Abstract
Recent studies have demonstrated a potential link between circulating cell-free mitochondrial DNA (mtDNA) content and cancers. However, there is no study evaluating the association between circulating mtDNA as a non-invasive marker of hepatocellular carcinoma (HCC) risk. We conducted a nested case-control study to determine circulating mtDNA content in serum samples from 116 HBV-related HCC cases and 232 frequency-matched cancer-free HBV controls, and evaluate the retrospective association between mtDNA content and HCC risk using logistic regression and their temporal relationship using a mixed effects model. HCC cases had significantly lower circulating mtDNA content than controls (1.06 versus 2.47, P = 1.7 × 10−5). Compared to HBV patients with higher mtDNA content, those with lower mtDNA content had a significantly increased risk of HCC with an odds ratio (OR) of 2.19 (95% confidence interval [CI] 1.28–3.72, P = 0.004). Quartile analyses revealed a significant dose-dependent effect (Ptrend = 0.001) for this association. In a pilot longitudinal sub-cohort of 14 matched cases-control pairs, we observed a trend of dramatically decreased mtDNA content in cases and slightly decreased mtDNA content in controls, with a significant interaction of case-control status with time (Pinteraction = 0.049). Our findings suggest that circulating mtDNA is a potential novel non-invasive biomarker of HCC risk in HBV patients.
Hepatocellular carcinoma (HCC) is a leading cause of cancer death, and chronic hepatitis B virus (HBV) infection represents the most common etiology of HCC worldwide1. In the United States, although hepatitis C virus (HCV) infection and alcohol and fat liver diseases are more common HCC etiological factors of HCC, HBV-related HCC has also grown significantly due to the influx of immigrants from HBV endemic regions such as Asia. It was reported that there were about 1.5 million chronic HBV-infected patients among the more than 40 million Americans born outside of the United States2. Moreover, over 60% of these HBV patients are relatively young, which may significantly increase the HBV-induced HCC risk and associated medical costs in the next a few decades3. These facts highlight the importance of risk prediction, close surveillance, and early diagnosis in HCC management.
Mitochondria are organelles in cytoplasm of the eukaryotic cells that play an important role in essential cellular functions such as energy metabolism, free radical generation, calcium homeostasis, and apoptosis4. Each cell contains several hundred to thousand mitochondria and each mitochondrion possesses its own genome5. Mitochondrial DNA (mtDNA) consists of an approximately 16.5 kb circular, maternally inherited, double-stranded extrachromosomal DNA6. Unlike nuclear DNAs, mtDNA does not have protective histones and sophisticated DNA repair mechanisms, which render it extremely susceptibility to oxidative stress and other genotoxic insults7. Moreover, high levels of reactive oxygen species (ROS) are generated around mtDNAs during metabolic events occurring in mitochondria. This environment of high oxidative stress also contributes to the high susceptibility of mtDNA to mutagenesis. As a defense mechanism, mitochondria increase their DNA copy numbers to mitigate the negative effects resulting from the high rates of mutations in their genomes8. The abnormal alteration of mtDNA copy number is well documented for numerous malignancies, including HCC9,10,11,12.
Biomarkers based on circulating DNA samples have unique advantages over other specimens due to their non-invasive nature as well as the capability of repetitive sampling13,14,15. Recently, it has been reported that the mtDNA content from plasma or serum samples are significantly associated with various diseases, such as Ewing’s sarcoma16, trauma17, and microbial infection18,19. It has also been associated with the mortality of patients in intensive care units20. However, as yet there is no reported study evaluating the association between circulating mtDNA content and HCC risk. In the current study, we sought to evaluate the association between circulating mtDNA content and the risk of HCC in patients with chronic HBV infection.
Results
Characteristics of study population
The patients’ characteristics were summarized in Table 1. This analysis included 116 HBV-related HCC cases and 232 cancer-free HBV controls that were 1:2 frequency-matched on age (mean ± standard deviation [SD], 55.7 ± 9.1 years vs. 54.1 ± 8.8 years, P = 0.123) and gender (P = 1.000). Additionally, no significant difference was observed in other major host characteristics, such as smoking status (P = 0.491), drinking status (P = 0.494), and family history of cancer (P = 0.742). As expected, a significantly higher percentage of cirrhotic patients was noted in HBV-related HCC cases (77.6%) than cancer-free HBV controls (39.7%) (P = 2.4 × 10−11).
Table 1. Summary of the study population.
| Variables | HBV Controls (n = 232, %) | HCC Cases (n = 116, %) | P value |
|---|---|---|---|
| Age (Mean ± SD) | 54.1 ± 8.8 | 55.7 ± 9.1 | 0.123 |
| Gender | |||
| Female | 26 (11.2) | 13 (11.2) | |
| Male | 206 (88.8) | 103 (88.8) | 1.000 |
| Smoking status | |||
| Never | 135 (58.2) | 63 (54.3) | |
| Ever | 97 (41.8) | 53 (45.7) | 0.491 |
| Drinking status | |||
| Never | 127 (54.7) | 59 (50.9) | |
| Ever | 105 (45.3) | 57 (49.1) | 0.494 |
| Family history of cancer | |||
| No | 160 (69.0) | 82 (70.7) | |
| Yes | 72 (31.0) | 34 (29.3) | 0.742 |
| Cirrhosis | |||
| No | 140 (60.3) | 26 (22.4) | |
| Yes | 92 (39.7) | 90 (77.6) | 2.4 × 10−11 |
Distributions of circulating mtDNA content in cases and controls by patients’ characteristics
The overall and stratified distributions of circulating mtDNA content between cases and controls were shown in Table 2. Overall, cases had significantly lower mtDNA content than controls (median, 1.06 [quartile range, 0.23–3.09] vs. 2.47 [0.90–5.15], P = 1.7 × 10−5). In stratified analyses, the significantly lower mtDNA content in cases was evident in all strata. In addition, we compared mtDNA differences between the strata of each variable in cases and controls separately. We observed a significantly lower mtDNA content in older than younger patients, and in males than females (P = 1.1 × 10−6 and 1.2 × 10−5, respectively) in controls. There was no significant difference between the strata of other variables in controls and all variables in cases. We further compared mtDNA content in the patients with different status of liver diseases by analyzing mtDNA content in three groups of patients: cancer-free HBV patients without liver cirrhosis, cancer-free HBV patients with liver cirrhosis, and HBV patients who were diagnosed as HCC. We found no statistically significant differences in the distributions of major patient characteristics among the three groups (Supp. Table 1). The median value (quartile range) of mtDNA content was 2.58 (1.12–5.21) in 140 HBV patients without cirrhosis, 2.21 (0.55–4.94) in 92 cirrhotic HBV patients, and 1.06 (0.23–3.09) in 116 HBV-HCC patients (P < 0.0001), showing an apparent decreasing trend in mtDNA content from non-cirrhotic HBV patients, to cirrhotic HBV patients, and then to HCC patients.
Table 2. Distributions of circulating mtDNA content by host characteristics in cases and controls.
| Variables | mtDNA: Median (Quartile range) | P† value | |
|---|---|---|---|
| HBV controls | HCC cases | ||
| Overall | 2.47 (0.90–5.15) | 1.06 (0.23–3.09) | 1.7 × 10−5 |
| Age, y | |||
| ≤54.3 | 3.37 (1.84–5.77) | 1.22 (0.30–3.92) | 1.5 × 10−4 |
| >54.3 | 1.36 (0.35–3.75) | 0.92 (0.23–2.75) | 0.049 |
| P‡ | 1.1 × 10−6 | 0.287 | |
| Gender | |||
| Female | 6.66 (2.76–10.94) | 1.20 (0.68–2.57) | 0.005 |
| Male | 2.18 (0.88–4.43) | 1.02 (0.22–3.28) | 3.1 × 10−4 |
| P‡ | 1.2 × 10−5 | 0.403 | |
| Smoking status | |||
| Never | 2.57 (0.95–5.17) | 0.92 (0.22–3.28) | 0.001 |
| Ever | 2.17 (0.88–4.75) | 1.20 (0.27–2.79) | 0.012 |
| P‡ | 0.438 | 0.905 | |
| Drinking status | |||
| Never | 2.56 (0.94–5.10) | 0.86 (0.19–3.28) | 0.001 |
| Ever | 2.27 (0.89–5.21) | 1.36 (0.29–2.84) | 0.011 |
| P‡ | 0.995 | 0.522 | |
| Family history of cancer | |||
| No | 2.23 (0.77–4.73) | 1.15 (0.27–3.49) | 0.005 |
| Yes | 2.71 (1.19–5.55) | 0.93 (0.18–2.75) | 3.2 × 10−4 |
| P‡ | 0.143 | 0.509 | |
| Cirrhosis | |||
| No | 2.58 (1.12–5.21) | 0.68 (0.18–1.95) | 0.001 |
| Yes | 2.21 (0.55–4.94) | 1.21 (0.27–3.49) | 0.018 |
| P‡ | 0.286 | 0.335 | |
P†, comparisons between cases and controls.
P‡, comparisons between the two strata of each individual variable.
Association between circulating mtDNA content and HCC risk
The association between circulating mtDNA content and HBV-related HCC risk was estimated using unconditional logistic regression model by analyzing the mtDNA content as a categorical variable based on the cutoff of median or quartile distribution of mtDNA content in controls. As shown in Table 3, patients with a lower level of serum mtDNA content (≤2.47) exhibited a significantly increased HCC risk with a crude OR of 2.22 (95% CI 1.39–3.56, P = 8.7 × 10−4) in univariate analysis and an adjusted OR of 2.19 (1.28–3.72, P = 0.004) in multivariate analysis adjusting for age, gender, smoking status, drinking status, family history of cancer, and cirrhosis, compared to those with a higher mtDNA content (>2.47). In quartile analysis, using the patients with the highest level of mtDNA content as reference, patients with lower levels of mtDNA content showed significantly increased HCC risk in a dose-dependent manner in both univariate and multivariate analyses (Pfor trend = 1.6 × 10−4, and 0.001, respectively). To analyze the confounding effect of alpha fetoprotein (AFP), the most commonly used HCC marker in clinics, we conducted a subset analysis in 252 patients with available AFP data. The results of the subset analysis showed that the association between mtDNA content and HCC risk remained significant (Pfor trend = 0.001) after adjusting for covariates including AFP (Table 3). Similar results were also noticed when major liver enzymes (e.g., alkaline phosphatase [ALP], aspartate aminotransferase [AST], alanine aminotransferase [ALT], gamma-glutamyltransferase [GGT]) were added to the multivariate analyses (Supp. Table 2). Moreover, we did not identify any correlation between mtDNA content and AFP level or tumor size (data not shown).
Table 3. The association of circulating mtDNA content with HCC risk.
| mtDNA | HBVcontrols | HCCcases | Univariate | Multivariate† | Multivariate‡ | Multivariate§ | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P value | OR (95% CI) | P value | OR (95% CI) | P value | OR (95% CI) | P value | |||
| By median | ||||||||||
| >2.47 | 116 | 36 | 1.00 | 1.00 | 1.00 | 1.00 | ||||
| ≤2.47 | 116 | 80 | 2.22 (1.39–3.56) | 8.7 × 10−4 | 2.19 (1.32–3.63) | 0.003 | 2.19 (1.28–3.72) | 0.004 | 2.95 (1.32–6.58) | 0.008 |
| By quartile | ||||||||||
| >4.93 | 58 | 19 | 1.00 | 1.00 | 1.00 | 1.00 | ||||
| 4.93– | 58 | 17 | 0.90 (0.42–1.89) | 0.771 | 0.95 (0.44–2.04) | 0.887 | 0.88 (0.39–1.98) | 0.758 | 2.94 (0.95–9.06) | 0.060 |
| 2.47– | 58 | 26 | 1.37 (0.68–2.74) | 0.376 | 1.42 (0.69–2.94) | 0.340 | 1.41 (0.66–3.03) | 0.376 | 3.46 (1.14–10.46) | 0.030 |
| ≤0.55 | 58 | 54 | 2.84 (1.50–5.37) | 0.001 | 2.96 (1.48–5.93) | 0.002 | 2.77 (1.33–5.76) | 0.006 | 9.03 (2.31–35.23) | 0.002 |
| Pfor trend | 1.6 × 10−4 | 4.8 × 10−4 | 0.001 | 0.001 | ||||||
†Adjusted for age, gender, smoking status, drinking status, and family history of cancer.
‡Adjusted for age, gender, smoking status, drinking status, family history of cancer, and cirrhosis.
§Adjusted for age, gender, smoking status, drinking status, family history of cancer, cirrhosis, and alpha fetoprotein.
Association between circulating mtDNA content and HCC risk stratified by patients’ characteristics
We further evaluated the association between circulating mtDNA content and HCC risk stratified by patients’ characteristics using multivariate analysis adjusted for age, gender, smoking status, drinking status, family history of cancer, and cirrhosis, where appropriate. We found that the significantly increased HCC risk conferred by decreased mtDNA content was evident in younger patients with an adjusted OR of 3.82 (1.69–8.60, P = 0.001) but not in older patients (P = 0.682) (Table 4). Moreover, a borderline statistically significant interaction effect was noted between age and mtDNA content on the risk of HBV-HCC (Pfor interaction = 0.062). An increased HCC risk associated with lower mtDNA content was also observed in both females (P = 0.022) and males (P = 0.044), never smokers (P = 0.011), never drinkers (P = 0.013), patients with a family history of cancer (P = 0.014), and patients with cirrhosis (P = 0.043). However, none of these variables exhibited a significant interaction with mtDNA content, except gender for which a borderline significant interaction was noted (Pfor interaction = 0.087) (Table 4).
Table 4. The associations of circulating mtDNA content with HCC risk stratified by patients’ characteristics.
| Variables | mtDNA | HBV controls | HCC cases | OR (95% CI)† | P value | Pfor interaction |
|---|---|---|---|---|---|---|
| Age | ||||||
| ≤ 54.3 | Higher | 76 | 16 | 1.00 | ||
| Lower | 40 | 31 | 3.82 (1.69–8.60) | 0.001 | ||
| > 54.3 | Higher | 40 | 20 | 1.00 | ||
| Lower | 76 | 49 | 1.16 (0.57–2.36) | 0.682 | 0.062 | |
| Gender | ||||||
| Female | Higher | 22 | 4 | 1.00 | ||
| Lower | 4 | 9 | 27.99 (1.61–486.69) | 0.022 | ||
| Male | Higher | 94 | 32 | 1.00 | ||
| Lower | 112 | 71 | 1.78 (1.02–3.10) | 0.044 | 0.087 | |
| Smoking status | ||||||
| Never | Higher | 70 | 19 | 1.00 | ||
| Lower | 65 | 44 | 2.60 (1.24–5.43) | 0.011 | ||
| Ever | Higher | 46 | 17 | 1.00 | ||
| Lower | 51 | 36 | 1.82 (0.83–4.02) | 0.136 | 0.414 | |
| Drinking status | ||||||
| Never | Higher | 65 | 17 | 1.00 | ||
| Lower | 62 | 42 | 2.68 (1.24–5.80) | 0.013 | ||
| Ever | Higher | 51 | 19 | 1.00 | ||
| Lower | 54 | 38 | 1.87 (0.88–3.96) | 0.101 | 0.575 | |
| Family history of cancer | ||||||
| No | Higher | 75 | 26 | 1.00 | ||
| Lower | 85 | 56 | 1.88 (0.99–3.57) | 0.053 | ||
| Yes | Higher | 41 | 10 | 1.00 | ||
| Lower | 31 | 24 | 3.97 (1.32–11.93) | 0.014 | 0.308 | |
| Cirrhosis | ||||||
| No | Higher | 73 | 6 | 1.00 | ||
| Lower | 67 | 20 | 2.78 (0.93–8.36) | 0.069 | ||
| Yes | Higher | 43 | 30 | 1.00 | ||
| Lower | 49 | 60 | 1.91 (1.02–3.59) | 0.043 | 0.184 | |
†ORs were adjusted for age, gender, smoking status, drinking status, family history of cancer, and cirrhosis, where appropriate.
Diagnostic value of mtDNA in combination with common demographic and clinical variables
We further explored the diagnostic value of mtDNA in both the overall dataset and the subsets of patients with available AFP or liver enzyme data. First, we found that in the overall dataset, when mtDNA content was added to multivariate analysis adjusting for age, gender, smoking status, drinking status, family history of cancer, and cirrhosis, the area under the curve (AUC) of receiver operating characteristic (ROC) curve significantly increased from 0.7133 (0.6582–0.7683) to 0.7511 (0.6981–0.8041) (P = 0.046) (Supp. Fig. 1A), indicating that mtDNA provided additional diagnostic value to the common demographic variables. Second, we conducted analyses in the subset of patients with available clinical values to compare the diagnostic performance of mtDNA with AFP or liver enzymes. There was no statistically significant difference in the AUCs between the model including mtDNA content and that including AFP (P = 0.527) or each of the liver enzyme (P = 0.149 for ALP, 0.216 for AST, 0.135 for ALT, and 0.545 for GGT) (Supp. Fig. 1B). Third, to investigate whether mtDNA content provided additional diagnostic value over AFP, we further compared the AUCs derived from the multivariate model including demographic variables plus AFP, and that including demographic variables plus both AFP and mtDNA. The AUC was 0.8020 (0.7400–0.8641) in the multivariate model including demographic variables plus AFP, and the AUC significantly increased to 0.8498 (0.7964–0.9032) after adding mtDNA to the model (P = 0.032, Supp. Fig. 2A). Similar results were found in the models including each of the liver enzymes (Supp. Fig. 2B–E). These new data demonstrated that mtDNA provided additional diagnostic value when jointly used with AFP or common liver enzymes.
Pilot longitudinal analysis
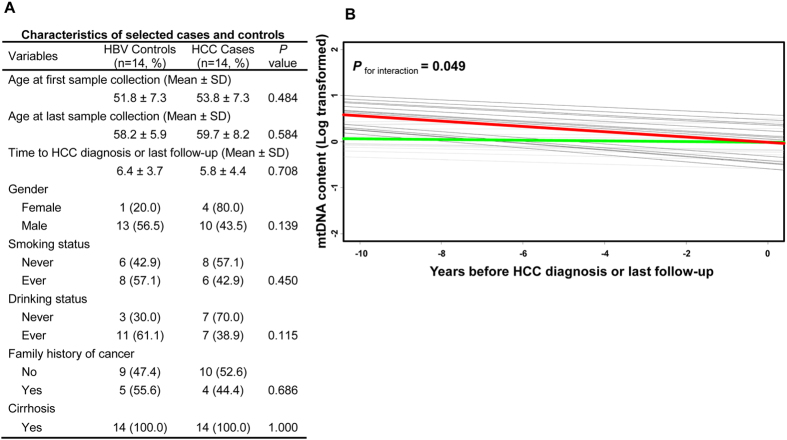
In order to illuminate the dynamic change of circulating mtDNA over time, we conducted a pilot longitudinal analysis in 14 pairs of cases and controls identified from the patients included in this analysis that had at least three samples available before HCC diagnosis (for cases) or last follow-up (for controls). Cases in this pilot analysis were those cancer-free HBV patients who developed HCC after at least one year of follow up. Controls were those HBV patients who remained cancer-free at last follow-up. The cases and controls were adequately matched on age at first sample collection (P = 0.484), age at last sample collection (P = 0.584), follow-up time (P = 0.708), gender (P = 0.139), smoking status (P = 0.450), drinking status (P = 0.115), and family history of cancer (P = 0.686). All cases and controls had cirrhosis at first sample collection (Fig. 1A). We measured mtDNA content in all available serum samples after study entry until HCC diagnosis (for cases) or last follow-up (for controls). Using a mixed effects model, we observed a trend of dramatically decreased mtDNA content in cases and a slightly decreased mtDNA content in controls during follow-up, with a significant interaction of case-control status with time (Pfor interaction = 0.049) (Fig. 1B).
Figure 1. Pilot longitudinal analysis of circulating mtDNA content and the risk of HBV-related HCC.
(A) Characteristics of 14 HBV-related HCC cases and 14 frequency-matched cancer-free HBV controls for longitudinal analysis. (B) Mixed effects model was used to analyze circulating mtDNA content and HCC risk through an interaction of case-control status with time in 14 case-control pairs. Grey lines, individual HBV patient who remained cancer-free (controls); black lines, individual HBV patient who developed HCC (cases); green lines, overall trend of mtDNA change in controls; red lines, overall trend of mtDNA change in cases.
Discussion
In the present study, we evaluated the association between circulating mtDNA from cell-free serum samples and HCC risk in a clinic-based cohort of HBV-infected patients. We found that a lower level of serum mtDNA content was significantly associated with an increased risk of HBV-related HCC in a dose-dependent manner, suggesting circulating mtDNA may be a novel non-invasive marker of HCC risk.
mtDNA exhibits about 10- to 200-fold higher rate of mutagenesis than that of nuclear DNA under an oxidative stress environment, due to the lack of protective histone proteins and the limited capacity of mitochondrial DNA repair system7,21. A growing body of evidence has described the alterations in the copy number of mtDNA in a cancer-specific manner9,10,11,12. The increase of mtDNA copy number, partially resulting from enhanced oxidative stress and feedback response that compensates for mitochondria with impaired respiratory function or elevated mtDNA mutations, has been found in a variety of malignancies, such as colorectal, esophageal, and lung cancers22,23,24,25. In comparison, studies in malignancies such as breast cancer, gastric cancer, renal cell carcinoma, Ewing’s sarcoma, and HCC reported a decreased mtDNA copy number26,27,28,29,30. It was proposed that point mutations adjacent to the replication region in the D-loop, the non-coding region regulating both replication and transcription of mitochondrial genome, may play an important role in the reduction of mtDNA copy number in HCC, breast cancer and Ewing’s sarcoma9,26,30. Genetic and genomic alterations in key players such as TP53 and mtDNA polymerases have also been linked to the decreased mtDNA copy number in colorectal and breast cancers31,32. In HCC, increased expression of nuclear DNA-encoded transacting factor, mitochondrial single strand DNA binding protein, which is important to mitochondrial biogenesis and mtDNA maintenance, was also associated with decreased copy number of mtDNA10. In the current study, we found that a lower level of mtDNA content in a serum sample was significantly associated with an increased HCC risk, which was consist with the findings in other HBV-HCC studies using tissues or peripheral blood leukocyte10,29,33. Injury to the respiratory machinery was considered a critical event in tumorigenesis resulting from mtDNA dysfunction34. The decrease in mtDNA content could decrease oxidative phosphorylation capacity and enhance production of ROS in aerobic metabolism, which is related to DNA damage, fast cellular growth, apoptosis resistance, and malignant cellular transformation33. Functional characterizations are needed to further elucidate the mechanisms underlying the association.
Although the origins of circulating DNAs in cell-free samples such as serum or plasma are not fully clear, it is commonly recognized that the majority of these nucleic acids are derived from necrotic and apoptotic cells from circulation system or target tissues35,36. This is especially relevant for HBV patients, in whom chronic inflammation further contributes to the generation and release of circulating nucleic acids through promoting repetitive liver damage37,38,39. Circulating DNAs have the unique advantage to serve as “liquid biopsy”, which may be used as cost-effective and non-invasive surrogates for tissue biopsies in cancer risk prediction and early detection40. Detection of aberrant changes of circulating mtDNA is becoming an increasingly important tool to assist early cancer diagnosis with some unique advantages over nuclear DNA15,41,42. For instance, the much shorter and more simply organized mitochondrial genome makes screening much easier and more cost-effective than for the nuclear genome. In addition, the higher number of mtDNA copy makes it easier to detect mtDNA content than nuclear DNAs in body fluid samples such as plasma and serum, in which the total DNA concentration is at very low abundance. Previous studies using peripheral blood leukocyte or tissue samples have shown an significant association between a lower mtDNA copy number variations and an elevated HCC risk in HBV patients29,33. Our study confirmed these findings and takes a step further to demonstrate that circulating mtDNA content in cell-free samples may be used as a novel cost-effective and non-invasive marker of HCC risk in HBV patients.
The concentration and components of circulating cell-free DNA vary in a wide range that is affected by physiological and pathological status of individuals14. Therefore, a single baseline circulating mtDNA measurement may not be reliable enough to yield robust findings. To help address this concern, we conducted a pilot longitudinal case-control analysis by measuring mtDNA content in all of the available serum samples collected before HCC diagnosis or last follow-up for each patient. In this analysis, we observed a slightly decreasing trend of mtDNA content along with increased age in HBV patients who did not develop HCC, which was not surprising because a negative correlation between mtDNA content and age has been documented43. Interestingly, a significant trend of reduction in mtDNA content was noticed in HBV patients who developed into HCC during follow up, consistent with the results of the main effects analysis of our study. Moreover, a significant interaction of case-control status with time was identified. These findings of the pilot longitudinal analysis established a temporal link and further substantiated the association between reduced circulating mtDNA content and increased HCC risk. However, due to the unplanned nature and small sample size, the results of the pilot longitudinal analysis need to be interpreted with caution and validated in future larger and well-designed longitudinal cohorts.
A major strength of our study is the unique and highly homogenous HBV patient population. Our study population is restricted to Korean Americans with HBV-infected patients only, which eliminates the potential confounding effects conferred by ethnicity and disease etiology. The strict matching criteria between cases and controls further minimized the confounding from other major risk factors. The non-invasive nature of measuring circulating mtDNA derived makes it possible for repetitive patient monitoring during follow-up and treatment. Furthermore, our findings were greatly strengthened by the pilot longitudinal analysis based on our unique serial collection of serum samples. There are also some limitations in our study. First, the restriction of the analysis to Korean HBV patients, although increasing study homogeneity, limited its generalizability. Second, we were not able to compare mtDNA content in serum with that in liver tissues (either HCC or normal tissue) due to unavailability of tissue samples in our study. Third, because the clinical data in our study were obtained through retrospective chart review, we were not able to conduct the multivariate analyses adjusting for all of the clinical variables in the same model due to the missing values of these variables. We also could not evaluate the correlations between mtDNA content and other clinicopathological variables such as tumor stage or vascular invasion, because these data were not available in our medical charts. In addition, although we detected a higher diagnostic sensitivity of Magnetic Resonance Imaging (MRI) compared to mtDNA alone (75.7% from multiple MRI examinations vs. 69.0% from single mtDNA measurement), the lack of MRI data in many of cancer-free HBV patients in our study made it impossible for us to compare the specificities of mtDNA with that of MRI. Last, owing to the retrospective case-control design, our study is also limited by the reverse-causation issue that is inherent in most retrospective studies, and cannot differentiate the causal relationship between circulating mtDNA content and HCC development. Further large-scale prospective studies are warranted to confirm our findings.
In conclusion, our study reports the first epidemiological evidence that lower circulating mtDNA content in cell-free serum samples is significantly associated with an increased risk of HCC in HBV patients. If validated, circulating mtDNA content may serve as a promising non-invasive biomarker for HCC diagnosis.
Methods
Study population
The subjects of this study were identified from an existing and ongoing clinic-based patient cohort. Patients were consecutively enrolled when they visited the Liver Disease Prevention Center at the Division of Gastroenterology and Hepatology in Thomas Jefferson University Hospital for the treatment of liver diseases such as HBV infection, HCV infection, cirrhosis, and HCC. Patient enrollment started in 1988 and is still ongoing. More details regarding this cohort were as previously described3. Since more than 90% of the patients in this cohort were of Korean ancestry and had HBV infection, to minimize the potential confounding effects from ethnicity and disease etiology, we restricted the study subjects in the current analysis to Korean HBV patients only. This study was approved by the Institutional Review Board of Thomas Jefferson University and the methods were carried out in accordance with the approved guidelines. A written informed consent was obtained from each patient.
Epidemiologic and clinical data collection
Demographic and clinical data were obtained for each patient through medical chart review and/or consulting with the treating physicians. Demographic variables collected in this study included age, gender, ethnicity, smoking status, drinking status, family history of cancer, and cirrhosis status. Individuals who had never smoked or had smoked less than 100 cigarettes in his or her lifetime were defined as a never smoker; otherwise as an ever smoker. Never drinker was defined as those who never consumed alcohol or consumed less than one drink per month; otherwise as an ever drinker. Liver cirrhosis and HCC were diagnosed by the combined use of clinical tests such as platelet counts and Child–Pugh scores and imaging studies especially magnetic resonance imaging, complemented by serum levels of AFP. Blood samples were drawn from patients when taking the clinical laboratory testing, and the extracted serum samples were stored at −80 °C for research purposes. The blood samples from most HCC patients were collected at the time of HCC diagnosis. For a few patients who had no blood samples collected at HCC diagnosis, we used the samples with collection dates close to the time of HCC diagnosis.
Measurement of serum mtDNA content
First, circulating DNA was isolated from 200 μL serum sample using QIAamp DNA Blood Mini kit (Qiagen, Carlsbad, CA) according to the manufacturer’s protocol. The relative mtDNA content was measured by quantitative real-time polymerase chain reaction (qRT-PCR) using a modified protocol described by previous studies28,33, in which the ratio of the copy number for mitochondrial ND1 gene to the copy of a human single copy gene 36B4 was used to determine the relative mtDNA content. Briefly, the 6 μL of qRT-PCR reaction for both ND1 and 36B4 genes consists of 1 X SYBR Green Master Mix (Applied Biosystems, CA, USA), 200 nmol/L each primer, and 2 μL of purified serum DNA sample. The primer sequences were as follows: Forward ND1 primer: 5′-CCCTAAAACCCGCCACATCT-3′; Reverse ND1 primer: 5′-GAGCGATGGTGAGAGCTAAGGT-3′; Forward 36B4 primer: 5′-CAGCAAGTGGGAAGGTGTAATCC-3′; Reverse 36B4 primer: 5′-CCCATTCTATCATCAACGGGTACAA-3′. The thermal cycling conditions for both genes were pre-denatured at 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 seconds and 58 °C for 1 minute with data collection. All samples were assayed in duplicate on a 384-well plate using ViiA 7 qRT-PCR system (Applied Biosystems, CA). The same negative control and calibrator DNA samples were incorporated into each plate for quality control and calibration of PCR efficiency. A reference DNA sample was used to construct a standard curve for mtDNA measurement in each palate. For each standard curve, the reference DNA sample was serially diluted by 1:2 to produce a ten-point standard curve with the initial concentration at 20 ng/μL. The R2 for each standard curve was ≥0.98, with acceptable standard deviation set at 0.25 for the cycle threshold (Ct) values. If the Ct value of a tested sample was out of the range of standard curve, the sample was repeated.
Statistical analyses
Statistical analyses were performed using the SAS software version 9.2 (SAS Institute, Cary, NC) or STATA 12.0 package (STATA Corp, College Station, TX). The differences in the distributions of patients’ characteristics between cases and controls were compared using Chi-square test for categorical variables, and student’s t test for continuous variables. The differences of serum mtDNA content between cases and controls or between dichotomized patients’ characteristics were compared by Wilcoxon rank-sum test. Serum mtDNA was analyzed as categorical variable by cutoffs at median or quartile values in the controls. The association between mtDNA and HCC risk was estimated by unconditional logistic regression model using univariate and multivariate analyses to determine the odds ratios (ORs) and 95% confidence intervals (95% CI). In subset analyses, AFP or serum liver enzymes were added into the multivariable analysis. Discrimination accuracy for HCC diagnosis was evaluated by AUC of ROC curve. The test for interaction between patients’ characteristics and mtDNA on HCC risk was performed by including a cross-product term into the logistic regression model. In the pilot longitudinal analysis to analyze the temporal relationship between mtDNA content and HCC risk, mtDNA content in 14 case-control pairs (matched on age at first sample collection, age at last sample collection, follow-up time, gender, smoking status, drinking status, family history of cancer, and cirrhosis ) were plotted after log transformation against follow-up time. A mixed effects model was used to analyze mtDNA content through an interaction of the case-control status with time. All statistical tests were two sided, and P < 0.05 was considered as the threshold of statistical significance.
Additional Information
How to cite this article: Li, L. et al. Cell-free circulating mitochondrial DNA content and risk of hepatocellular carcinoma in patients with chronic HBV infection. Sci. Rep. 6, 23992; doi: 10.1038/srep23992 (2016).
Supplementary Material
Acknowledgments
The work was supported by National Cancer Institute grants CA153099 and CA152703, American Cancer Society grants 123741-RSG-13-003-01-CCE and IRG-08-060-01, and a Tobacco Grant from Pennsylvania Department of Health.
Footnotes
Author Contributions H.S.Y., L.L., H.W.H. and S.G.W. conceived and designed the study. S.G.W. and Z.Y. conducted the experiments. L.L., H.W.H., S.G.W., R.S.H. and X.S.Y. collected clinical data. L.L., C.W., Y.Z.L. and B.S.L. analyzed the data. H.W.H., R.E.M. and J.L.X. contributed reagents/materials/analysis tools. H.S.Y., L.L., H.W.H., S.G.W., A.E. and R.E.M. wrote the manuscript. All authors reviewed the manuscript.
References
- Jemal A. et al. Global cancer statistics. CA Cancer J Clin. 61, 69–90 (2011). [DOI] [PubMed] [Google Scholar]
- Kim W. R. Epidemiology of hepatitis B in the United States. Hepatology. 49, S28–34 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X. et al. Relative telomere length: a novel non-invasive biomarker for the risk of non-cirrhotic hepatocellular carcinoma in patients with chronic hepatitis B infection. Eur J Cancer. 48, 1014–1022 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppenol W. H., Bounds P. L. & Dang C. V. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer. 11, 325–337 (2011). [DOI] [PubMed] [Google Scholar]
- Chen X. J. & Butow R. A. The organization and inheritance of the mitochondrial genome. Nat Rev Genet. 6, 815–825 (2005). [DOI] [PubMed] [Google Scholar]
- Wallace D. C. Mitochondrial DNA mutations in disease and aging. Environ Mol Mutagen. 51, 440–450 (2010). [DOI] [PubMed] [Google Scholar]
- Larsen N. B., Rasmussen M. & Rasmussen L. J. Nuclear and mitochondrial DNA repair: similar pathways? Mitochondrion. 5, 89–108 (2005). [DOI] [PubMed] [Google Scholar]
- Taylor R. W. & Turnbull D. M. Mitochondrial DNA mutations in human disease. Nat Rev Genet. 6, 389–402 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. C. et al. Somatic mutations in the D-loop and decrease in the copy number of mitochondrial DNA in human hepatocellular carcinoma. Mutat Res. 547, 71–78 (2004). [DOI] [PubMed] [Google Scholar]
- Yin P. H. et al. Alteration of the copy number and deletion of mitochondrial DNA in human hepatocellular carcinoma. Br J Cancer. 90, 2390–2396 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada S. et al. Correlation between copy number of mitochondrial DNA and clinico-pathologic parameters of hepatocellular carcinoma. Eur J Surg Oncol. 32, 303–307 (2006). [DOI] [PubMed] [Google Scholar]
- Vivekanandan P., Daniel H., Yeh M. M. & Torbenson M. Mitochondrial mutations in hepatocellular carcinomas and fibrolamellar carcinomas. Mod Pathol. 23, 790–798 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischhacker M. & Schmidt B. Circulating nucleic acids (CNAs) and cancer–a survey. Biochim Biophys Acta. 1775, 181–232 (2007). [DOI] [PubMed] [Google Scholar]
- Schwarzenbach H., Hoon D. S. & Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 11, 426–437 (2011). [DOI] [PubMed] [Google Scholar]
- Gonzalez-Masia J. A., Garcia-Olmo D. & Garcia-Olmo D. C. Circulating nucleic acids in plasma and serum (CNAPS): applications in oncology. Onco Targets Ther. 6, 819–832 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M., Wan Y. F. & Zou Q. H. Cell-free circulating mitochondrial DNA in the serum: a potential non-invasive biomarker for Ewing’s sarcoma. Arch Med Res. 43, 389–394 (2012). [DOI] [PubMed] [Google Scholar]
- Zhang Q. et al. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature. 464, 104–107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossarizza A. et al. Increased plasma levels of extracellular mitochondrial DNA during HIV infection: a new role for mitochondrial damage-associated molecular patterns during inflammation. Mitochondrion. 11, 750–755 (2011). [DOI] [PubMed] [Google Scholar]
- Sursal T. et al. Plasma bacterial and mitochondrial DNA distinguish bacterial sepsis from sterile systemic inflammatory response syndrome and quantify inflammatory tissue injury in nonhuman primates. Shock. 39, 55–62 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahira K. et al. Circulating mitochondrial DNA in patients in the ICU as a marker of mortality: derivation and validation. PLoS Med. 10, e1001577 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi N. O., Bianchi M. S. & Richard S. M. Mitochondrial genome instability in human cancers. Mutat Res. 488, 9–23 (2001). [DOI] [PubMed] [Google Scholar]
- Chen T. et al. The mitochondrial DNA 4,977-bp deletion and its implication in copy number alteration in colorectal cancer. BMC Med Genet. 12, 8 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C. S. et al. The role of mitochondrial DNA alterations in esophageal squamous cell carcinomas. J Thorac Cardiovasc Surg. 139, 189–197 e184 (2010). [DOI] [PubMed] [Google Scholar]
- Bonner M. R. et al. Mitochondrial DNA content and lung cancer risk in Xuan Wei, China. Lung Cancer. 63, 331–334 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. C. & Wei Y. H. Mitochondrial DNA instability and metabolic shift in human cancers. Int J Mol Sci. 10, 674–701 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M. et al. Reduced mitochondrial DNA copy number is correlated with tumor progression and prognosis in Chinese breast cancer patients. IUBMB Life. 59, 450–457 (2007). [DOI] [PubMed] [Google Scholar]
- Liao L. M. et al. Mitochondrial DNA copy number and risk of gastric cancer: a report from the Shanghai Women’s Health Study. Cancer Epidemiol Biomarkers Prev. 20, 1944–1949 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing J. et al. Mitochondrial DNA content: its genetic heritability and association with renal cell carcinoma. J Natl Cancer Inst. 100, 1104–1112 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu C. C., Lee H. C. & Wei Y. H. Mitochondrial DNA alterations and mitochondrial dysfunction in the progression of hepatocellular carcinoma. World J Gastroenterol. 19, 8880–8886 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M., Wan Y. & Zou Q. Decreased copy number of mitochondrial DNA in Ewing’s sarcoma. Clin Chim Acta. 411, 679–683 (2010). [DOI] [PubMed] [Google Scholar]
- Chang S. C. et al. Mitochondrial D-loop mutation is a common event in colorectal cancers with p53 mutations. Int J Colorectal Dis. 24, 623–628 (2009). [DOI] [PubMed] [Google Scholar]
- Singh K. K., Ayyasamy V., Owens K. M., Koul M. S. & Vujcic M. Mutations in mitochondrial DNA polymerase-gamma promote breast tumorigenesis. J Hum Genet. 54, 516–524 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S. et al. Association of mitochondrial DNA content in peripheral blood leukocyte with hepatitis B virus-related hepatocellular carcinoma in a Chinese Han population. Cancer Sci. 102, 1553–1558 (2011). [DOI] [PubMed] [Google Scholar]
- Warburg O. On the origin of cancer cells. Science. 123, 309–314 (1956). [DOI] [PubMed] [Google Scholar]
- Jahr S. et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 61, 1659–1665 (2001). [PubMed] [Google Scholar]
- Kanduc D. et al. Cell death: apoptosis versus necrosis (review). Int J Oncol. 21, 165–170 (2002). [PubMed] [Google Scholar]
- Brenner C., Galluzzi L., Kepp O. & Kroemer G. Decoding cell death signals in liver inflammation. J Hepatol. 59, 583–594 (2013). [DOI] [PubMed] [Google Scholar]
- Dunn C. et al. Cytokines induced during chronic hepatitis B virus infection promote a pathway for NK cell-mediated liver damage. J Exp Med. 204, 667–680 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida M. et al. Relation between serum levels of cell-free DNA and inflammation status in hepatitis C virus-related hepatocellular carcinoma. Oncology reports. 20, 761–765 (2008). [PubMed] [Google Scholar]
- Diaz L. A. Jr. & Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol. 32, 579–586 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler C. et al. Levels of plasma circulating cell free nuclear and mitochondrial DNA as potential biomarkers for breast tumors. Mol Cancer. 8, 105 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachariah R. R. et al. Levels of circulating cell-free nuclear and mitochondrial DNA in benign and malignant ovarian tumors. Obstet Gynecol. 112, 843–850 (2008). [DOI] [PubMed] [Google Scholar]
- Dimmock D., Tang L. Y., Schmitt E. S. & Wong L. J. Quantitative evaluation of the mitochondrial DNA depletion syndrome. Clin Chem. 56, 1119–1127 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.