Abstract
Segregation of embryonic endomesoderm into separate endoderm and mesoderm fates is not well understood in deuterostomes. Using sea urchin embryos, we show that Notch signaling initiates segregation of the endomesoderm precursor field by inhibiting expression of a key endoderm transcription factor in presumptive mesoderm. The regulatory circuit activated by this transcription factor subsequently maintains transcription of a cWnt ligand only in endoderm precursors. This cWnt ligand reinforces the endoderm state, amplifying the distinction between emerging endoderm and mesoderm. Before gastrulation, Notch-dependent nuclear export of an essential β-catenin transcriptional coactivator from mesoderm renders it refractory to cWnt signals, insulating it against an endoderm fate. Thus we report that endomesoderm segregation is a progressive process, requiring a succession of regulatory interactions between cWnt and Notch signaling.
Early endomesoderm induction and subsequent segregation of endoderm from mesoderm are fundamental processes in animal development. While initial endomesoderm specification has been studied extensively (1, 2), its separation in deuterostomes is poorly understood. In several deuterostomes including vertebrates like zebrafish and Xenopus and echinoderms such as sea urchins and sea stars (3–7), Notch signaling induces the expression of endoderm- or mesoderm-specific markers within an endomesoderm field. Although Notch might regulate endomesoderm segregation, it is unknown whether it alters the early endomesoderm signaling milieu, a change that is likely required to stabilize lineage identities. cWnt signaling probably establishes that precursor environment given its ancestral role in specifying early endomesoderm (8). Elucidating the mechanisms underlying such a Notch-cWnt interaction would significantly advance our understanding of the progressive specialization of the endomesoderm.
Like other deuterostomes, the cWnt signaling effector, nuclear β-catenin (nβ-catenin) is enriched asymmetrically in sea urchin embryos (9), where it specifies endomesoderm precursors in three ways. First, it establishes an early endoderm regulatory state in a tier of vegetal blastomeres (veg2, Fig. 1A) at cleavage stages (9–11). Second, in micromere descendants located immediately adjacent to the veg2 tier (Fig. 1A), nβ-catenin induces expression of the ligand, Delta (12–14), which signals through the Notch receptor in veg2 blastomeres and activates mesoderm gene expression (5, 6, 13, 15). Third, cWnt also makes veg2 cells competent to receive the micromere Delta signal (12). Thus, specification of both endoderm and mesoderm is initiated by blastula stage throughout veg2 blastomeres (Fig. 1A) (6, 9, 11, 16, 17).
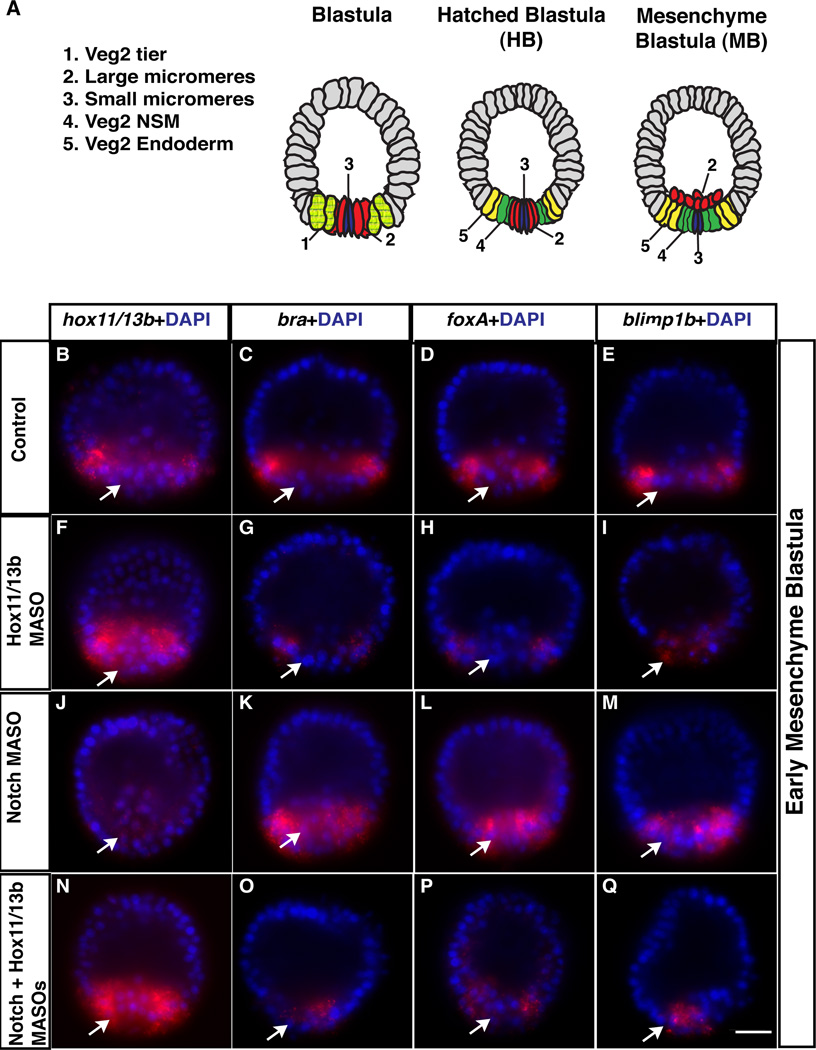
Figure 1. Notch inhibits a Hox11/13b-dependent regulatory circuit in mesoderm.
(A) Diagram of endomesoderm blastomeres: Large (red) micromere progeny are adjacent to veg2 cells in which early cWnt activates endoderm and mesoderm specification (hatched light green) in early blastulae. By HB stage, veg2 descendants form outer (presumptive endoderm, yellow) and inner (presumptive mesoderm, green) rings. Large micromere progeny (red) ingress by MB stage. (B to Q) Endodermal brachyury (bra), foxa and blimp1b transcripts are downregulated in Hox11/13b morphants (MASO). (B, F, N) hox11/13b mRNA is upregulated in Hox11/13b morphants, because Hox11/13b represses its own transcription (11). (K to M) Ectopic expression of brachyury (bra), foxa and blimp1b mRNA occurs in inner veg2 cells of Notch morphants. hox11/13b mRNA concentration is low through auto-repression by ectopic Hox11/13b protein (11) (J). (O to Q) In Notch+Hox11/13b double morphants, ectopic expression of Hox11/13b target genes in inner veg2 cells (arrows) is significantly lower (Q). Scale bar=20 µm.
By hatching blastula (HB) stage, veg2 progeny form outer and inner rings of cells (Fig. 1A). Only inner veg2 daughters adjacent to Delta-expressing micromere progeny can transduce the cell contact-dependent Notch signal and continue expressing mesoderm markers (17). Transcripts encoded by endoderm regulatory genes, in turn, are detected in outer veg2 daughters by this time (11, 17). Notch is required for this restriction, since it inhibits expression of the endoderm markers, foxa, blimp1b and dac in inner veg2 daughters (17–19). During cleavage and HB stages, nβ-catenin is detected throughout the veg2 endomesoderm precursor field (9). By mesenchyme blastula (MB) stage, 6–8 hours after endoderm marker expression first clears from inner veg2 daughters, nβ-catenin is down-regulated in these mesoderm precursors through a process requiring Notch (9, 20, 21). Thus, Notch likely plays a substantial role in endomesoderm segregation beyond merely activating mesoderm regulatory genes. However, several major questions remain unanswered. First, how does Notch restrict endoderm fate to a subset of the endomesoderm progenitor field? Second, how does Notch inhibit endomesoderm-inducing cWnt in presumptive mesoderm (9, 21) 6–8 hours after initial endoderm marker expression has disappeared? Third, are these Notch-dependent events mechanistically linked?
To understand initial Notch-dependent restriction of endoderm potential from mesoderm, we used Notch-deficient embryos to systematically assess the expression of each gene in the Early Endoderm Gene Regulatory Network (GRN) (fig. S1A), which represents cWnt-induced endoderm specification until the HB stage (10, 11). We found that hox11/13b, brachyury, blimp1b and foxA transcripts accumulated ectopically in inner veg2 descendants when Notch was knocked down (fig. S1B to E and I to L). Since only part of the cWnt-activated Early Endoderm GRN is thus subject to Notch inhibition (fig. S1F to H and M to O), Notch does not initially modulate upstream cWnt signaling. Instead, it probably inhibits an intermediate endoderm GRN factor(s) activated in endomesoderm precursors by the earlier cWnt input. Such a mechanism would also allow mesoderm induction in veg2 progeny, which require cWnt for competence to transduce the Notch signal (12). Furthermore, since brachyury, blimp1b and foxA are Hox11/13b target genes (Fig.1, B to I) (11), we hypothesized that Notch could initiate endoderm restriction simply by suppressing hox11/13b expression in mesoderm. We confirmed this through two observations. First, brachyury, foxa and blimp1b are not expressed ectopically in inner veg2 descendants (Fig. 1K to M vs O to Q) in Notch-deficient embryos that also lack Hox11/13b. Second, Notch-suppressed endoderm genes still accumulate ectopically in inner veg2 descendants in double morphants that lack both Notch and any one of Brachyury, FoxA or Blimp1b (figs. S2, S3 and S4). Thus, Notch initially suppresses mesodermal accumulation of a single endoderm transcription factor, Hox11/13b. This prevents ectopic activation of the Hox11/13b-dependent regulatory circuit, consisting of Brachyury, Blimp1b and FoxA, despite the presence of nβcatenin in the mesoderm (9) (step 1 in Fig. 3J).
We hypothesized that Notch clears nβ-catenin from mesoderm by the MB stage (9, 21) by inhibiting expression of a cWnt ligand in this lineage and identified wnt1 as a candidate through two observations: First, like nβ-catenin, wnt1 mRNA is detected throughout veg2 endomesoderm (fig. S5) between HB and MB stages, and is down-regulated through Notch in mesoderm by MB stage (Fig. 2A to D) (9, 21). Second, at MB stage, Wnt1 stimulates nβ-catenin activity, as revealed by cWnt-dependent TOPFLASH (22) luciferase reporter assays (Fig. 2E). Since wnt1 expression is restricted to the endoderm 6–8 hours after Notch confines expression of Hox11/13b-activated regulatory genes to this lineage, we assessed the potential linkage between these events. Both endodermal wnt1 expression in normal embryos at MB stage and its persistence in mesoderm in the absence of Notch require Hox11/13b and its target, Brachyury (Fig. 2F to K). These wnt1 regulators are also essential for normal levels of cWnt activity at this time (Fig. 2E). Thus, by inhibiting mesodermal hox11/13b expression at the HB stage, Notch indirectly restricts wnt1 accumulation to the endoderm, where it likely contributes to maintaining cWnt activity (step 2 in Fig. 3J).
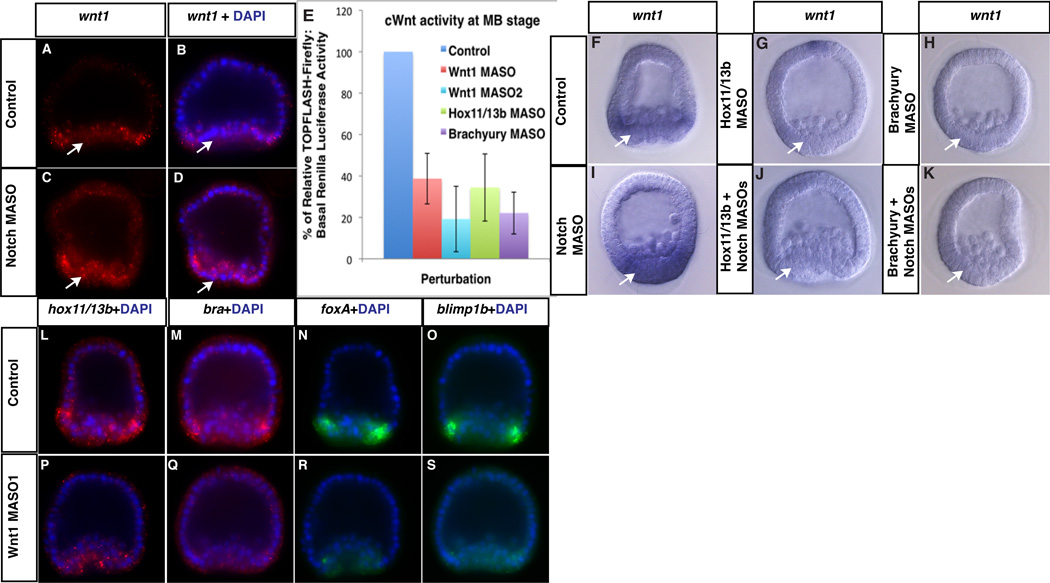
Figure 2. Through the Hox11/13b circuit, Notch indirectly restricts wnt1 to endoderm, reinforcing its differentiation.
(A to D) In Notch morphants, ectopic wnt1 mRNA accumulates in inner veg2 descendants (arrows). (E) cWnt signaling activity (TOPFLASH-see Methods) is strongly reduced in Wnt1 (MASO or MASO2), Hox11/13b or Brachyury morphants [error bars indicate ±SD from the mean (n=3)]. (F to H) wnt1 mRNA accumulation in control MB endoderm and (I to K) its ectopic expression in inner veg2 cells (arrows) of Notch morphants requires Hox11/13b and Brachyury. (L to S) Endodermal MB expression of hox11/13b, brachyury (bra), foxa and blimp1b mRNA is reduced in Wnt1 morphants.
Since the Endoderm GRN depends on cWnt (11, 18, 23), we hypothesized that ligands activating cWnt signaling stabilize the endoderm regulatory state and amplify its distinction from mesoderm. Consistent with this idea, two different Wnt1 morpholinos interfere with endodermal expression of hox11/13b, brachyury, foxa and blimp1b (Fig. 2L to S; fig. S6) at the MB stage and significantly delay gastrulation (fig. S7). Additionally, Notch-mediated wnt1 down regulation could help deplete nβ-catenin in the mesoderm (9), leading to repression of cWnt-sensitive endoderm genes through its transcriptional binding partner, TCF (24). This would further reinforce endomesoderm segregation, since TCF represses at least one endoderm factor, FoxA, in the mesoderm at MB stage (18). Other Wnt ligands, including Wnt6 and Wnt8 that are required for endoderm specification (25–27), might also contribute to endomesoderm separation by increasing cWnt signaling. Consistent with the timing of its restriction to endoderm, Wnt1 reinforces endoderm segregation only at MB stage. It does not affect its initial restriction at HB stage because Notch-deficient embryos lacking Wnt1 protein still show ectopic expression of Hox11/13b target genes in inner veg2 descendants (fig. S8).
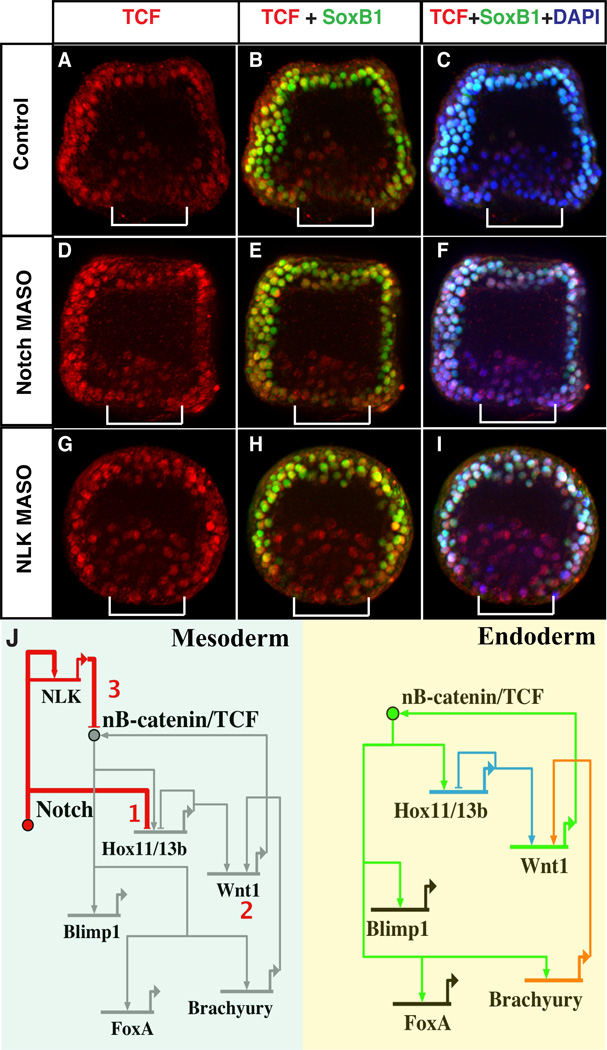
Delta-Notch signaling can also inhibit cWnt activity in sea urchin embryos by activating Nemo-like Kinase (NLK) expression in veg2 descendants (28). In other systems, NLK, a MAP kinase, modulates cWnt signaling by phosphorylating β-catenin’s essential transcriptional coactivator, TCF, and promoting its nuclear export (29). The loss of TCF could eliminate cWnt signaling via any Wnt ligand in the mesoderm. We detected TCF protein in all nuclei until late MB stage (fig. S9A to C). However, just before gastrulation begins, 4 hours after MB stage, TCF clears specifically from mesoderm nuclei (fig. S9) in a Notch- and NLK-dependent manner (Fig. 3A to I), which strongly antagonizes cWnt in these cells (step 3 in Fig. 3J).
Figure 3. Notch down regulates nuclear TCF in mesoderm.
(A to C) Nuclear TCF clears from mesoderm (brackets; SoxB1-negative; see fig. S9 for lineage identification using SoxB1) in control embryos. (D to F) Without Notch or (G to I) NLK, nuclear TCF persists in inner veg2 descendants (brackets). (J) Summary of endomesoderm segregation steps. (Step 1) Notch inhibits the Hox11/13b regulatory circuit in mesoderm (green box) by HB stage to initiate segregation. The Hox11/13b circuit stays active in endoderm (yellow). (Step 2) Consequently, by MB stage, Hox11/13b- and Brachyury-dependent expression of wnt1 is restricted to endoderm, where it reinforces cWnt signaling and endoderm fate. (Step 3) By pregastrula stage, a Notch-NLK pathway clears nuclear TCF from mesoderm, making it unresponsive to cWnt. The endoderm retains nuclear TCF and cWnt sensitivity.
In this study, we elucidate sequential Notch-dependent mechanisms that precisely regulate endomesoderm segregation. Notch initially restricts a transcription factor(s) to the endodermal daughters of endomesoderm precursors, where it subsequently activates cWnt. Such a mechanism could be shared with other deuterostomes because Notch affects expression of brachyury and foxA during endomesoderm segregation in both echinoderm and vertebrate embryos (3, 4, 7, 17–19). The regulatory device through which Notch indirectly modifies the early endomesoderm cWnt signaling state could also be conserved because Brachyury maintains transcription of cWnt ligands in both zebrafish and sea urchin embryos (30). Restricting Brachyury expression to either endoderm or mesoderm would also confine cWnt signaling. This could, in turn, reinforce lineage segregation, as seen with the Brachyury>Wnt1>endoderm and the Brachyury>cWnt>posterior mesoderm pathways in sea urchins and zebrafish, respectively (30). More generally, similar Notch-dependent mechanisms could modulate additional pathways such as Nodal/TGFβ that induce endomesoderm in vertebrate embryos (31). Finally, it is unknown whether Notch also insulates mesoderm or endoderm from incident cWnt signals through NLK activity in vertebrate embryos. We thus uncover a remarkable timing buffer that uses a cell contact-dependent signal to separate regulatory states within a broadly induced endomesoderm field without immediately altering its signaling milieu. This preserves the competence of each lineage and correctly institutes its specification. Individual lineage choices are then reinforced and cemented through successive signaling state changes.
Supplementary Material
Acknowledgments
The authors thank D. Adams and R. Benabentos for their comments. Funding was provided by the Intramural Program at the National Institute of Dental and Craniofacial Research, NIH.
Footnotes
figures S1-S9
Materials and Methods
References (32–36) [Note: The numbers refer to any additional references cited only within the SOM]
References
- 1.Kimelman D, Griffin KJ. Curr. Opin. Genet. Dev. 2000;10:350. doi: 10.1016/s0959-437x(00)00095-2. [DOI] [PubMed] [Google Scholar]
- 2.Rodaway A, Patient R. Cell. 2001;105:169. doi: 10.1016/s0092-8674(01)00307-5. [DOI] [PubMed] [Google Scholar]
- 3.Kikuchi Y, et al. Dev Dyn. 2004;229:756. doi: 10.1002/dvdy.10483. [DOI] [PubMed] [Google Scholar]
- 4.Revinski DR, Paganelli AR, Carrasco AE, Lopez SL. Dev. Biol. 2010;339:477. doi: 10.1016/j.ydbio.2010.01.010. [DOI] [PubMed] [Google Scholar]
- 5.Sherwood DR, McClay DR. Development. 1997;124:3363. doi: 10.1242/dev.124.17.3363. [DOI] [PubMed] [Google Scholar]
- 6.Sherwood DR, McClay DR. Development. 1999;126:1703. doi: 10.1242/dev.126.8.1703. [DOI] [PubMed] [Google Scholar]
- 7.Hinman VF, Davidson EH. Proc Natl Acad Sci U S A. 2007;104:19404. doi: 10.1073/pnas.0709994104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petersen CP, Reddien PW. Cell. 2009;139:1056. doi: 10.1016/j.cell.2009.11.035. [DOI] [PubMed] [Google Scholar]
- 9.Logan CY, Miller JR, Ferkowicz MJ, McClay DR. Development. 1999;126:345. doi: 10.1242/dev.126.2.345. [DOI] [PubMed] [Google Scholar]
- 10.Davidson EH, et al. Dev. Biol. 2002;246:162. doi: 10.1006/dbio.2002.0617. [DOI] [PubMed] [Google Scholar]
- 11.Peter IS, Davidson EH. Dev. Biol. 2010;340:188. doi: 10.1016/j.ydbio.2009.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McClay DR, Peterson RE, Range RC, Winter-Vann AM, Ferkowicz MJ. Development. 2000;127:5113. doi: 10.1242/dev.127.23.5113. [DOI] [PubMed] [Google Scholar]
- 13.Sweet HC, Gehring MC, Ettensohn CA. Development. 2002;129:1945. doi: 10.1242/dev.129.8.1945. [DOI] [PubMed] [Google Scholar]
- 14.Oliveri P, Carrick DM, Davidson EH. Dev Biol. 2002;246:209. doi: 10.1006/dbio.2002.0627. [DOI] [PubMed] [Google Scholar]
- 15.Sweet HC, Hodor PG, Ettensohn CA. Development. 1999;126:5255. doi: 10.1242/dev.126.23.5255. [DOI] [PubMed] [Google Scholar]
- 16.Wikramanayake AH, Huang L, Klein WH. Proc Natl Acad Sci U S A. 1998;95:9343. doi: 10.1073/pnas.95.16.9343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Croce JC, McClay DR. Development. 2010;137:83. doi: 10.1242/dev.044149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de-Leon SB, Davidson EH. Proc Natl Acad Sci U S A. 2010;107:10103. doi: 10.1073/pnas.1004824107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peter IS, Davidson EH. Nature. 2011;474:635. doi: 10.1038/nature10100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruffins SW, Ettensohn CA. Development. 1996;122:253. doi: 10.1242/dev.122.1.253. [DOI] [PubMed] [Google Scholar]
- 21.Sherwood DR, McClay DR. Development. 2001;128:2221. doi: 10.1242/dev.128.12.2221. [DOI] [PubMed] [Google Scholar]
- 22.Korinek V, et al. Science. 1997;275:1784. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 23.Smith J, Kraemer E, Liu H, Theodoris C, Davidson E. Dev Biol. 2008;313:863. doi: 10.1016/j.ydbio.2007.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoppler S, Kavanagh CL. J. Cell Sci. 2007;120:385. doi: 10.1242/jcs.03363. [DOI] [PubMed] [Google Scholar]
- 25.Croce J, et al. Development. 2011;138:3297. doi: 10.1242/dev.058792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wikramanayake AH, et al. Genesis. 2004;39:194. doi: 10.1002/gene.20045. [DOI] [PubMed] [Google Scholar]
- 27.Smith J, Theodoris C, Davidson EH. Science. 2007;318:794. doi: 10.1126/science.1146524. [DOI] [PubMed] [Google Scholar]
- 28.Röttinger E, et al. Development. 2006;133:4341. doi: 10.1242/dev.02603. [DOI] [PubMed] [Google Scholar]
- 29.Lo MC, Gay F, Odom R, Shi Y, Lin R. Cell. 2004;117:95. doi: 10.1016/s0092-8674(04)00203-x. [DOI] [PubMed] [Google Scholar]
- 30.Martin BL, Kimelman D. Dev. Cell. 2008;15:121. doi: 10.1016/j.devcel.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schier AF. Annu. Rev. Cell Dev. Biol. 2003;19:589. doi: 10.1146/annurev.cellbio.19.041603.094522. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.