Abstract
Amide-linked conjugates of indole-3-acetic acid (IAA) have been identified in most plant species. They function in storage, inactivation or inhibition of the growth regulator auxin. We investigated how the major known endogenous amide-linked IAA conjugates with auxin-like activity act in auxin signaling and what role ILR1-like proteins play in this process in Arabidopsis. We used a genetically encoded auxin sensor to show that IAA-Leu, IAA-Ala and IAA-Phe act through the TIR1-dependent signaling pathway. Furthermore, by using the sensor as a free IAA reporter, we followed conjugate hydrolysis mediated by ILR1, ILL2 and IAR3 in plant cells and correlated the activity of the hydrolases with a modulation of auxin response. The conjugate preferences that we observed are in agreement with available in vitro data for ILR1. Moreover, we identified IAA-Leu as an additional substrate for IAR3 and showed that ILL2 has a more moderate kinetic performance than observed in vitro. Finally, we proved that IAR3, ILL2 and ILR1 reside in the endoplasmic reticulum, indicating that in this compartment the hydrolases regulate the rates of amido-IAA hydrolysis which results in activation of auxin signaling.
The cellular pool of the plant hormone auxin is represented by free indole-3-acetic acid (IAA), IAA precursors and IAA conjugated forms. Conjugated forms of IAA account for a significant amount of total IAA within most plant tissues and they include amide-linked conjugates such as amino acids and peptides and ester-linked conjugates such as myo-inositol and myo-inositol-sugars1,2. Ester-linked conjugates are found in endosperm tissues of monocots and dicots, whereas amide-linked IAA-l-amino acids conjugates (IAA-aa) predominate in mature dicot seeds and light grown vegetative tissues of most plants, both monocots and dicots3. Various IAA-aa conjugates have been identified in different plant species and include IAA-leucine (IAA-Leu), IAA-alanine (IAA-Ala), IAA-aspartate (IAA-Asp), IAA-glutamate (IAA-Glu), and IAA-tryptophan (IAA-Trp)4,5. The occurrence of other conjugates such as IAA-valine (IAA-Val) and IAA-phenylalanine (IAA-Phe) has been postulated based on the identification of oxidative metabolites in Arabidopsis6. The different IAA-aa conjugates have been implicated in a variety of biological processes. For example, IAA-Glu and IAA-Asp, which are readily synthesized upon incubation of plants with high dosage of IAA4, are considered precursors for auxin degradation. IAA-Trp functions as an endogenous inhibitor which interferes with several physiological responses to auxin7. IAA-Trp requires the auxin receptor TIR1 for full activity though reaction pathways are not yet elucidated7. Other conjugates such as IAA-Ala, -Leu, -Phe, -Val are biologically active and mimic the effect of free IAA in inducing plant developmental responses like root and hypocotyl elongation inhibition8.
Early studies showed that the biological activity of IAA-aa conjugates was related to the amount of free IAA detected in the stem tissue resulting from their hydrolysis9. This demonstrated, for the first time, a direct link between the activity of the conjugates and their hydrolysis.
Mutant screens based on reduced sensitivity to biologically active IAA-aa in root growth inhibition assays led to the identification of a specific group of amidohydrolases8,10. IAA-Leu-resistant1 (ilr1) was the first mutant to be identified; the gene encodes an amidohydrolase with high affinity for IAA-Leu and IAA-Phe8. In Arabidopsis, the ILR1-like (ILL) family consists of 7 members: ILR1, ILL1, ILL2, ILL3, IAR3 (ILL4), ILL5 and ILL6. The best characterized are ILR1, ILL1, ILL2, and IAR3 for which IAA-amino acids cleavage activity and substrate specificity have been shown by in vitro enzymatic assays10,11. IAR3 and ILL2 show highest catalytic activity with IAA-Ala as a substrate, while ILR1 is most efficient in hydrolyzing IAA-Leu and IAA-Phe. ILL3 and ILL6 show no activity on IAA-aa in vitro10 or very little activity11. ILL5 is an apparent pseudogene10,11. IAA-Asp and IAA-Glu are not efficiently hydrolyzed by any of the Arabidopsis amidohydrolases10. Single mutants of the ILR-1 like family such as ilr1, ill2 and iar3, display a reduced sensitivity to biologically active IAA-aa conjugates, while plants overexpressing the hydrolases show higher sensitivity to certain IAA-aa conjugates12. The triple mutant ilr1iar3ill2 is less sensitive to IAA-Phe and IAA-Ala and essentially insensitive to IAA-Leu13. These genetic data correlate with hydrolysis rates for each IAA-aa conjugate reported from in vitro enzymatic assays10, suggesting that the hydrolase activity of ILR1-like proteins is directly linked to the biological activity of the IAA-aa conjugates.
This study evaluates how the major endogenous IAA-aa conjugates with auxin-like activity act in auxin signaling and how this activity is modulated by the amidohydrolases. We applied a genetically encoded auxin sensor which is based on the TIR1-mediated auxin-dependent degradation of Aux/IAAs14 and showed that the biological activity of IAA-Ala, IAA-Leu and IAA-Phe is, at least in part, mediated by the TIR1-dependent auxin signaling pathway. Additionally, by using the sensor as a reporter of free IAA, we investigated the hydrolysis activity of members of ILR1-like family in a single plant cell system. Thereby we positively correlated conjugate preference for ILR1 and IAR3 with in vitro data10. Furthermore, we identified IAA-Leu as an additional substrate for IAR3 and found a different conjugate preference and a more moderate kinetic performance for ILL2. Finally, we experimentally showed that IAR3, ILL2 and ILR1 are localized in the endoplasmic reticulum (ER). This defines the ER as the compartment where the amidohydrolases modulate the rate of IAA-aa hydrolysis which results in activation of auxin signaling.
Results and Discussion
Correlating amide-conjugates IAA-Ala, IAA-Leu and IAA-Phe with auxin signaling
IAA-aa conjugates IAA-Ala, IAA-Leu and IAA-Phe mimic the effect of free IAA in inducing plant developmental responses in Arabidopsis13. Genetic and biochemical studies indicate a direct relationship between the biological activity of these conjugates and their hydrolysis rates mediated by amidohydrolases from the ILR1-like family. The underlying molecular basis of IAA-like activity however has not yet been elucidated. To address this question, we investigated how IAA-aa and their hydrolysis act in term of auxin perception and activation of auxin-signaling pathway.
Auxin signaling is initiated through binding of IAA to the TRANSPORT INHIBITOR RESPONSE1/AUXIN SIGNALING F-BOX PROTEIN (TIR1/AFB) and AUXIN/INDOLE ACETIC ACID (Aux/IAA) protein co-receptors and the consequent targeting of the Aux/IAA proteins for degradation15,16,17. Upon Aux/IAA degradation, repression of AUXIN RESPONSIVE FACTORS (ARFs) transcription factors is released and transcription of auxin-regulated genes takes place, thereby initiating auxin signaling. In light of this, we investigated how biologically active IAA-aa conjugates are involved in the TIR1/AFBs-Aux/IAAs-ARFs pathway by employing a genetically encoded ratiometric auxin sensor14. The functional principle of this sensor is based upon the auxin-dependent formation of a TIR1/AFB-Aux/IAA-reporter like complex. The sensor comprises two modules: an auxin responsive module and an auxin-insensitive module. The auxin responsive module consists of firefly luciferase translationally fused with the conserved degron motive of Aux/IAAs. The auxin-insensitive module consists of renilla luciferase and serves as normalization of response. Thus, upon expression of the sensor in plant cells, auxin-dependent degradation of the sensor is monitored as a decrease in firefly (responsive module) relative to renilla (normalization module) luminescence. Notably, this degradation-based sensor is different from widely used DR5- based auxin inducible reporters whose functional principle relies on ARF’s binding sites and activation of auxin signalling18,19.
Previously, it was shown that the sensor enables quantitative monitoring of changes in cellular levels of IAA as well as of synthetic compounds with auxin-like activity14. We first explored whether the biologically active IAA conjugates led to TIR1-dependent sensor degradation. Arabidopsis leaf protoplast cells expressing the sensor were incubated with each conjugate at various concentrations for 1 hour and then the ratiometric determinations of firefly and renilla luminescence (F/R) were performed (Fig. 1a). All conjugates induced sensor degradation starting as low as 1 μM and resulted in ~50% of sensor degradation at 10 μM. An even stronger effect was observed at higher concentrations (Fig. 1a). These results indicate that biologically active conjugates may trigger the sensor degradation either by direct binding to TIR1/AFBs and subsequent ubiquitination and degradation of the sensor or it is free IAA, released upon hydrolysis, acting as the effector molecule.
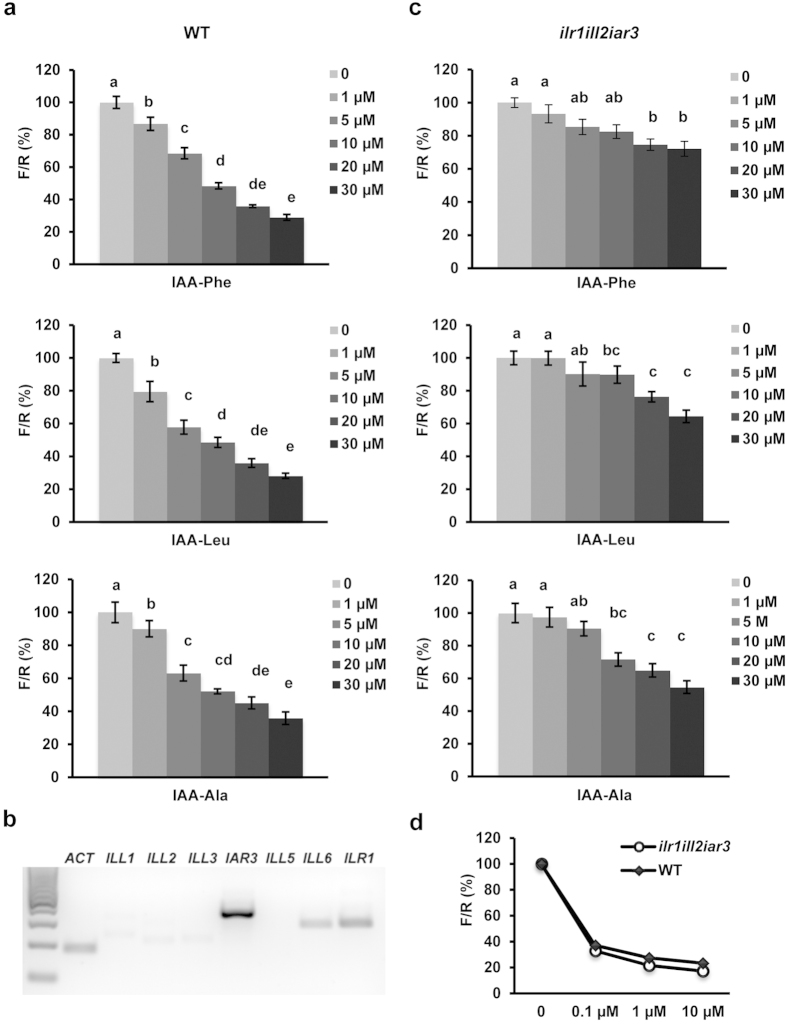
Figure 1. Correlation of amide-conjugates IAA-Ala, IAA-Leu and IAA-Phe with TIR1/AFBs signaling pathway using ratiometric auxin sensor in plant cells.
(a,c,d) Sensitivity of the auxin sensor towards IAA-Ala, IAA-Leu, IAA-Phe and IAA, in plant cells. Protoplasts from Col-0 wild type (a) ilr1ill2iar3 triple mutant (c) or both (d) were transformed with the sensor construct. After 20 h incubation in hormone-free medium, 0, 1, 5, 10, 20, 30 μM each of IAA-Ala, IAA-Leu and IAA-Phe (a,c) or 0, 0.1, 1, 10 μM IAA (d) were added to the samples. After 1 h treatment, the ratiometric determinations of firefly/renilla (F/R) were performed. Results are represented as percentile of the control samples (no auxin treatment). Results are means ± s.e.m. (n = 16). Statistical significances for each conjugates are indicated with lower case letters (the same letters indicate no statistical significant difference between the samples, one-way ANOVA, P < 0.001). (b) Expression analysis of ILL1, ILL2, ILL3, IAR3, ILL5, ILL6 and ILR1 in Arabidopsis Col-0 protoplast-derived cells. ACTIN (ACT) was included as a positive control.
Therefore, to understand whether the sensor degradation observed upon incubation with the IAA conjugates (Fig. 1a) requires the activity of the hydrolases, we analyzed the expression levels of members of the ILR1-like genes in Arabidopsis leaf protoplasts. RT-PCR analysis showed that ILR1, ILL6 and IAR3 were expressed in Arabidopsis cells (Fig. 1b). Expression level of ILL1, ILL2 and ILL3 was at the limit of detection, indicating low abundance of these gene transcripts. ILL5 could not be amplified. The presence of ILR1, ILL6 and IAR3 in protoplasts indicates that the sensor degradation might indeed reflect the accumulation of free IAA in the cells upon conjugate hydrolysis. To validate this suggestion, we investigated mutant plants defective of ILR1, IAR3 and ILL2, the hydrolases which showed the highest in vitro hydrolysis performance towards IAA-amido conjugates13.
The degradation kinetic analysis of the auxin sensor was compared in WT and ilr1ill2iar3 triple mutant cells20. Impaired hydrolysis resulted in a significant reduction of sensitivity towards the three conjugates (Fig. 1c). Treatment with up to 5 μM of either conjugate resulted in statistically insignificant sensor degradation in ilr1iar3ill2 cells. At the highest concentration (30 μM), sensor degradation was strongly decreased: 30% vs. 71% (IAA-Phe), 40% vs. 72% (IAA-Leu), and 50% vs. 65% (IAA-Ala) in triple mutant and WT cells, respectively. To test whether auxin response was in general affected in the triple mutant, treatment with free IAA in WT and ilr1iar3ill2 cells was performed (Fig. 1d). The sensor was degraded in the triple mutant to the same extent as in WT indicating that the TIR1/AFBs pathway is not affected in the triple mutant.
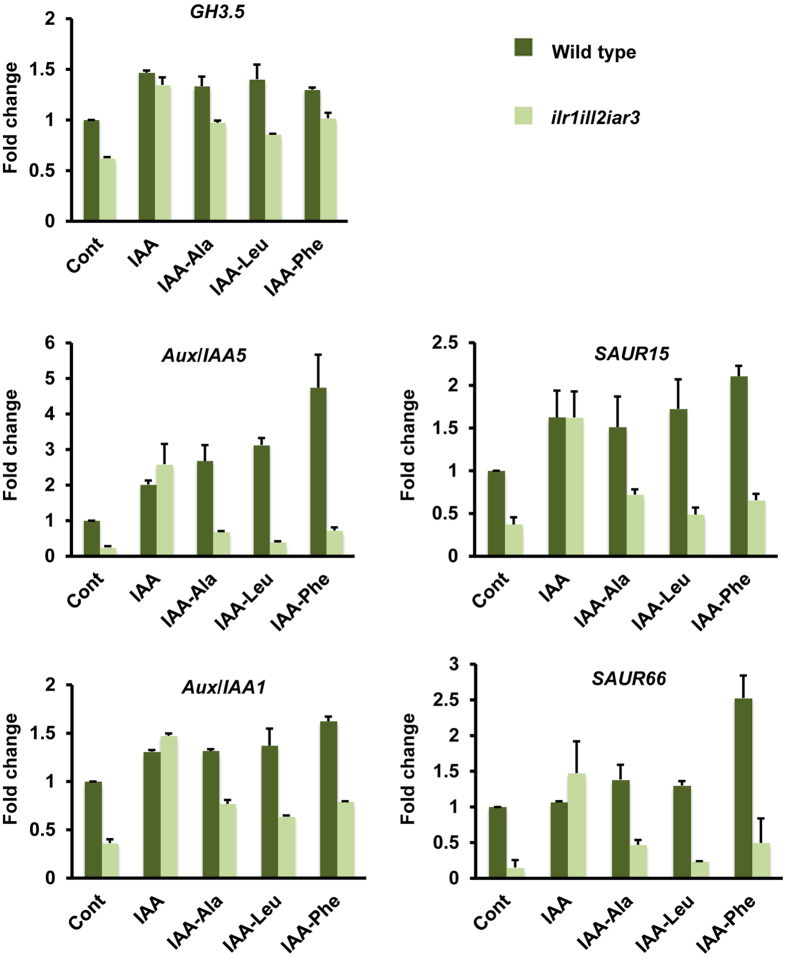
We further analyzed the effect of IAA-aa conjugates and their hydrolysis on transcriptional activation of auxin responsive genes. The best characterized early auxin response genes are represented by three gene families21: Aux/IAAs, SMALL AUXIN UP RNAs (SAURs), and GRETCHEN HAGEN3s (GH3s). We therefore tested the expression of representative members of these gene families, i.e. Aux/IAA1, Aux/IAA5, SAUR15, SAUR66 and GH3.5, upon treatment with IAA-aa conjugates in Arabidopsis WT and ilr1ill2iar3 seedlings. Free IAA was used as a positive control. Quantitative real time PCR (qPCR) analysis (Fig. 2) revealed a lower basal expression level of auxin responsive genes in the triple mutant. This result is in accordance with the developmental phenotypes and a reduced free IAA level measured in ilr1ill2ir3 triple mutant13. Analysis of gene expression upon treatment with free IAA showed similar expression levels of Aux/IAA1, Aux/IAA5, SAUR15, SAUR66 and GH3.5 in both, triple mutant and WT seedlings. On the other hand, analysis of gene expression upon treatment with IAA-Ala, IAA-Leu and IAA-Phe showed that induction of auxin regulated genes was affected in the mutant (Fig. 2).
Figure 2. Expression analysis of early auxin induced genes in Arabidopsis Col-0 wild type and ilr1ill2air3 triple mutant seedlings.
The seedlings were incubated for 1 hour in liquid culture medium (CONT), or supplied with 10 μM each of IAA-Ala, IAA-Leu, IAA-Phe or 100 nM IAA. Expression analysis of Aux/IAA1, Aux/IAA5, GH3.5, SAUR15, SAUR 66 (auxin regulated genes) was performed using quantitative real time PCR. Transcript levels of target genes were normalized to expression of ACTIN (housekeeping gene). Expression is shown relative to wild type (CONT). Results are means ± s.e.m. (n = 2).
Collectively these results indicate that IAA-mediated auxin signaling machinery per se is not impaired in the triple mutant and that activation of auxin signaling cascade by IAA-aa conjugates requires the activity of ILR1-like hydrolases.
Application of the auxin sensor to monitor IAA amide conjugate hydrolysis in plant cells
The ratiometric auxin sensor assays showed the relevance of the hydrolases to modulate intracellular free IAA levels. Furthermore, these assays provide an ex vivo experimental system to study the hydrolase activity, which so far has only been characterized in vitro10,11. Therefore we applied the auxin sensor to study conjugate hydrolysis mediated by ILR1, ILL2 and IAR3 in plant cells. The sensor was co-expressed with ILR1, IAR3, ILL2 or GFP (a protein not involved in auxin metabolism) as a negative control. Comparable expression of the hydrolases was confirmed at transcript and protein levels (Supplementary Fig. S1). To highlight the specific activity of each hydrolase and to minimize the effect of endogenous enzymes, the assays were performed in ilr1ill2iar3 cells. 20 hours after transformation, the cells were incubated with either IAA-Leu, IAA-Phe, or IAA-Ala, for 1 hour. For all tested conjugates, increased sensor degradation was observed upon over expression of the hydrolases (Fig. 3). However, the conjugate preference and the relative activity differed among the three enzymes. IAA-Phe was hydrolyzed by ILR1 and ILL2. IAA-Leu was preferentially hydrolyzed by ILR1 and to lesser extent by ILL2 and IAR3. IAA-Ala, which retained most activity in the triple mutant cells, was hydrolyzed by all three hydrolases but to lesser extent than IAA-Leu and IAA-Phe. These results show that the three hydrolases are involved in activation of auxin signaling by hydrolysis of IAA-Ala, -Leu and -Phe into biologically active IAA. Particularly, ILR1 is primarily involved in mediating plant responses to IAA-Leu and IAA-Phe, IAR3 to IAA-Ala and IAA-Leu and ILL2 to IAA-Phe and IAA-Ala.
Figure 3. Application of the auxin sensor to monitor the modulation of intracellular auxin pool mediated by ILR1, ILL2 and IAR3, in plant cells.
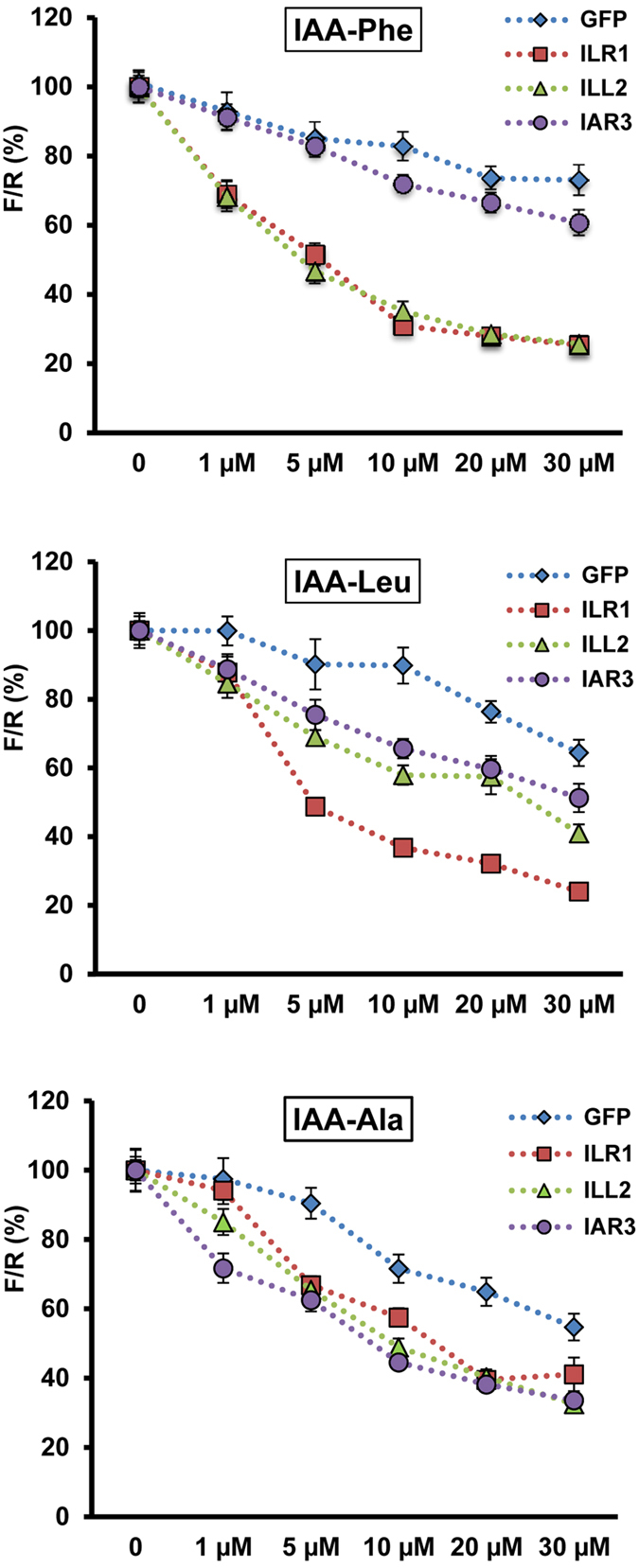
The sensor was co-expressed with either ILR1, or ILL2 or IAR3 in triple mutant ilr1ill2iar3 protoplasts. GFP was also co-expressed as a control. After 20 h incubation in hormone-free medium, 0, 1, 5, 10, 20, 30 μM each of IAA-Ala, IAA-Leu and IAA-Phe were supplemented to the culture medium. After 1 h treatment, the ratiometric determinations (F/R) were performed. Results are represented as percentile of the control samples (no auxin treatment). Results are means ± s.e.m. (n = 16).
These findings are in positive correlation with physiological assays performed with the hydrolase mutants and partly in agreement with the reported hydrolase activity and substrate specificities observed in vitro. For example, ILR1-mediated in vitro hydrolysis activity was reported for IAA-Phe, IAA-Leu and IAA-Ala10. Similarly, in root elongation inhibition assays on single, double and triple mutants, ilr1 contributed to resistance to IAA-Leu, IAA-Phe and to a minor extent to IAA-Ala13. Our data on ILR1 are in agreement with these results. On the other hand, IAR3 showed in vitro hydrolysis activity with IAA-Ala, but no activity with IAA-Leu10. Our assays confirmed IAA-Ala as a substrate of IAR3, but also identified IAA-Leu as an additional substrate. This finding can now explain the contribution of iar3 to IAA-Leu resistance observed in physiological assays when combined with ilr1ill2 mutant13. The least agreement between our and in vitro data was found for ILL2. ILL2 showed highest amidohydrolase activity in vitro, with the broadest range of substrate specificity and highest hydrolysis activity with IAA-Ala10. In our assays, ILL2 showed highest hydrolysis activity with IAA-Phe, with comparable sensor degradation kinetics as ILR1, while IAR3 showed higher hydrolysis activity with IAA-Ala than ILL2. The reasons for the discrepancy between our data and in vitro characterization could be diverse. Multiple factors which are absent in standardized in vitro conditions might influence the activity of an enzyme in vivo. For example, while determining optimal conditions for hydrolysis activity it was shown that different pH and metal ion cofactors influenced the hydrolysis rates10. Therefore metal ion homeostasis in the cell may modulate the activity of the hydrolases differently. Accordingly, the ilr2 mutant, which is resistant to IAA-Phe and IAA Leu, exhibited altered metal transport22.
Interestingly, the conjugate preference and a more moderate hydrolysis activity of ILL2 observed in the plant cell assays are in agreement with genetic data. Albeit ILL2 was the most active hydrolases in vitro10, no ill2 alleles were isolated in genetic screens for conjugate resistant seedlings (in contrast to ilr1 and iar3 alleles)23. Additionally, when ill2 allele was combined in double and triple mutants with ilr1 and iar3, it mainly contributed to resistance to IAA-Phe and to lesser extent to IAA-Ala13. Our sensor-based assays also showed that IAA-Ala was the least hydrolyzed by ILR1, IAR3 or ILL2. This suggests that other proteins might be involved in hydrolysis of IAA-Ala. In protoplasts at least one more member of ILR1-like family is expressed: ILL6 (Fig. 1b). However, ILL6 showed no10 or little in vitro hydrolysis activity, around 10 fold less than IAR3 with IAA-Ala as a substrate11. Higher order hydrolase mutants will help in future to discriminate the contribution of other hydrolases for IAA-Ala sensitivity.
Our experimental approach proved efficient to study the auxin conjugate hydrolase activity in Arabidopsis single cells, thus representing a valuable tool to complement in vitro studies. Moreover, this system could be adapted to the analysis of other genes and physiologically active compounds involved in auxin homeostasis and signaling networks in Arabidopsis and other plant species.
Subcellular localization of ILL2, ILR1 and IAR3
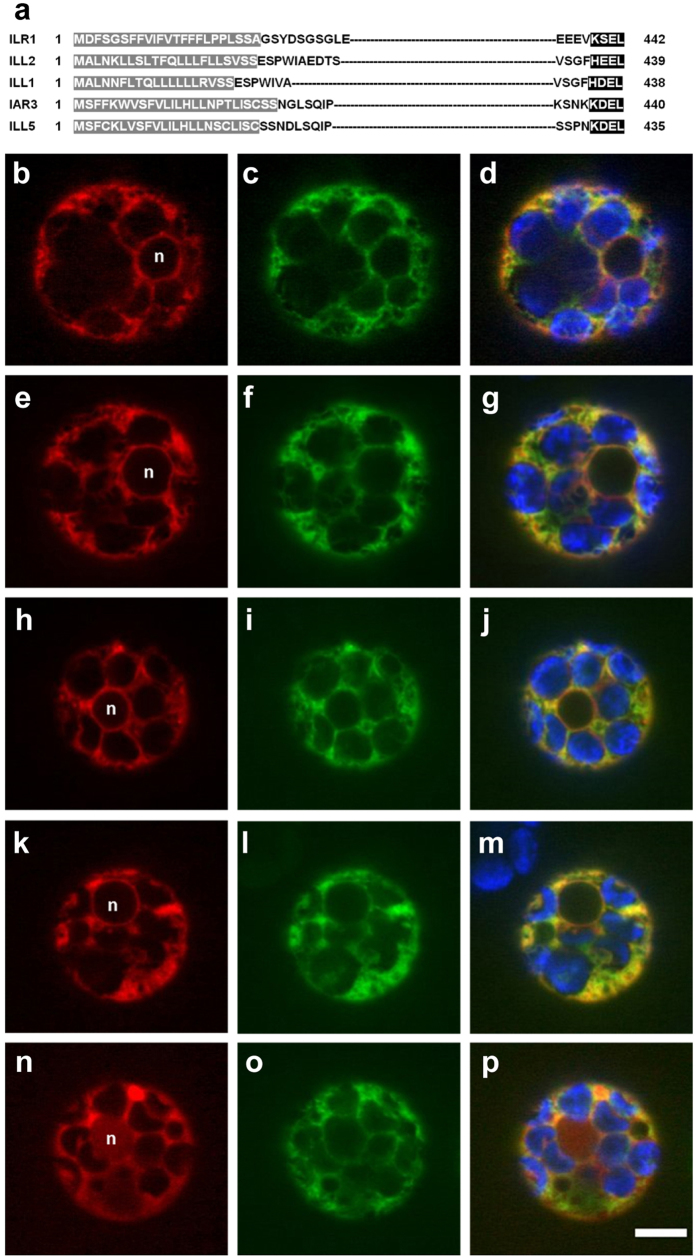
We next analyzed in what cellular compartment the hydrolases function. Except for ILL6 and ILL3, the other five members of the ILR1-like family have both a predicted amino terminal signal sequence and a carboxy terminal peptide that might serve as an ER retrieval tag24. In particular, ILL1, IAR3 and ILL5 have a K/HDEL carboxy-terminal peptide (Fig. 4a), a well-known ER retention signal for soluble proteins25. ILR1 and ILL2 have the variant motives (KSEL and HEEL, respectively), for which there is no evidence of ER retention in plants. Based on these in silico analysis, it has long been assumed that some of these hydrolases are ER-localized. This, in combination with the relatively recent characterization of auxin carriers localized at the ER26,27,28, has led to the suggestion of a possible role of the ER in the conjugation-based metabolism of auxin29,30. However, the subcellular localization of the ILR1-like amidohydrolases has not been validated until now. We therefore decided to experimentally test the in silico predictions. We generated translational fusions of IAR3, ILL2 and ILR1 with the fluorescent protein GFP inserted at the N-terminal end of the proteins after the predicted signal peptide (Fig. 4a). The fusion proteins were transiently co-expressed with an ER-mCherry marker in Arabidopsis protoplasts and visualized using spinning disk confocal microscopy (Fig. 4b–j). The overlap of the fluorescent signals of ILR1-GFP, ILL2 and IAR3-GFP with the ER-mCherry marker confirmed the ER localization of the ILR1-like amidohydrolases (Fig. 4d,g,j). We further studied the in silico predicted ER retention signals and showed that the KSEL motif is essential for the retention of ILR1 in the ER. Upon removal of the tetrapeptide, the mutated ILR1-mCherry protein showed a cytoplasm-like localization and could be detected in the nucleus (Fig. 4n). On the other hand, removal of HEEL from ILL2 (Fig. 4l) did not affect the localization pattern suggesting that other motives/interactions are sufficient to retain ILL2 in the ER.
Figure 4. Subcellular localization of ILR1, ILL2 and IAR3.
(a) N- and C-terminal protein sequences of Arabidopsis ILR1, ILL2, IAR3, ILL1 and ILL5. The predicted signal peptides, according to TargetP40, are highlighted in grey; the potential retention signals are highlighted in black. (b–j) Subcellular localization of ILR1, ILL2 and IAR3. Confocal images of Arabidopsis leaf protoplasts co-expressing the mCherry-HDEL ER marker (b,e,h) and either ILR1 (c) ILL2 (f) or IAR3 (i) fused to eGFP. (k–p) Co-localization analysis of ILL2 depleted of the potential retention signal HEEL (l) with the ER mCherry marker (k) or ILR1 depleted of the potential retention signal KSEL (n) with ILR1 fused to eGFP (o). Merged signal of red and green channels (d,g,j,m,p) indicates co-localization. In the merged images the chloroplasts are visualized in blue and in the red channel images the nucleus position is indicated (n). The scale bar corresponds to 5 μM.
All together our findings indicate that in Arabidopsis hydrolysis of amide conjugates with auxin-like activity indeed occurs in the ER. These results provide first experimental evidence to a current model of subcellular compartmentation of auxin metabolism1, according to which conjugate hydrolysis takes place preferably in the ER, whilst the synthesis of IAA-aa conjugates is most likely happening in the cytosol. Besides Arabidopsis, auxin conjugate hydrolases from other species have ER retention sequences. Hydrolases with a typical retention signal have been characterized in Brassica rapa31. Additionally, phylogenetic analysis of ILR1-like family orthologues across the plant kingdom32 identified two monocot clades that evolved from the dicot orthologues: one possessing putative ER localization signals and one lacking them. The inverse reaction, the synthesis of IAA-aa conjugates, is catalyzed by members of the GH3 family33. So far, GH3 subcellular location has been studied only in moss Physcomitrella patens, where GH3-1 was experimentally shown to be cytoplasmic and GH3-2 was predicted to be in the cytosol34. Therefore, further experimental data are needed to understand the interplay between the ER and the cytosol located proteins involved in synthesis and hydrolysis of amide-linked IAA-l-amino acids conjugates.
Direct impact of IAA-aa hydrolysis on activation of auxin signaling suggests that the ER is not only a site of auxin storage. Gain of function mutants of ER localized auxin carriers PIN5, PIN8 and PIL5 resulted in altered auxin response26,27,28 supporting the idea that the IAA content in the ER indeed plays an important role in auxin signaling. Recently, enzymes involved in auxin biosynthesis and catalysing adjacent steps in YUCCA-dependent biosynthesis were also shown to be localized to the ER and this led to the suggestion that the relative concentration of auxin in different subcellular compartments might represent an additional cellular strategy to regulate auxin action35. Interestingly, IAR3 is also involved in the hydrolysis of jasmonyl-L-isoleucine (JA-Ile), which is the critical molecule to activate the jasmonate pathway11. Thus, the ER might also represent a site of metabolic/signaling crosstalk between auxin and jasmonate pathways.
Methods
Plant material
Arabidopsis thaliana wild type (Col-0) and amidohydrolase single and triple mutants ilr1, iar3 and ilr1ill2iar320 plant cultures were as described in14. ilr1, iar3 and ilr1ill2iar3 seeds were kindly provided by Dr. Bethany Zolman.
Constructs
Total RNA isolated from cauline leaves or inflorescence of Arabidopsis thaliana Col-0 was used as template to amplify the cDNAs of ILL2 (AT5G56660), ILR1 (AT3G02875) and IAR3 (AT1G51760) using the primer pairs ILL2 for/ILL2 rev, ILR1 for/ILR1 rev and IAR3 for/IAR3 rev, respectively. The obtained fragments were cloned in the pENTR/D-TOPO plasmid (Invitrogen, Germany) to generate the plasmids pENTR-ILL2, pENTR-ILR1 and pENTR-IAR3. pENTR-ILR1 was used as template to amplify the clone ILR1-wo-KSEL using the primer pairs ILR1 for/ILR1woKSEL rev. The CDS ILR1woKSEL was further cloned in the pENTR/D-TOPO plasmid. Mistake-free clones were subsequently introduced by Gateway cloning into the pMIR vector containing the L2 min17-Luc auxin sensor and an additional 35S driven expression cassette14 to generate the plasmids: pMIR-L2 min17-Luc-ILL2, pMIR-L2 min17-Luc-ILR1and pMIR-L2 min17-Luc-IAR3. Control auxin sensor plasmid containing the GFP expression cassette was used according to14. Fluorescently tagged ILL2, ILR1 and IAR3 were generated by translational fusion with eGFP (GenBank AFA52654.1), with eGFP inserted after the amino acid position 33, 34 and 30 from the start codon (ATG), for ILL2, ILR1 and IAR3, respectively. GFP was inserted using the Gibson assembly method. To assemble the plasmid pENTR-ILL2eGFP, three PCR fragments were combined: one amplified with the primers ILL2-eGFP for/eGFP-ILL2 rev and eGFP’s cDNA as template; one amplified with the primers eGFP-ILL2 for/pENTR Kan and pENTR-ILL2 as template; and one amplified with the primers pENTR Kan for/ILL2-eGFP rev and pENTR-ILL2 as template. To assemble the plasmid pENTR-ILR1eGFP, three PCR fragments were combined: one amplified with the primers ILR1-eGFP for/eGFP-ILR1 rev and eGFP’s cDNA as template; one amplified with the primers eGFP-ILR1 for/pENTR Kan and pENTR-ILR1 as template; and one amplified with the primers pENTR Kan for/ILR1-eGFP rev and pENTR-ILR1 as template. To assemble the plasmid pENTR-ILR1eGFPwoKSEL, the same primers employed to generate the clone pENTR-ILR1eGFP were used but pENTR-ILR1woKSEL was used as PCR template instead of pENTR -ILR1. To assemble the plasmid pENTR-IAR3eGFP, three PCR fragments were combined: one amplified with the primers IAR3-eGFP for/eGFP-IAR3 rev and eGFP’s cDNA as template; one amplified with the primers eGFP-IAR3 for/pENTR Kan and pENTR-IAR3 as template; and one amplified with the primers pENTR Kan for/IAR3eGFP rev and pENTR/D-TOPO-IAR3 as template. To assemble the plasmid pENTR-ILL2eGFPwoHEEL, three PCR fragments were combined: one amplified with the primers ILL2-eGFP for/ILL2woHEEL rev; one amplified with the primers ILL2woHEEL for/pENTR Kan rev; and one amplified with the primers pENTR Kan for/ILL2-eGFP rev. For all the three fragments pENTR-ILL2eGFP was used as PCR template. pENTR-ILL2eGFP, pENTR-ILR1eGFP, pENTR-IAR3eGFP, pENTR-ILL2eGFPwoHEEL and pENTR-ILR1eGFPwoKSEL were subsequently used for Gateway cloning with the plant expression vector p2GW7.036. The sequences of all the primers used are given in Table 1.
Table 1. Oligonucleotide sequences.
| Primer name | Sequence (5’ to 3’) | Use |
|---|---|---|
| ILL2 for | CACCATGGCTCTAAACAAGCTCCTCAGT | cDNA amplification |
| ILL2 rev | TTAGAGTTCTTCATGAAAGCCTGA | |
| IAR3 for | CACCAGAGATAAGTCATGAGTTTCTTC | |
| IAR3 rev | AGAAGCCAATGTTTGTTGGCA | |
| ILR1 for | CACCATGGATTTCTCAGGGAGCTTCT | |
| ILR1 rev | AGCTGATTTTCTCCCAACACC | |
| RT-IAR3 for | AAGTGAGCTCGAGAGAGGGT | RT-PCR/qPCR |
| RT-IAR3 rev | CATATTCACGCTCGCTTGCC | |
| RT-ILL1 for | TTGGTGGCATCGGTTGGTTA | |
| RT-ILL1 rev | CGTGAAACTGACCCTTCGGA | |
| RT-ILL2 for | AAGGGAATCCGGGCAGAAAG | |
| RT-ILL2 rev | CATGCTTGCGGACATGATGG | |
| RT-ILL3 for | TGCTCCTTGGTGCTGCTAAA | |
| RT-ILL3 rev | CCAAAGCAGGACCCGAGATT | |
| RT-ILL5 for | AGGATCCACGAGAACCCAGA | |
| RT-ILL5 rev | CTCAAAGCAACAAAGGGGGC | |
| RT-ILL6 for | CTTTGGATGACGTGGAGGCT | |
| RT-ILL6 rev | GCGCCACATCGAGACTATGA | |
| RT-ILR1 for | CACGGTTCACGGTCAAGGT | |
| RT-ILR1 rev | ACCGATGCTTGTGCCTCTG | |
| RT-IAA1 for | ATGGAAGTCACCAATGGGCTTAACCT | |
| RT-IAA1 rev | TCATAAGGCAGTAGGAGGAGCTTCGGATC | |
| RT-IAA5 for | TCCGCTCTGCAAATTCTGTTCGG | |
| RT-IAA5 rev | CCCAAGGAACATCTCCAGCAAG | |
| RT-SAUR15 for | ATGGCTTTTTTGAGGAGTTTCTTGGG | |
| RT-SAUR15 rev | TCATTGTATCTGAGATGTGACTGTG | |
| RT-SAUR66 for | CACAAAGAAACTCATGAAGATGG | |
| RT-SAUR66 rev | GAATCGAATCGAATGGCAAC | |
| RT-GH3.5 for | AGCCCTAACGAGACCATCCT | |
| RT-GH3.5 rev | AAGCCATGGATGGTATGAGC | |
| RT-ACT for | TGCTGGACGTGACCTTACTG | |
| RT-ACT rev | TCTCGATGGAAGAGCTGGT | |
| ILR1-eGFP for | GGTTCGGGTCTCGAGTCAGTGAGCAAGGGCGAGGAGCTGT | Cloning |
| eGFP-ILR1 rev | CCCGCGAGCGAGGCCGCCCTTGTACAGCTCGTCCATGCCG | |
| eGFP-ILR1 for | GACGAGCTGTACAAGGGCGGCCTCGCTCGCGGGATGCTTCATTC | |
| pENTR Kan rev | CCATACAAGCGATAGATTGTCG | |
| pENTR Kan for | ATAATGTCGGGCAATCAGGTG | |
| ILR1-eGFP rev | CTCCTCGCCCTTGCTCACTGACTCGAGACCCGAACCAGAA | |
| IAR3-eGFP for | TCTAATGGGTTATCTCAAGTGAGCAAGGGCGAGGAGCTGT | |
| eGFP-IAR3 rev | CTTTGAAGGTATGCCGCCCTTGTACAGCTCGTCCATGCCG | |
| eGFP-IAR3 for | GAGCTGTACAAGGGCGGCATACCTTCAAAGTTTCTTACTT | |
| IAR3-eGFP rev | CTCCTCGCCCTTGCTCACTTGAGATAACCCATTAGAGGAA | |
| ILL2-eGFP for | GCCGAAGATACGTCTCAAGTGAGCAAGGGCGAGGAGCTGT | |
| eGFP-ILL2 rev | CTTCGTCTGGATGCCGCCCTTGTACAGCTCGTCCATGCCG | |
| eGFP-ILL2 for | GAGCTGTACAAGGGCGGCATCCAGACGAAGCTCCTCGAAT | |
| ILL2-eGFP rev | CTCCTCGCCCTTGCTCACTTGAGACGTATCTTCGGCGATC | |
| ILR1woKSEL rev | CTAAACCTCTTCTTCATGGCTATGAC | |
| ILL2woHEEL for | GTCTCAGGCTTTTAACGACCCAGCTTTCTTGTACAAAGTT | |
| ILL2woHEEL rev | CAAGAAAGCTGGGTCGTTAAAAGCCTGAGACAGAACCTTTAG |
Gene expression analysis
For expression analysis of auxin induced genes in Arabidopsis seedlings, 10-day old Col-0 and ilr1ill2iar3 seedlings were transferred to liquid SCA medium37 supplied with 10 μM each of IAA-Ala, IAA-Leu, IAA-Phe or 100 nM IAA. After 1 hour, 3–4 seedlings/condition were frozen in liquid nitrogen and further used for total RNA isolation. For expression analysis of ILR1-like genes in Arabidopsis Col-0 protoplasts and for expression analysis of ILR1, ILL2 and IAR3 in transiently transformed ilr1ill2iar3 protoplasts, 5∙105cells/sample were harvested 20 h after isolation/transformation, and used for total RNA isolation. Total RNA isolation was performed with RNeasy Plant mini kit (Qiagen, Germany) according to the manufacture instructions, with an additional DNA elimination step. 250 ng of purified RNA was used for cDNA synthesis with Maxima Reverse Transcriptase (Fermentas, Germany) using a combination of oligo(dT) and random primers and following the manufacture instructions. For RT-PCR analysis, 1 μl for each reaction was used for subsequent PCR amplification using the DreamTaq polymerase (Fermentas, Germany) according to the manufacture instructions. 25 amplification cycles were used. Equal amount of each reactions were loaded on 2% agarose gel. For qPCR analysis, DyNAmo Flash SYBR Green qPCR Kit (Thermo Scientific, Germany) was used. Real-time was performed using LightCycler 480 (Roche, Germany). For expression analysis of auxin induced genes, relative quantification analysis was applied. For expression analysis of ILR1, ILL2 and IAR3 in transient transformed protoplast, absolute quantification using a standard curve was applied. Primer pairs used for RT-PCR and qPCR are given in Table 1.
Protein expression analysis
For protein accumulation analysis of ILR1, ILL2 and IAR3, triple mutant protoplasts were transformed using the plasmids p2GW7-ILR1eGFP, ILL2eGFP and IAR3eGFP, respectively. 20 h after transformation, 1*106 cells were collected and used for microsomal enriched protein isolation according to38. Membrane pellets were suspended in 50 μl sample buffer38. Half sample was used for Western analysis, another half for Coomassie staining. Gels were blotted, stained and Western analysis was performed using standard procedures. For Western analysis GFP antibody (Roche) was used. Chemi-luminescent signal was recorded using Fusion Capt Advance SL4 software (Fusion SL, Peqlab).
Protoplast isolation, transformation and luminescence analysis
For transient expression assays and for luminescence measurement assays using the auxin sensor, isolation of Arabidopsis protoplasts, PEG-mediated DNA uptake, auxin treatments and luminescence measurements were performed as described in14. For each luminescent measurement, ~1.8*104 protoplasts/sample were used. For each trial 6–8 samples were analysed. Data are presented as average of two independent trials and a total of 12–16 samples. The ratiometric values were used for statistical analysis performed using one-way ANOVA method in Minitab software (Minitab, Ltd, UK). Grouping was performed using Tukey’s multiple comparisons with 95% simultaneous confidence intervals. Statistical significances are indicated with lower case letters, means that do not share a letter are significantly different. Auxin treatment was performed 19-20 hours after DNA transfection, for 1 hour. IAA (Dushefa Biochemie, The Netherlands) was prepared as 10 mg/ml stock; IAA-L-Phe (AldrichCPR, Germany MAR000005), IAA-L-Leu (Santa Cruz Biotechnologie, Germany CAS 57105-39-2), IAA-L-Ala (Santa Cruz Biotechnologie, Germany sc-257809) were prepared as 10 mM stock. All stocks were prepared in 90% Ethanol and stored at −20 °C.
For subcellular localization analysis the following plasmids were used: p2GW7.0-ILL2eGFP, p2GW7.0-ILR1eGFP, p2GW7.0-ILR1woKSELmCherry, p2GW7.0-ILL2woHEELeGFP, p2GW7.0-IAR3eGFP and ER-mCherry39.
Microscopy
Spinning disk Andromeda microscope (Till Photonics GmbH, Germany) was used in the analysis of the subcellular localization of the fluorescently tagged amidohydrolases and the ER compartment. Image acquisition was performed 20 h after protoplast transformation using LA software (Till Photonics GmbH, Germany). The GFP-tagged proteins were excited with 488 nm diode laser and detected with BP 525/50 emission filter. The mCherry-tagged proteins were excited with 561 nm diode laser and detected with BP 593/40 emission filter. Chlorophyll fluorescence of the chloroplasts was detected using 488 nm diode laser for excitation and quad-band BP FF01-446/523/600/677-25 filter for emission. Images were acquired as Z-stacks with 0.35 nm step size using 63xNA 1.3 Immersion-water objective (Carl Zeiss, Jena). Representative confocal sections through the nucleus area were used for image processing of the detected channels using Fiji version of ImageJ software (http://fiji.sc/Fiji).
Additional Information
How to cite this article: Sanchez, A. P. et al. Hydrolases of the ILR1-like family of Arabidopsis thaliana modulate auxin response by regulating auxin homeostasis in the endoplasmic reticulum. Sci. Rep. 6, 24212; doi: 10.1038/srep24212 (2016).
Supplementary Material
Acknowledgments
The authors wish to thank Sean Walsh for the critical reading and editing of the manuscript; and Stefanie Wrobel and Katja Rapp for technical assistance. We are grateful to Dr. Bethany Zolman for providing the mutant line ilr1ill2iar3 and related information. This work was supported by Bundesministerium für Bildung und Forschung (BMBF SYSBRA, SYSTEC Microsystems). A.S. acknowledges support from DAAD (bi-lateral program “A new passage to India”).
Footnotes
Author Contributions C.D.B., A.D. and K.P. conceived the project; C.D.B. designed and supervised the experiments; A.P.S.C., A.S., K.S., C.D.B. and A.D. performed the experiments and analyzed the data; C.D.B. wrote the article; A.D. and K.P. complemented the writing.
References
- Ludwig-Müller J. Auxin conjugates: their role for plant development and in the evolution of land plants. J. Exp. Bot. 62, 1757–1773 (2011). [DOI] [PubMed] [Google Scholar]
- Bajguz A. & Piotrowska A. Conjugates of auxin and cytokinin. Phytochemistry 70, 957–969 (2009). [DOI] [PubMed] [Google Scholar]
- Normanly J., Slovin J. P. & Cohen J. D. In Plant hormones: biosynthesis, signal transduction, action! revised 3d edn, (eds Davies P. J.) Ch. B1, 36–62 (Springer Science+Business Media B.V., 2010). [Google Scholar]
- Östin A., Kowalyczk M., Bhalerao R. P. & Sandberg G. Metabolism of Indole-3-Acetic Acid in Arabidopsis. Plant Physiol. 118, 285–296 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk M. & Sandberg G. Quantitative Analysis of Indole-3-Acetic Acid Metabolites in Arabidopsis. Plant Physiol. 127, 1845–1853 (2001). [PMC free article] [PubMed] [Google Scholar]
- Kai K., Horita J., Wakasa K. & Miyagawa H. Three oxidative metabolites of indole-3-acetic acid from Arabidopsis thaliana. Phytochemistry 68, 1651–1663 (2007). [DOI] [PubMed] [Google Scholar]
- Staswick P. E. The Tryptophan Conjugates of Jasmonic and Indole-3-Acetic Acids Are Endogenous Auxin Inhibitors. Plant Physiol. 150, 1310–1321 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel B. & Fink G. R. ILR1, an amidohydrolase that releases active indole-3-acetic acid from conjugates. Science 268, 1745–1748 (1995). [DOI] [PubMed] [Google Scholar]
- Bialek K., Meudt W. J. & Cohen J. D. Indole-3-acetic Acid (IAA) and IAA conjugates applied to bean stem sections: IAA content and the growth response. Plant Physiol. 73, 130–134 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeClere S., Tellez R., Rampey R. A., Matsuda S. P. & Bartel B. Characterization of a family of IAA- amino acid conjugate hydrolases from Arabidopsis. J. Biol. Chem. 277, 20446–20452 (2002). [DOI] [PubMed] [Google Scholar]
- Widemann E. et al. The Amidohydrolases IAR3 and ILL6 Contribute to Jasmonoyl-Isoleucine Hormone Turnover and Generate 12-Hydroxyjasmonic Acid Upon Wounding in Arabidopsis Leaves. J. Biol. Chem. 288, 31701–31714 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies R. T., Goetz D. H., Lasswell J., Anderson M. N. & Bartel B. IAR3 encodes an auxin conjugate hydrolase from Arabidopsis. Plant Cell 11, 365–376 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampey R. A. et al. A family of auxin-conjugate hydrolases that contributes to free indole-3-acetic acid levels during Arabidopsis germination. Plant Physiol. 135, 978–988 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wend S. et al. A quantitative ratiometric sensor for time-resolved analysis of auxin dynamics. Sci. Rep. 3, 2052 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri N., Dharmasiri S. & Estelle M. The F-box protein TIR1 is an auxin receptor. Nature 435, 441–445 (2005). [DOI] [PubMed] [Google Scholar]
- Kepinski S. & Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435, 446–451 (2005). [DOI] [PubMed] [Google Scholar]
- Calderon Villalobos L. I. et al. A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat. Chem. Biol. 8, 477–485 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T., Hagen G. & Guilfoyle T. J. Activation and repression of transcription by auxin-response factors. Proc. Natl. Acad. Sci. USA 96, 5844–5849 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao C.-Y. et al. Reporters for sensitive and quantitative measurement of auxin response. Nat. Methods 12, 207–210 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiess G. M. et al. Auxin input pathway disruptions are mitigated by changes in auxin biosynthetic gene expression in Arabidopsis. Plant Physiol. 165, 1092–1104 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen G. & Guilfoyle T. Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol. Biol. 49, 373–385. (2002). [PubMed] [Google Scholar]
- Magidin M., Pittman J. K., Hirschi K. D. & Bartel B. ILR2, a novel gene regulating IAA conjugate sensitivity and metal transport in Arabidopsis thaliana. Plant J. 35, 523–534 (2003). [DOI] [PubMed] [Google Scholar]
- Woodward A. W. & Bartel B. Auxin: regulation, action, and interaction. Ann. Bot. 95, 707–735 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitto. E. et al. X-ray structure of ILL2, an auxin-conjugate amidohydrolase from arabidopsis thaliana. Proteins 74, 61–71 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napier R. M., Fowke L. C., Hawes C., Lewis M. & Pelham H. R. Immunological evidence that plants use both HDEL and KDEL for targeting proteins to the endoplasmic reticulum. J. Cell Sci. 102, 261–271 (1992). [DOI] [PubMed] [Google Scholar]
- Mravec J. et al. Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature 459, 1136–1140 (2009). [DOI] [PubMed] [Google Scholar]
- Dal Bosco C. et al. The endoplasmic reticulum localized PIN8 is a pollen-specific auxin carrier involved in intracellular auxin homeostasis. Plant J. 71, 860–870 (2012). [DOI] [PubMed] [Google Scholar]
- Barbez E. et al. A novel putative auxin carrier family regulates intracellular auxin homeostasis in plants. Nature 485, 119–122 (2012). [DOI] [PubMed] [Google Scholar]
- Rosquete M. R., Barbez E. & Kleine-Vehn J. Cellular auxin homeostasis: gatekeeping is housekeeping. Mol. Plant 5, 772–786 (2012). [DOI] [PubMed] [Google Scholar]
- Barbez E. & Kleine-Vehn J. Divide Et Impera-cellular auxin compartmentalization. Curr. Opin. Plant Biol. 16, 78–84 (2013). [DOI] [PubMed] [Google Scholar]
- Schuller A. & Ludwig-Müller J. A family of auxin conjugate hydrolases from Brassica rapa: characterization and expression during clubroot disease. New Phytol. 171, 145–158 (2006). [DOI] [PubMed] [Google Scholar]
- Campanella J. J., Larko D. & Smalley J. A molecular phylogenomic analysis of the ILR1-like family of IAA amidohydrolase genes. Comp. Funct. Genomics 4, 584–600 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick P. E. et al. Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid Plant Cell 17, 616–627 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig-Müller J., Jülke S., Bierfreund N. M., Decker E. L. & Reski R. Moss (Physcomitrella patens) GH3 proteins act in auxin homeostasis. New Phytol. 181, 323–338 (2009). [DOI] [PubMed] [Google Scholar]
- Kriechbaumer V., Seo H., Park W. J. & Hawes C. Endoplasmic reticulum localization and activity of maize auxin biosynthetic enzymes. J. Exp. Bot. 66, 6009–6020 (2015). [DOI] [PubMed] [Google Scholar]
- Karimi M., Inzé D. & Depicker A. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7, 193–195 (2002). [DOI] [PubMed] [Google Scholar]
- Dovzhenko A., Dal Bosco C., Meurer J. & Koop H. U. Efficient regeneration from cotyledon protoplasts in Arabidopsis thaliana. Protoplasma 222, 107–111 (2003). [DOI] [PubMed] [Google Scholar]
- Abas L. & Luschnig C. Maximum yields of microsomal-type membranes from small amounts of plant material without requiring ultracentrifugation. Anal. Biochem. 401, 217–227 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson B. K., Cai X. & Nebenführ A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51, 1126–1136 (2007). [DOI] [PubMed] [Google Scholar]
- Emanuelsson O., Brunak S., von Heijne G. & Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2, 953–971 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.