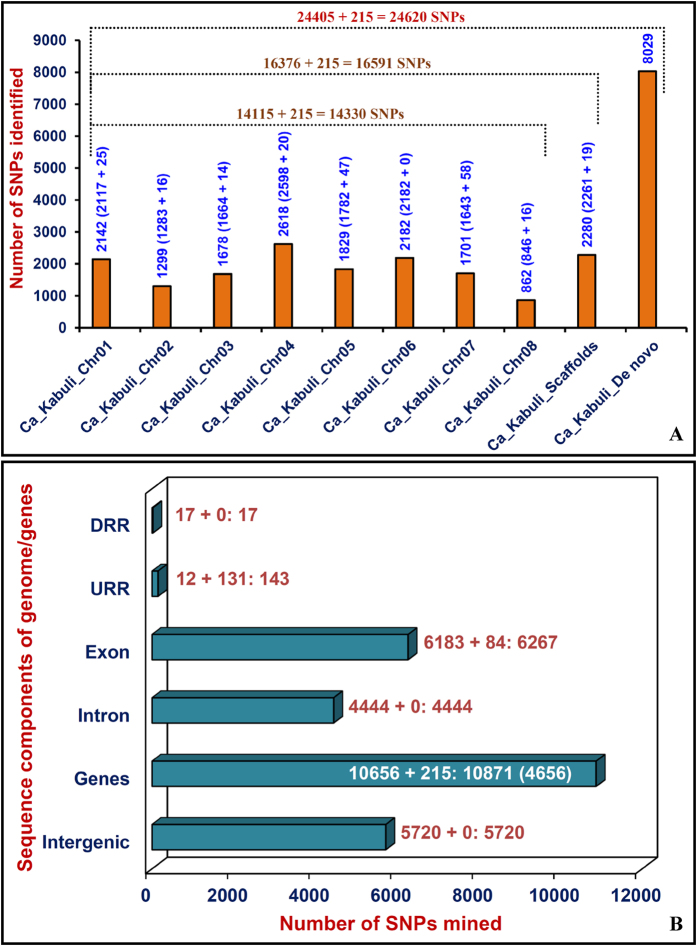
The kabuli reference genome (16376)- and de novo (8029)-based GBS (genotyping-by-sequencing) assays were employed to mine 24405 high-quality SNPs from 92 desi and kabuli chickpea accessions (Table S1)44. The GBS assay is advantageous in rapid large-scale mining and high-throughput genotyping of high-quality accurate genome-wide SNPs simultaneously for high-resolution genetic and association mapping37,38,39,44,45. Henceforth, this NGS-based strategy has potential to identify genes/QTLs regulating important agronomic traits in chickpea. A diverse array of known cloned genes involved in homeostasis, including uptake, translocation, regulation, signaling, storage and compartmentalization of Fe and Zn-contents have been isolated and characterized in multiple crop plants, including dicots46,47,48. Therefore, it would be interesting to assess the association potential of these known cloned genes for seed-Fe and Zn contents in chickpea. This can be simply achieved by large-scale genotyping of novel coding and regulatory SNP allelic variants mined from the seed-Fe and Zn contents-related known cloned genes in a diverse set of phenotypically well-characterized natural germplam lines. With a prime objective to discover gene-based SNPs, the high-quality cloned amplicon sequences of 13 known Fe and Zn content-regulating gene orthologs of chickpea (involved in transport and signalling pathways of Fe and Zn contents in crop plants) were compared initially among 12 representative desi and kabuli accessions. The subsequent high-throughput genotyping of these mined SNPs in all 92 desi and kabuli accessions by Illumina GoldenGate assay identified 215 coding and regulatory SNPs in 13 genes (46421 bp) with a mean density of one SNP per 215.9 bp (Table 1). Altogether, kabuli reference genome- and de novo-based GBS (24405) assay as well as known gene-derived (215) SNP genotyping assay discovered 24620 SNPs (Fig. 1A). This includes 16591 reference genome-based SNPs, of which 14330 and 2280 SNPs were physically mapped across eight chromosomes (mean map density: 24.2 kb) and unanchored scaffolds of kabuli genome, respectively.
Table 1. Thirteen chickpea gene orthologs known to regulate signalling and transport of Fe and Zn contents in crop plants selected for trait association mapping.
| Characterized known cloned genes |
Kabuli chickpea gene homologs |
||||||
|---|---|---|---|---|---|---|---|
| Cropplants | Gene identities with functions | Geneaccession IDs | Geneaccession IDs | Chromosomes | Sequenced geneamplicon size (bp) | Coding and upstreamregulatory SNPs mined | |
| Arabidopsis | AtNRAMP1 | Natural resistance-associated macrophage protein 1 | At1g80830 | Ca02923 | Ca_Kabuli_Chr01 | 3485 | 13 |
| Arabidopsis | AtZIP2 | Zinc/iron permease Zinc/iron regulated transporter-related protein 2 | At5g59520 | Ca02630 | Ca_Kabuli_Chr01 | 3005 | 12 |
| Arabidopsis | AtFER3 | Ferritin 3 | At3g56090 | Ca12453 | Ca_Kabuli_Chr02 | 2765 | 16 |
| Arabidopsis | AtFER1 | Ferritin 1 | At5g01600 | Ca09416 | Ca_Kabuli_Chr03 | 2777 | 14 |
| Arabidopsis | AtYSL1 | Yellow stripe-like 1 Oligopeptide transporter OPT superfamily | At4g24120 | Ca03842 | Ca_Kabuli_Chr04 | 3923 | 20 |
| Arabidopsis | AtIRT3 | Iron-regulated transporter 3 | At1g60960 | Ca01633 | Ca_Kabuli_Chr05 | 3224 | 15 |
| Arabidopsis | AtZIP1 | Zinc/iron permease Zinc/iron regulated transporter-related protein 1 | At3g12750 | Ca07626 | Ca_Kabuli_Chr05 | 3044 | 14 |
| Arabidopsis | AtYSL2/3 | Yellow stripe-like 2/3 Oligopeptide transporter OPT superfamily | At5g24380 | Ca04020 | Ca_Kabuli_Chr05 | 3863 | 18 |
| Arabidopsis | AtHMA2/4 | Heavy metal ATPases 2/4 ATPase, P-type, H+ transporting proton pump | At4g30110 At2g19110 | Ca03227 | Ca_Kabuli_Chr07 | 5099 | 25 |
| Arabidopsis | AtNRAMP3 | Natural resistance-associated macrophage protein 3 | At2g23150 | Ca09322 | Ca_Kabuli_Chr07 | 3533 | 12 |
| Arabidopsis | AtFRO2 | Ferric chelate reductase 2 FAD-binding 8 | At1g01580 | Ca13723 | Ca_Kabuli_Chr07 | 4151 | 21 |
| Arabidopsis | AtNRAMP4 | Natural resistance-associated macrophage protein 4 | At5g67330 | Ca02191 | Ca_Kabuli_Chr08 | 3521 | 16 |
| Arabidopsis | AtYSL4/6 | Yellow stripe-like 4/6 Oligopeptide transporter OPT superfamily | At5g41000 At3g27020 | Ca27338 | Ca_Kabuli_Scaffold637 | 4031 | 19 |
Figure 1.
(A) Genomic distribution of 24620 SNPs discovered from the reference kabuli genome (eight chromosomes and scaffolds)- and de novo-based GBS (24405 SNPs) assays as well as by combining the Fe and Zn contents-related known cloned chickpea gene orthologs-led amplicon resequencing assay with a Illumina GoldenGate (215 SNPs) SNP genotyping assay. (B) Structural annotation of SNPs in the diverse non-coding (intron, URR and DRR) and coding (synonymous and non-synonymous) sequence components of genes and intergenic regions of kabuli genome. The CDS (coding sequences), URR (upstream regulatory region) and DRR (downstream regulatory region) of genes (mentioned in the Parentheses) were demarcated according to gene annotation information of reference kabuli genome (Varshney et al.35). The digits mentioned within/above the bars indicate the structural annotation of genome-wide GBS and candidate gene-based SNPs.
The structural annotation of 16591 SNPs, including 16376 kabuli reference genome- and 215 gene-based SNPs was performed. This revealed presence of 5720 and 10871 SNPs in the intergenic regions and 4656 genes, respectively (Fig. 1B). The annotation of SNPs within the genes exhibited the occurrence of a highest and lowest number of 6267 (6183 genome-wide and 84 gene-based SNPs) and 17 SNPs in the exons/CDS (coding sequence) and DRRs (downstream regulatory regions), respectively. The coding SNPs comprised of 4259 synonymous and 2008 non-synonymous (missense and nonsense) SNPs. The regulatory SNPs (143, including 12 genome-wide and 131 gene-based SNPs) mostly derived from the URRs (upstream regulatory regions) of kabuli chickpea genes (Fig. 1B). The GBS-based genome-wide and known gene-derived novel SNP allelic variants discovered among a diverse set of desi and kabuli chickpea accessions can be deployed for various high-throughput genetic analysis particularly in rapid identification of genes (alleles)/QTLs governing complex Fe and Zn-content traits in chickpea.
Construction of a high-density intra-specific chickpea genetic linkage map
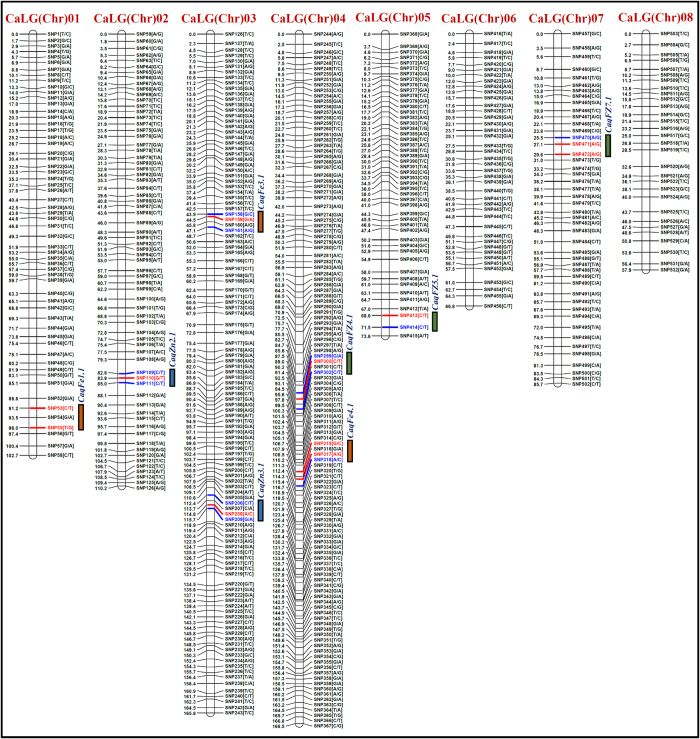
For generating an intra-specific high-resolution genetic linkage map, 536 SNPs (529 genome-wide GBS- and 7 known gene-based SNPs) exhibiting polymorphism between parental accessions (desi accession ICC 4958 and kabuli accession ICC 8261) were genotyped in 277 individuals of a RIL (recombinant inbred line) mapping population (ICC 4958 x ICC 8261). The use of these SNP genotyping data in linkage analysis mapped 533 SNPs onto eight LGs (linkage groups) of an intra-specific genetic map of chickpea (Table S2, Fig. 2). The genetic map consisting of eight LGs covered a total map length of 831.2 cM with a mean inter-marker distance of 1.56 cM. The LG4 had longest map length spanning 168.5 cM, whereas LG8 exhibited shortest map length of 57.9 cM (Table S2, Fig. 2). A highest number of SNPs were mapped on LG4 (124 markers), followed by LG3 (118) and lowest on LG8 (30). The most saturated genetic map was observed in LG4 (a mean inter-marker distance of 1.36 cM), while LG8 (1.93 cM) had the least saturated map (Table S2, Fig. 2). Overall, an intra-specific genetic linkage map constructed in our study had a comparable/higher mean map-density (an average inter-marker distance of 1.56 cM) than that recently documented in diverse microsatellite and SNP markers-anchored intra- and inter-specific genetic maps of chickpea35,36,38,39,41,49,50,51,52,53,54,55.
Figure 2. Eight major genomic regions harbouring eight robust QTLs (16.9–23.6% PVE) associated with seed-Fe and Zn concentrations were identified and mapped on six chromosomes (LOD > 6.0) using a 277 RIL mapping population (ICC 4958 × ICC 8261) of chickpea.
The genetic distance (cM) and identity of the marker loci integrated on the chromosomes are indicated on the left and right side of the chromosomes, respectively. The markers flanking and tightly linked to the QTLs are indicated with blue and red coloured lines, respectively. Orange, blue and green boxes indicate the QTLs governing seed-Fe, seed-Zn and both seed-Fe and Zn concentrations, respectively mapped on the chromosomes of a high-density genetic map.
Molecular mapping of QTLs associated with seed-Fe and Zn concentrations
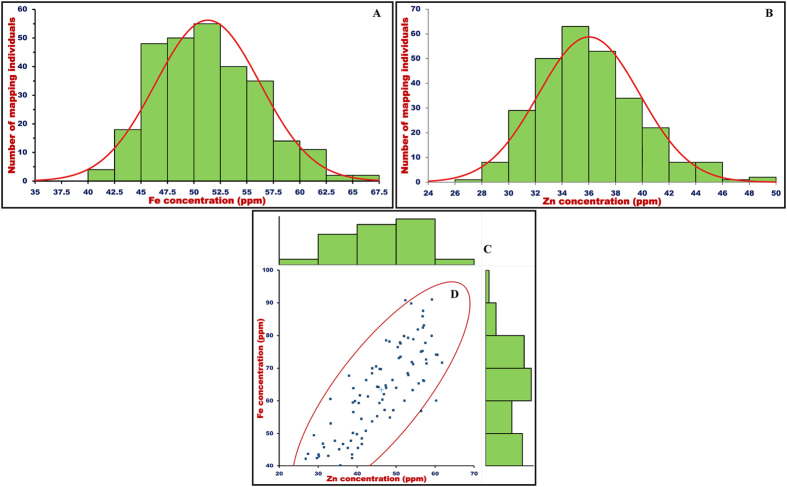
A highly significant difference (P < 0.001) of seed-Fe (51.3 ± 5.0, 40.3–67.0 ppm with 81% H2) and Zn (36.0 ± 3.8, 27.9–48.5 ppm with 83% H2) concentrations in two parental accessions and 277 individuals of a RIL mapping population [ICC 4958 (Fe: 43.0 ± 2.9 and Zn: 30.2 ± 2.5 ppm) × ICC 8261 (Fe: 64.8 ± 3.3 and Zn: 42.0 ± 2.7 ppm)] across two years based on ANOVA was observed (Table S3, Fig. 3A,B). Especially, ANOVA results inferred a highly significant difference (P < 0.001) among RIL individuals for both seed-Fe and Zn concentrations despite significant environmental (years and geographical locations) effects on these traits. A significant interaction between genotypes (G)/accessions and environment (E) for seed-Fe and Zn concentration traits was evident. However, G × E and E-variances (% of total mean squares) was found to be almost equal between seed-Fe and Zn concentrations at a significant level of P < 0.001. The normal frequency distribution, including bi-directional transgressive segregation of two target traits among mapping individuals and parental accessions was evident (Fig. 3A,B). The Pearson’s correlation coefficient estimation revealed a significant positive correlation (0.53–0.68 with mean r: 0.61, P < 0.0001) between seed-Fe and Zn contents among individuals of a mapping population.
Figure 3.
Frequency distribution of seed-Fe and Zn concentration traits variation estimated among 277 individuals and parental accessions of an intra-specific F7 RIL mapping population (ICC 4958 × ICC 8261) (A , B) as well as in a structured population of 92 desi and kabuli chickpea accessions (C) depicted a goodness of fit to the normal distribution. (D) Correlation based on Pearson’s coefficient estimated between seed-Fe and Zn concentrations among 92 chickpea accessions.
The use of genotyping data of 533 SNPs mapped on an intra-specific genetic linkage map and phenotyping data of 277 RIL mapping individuals in QTL mapping identified eight major genomic regions underlying eight robust QTLs (6.0–8.8 LOD) significantly associated with seed-Fe and Zn concentrations in chickpea (Table 2, Fig. 2). These validated QTLs exhibiting consistent phenotypic expression (>10% phenotypic variation explained49) across environments (years and geographical locations) at a higher LOD (>6.0) and H2 (>90%) were considered as robust QTLs governing chickpea seed-Fe and Zn contents. Eight major genomic regions harbouring eight robust QTLs (CaqFe1.1, CaqZn2.1, CaqFe3.1 CaqZn3.1 CaqFZ4.1 CaqFe4.1 CaqFZ5.1 and CaqFZ7.1) were mapped on six chromosomes (except chromosomes 6 and 8) (Table 2, Fig. 2). These QTL intervals spanning in the range from 2.2 cM on chromosome 2 to 4.8 cM on chromosome 1 covered with a total of 27 mapped SNP loci. The proportion of phenotypic variation explained (PVE) by individual QTL ranged from 16.9 to 23.6%. The combined PVE measured for all eight robust QTLs was 39.4%.
Table 2. Significant QTLs associated with seed-iron and zinc concentrations identified and mapped on the chromosomes of a high-density intra-specific chickpea genetic linkage map (ICC 4958 × ICC 8261).
| QTLs | LGs/chromosomes | SNP marker intervals with geneticpositions (cM) | Traits associated | SNP markers tightlylinked with QTLs | 2012 |
2013 |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| LOD | PVE(%) | A | LOD | PVE(%) | A | |||||
| CaqFe1.1 | Ca_Kabuli_Chr01 | SNP53(91.2)-SNP55(96.0) | Seed-Fe concentration | SNP53 and SNP55 | 8.6 | 23.6 | 6.5 | 8.5 | 22.6 | 6.8 |
| CaqZn2.1 | Ca_Kabuli_Chr02 | SNP109(82.8)-SNP111(85.0) | Seed-Zn concentration | SNP110 | 8.3 | 21.8 | 5.4 | 8.2 | 20.5 | 5.5 |
| CaqFe3.1 | Ca_Kabuli_Chr03 | SNP158(43.9)-SNP161(47.1) | Seed-Fe concentration | SNP159 | 8.5 | 20.8 | 4.8 | 8.0 | 21.1 | 4.3 |
| CaqZn3.1 | Ca_Kabuli_Chr03 | SNP206(112.4)-SNP209(115.7) | Seed-Zn concentration | SNP208 | 7.3 | 19.6 | 5.1 | 7.0 | 18.7 | 5.5 |
| CaqFZ4.1 | Ca_Kabuli_Chr04 | SNP299(87.5)-SNP302(91.4) | Seed-Fe and Zn concentration | SNP300 | 6.5 | 17.7 | 4.3 | 6.7 | 18.5 | 4.7 |
| 7.1 | 18.6 | 4.1 | 7.4 | 17.8 | 4.0 | |||||
| CaqFe4.1 | Ca_Kabuli_Chr04 | SNP315(106.7)-SNP318(110.2) | Seed-Fe concentration | SNP315 and SNP317 | 8.8 | 22.6 | 6.7 | 8.6 | 23.4 | 6.5 |
| CaqFZ5.1 | Ca_Kabuli_Chr05 | SNP413(68.6)-SNP414(71.5) | Seed-Fe and Zn concentration | SNP413 | 7.1 | 19.5 | 7.4 | 7.3 | 18.6 | 7.0 |
| 6.8 | 23.7 | 8.3 | 7.6 | 20.5 | 8.0 | |||||
| CaqFZ7.1 | Ca_Kabuli_Chr07 | SNP470(25.5)-SNP472(29.6) | Seed-Fe and Zn concentration | SNP471 and SNP472 | 6.3 | 16.9 | 8.5 | 6.0 | 17.8 | 8.7 |
| 6.5 | 19.5 | 8.3 | 6.3 | 18.9 | 8.4 | |||||
*CaqFe1.1 (Cicer arietinum QTL for seed-iron concentration on chromosome 1 number 1), CaqZn2.1 (Cicer arietinum QTL for seed-zinc concentration on chromosome 2 number 1) and CaqFZ4.1 (Cicer arietinum QTL for seed-iron and zinc concentrations on chromosome 4 number 1). PVE: Phenotypic variation explained. A: Additive effect of alleles from ICC 8261 with high seed-iron and zinc concentrations. Details regarding SNPs are provided in the Table S1.
For seed-Fe concentration, three major genomic regions harbouring three robust QTLs (21.1–23.4% PVE with 8.0–8.8 LOD) were mapped on three chromosomes (1, 3 and 4) (Table 2, Fig. 2). The combined PVE of all three robust QTLs was 34.6%. These QTL regions covered (3.2 cm on chromosome 3 to 4.8 cM on chromosome 1) with 11 SNPs, were genetically mapped on chromosomes. The identified major QTLs exhibited positive additive gene effect for increasing seed-Fe concentration with higher effective allelic contribution from ICC 8261. For seed-Zn concentration, two major genomic regions underlying two robust QTLs were identified and mapped on two chromosomes (2 and 3) with 18.7–21.8% PVE (7.0–8.3 LOD) (Table 2, Fig. 2). These QTL intervals covered (2.2 cM on chromosome 2 to 3.3 cM on chromosome 3) with seven SNPs, were genetically mapped on chromosomes. The combined PVE for all two robust QTLs was 33.9%. These QTLs revealed positive additive gene effect for increasing seed-Zn concentration with major allelic contributions from ICC 8261. For both seed-Fe and Zn concentrations, three major genomic regions harbouring three robust QTLs were identified and mapped on three chromosomes (4, 5 and 7) with 16.9–18.5% PVE (6.0–6.7 LOD) (Table 2, Fig. 2). The combined PVE of all three robust QTLs was 36.4%. These QTL regions covered (2.9 cm on chromosome 5 to 4.1 cM on chromosome 7) by nine genetically mapped SNP loci, were exhibited positive additive gene effect for increasing both seed-Fe and Zn concentrations with maximum effective allelic contribution from ICC 8261. All 11 informative SNP loci tightly linked to eight QTLs regulating seed-Fe and Zn concentrations are provided in the Table 2.
The 11 SNP loci linked to the robust eight major QTLs governing seed-Fe and Zn concentrations were compared/correlated with that of an only previous association mapping study on similar traits available till date in chickpea14. This revealed correspondence of two SNP loci (SNP53 and SNP317) in the genes (encoding elongation factor and I-Q domain like protein) linked tightly with two major QTLs (CaqFe1.1 and CaqFe4.1) between previous and present study. Molecular mapping of QTLs associated with any of such traits pertaining to seed-Fe and Zn concentrations has not been reported so far in chickpea. In this context, the informative SNPs especially derived from the major known/candidate genes identified and mapped at robust seed-Fe and Zn concentrations-associated QTL regions in our study seem quite relevant. The gene-based SNPs tightly linked with major QTLs identified, can be utilized in marker-assisted genetic improvement for developing cultivars with high level of Fe and Zn contents in the seeds of chickpea.
Association mapping of seed-Fe and Zn concentrations
The implication towards integrating GWAS with candidate gene-based association analysis for efficient quantitative dissection of diverse complex agronomic traits have been realized currently in chickpea38,39,56,57. Considering this, an integrated high-resolution association mapping strategy involving GWAS and candidate gene-based association analysis has been deployed in our study to narrow-down the potential genomic loci/genes governing quantitative traits of seed-Fe and Zn concentrations at a genome-wide scale in chickpea. For GWAS and candidate gene-based association mapping, the genotyping information of 16591 informative SNPs (differentiating the 92 desi and kabuli chickpea accessions) mined through genome-wide GBS (16376 SNPs)- and known/candidate gene (215 SNPs)-based SNP genotyping assays were utilized (Fig. 1A). The classification of 92 chickpea accessions into two distinct population groups (POP I and POP II) based on neighbour-joining phylogenetic tree, high-resolution population genetic structure and principal component analysis (PCA) using 16591 SNP genotyping data was apparent37. The determination of LD pattern using the genotyping data of 14330 genome-wide SNPs (physically mapped on eight kabuli chromosomes) in 92 desi and kabuli chickpea accessions exhibited 200–250 kb LD decay across chromosomes. The normal frequency distribution along with a wider phenotypic variation for seed-Fe (63.3 ± 13.3, 40.2–91.0 ppm with 80% H2) and Zn (46.2 ± 9.1, 26.8–61.8 ppm with 82% H2) concentrations among 92 desi and kabuli chickpea accessions was observed (Table S3, Fig. 3C). A higher significant positive Pearson correlation (0.71–0.86 with mean r: 0.80, P < 0.0001) between these two target traits in 92 chickpea accessions was apparent (Fig. 3D). By comparing the multi-location field phenotyping information of seed-Fe and Zn concentrations observed in our constituted natural and mapping populations, a broader target trait variation in 92 natural germplasm lines than that of 277 RIL individuals of an intra-specific mapping population (ICC 4958 × ICC 8261) was observed. This infers that the natural germplasm lines representing 16 diverse geographical regions of the world selected by us are rich in allelic diversity for seed-Fe and Zn content traits. Therefore, the screened germplasm lines can serve as a useful genetic resource for mining novel functional allelic variants to accelerate trait association mapping at a genome-wide and/or gene-level in chickpea. This will essentially expedite the identification of potential genes/alleles regulating seed-Fe and Zn concentrations in domesticated desi and kabuli chickpea.
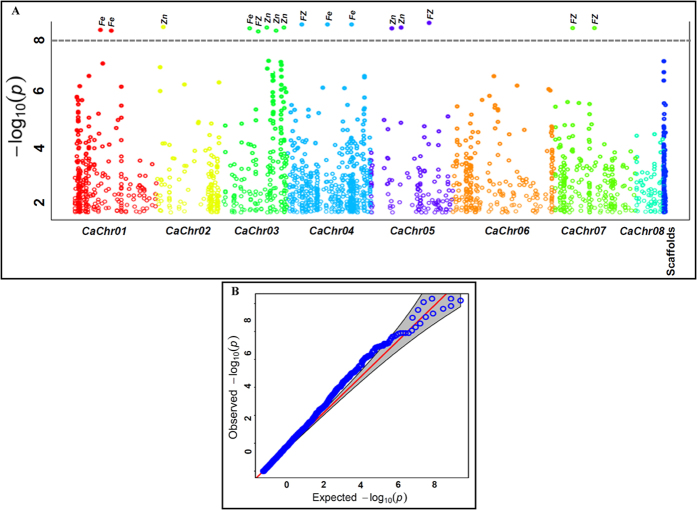
The CMLM and P3D/EMMAX model-based association mapping approach (FDR cut-off ≤0.05) identified 16 genomic loci (gene-based SNPs) exhibiting significant association (P ≤ 10−7) with seed-Fe and Zn concentrations in chickpea (Table 3, Fig. 4). Twelve SNPs of these, were scanned from the genome-wide GBS information, whereas four SNPs derived from the known cloned kabuli chickpea genes regulating Fe and Zn contents. All 16 trait-associated SNPs were physically mapped on six chromosomes (except chromosomes 6 and 8) of kabuli genome (Table 3, Fig. 4). A highest number of five trait-associated genomic loci were mapped on chromosome 3. Nine and seven of 16 trait-associated genomic loci was represented from diverse coding (five synonymous and four non-synonymous SNPs) and non-coding (four intronic and three URRs) sequence components of 16 genes, respectively (Table 3, Fig. 4). The phenotypic variation for seed-Fe and Zn concentrations explained by 16 known/candidate gene-derived maximum effect genomic SNP loci varied from 6 to 14% (P: 1.2 × 10−8 to 1.0 × 10−9) among 92 chickpea accessions (Table 3, Fig. 4). The combined phenotypic variation for seed-Fe and Zn concentrations explained by all significant 16 genic SNP loci was 29% (Table 3, Fig. 4). Specifically, six gene-based SNP loci, including one of each non-synonymous and regulatory SNPs were significantly associated (6–12% R2 with P: 3.2 × 10−8 to 1.3 × 10−9) with seed-Fe concentration in chickpea (Table 3, Fig. 4). For seed-Zn concentration, six, including two non-synonymous genic SNPs revealing significant association (7–14% R2 with P: 1.0 × 10−7 to 1.0 × 10−9) with target trait were detected. In addition, four gene-derived SNPs (including one non-synonymous and two URR-SNPs) associated (6–12% R2 with P: 1.2 × 10−7 to 1.0 × 10−9) with both seed-Fe and Zn concentrations were identified (Table 3, Fig. 4). The non-synonymous coding and regulatory SNPs in the known/candidate genes associated with seed-Fe and Zn concentrations delineated by us combining both GWAS and candidate gene-based association mapping in our study have functional significance towards quantitative dissection of these complex seed quality component traits in chickpea. These inputs further can essentially be utilized for establishing the rapid marker-trait linkages and efficient identification of potential genes/QTLs governing seed-Fe and Zn concentrations in chickpea.
Table 3. Sixteen genes with SNPs regulating seed-Fe and Zn concentrations delineated by integrating GWAS and candidate gene-based association analysis with QTL mapping and expression profiling.
| SNP IDs | Kabulichromosomes | SNP physical positions (bp) | SNPs | Gene accession IDs | Structural annotation | Known/putative functions | Association analysis |
||
|---|---|---|---|---|---|---|---|---|---|
| P | R2 (%) | Traits associated | |||||||
| *,#CakSNP1426/SNP53 | Ca_Kabuli_Chr01 | 16379258 | [C/T] | Ca06927 | Intron | Ubiquitin-associated/translation elongation factor EF1B | 1.5 × 10−8 | 10 | Seed Fe concentration |
| *,#CakSNP1628/SNP55 | Ca_Kabuli_Chr01 | 24071014 | [T/G] | Ca19289 | Intron | AUX/IAA (auxin/indole-3-acetic acid) protein | 1.9 × 10−8 | 10 | Seed Fe concentration |
| *,#CakSNP14941/SNP110 | Ca_Kabuli_Chr02 | 96532 | [G/T] | Ca27126 | Intron | Vacuolar protein sorting-associated protein | 1.0 × 10−9 | 14 | Seed Zn concentration |
| *,#SNP159 | Ca_Kabuli_Chr03 | 20101590 | [G/A] | Ca09416 | URR | Ferritin 1 (FER1) | 2.0 × 10−8 | 10 | Seed Fe concentration |
| #CakSNP4409/ SNP186 | Ca_Kabuli_Chr03 | 30837284 | [C/T] | Ca12181 | CDS(Nsyn) | Domain of unknown function 828 (DUF828) | 1.2 × 10−7 | 6 | Seed Fe and Zn concentrations |
| *,#CakSNP4476/SNP208 | Ca_Kabuli_Chr03 | 31631451 | [A/C] | Ca12262 | CDS(Nsyn) | RNA recognition motif domain | 2.0 × 10−8 | 12 | Seed Zn concentration |
| #CakSNP4678/SNP216 | Ca_Kabuli_Chr03 | 36050319 | [T/C] | Ca00903 | CDS(Syn) | SWAP (suppressor-of-white-apricot)/Surp domain-containing protein | 1.2 × 10−7 | 9 | Seed Zn concentration |
| #CakSNP4740/SNP229 | Ca_Kabuli_Chr03 | 36387718 | [C/T] | Ca00947 | CDS(Syn) | WD40 [tryptophan-aspartic acid (W-D) dipeptide] repeat-containing protein | 1.0 × 10−7 | 8 | Seed Zn concentration |
| *,#SNP300 | Ca_Kabuli_Chr04 | 4138693 | [C/T] | Ca03842 | URR | Yellow stripe-like 1 (YSL1) | 1.0 × 10−9 | 12 | Seed Fe and Zn concentrations |
| *,#CakSNP6693/SNP315 | Ca_Kabuli_Chr04 | 30258596 | [G/C] | Ca14189 | CDS(Syn) | Late embryogenesis abundant (LEA) protein | 1.3 × 10−9 | 12 | Seed Fe concentration |
| *,#CakSNP7405/SNP317 | Ca_Kabuli_Chr04 | 44814982 | [A/G] | Ca09146 | Intron | IQ-domain like protein | 2.5 × 10−9 | 11 | Seed Fe concentration |
| #CakSNP15585/SNP380 | Ca_Kabuli_Chr05 | 43372 | [C/T] | Ca22196 | CDS(Syn) | Ubiquitin | 1.1 × 10−7 | 8 | Seed Zn concentration |
| #CakSNP15145/SNP396 | Ca_Kabuli_Chr05 | 82342 | [C/T] | Ca24399 | CDS(Nsyn) | Unknown expressed gene | 1.3 × 10−7 | 7 | Seed Zn concentration |
| *,#CakSNP9048/SNP413 | Ca_Kabuli_Chr05 | 41080035 | [C/T] | Ca07626 | CDS(Syn) | Zinc/iron permease (AtZIP1) | 1.0 × 10−8 | 11 | Seed Fe and Zn concentrations |
| *,#CakSNP11677/SNP471 | Ca_Kabuli_Chr07 | 781409 | [A/G] | Ca03400 | CDS(Nsyn) | Octicosapeptide/Phox/Bem1p (PB1) domain-containing protein | 3.2 × 10−7 | 6 | Seed Fe concentration |
| *,#SNP472 | Ca_Kabuli_Chr07 | 2515209 | [A/G] | Ca03227 | URR | Heavy metal ATPases 2/4 (HMA2/4) | 3.5 × 10−7 | 10 | Seed Fe and Zn concentrations |
*Validated by QTL mapping and
#differential expression profiling.
Figure 4.
(A) GWAS-based Manhatton plot exhibiting significant P-values (measured by CMLM and P3D/EMMAX model) associated with seed-Fe and Zn concentrations using 16591 genome-wide GBS- and candidate gene-based SNPs in chickpea. The x-axis illustrates the relative density of reference genome-based SNPs physically mapped on eight chromosomes and scaffolds of kabuli genome. The y-axis displays the -log10 (P)-value for significant association of 16 SNP loci with seed-Fe and Zn concentrations. The SNPs revealing significant association with seed-Fe and Zn concentrations at cut-off P value ≤10−8 are marked with dotted lines. (B) Quantile-quantile plot illustrating the comparison between expected and observed -log10(P)-values with a FDR cut-off <0.05 to detect significant genomic loci (genes) associated with seed-Fe and Zn concentrations in chickpea.
Interestingly, we scaled-down 11, including six, two and three genomic SNP loci in 11 known/candidate genes showing strong association with seed-Fe, Zn and both Fe-Zn concentrations, respectively based on association mapping. Interestingly, these gene-based SNP loci were also found to be tightly linked with the robust nine major QTLs governing respective seed-Fe and Zn content traits (detected by QTL mapping in the present study) in chickpea (Tables 2 and 3). Of these, strong association of SNPs derived from three known/candidate genes [encoding late embryogenesis abundant (LEA) protein (12% R2 with P: 2.3 × 10−9), vacuolar protein sorting-associated protein (14% R2 with P: 1.0 × 10−9) and yellow stripe-like 1 (YSL1) protein (12% R2 with P: 1.0 × 10−9)] localized at three major target robust QTLs (17.7–23.4% R2 at 6.5–8.8 LOD) governing seed-Fe, Zn and both Fe-Zn concentrations, respectively was evident (Tables 2 and 3). Henceforth, 11 known/candidate gene-derived SNPs validated by both association and QTL mapping, were further selected as potential candidates regulating seed-Fe and Zn concentrations in chickpea.
Validation of trait-associated genes through expression profiling
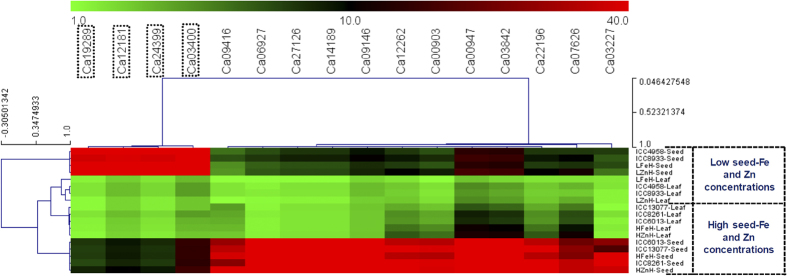
A set of 16 trait-associated known/candidate genes with SNPs, including 11 validated by both QTL and association mapping were analysed to determine the differential expression profile and functional regulatory pattern of these genes governing seed-Fe and Zn concentrations in chickpea. The differential expression analysis (by quantitative RT-PCR assay) of these trait-associated genes in the leaves and seed maturation developmental stages of five chickpea accessions/parents and four homozygous mapping individuals with contrasting level of low (ICC 4958 and ICC 8933 with 42–43 ppm Fe and 27–30 ppm Zn) and high (ICC 8261, ICC 6013 and ICC 13077 with 65–90 ppm Fe and 42–62 ppm Zn) seed-Fe and Zn concentrations was performed. All 16 trait-associated known/candidate genes (11 genes validated by QTL mapping) exhibited seed-specific expression (>4-fold) as compared to leaf tissues of parental accessions and homozygous mapping individuals with low and high level of Fe and Zn contents (Fig. 5). Of these, twelve and four genes gave pronounced differential up (~7-fold, P ≤ 0.0001)- and down (~5-fold, P ≤ 0.0001)-regulation, respectively in the seeds of parental accessions/mapping individuals with high level of Fe and Zn concentrations as compared to that of parental accessions/individuals with low Fe and Zn contents (Fig. 5). Collectively, 16 differentially expressed potential genes with SNPs governing seed-Fe and Zn concentrations by integrating GWAS and candidate gene-based association analysis with expression profiling and 11 genes of these, by inclusion of QTL mapping with aforesaid strategy, were delineated in chickpea (Table 3).
Figure 5. Hierarchical cluster display depicting the differential expression profiles of 16 seed-Fe and Zn concentrations-associated genes (including 11 validated by QTL mapping) with SNPs in the leaves and seed maturation developmental stages of five chickpea accessions/parents and four representative homozygous mapping individuals (ICC 4958 × ICC 8261) with contrasting level of low (ICC 4958 and ICC 8933) and high (ICC 8261, ICC 6013 and ICC 13077) seed-Fe and Zn concentrations.
The average log signal expression value of genes in leaves and seed developmental stages are represented at the top with a colour scale; in which green, black and red color denote low, medium and high level of expression, respectively. The genes showing seed-specific expression, including pronounced differential down-regulation are highlighted with black boxes, respectively. The structural and functional annotation of 16 genes with SNPs are mentioned in the Table 3. The tissues/stages and genes utilized for expression profiling are provided on the right and upper side of expression map, respectively. Digits mentioned in the vertical and horizontal bars illustrate the range (minimum, optimum and maximum) of correlation coefficient varying among tissues/stages of diverse contrasting accessions/mapping individuals and across trait-associated genes, respectively. LFeH: low seed-Fe concentration homozygous mapping individual and LZnH: low seed-Zn concentration homozygous mapping individual. HFeH: high seed-Fe concentration homozygous mapping individual and HZnH: high seed-Zn concentration homozygous mapping individual.
The seed-specific expression, including a pronounced upregulation and enhanced transcript accumulation of three strong seed-Fe and Zn contents-associated known/candidate genes [encoding LEA, YSL1 and vacuolar protein sorting-associated proteins] have ascertained significant involvement of these genes in storing high level of Fe and Zn contents in the maturing seeds of multiple crop (legumes) accessions46,47,48,58,59,60. These genes with useful properties of Fe/Zn-binding, are known to be involved in micronutrients trafficking, signalling and storage, especially overall homeostasis of Fe and Zn contents in seeds of crop plants. Recently, novel functional allelic variants scanned from the selected candidate genes that encode elongation factor, ubiquitin family [SWAP (suppressor-of-white-apricot)/Surp domain-containing protein] and IQ-domain like protein as well as unknown expressed protein, are found to be associated with seed-Fe and Zn concentrations based on 1536 SNP markers-led trait association mapping in chickpea14. A number of DUF 828 (domain of unknown function 828), WD40 [tryptophan-aspartic acid (W-D) dipeptide] repeat and RNA recognition motif domain-containing protein-coding genes belonging to Zn-finger family identified in our study are known to reflect molecular response towards Zn homeostasis in plant species48. The essential role of a diverse array of vacuolar protein sorting genes, including HMA2/4 (heavy metal ATPases 2/4) (ATPase/P-type/H+ transporting proton pump) in regulation of seed Fe and Zn-contents suggests the critical involvement of these genes in translocation, remobilization and storage of Fe and Zn-contents in crop plants61. These vacuolar protein-encoding genes thus can be selected as potential candidates in deciphering the functional regulatory pathways underlying Fe and Zn content-accumulation in seeds of chickpea. Additionally, an YSL gene representing oligopeptide transporter OPT superfamily and ferritin (FER) gene regulating Fe translocation and storage, respectively have been isolated and characterized in higher plants46,47,48. A zinc/iron permease (ZIP1) gene encodes zinc/iron regulated transporter-related protein 1 belonging to a novel metal transporter ZIP gene family, is known to regulate transport of a wide-range of cations, including Fe and Zn for retaining their cellular homeostasis in plants47,48,62.
The aforementioned previous reports altogether suggests that the novel functional SNP allelic variants in the known/candidate genes and high-resolution major QTLs regulating seed-Fe and Zn concentrations delineated in our study, by deploying a combinatorial genomics-assisted breeding approach (QTL mapping, GWAS, candidate gene-based association analysis and differential expression profiling) seems much relevant towards rapid quantitative dissection of complex seed quality component target traits in chickpea. These informative molecular tags thus have potential to expedite marker-assisted genetic enhancement for developing iron/zinc-biofortified cultivars of chickpea.
Methods
Discovery and genotyping of SNPs
The structural/functional annotation, including genotyping information of kabuli reference genome- and de novo-based SNPs discovered from the sequenced 92 (39 desi and 53 kabuli) diverse chickpea accessions through a GBS-assay, were acquired37. For large-scale discovery and genotyping of gene-derived SNPs in chickpea, a selected set of 30 cloned genes known to be involved in regulation, transport and signalling pathways of Fe and Zn contents in crop plants, including dicots46,47,48 were obtained. To find out the best possible chickpea gene orthologs, the coding sequence of these known genes were BLAST homology searched (cut-off E-value ≤ 1e–41 and bit score ≥ 500) against the CDS of kabuli genes. The CDS and 2000-bp URRs of these identified chickpea gene orthologs were retrieved. These coding and regulatory sequences were used to design multiple overlapping forward and reverse primers with expected amplification product size of 400 to 500 bp per primer. The gene-based primers were amplified using the genomic DNA of 12 chickpea accessions (selected from aforesaid 92 accessions). The amplified PCR product was purified, cloned and sequenced twice in both forward and reverse directions by automated 96 capillary ABI 3730xl DNA Analyzer (Applied Biosystems, USA). The high-quality SNPs detected from the gene sequences were further genotyped in the genomic DNA of 92 chickpea accessions using Illumina GoldenGate assay. Subsequently, the genotyping information of SNPs generated among accessions was utilized for trait association mapping (following Bajaj et al.41).
Phenotyping for seed-Fe and Zn contents
During normal crop season, 92 desi and kabuli chickpea accessions, and 277 individuals of an intra-specific F7 RIL mapping population (desi traditional cultivar/landrace ICC 4958 x kabuli landrace ICC 8261) were grown in the field (following randomised complete block design with at least two replications) for two consecutive years (2012 and 2013) at two diverse geographical locations (New Delhi: 28° 4′N/77° 2′E and Patancheru, Hyderabad: latitude 17° 3′N/longitude 77° 2′E) of India. The mature seeds of each accession and mapping individual (2–4 representative plants from each accession/individual) were collected separately during harvesting stage. The healthy mature seeds (20 g) of chickpea accessions were analyzed at Central Analytical Services Laboratory, ICRISAT, Patancheru to measure their Fe and Zn contents. Precautions were taken to avoid the contamination of seeds with dust and metal particles during harvest and while preparing them for analysis.
Further, the seed samples were washed with distilled water and oven-dried at 60 °C for 48 h before grinding. The dried-seed samples (20 g) from each accession were then powdered in a mill with Teflon chambers and ground samples kept overnight in an oven at 60 °C. The standards and samples were digested simultaneously with appropriate blanks in duplicate (two independent analyses). One g of ground sample was transferred to a digestion tube [75 ml capacity and 10 ml of tri-acid mixture consisting of nitric acid, sulfuric and perchloric acid in the ratio of 10:0.5:2 (v/v)] and the contents left overnight for cold digestion in a digestion chamber. The samples were digested initially at 120 °C for one h, followed by digestion at 230 °C for about 2 h to get clear and colourless digests. Following cooling of the digests, the contents were dissolved in distilled water and volume made up to 75 ml, and shaken well. Aliquots were taken from the digests and analyzed for Fe and Zn concentrations (mg kg−1) by atomic absorption spectrophotometry (Varian Spectra AA 2063). The concentrations of Fe and Zn were estimated and expressed as mg kg−1 (ppm) seed. The seed-Fe and Zn contents of accessions were measured on a plot basis and their mean values were used for statistical analysis.
The diverse statistical parameters, including mean, standard deviation, coefficient of variation (CV), analysis of variance (ANOVA), frequency distribution and Pearson’s correlation coefficient of seed-Fe and Zn concentrations among accessions and RIL mapping individuals were measured using SPSSv17.0 as per Bajaj et al.41. The inheritance pattern of seed-Fe and Zn concentration traits was determined by estimating the effect of genotypes (accessions) (G), environments (experimental years and/or geographical locations) (E) and G × E interactions based on two-way ANOVA. The broad-sense heritability [H2 = σ2g/(σ2g + σ2ge/n + σ2e/nr)] of these two target traits was measured individually using the optimum criteria such as σ2g (genetic), σ2ge (G × E) and σ2e (error) variance with n (number of experimental years/environments) =2 and r (number of replicates) =2.
QTL mapping
The multi-location/years replicated field phenotyping data for seed-Fe and Zn contents (ppm) of parental accessions and 277 individuals derived from a RIL mapping population (ICC 4958 × ICC 8261) was obtained. A selected set of parental polymorphic GBS-based (obtained from Kujur et al.37) and known gene-derived SNPs physically mapped on eight chromosomes was genotyped in 277 RIL mapping individuals and parental accessions using an Illumina GoldenGate SNP genotyping assay (as per Bajaj et al.41). The SNP genotyping data of a RIL mapping population were utilized to construct an intra-specific high-density genetic linkage map. Subsequently, this was correlated with the aforementioned phenotyping data of RIL mapping individuals and parents for QTL mapping [composite interval mapping at significant LOD (logarithm of odds) threshold >4.0 at a p < 0.05] as per Bajaj et al.41 and Saxena et al.42. Accordingly, identification and molecular mapping of the major genomic regions underlying robust QTLs associated with seed-Fe and Zn concentrations were performed in chickpea.
Association mapping
For GWAS, the information regarding the genetic diversity (phylogenetic tree, PCA and population structure) and genome-wide/population-specific LD patterns (LD estimates and LD decay) among 92 chickpea accessions were obtained from the previous study37. The population structure ancestry coefficient (Q), kinship matrix (K) and PCA (P) data as well as the genome-wide and candidate gene-based SNP genotyping and phenotyping information (seed-Fe and Zn concentrations) of 92 accessions were analysed through mixed model (P + K, K and Q + K)-based CMLM (compressed mixed linear model) and P3D (population parameters previously determined64)/EMMAX (efficient mixed model association expedited65) approaches of GAPIT (following38,66). To ascertain the accuracy and robustness of SNP marker-trait association, the comparison of observed and expected -log10(P)-value relative distribution for each trait-associated genomic loci based on quantile-quantile plot and correction of their adjusted P-value threshold of significance for multiple comparison by false discovery rate (FDR cut-off ≤0.05, Benjamini and Hochberg 1995) were performed following Kujur et al.38 and Kumar et al.66. The degree of association of SNP loci with seed-Fe and Zn concentration traits was estimated by the R2 using a model with the SNP and adjusted P-value following a FDR-controlling method. The SNP loci in the genomic (gene) regions revealing significant association with seed-Fe and Zn concentrations at the lowest FDR adjusted P-values (threshold P < 10−7) and highest R2 (degree of SNP marker-trait association) were identified in chickpea.
Expression profiling
To determine the regulatory pattern of candidate genes associated (validated by QTL mapping and association analysis) with seed-Fe and Zn concentrations, the differential expression profiling of these genes was performed using the quantitative RT-PCR assay. The leaves and late seed maturation developmental stages [21–30 days after podding (DAP)67] of five chickpea accessions as well as parents and four homozygous individuals of a RIL mapping population (ICC 4958 × ICC 8261) with contrasting level of low and high seed-Fe and Zn contents (ppm), were used for RNA isolation. In the RT-PCR assay, RNA isolated from three independent biological replicates of each sample and two technical replicates of each biological replicate with no template and primer as control were utilized. In addition, an internal control gene elongation factor 1-alpha (EF1α) for normalization of expression value and gene-specific primers for determining the differential gene expression profile across tissues of accessions and mapping parents/individuals were used (as per Bajaj et al.67). The significant difference of gene expression among accessions and mapping parents/individuals was determined by constructing a heat map through TIGR MultiExperiment Viewer (MeV, http://www.tm4.org/mev).
Additional Information
How to cite this article: Upadhyaya, H. D. et al. Genetic dissection of seed-iron and zinc concentrations in chickpea. Sci. Rep. 6, 24050; doi: 10.1038/srep24050 (2016).
Supplementary Material
Acknowledgments
The authors gratefully acknowledge the financial support for this study provided by a research grant from the Department of Biotechnology (DBT), Government of India (102/IFD/SAN/2161/2013-14). Shouvik Das acknowledges the DBT for Senior Research Fellowship award.
Footnotes
Author Contributions H.D.U., D.B. and S.D. conducted experiments and drafted the manuscript. D.B., S.D. and V.K. involved in data analysis. H.D.U., C.L.L.G. and S.S. helped in constitution of association panel and performed phenotyping. S.K.P. and A.K.T. conceived and designed the study, guided data analysis and interpretation, participated in drafting and correcting the manuscript critically and gave the final approval of the version to be published. All authors have read and approved the final manuscript.
References
- Godfray H. C. J. Food security: the challenge of feeding 9 billion people. Science 327, 812–818 (2010). [DOI] [PubMed] [Google Scholar]
- Bohra A. et al. Genetics-and genomics-based interventions for nutritional enhancement of grain legume crops: status and outlook. J. Appl. Genet. 56, 151–161 (2015). [DOI] [PubMed] [Google Scholar]
- Welch R. M. Breeding strategies for biofortified staple plant foods to reduce micronutrient malnutrition globally. J. Nutr. 132, 495S–499S (2002). [DOI] [PubMed] [Google Scholar]
- Welch R. M. & Graham R. D. Breeding for micronutrients in staple food crops from a human nutrition perspective. J. Exp. Bot. 55, 353–364 (2004). [DOI] [PubMed] [Google Scholar]
- Thavarajah P. Evaluation of chickpea (Cicer arietinum L.) micronutrient composition: Biofortification opportunities to combat global micronutrient malnutrition. Food Res. Int. 49, 99–104 (2012). [Google Scholar]
- Fletcher R. J., Bell I. P. & Lambert J. P. Public health aspects of food fortification: a question of balance. Proc. Nutr. Soc. 63, 605–614 (2004). [DOI] [PubMed] [Google Scholar]
- Welch R. M. & Graham R. D. A new paradigm for world agriculture: meeting human needs: productive, sustainable, nutritious. Field Crops Res. 60, 1–10 (1999). [Google Scholar]
- Cichy K. A., Caldas G. V., Snapp S. S. & Blair M. W. QTL analysis of seed iron, zinc, and phosphorus levels in an Andean bean population. Crop Sci. 49, 1742–1750 (2009). [Google Scholar]
- Blair M. W. et al. QTL for seed iron and zinc concentration and content in a Mesoamerican common bean (Phaseolus vulgaris L.) population. Theor. Appl. Genet. 121, 1059–1070 (2010). [DOI] [PubMed] [Google Scholar]
- Brown K. H., Peerson J. M., Rivera J. & Allen L. H. Effect of supplemental zinc on the growth and serum zinc concentrations of prepubertal children: a meta-analysis of randomized controlled trials. Am. J. Clin. Nutr. 75, 1062–1071 (2002). [DOI] [PubMed] [Google Scholar]
- White P. J. & Broadley M. R. Biofortifying crops with essential mineral elements. Trends Plant Sci. 10, 586–593 (2005). [DOI] [PubMed] [Google Scholar]
- Mayer J. E., Pfeiffer W. H. & Beyer P. Biofortified crops to alleviate micronutrient malnutrition. Curr. Opin. Plant Biol. 11, 166–170 (2008). [DOI] [PubMed] [Google Scholar]
- Blair M. W., Astudillo C., Grusak M. A., Graham R. & Beebe S. E. Inheritance of seed iron and zinc concentrations in common bean (Phaseolus vulgaris L.). Mol. Breed. 23, 197–207 (2009). [Google Scholar]
- Diapari M. et al. Genetic diversity and association mapping of iron and zinc concentrations in chickpea (Cicer arietinum L.). Genome 57, 459–468 (2014). [DOI] [PubMed] [Google Scholar]
- Uauy C., Distelfeld A., Fahima T., Blechl A. & Dubcovsky J. A. NAC Gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 314, 1298–1301 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K. et al. Quantitative trait loci controlling Cu, Ca, Zn, Mn and Fe content in rice grains. J. Genet. 87, 305–310 (2008). [DOI] [PubMed] [Google Scholar]
- Tiwari V. K. et al. Mapping of quantitative trait Loci for grain iron and zinc concentration in diploid A genome wheat. J. Hered. 100, 771–776 (2009). [DOI] [PubMed] [Google Scholar]
- Anuradha K. et al. Mapping QTLs and candidate genes for iron and zinc concentrations in unpolished rice of Madhukar × Swarna RILs. Gene 508, 233–240 (2012). [DOI] [PubMed] [Google Scholar]
- Jin T. et al. The genetic architecture of zinc and iron content in maize grains as revealed by QTL mapping and meta-analysis. Breed. Sci. 63, 317–324 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankaran R. P., Huguet T. & Grusak M. A. Identification of QTL affecting seed mineral concentrations and content in the model legume Medicago truncatula. Theor. Appl. Genet. 119, 241–253 (2009). [DOI] [PubMed] [Google Scholar]
- Thavarajah P., Thavarajah D. & Vandenberg A. Low phytic acid lentils (Lens culinaris L.): a potential solution for increased micronutrient bioavailability. J. Agric. Food Chem. 57, 9044–9049 (2009). [DOI] [PubMed] [Google Scholar]
- Blair M. W. et al. Variation and inheritance of iron reductase activity in the roots of common bean (Phaseolus vulgaris L.) and association with seed iron accumulation QTL. BMC Plant Biol. 10, 215 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair M. W., Astudillo C., Rengifo J., Beebe S. E. & Graham R. QTL analyses for seed iron and zinc concentrations in an intra-genepool population of Andean common beans (Phaseolus vulgaris L.). Theor. Appl. Genet. 122, 511–521 (2011). [DOI] [PubMed] [Google Scholar]
- Shunmugam A. S. K. et al. Mapping seed phytic acid concentration and iron bioavailability in a pea recombinant inbred line population. Crop Sci. 55, 828–836 (2014). [Google Scholar]
- Kumar A., Choudhary A. K., Solanki R. K. & Pratap A. Towards marker-assisted selection in pulses: a review. Plant Breed. 130, 297–313 (2011). [Google Scholar]
- Varshney R. K. et al. Achievements and prospects of genomics-assisted breeding in three legume crops of the semi-arid tropics. Biotechnol. Adv. 31, 1120–1134 (2013). [DOI] [PubMed] [Google Scholar]
- Jain M. et al. A draft genome sequence of the pulse crop chickpea (Cicer arietinum L.). Plant J. 74, 715–729 (2013). [DOI] [PubMed] [Google Scholar]
- Varshney R. K. et al. Draft genome sequence of chickpea (Cicer arietinum) provides a resource for trait improvement. Nat. Biotechnol. 31, 240–246 (2013). [DOI] [PubMed] [Google Scholar]
- Parween S. et al. An advanced draft genome assembly of a desi type chickpea (Cicer arietinum L.). Sci. Rep. 5, 12806 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukanti A. K., Gaur P. M., Gowda C. L. L. & Chibbar R. N. Nutritional quality and health benefits of chickpea (Cicer arietinum L.): a review. Brit. J. Nut. 108, S11–S26 (2012). [DOI] [PubMed] [Google Scholar]
- Konishi S. et al. SNP caused loss of seed shattering during rice domestication. Science 312, 1392–1396 (2006). [DOI] [PubMed] [Google Scholar]
- Tian Z. et al. Allelic diversities in rice starch biosynthesis lead to a diverse array of rice eating and cooking qualities. Proc. Natl. Acad. Sci. USA 106, 21760–21765 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharabian-Masouleh A., Waters D. L. E., Reinke R. F., Ward R. & Henry R. J. SNP in starch biosynthesis genes associated with nutritional and functional properties of rice. Sci. Rep. 2, 557 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parida S. K., Mukerji M., Singh A. K., Singh N. K. & Mohapatra T. SNPs in stress-responsive rice genes: validation, genotyping, functional relevance and population structure. BMC Genomics 13, 426 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujur A. et al. Functionally relevant microsatellite markers from chickpea transcription factor genes for efficient genotyping applications and trait association mapping. DNA Res. 20, 355–374 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujur A. et al. An efficient and cost-effective approach for genic microsatellite marker-based large-scale trait association mapping: identification of candidate genes for seed weight in chickpea. Mol. Breed. 34, 241–265 (2014). [Google Scholar]
- Kujur A. et al. Employing genome-wide SNP discovery and genotyping strategy to extrapolate the natural allelic diversity and domestication patterns in chickpea. Front. Plant Sci. 6, 162 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujur A. et al. A genome-wide SNP scan accelerates trait-regulatory genomic loci identification in chickpea. Sci. Rep. 5, 11166 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujur A. et al. Ultra-high density intra-specific genetic linkage maps accelerate identification of functionally relevant molecular tags governing important agronomic traits in chickpea. Sci. Rep. 5, 9468 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrao S. New allelic variants found in key rice salt-14 tolerance genes: an association study. Plant Biotech. J. 11, 87–100 (2013). [DOI] [PubMed] [Google Scholar]
- Bajaj D. et al. combinatorial approach of comprehensive QTL-based comparative genome mapping and transcript profiling identified a seed weight-regulating candidate gene in chickpea. Sci. Rep. 5, 9264 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena M. S. et al. An Integrated genomic approach for rapid delineation of candidate genes regulating agro-morphological traits in chickpea. DNA Res. 21, 695–710 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J. & Li J. Molecular genetic dissection of quantitative trait loci regulating rice grain size. Annu. Rev. Genet. 48, 99–118 (2014). [DOI] [PubMed] [Google Scholar]
- Deokar A. A. et al. Genome wide SNP identification in chickpea for use in development of a high density genetic map and improvement of chickpea reference genome assembly. BMC Genomics 15, 708 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaganathan D. et al. Genotyping-by-sequencing based intra-specific genetic map refines a “QTL-hotspot” region for drought tolerance in chickpea. Mol. Genet. Genomics 290, 559–571 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curie C. & Briat J. F. Iron transport and signaling in plants. Annu. Rev. Plant Biol. 54, 183–206 (2003). [DOI] [PubMed] [Google Scholar]
- Kobayashi T. & Nishizawa N. K. Iron uptake, translocation, and regulation in higher plants. Annu. Rev. Plant Biol. 63, 131–152 (2012). [DOI] [PubMed] [Google Scholar]
- Bouain N. et al. Phosphate and zinc transport and signalling in plants: toward a better understanding of their homeostasis interaction. J. Exp. Bot. 65, 5725–5741 (2014). [DOI] [PubMed] [Google Scholar]
- Nayak S. N. et al. Integration of novel SSR and gene-based SNP marker loci in the chickpea genetic map and establishment of new anchor points with Medicago truncatula genome. Theor. Appl. Genet. 120, 1415–1441 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gujaria N. et al. Development and use of genic molecular markers (GMMs) for construction of a transcript map of chickpea (Cicer arietinum L.). Theor. Appl. Genet. 122, 1577–1589 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiremath P. J. et al. Large-scale transcriptome analysis in chickpea (Cicer arietinum L.), an orphan legume crop of the semi-arid tropics of Asia and Africa. Plant Biotech. J. 9, 922–931 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thudi M. et al. Novel SSR markers from BAC-end sequences, DArT arrays and a comprehensive genetic map with 1,291 marker loci for chickpea (Cicer arietinum L.). PLoS One 6, e27275 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur R. et al. High-throughput SNP discovery and genotyping for constructing a saturated linkage map of chickpea (Cicer arietinum L.). DNA Res. 19, 357–373 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabbavarapu M. M. et al. Molecular mapping of QTLs for resistance to Fusarium wilt (race 1) and Ascochyta blight in chickpea (Cicer arietinum L.). Euphytica 193, 121–133 (2013). [Google Scholar]
- Varshney R. K., Thudi M. & Nayak S. N. Genetic dissection of drought tolerance in chickpea (Cicer arietinum L.). Theor. Appl. Genet. 127, 445–462 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale D. B. & Savolainen O. Association genetics of complex traits in conifers. Trends Plant Sci. 9, 325–330 (2004). [DOI] [PubMed] [Google Scholar]
- Varshney R. K. et al. Genome wide association analyses for drought tolerance related traits in barley (Hordeum vulgare L.). Field Crops Res. 126, 171–180 (2012). [Google Scholar]
- Kim S. A. & Guerinot M. L. Mining iron: iron uptake and transport in plants. FEBS Letters 581, 2273–2280 (2007). [DOI] [PubMed] [Google Scholar]
- Liu G., Xu H., Zhang L. & Zheng Y. Fe binding properties of two soybean (Glycine max L.) LEA4 proteins associated with antioxidant activity. Plant Cell Physiol. 52, 994–1002 (2011). [DOI] [PubMed] [Google Scholar]
- Singh S. P. et al. Comparative transcriptional profiling of two wheat genotypes, with contrasting levels of minerals in grains, shows expression differences during grain filling. PLoS One 9, e111718 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santi S. & Schmidt W. Laser microdissection-assisted analysis of the functional fate of iron deficiency induced root hairs in cucumber. J. Exp. Bot. 59, 697–704 (2008). [DOI] [PubMed] [Google Scholar]
- Guerinot M. L. The ZIP family of metal transporters. Biochim. Biophys. Acta 1465, 190–198 (2000). [DOI] [PubMed] [Google Scholar]
- Sahrawat K. L., Ravi Kumar G. & Rao J. K. Evaluation of triacid and dry ashing procedures for determining potassium, calcium, magnesium, iron, zinc, manganese and copper in plant materials. Commu. Soil Sci. Plant Anal. 33, 95–102 (2002). [Google Scholar]
- Kumar V. et al. Genome-wide association mapping of salinity tolerance in rice (Oryza sativa). DNA Res. 22, 133–145 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z. et al. Mixed linear model approach adapted for genome-wide association studies. Nat. Genet. 42, 355–360 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H. M. et al. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 42, 348–354 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajaj D. et al. Genome-wide high-throughput SNP discovery and genotyping for understanding natural (functional) allelic diversity and domestication patterns in wild chickpea. Sci. Rep. 5, 11627 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.