Abstract
Many organisms accumulate a pool of germline stem cells during development that is maintained in later life. The dynamics of establishment, expansion and homeostatic maintenance of this pool are subject to both developmental and physiological influences including the availability of a suitable niche microenvironment, nutritional status, and age. Here, we investigated the dynamics of germline proliferation during stages of expansion and homeostasis, using the C. elegans germ line as a model. The vast majority of germ cells in the proliferative zone are in interphase stages of mitosis (G1, S, G2) rather than in the active mitotic (M) phase. We examined mitotic index and DNA content, comparing different life stages, mutants, and physiological conditions. We found that germ cells in larval stages cycle faster than in adult stages, but that this difference could not be attributed to sexual fate of the germ cells. We also found that larval germ cells exhibit a lower average DNA content compared to adult germ cells. We extended our analysis to consider the effects of distance from the niche and further found that the spatial pattern of DNA content differs between larval and adult stages in the wild type and among mutants in pathways that interfere with cell cycle progression, cell fate, or both. Finally, we characterized expansion of the proliferative pool of germ cells during adulthood, using a regeneration paradigm (ARD recovery) in which animals are starved and re-fed. We compared adult stage regeneration and larval stage expansion, and found that the adult germ line is capable of rapid accumulation but does not sustain a larval-level mitotic index nor does it recapitulate the larval pattern of DNA content. The regenerated germ line does not reach the number of proliferative zone nuclei seen in the continuously fed adult. Taken together, our results suggest that cell cycle dynamics are under multiple influences including distance from the niche, age and/or maturation of the germ line, nutrition and, possibly, latitude for physical expansion.
Keywords: Cell fate, Irises, Notch, Insulin, S6 Kinase, Adult reproductive diapause
1. Introduction
When considering the accumulation and/or maintenance of a pool of proliferating cells, it can be difficult to distinguish between the effects of cell fate (undifferentiated versus differentiated) and cell cycle (the rate of cell division). For example, interfering with signaling pathways necessary to maintain the proliferative fate of cells within a pool may reduce the number of proliferative cells, but may or may not interfere with the rate of cell cycle progression. Similarly, quiescent or slow-cycling cells may retain a “proliferative-competent” state but lack critical cues to activate a robust mitotic cell cycle. Moreover, in circumstances where the spatial distribution of proliferative cells is important (e.g., in response to signaling from a niche), the features of the cell cycle may differ depending on proximity to the niche. Finally, the states of expansion and establishment of a stem cell pool may differ from those in homeostasis.
The C. elegans germ line is a relatively simple paradigm for studying the cellular and molecular underpinnings of the influences of signaling and nutrition on a proliferating pool of cells, such as stem and progenitor cells. In the C. elegans hermaphrodite, a single cell, the distal tip cell (DTC) acts as a niche. A DTC caps each of two gonad arms and is required to establish and maintain the population of proliferative germ cells adjacent to it (Kimble and White, 1981). Ligands produced by the DTC interact with and activate GLP-1, a Notch family receptor present on the surface of distal germ cells, to prevent differentiation (Hansen and Schedl, 2013; Kershner et al., 2013). Additionally, the proliferative germ cell pool is sensitive to optimal nutrition and is regulated by nutritionally sensitive pathways such as Insulin/IGF and TOR/S6-Kinase (S6K) (Hubbard et al., 2012).
During the germline expansion phase of the third (L3) and fourth (L4) larval stages, the pool of distal proliferative germ cells accumulates rapidly from approximately 30 to over 200 cells in each of the two arms of the hermaphrodite gonad. Meiotic entry begins in proximal germ cells, those farthest from the DTC, at the mid-L3 stage (Hansen et al., 2004; Hirsh et al., 1976). Therefore, while the number of proliferative germ cells provides a convenient estimate of the expansion of the proliferative zone, it underestimates the number of cells that are produced after the mid-L3 since the pool is continuously donating cells to the meiotic pathway. The position relative to the DTC at which meiotic entry occurs ranges from ~ 13 cell diameters (CD) at the time of initial meiosis in the L3 stage to 20–25 CD in the adult. Characteristic crescent-shaped nuclear morphology of leptotene and zygotene stages of prophase of meiosis I indicate meiotic entry in the ‘transition zone (TZ)’ (Hansen et al., 2004; Hirsh et al., 1976). By convention, the “proliferative zone” (or “mitotic region”) is defined as the cells between the distal tip and the first row of germ cells containing 2 or more crescent shaped nuclei (Crittenden et al., 2006). In the adult, the proliferative zone also contains a large fraction of cells in meiotic S phase (Fox et al., 2011).
While the precise relationship between Notch signaling and cell cycle is unknown for the germ line, a recent model suggests that cells within the proliferative zone which enter a sub-thresh-old region of GLP-1 activity (~ 10 cell diameters from the distal tip in the adult) complete one final mitotic division prior to meiotic entry (Fox and Schedl, 2015). In addition, precedent exists for cell-cycle gating of Notch receptor activity in vulval precursor cells, where sequential LIN-12/Notch signaling in the G1 and again in the G2 is thought to direct different cell fate decisions (Ambros, 1999). However, cell cycle components may also promote or inhibit the activity of LIN-12 in the G1 and G2, respectively (Nusser-Stein et al., 2012). In the germ line, the cell cycle is more difficult to follow due to its duration and due to the extreme sensitivity of the germ line to manipulation (Gerhold et al., 2015; Michaelson et al., 2010), both of which hinder informative long-term live imaging.
In addition to Notch mediated signaling that is required to maintain the proliferative pool in all stages, robust larval expansion of the proliferative pool requires adequate nutrition and the activity of nutrition-sensitive signaling pathways (Hubbard et al., 2012). In contrast, maintenance of the steady number of cells in the proliferative pool during early adult homeostasis is less sensitive to the activity of these nutrition-sensitive pathways (Korta et al., 2012; Michaelson et al., 2010). Thus, reducing the activity of either the Notch, Insulin, or TOR/ S6K pathways can interfere with the accumulation proliferative cells, but by different mechanisms (Hubbard et al., 2012). Consistent with its role as an arbiter of undifferentiated versus differentiated fate, reducing GLP-1/Notch signaling does not alter the mitotic index of larval progenitors. Rather, they differentiate at a position closer to the DTC and thereby have fewer undifferentiated cells in the proliferative zone. By contrast, Insulin pathway mutants reduce the cell cycle rate but do not alter cell fate (Michaelson et al., 2010). Finally, S6K, one of two well-characterized downstream effectors of TORC1 signaling, interferes with both (Korta et al., 2012).
Separable or inseparable cell fate (mitosis/meiosis) versus cell cycle control is also evident among mutants that cause more severe proliferation defects. For example, animals bearing mutations in some genes exhibit severely reduced germ cell numbers but these cells can still undergo differentiation in the absence of glp-1 (e.g., nst-1 (Kudron and Reinke, 2008), mex-3; puf-8 (Ariz et al., 2009)), while germ cells in animals bearing mutations in other genes that cause severely reduced germ cell numbers fail to differentiate, even in the absence of GLP-1/Notch activity (e.g., glp-4 (Beanan and Strome, 1992), rpl-11.1 (Maciejowski et al., 2005), glp-3 (Kadyk et al., 1997)). Indeed, bona fide cell cycle proteins such as cyclin E (cye-1) have roles in both cell cycle progression and in preventing premature differentiation (Fox et al., 2011).
To examine more closely the cell cycle features within the proliferative pool during expansion and homeostasis, we examined the profiles of mitosis and cycling interphase (G1, S, G2) stages using a combination of standard markers and Irises software we developed to measure DNA content (ploidy) in situ (Vogel et al., 2014). We have applied these approaches to both expansion and homeostasis phases of the pool of proliferative germ cells in wild type and in selected mutants that we have previously characterized. We found that the proportion of nuclei in early and late stages of the cell cycle (low versus high DNA content) differs between larval and adult stages. We also found that in the adult these proportions differ spatially between distal and proximal nuclei in the proliferative zone. Since the first germ cells to differentiate become sperm (Hansen and Pilgrim, 1999), we tested the hypothesis that the stage-specific differences in mitotic index could be attributed to the sexual fate of the cells, and found that it could not. Further, we perturbed pathways that govern cell fate, cell cycle or both and examined the effects on trends among temporal and spatial patterns of DNA content. Finally, using a post-starvation re-feeding assay (Angelo and Van Gilst, 2009; Seidel and Kimble, 2011) to regenerate the germ line during adult stages, we tested the hypothesis that developmental stage constrains these patterns and found that it constrains the size of the pool, but not the accumulation rate. Taken together, our results point to a complex combination of driving factors and constraints – including stage, nutrition and possibly space – to account for differences in cell cycle dynamics between expansion and homeostatic stages.
2. Materials and methods
2.1. Worm handling and strains
Strains were derived from Bristol N2 wild type and maintained using standard methods (Brenner, 1974). Mutant strains: reduction of function (rf) temperature sensitive alleles glp-1(e2141) and daf-2 (e1370), the null alleles rsks-1 (sv31) and fog-2(oz40). Unless otherwise indicated, worms were grown on standard OP-50 E. coli bacteria at 20 °C, which is a semi-permissive temperature for glp-1(e2141) and daf-2(e1370). For experiments conducted under well-fed conditions, worms were synchronized by hatch-off within a 2–3 h window and developmental stage was monitored by vulval morphology as described previously (Michaelson et al., 2010; Pepper et al., 2003; Seydoux et al., 1993). For the “L4/YA molt” time point, from populations of synchronized worms in which 50% or more were adults (and the remaining late L4), only worms with adult vulval morphology and no oocytes were scored. The “YA stage” was 12 h post-mid-L4 when worms contained oocytes and up to a few embryos. Whole worms were ethanol fixed, DAPI stained and imaged as described previously (Michaelson et al., 2010).
2.2. Induction of Adult Reproductive Diapause (ARD) and Re-feeding
Preparation of worms was modified from Seidel and Kimble (2011). Gravid wild-type worms were treated with 12:2:1 of M9 buffer: sodium hypochlorite (Sigma-Aldrich #425044): 5 N sodium hydroxide solution to release the eggs (modified from Stiernagle, 2006). Eggs were washed 3 times in M9 solution and allowed to hatch in the absence of food in S-Basal buffer solution overnight. Synchronized L1 larvae were then placed on OP-50 E. coli and allowed to develop to the early L4 larval stage. L4 larvae were washed with M9 solution, allowed to settle by gravity for 15–20 min, and then washed another 4–5 times to allow the larvae to defecate and to ensure food within and outside the animals was eliminated. These animals were then placed on high-agar (2.5%) NGM plates and starved for 5 days. For re-feeding, 5 day starved animals were placed onto the original OP-50 E. coli culture and were collected at 2, 6, 24, 48, 72 and 96 hrs post re-feeding for analysis.
2.3. Germ cell nuclei counts and mitotic index
Total number of germ cell nuclei in the proliferative zone was determined by a semi-automated method using a modified ImageJ plug-in (Korta et al., 2012). The mitotic index was defined as the number of metaphase and anaphase figures over the total number of nuclei in the proliferative zone and is represented as a percent value (Maciejowski et al., 2006).
2.4. Analysis of DNA Content by Genotype, CD Position, and Age (Larval/Adult)
DNA quantification was performed with the Irises software as described (Vogel et al., 2014) along with spot checking by manual quantification (Michaelson et al., 2010) to confirm that both methods gave equivalent results.
Further, here, we introduce an extension to Irises (Vogel et al., 2014) “Spatial_Analysis_of_DNA_Content” to facilitate analysis of DNA content on a per-nucleus basis over the distal-to-proximal axis. A position was assigned to each nucleus by a semi-automated process that effectively linearizes the gonad. A series of connected line segments are manually drawn through the center of gonad in 2D, to define the central gonad axis. Then, each nucleus was assigned to an individual line segment of the gonad axis to minimize the distance between the center of the nucleus and it's projection onto the line segment. On curved specimens with multiple line segments, the projection of the nucleus onto its assigned segment marks the nucleus’ location relative to the curved distal-proximal axis of the gonad. Once each nucleus is assigned to a segment, a relative distance from the distal end is computed as the sum of all distal segment lengths plus the length of the part of the assigned segment between its distal end and the projection of the nucleus onto that segment. In practice, for most gonads used in this analysis, one line segment was sufficient to define the center since the arms were essentially straight. The total distance from the distal end is normalized by an estimate of CD (in pixels) given by the user. This computation ultimately provides the distance in CD of the nucleus from the distal end of the gonad. Irises contains the X, Y, Z coordinates and relative fluorescence intensity (a proxy for DNA content) for each nucleus. The extension program then sorts each nucleus’ fluorescence intensity as calculated in Irises by distance from the distal tip. The output is in the form of .csv text file that includes for each nucleus, a “distance diameter” in CD, “distance um” in µm, the fluorescence intensity, the X Y Z position, and the calculated DNA content “N values”. Note, the original Irises output includes a column labeled “Cell Diameter” that refers to the size of the nucleus, not the CD position. This column does not appear in the output from the extension. Please see https://sourceforge.net/projects/irises/files/?source=navbar for files providing code and instructions on running the “Spatial_Analysis_of_DNA_-Content” extension to Irises.
For each gonad arm analyzed, DNA content was calculated for all nuclei at each CD position from the distal tip to the CD position third from the TZ, thus excluding the last two CD of the proliferative zone. The excluded CDs contained nuclei that are virtually all in pre-meiotic S.
Table S1 provides the numbers of animals and nuclei included in the DNA content analysis by genotype, age, and CD position. The number of scoreable nuclei per individual animal varied due to imaging constraints: deeper nuclei in the preparations were avoided to minimize variation in fluorescence. Normalization was done with nuclei in the same image (see Vogel et al., 2014). These data are presented in several figures and tables. In Figs. 1 and 2, “Pooled Average DNA Content” is the average DNA content of all nuclei in all animals per age group per genotype, without regard for number of animals or number of nuclei measured per animal. For these data, pairwise significance was calculated using nonparametric Mann–Whitney U test. “Binned DNA Content” (Fig. 2), presents the pooled DNA content (by age group and genotype) binned into categories of “Low”, “Mid” and “High” DNA content (< 2.5 = Low; ≥ 2.5 and ≤ 3.5 = Mid, and > 3.5 High). These categories roughly correspond to G1/early S, S phase, and late S/G2. However, since some of fluorescence values calculated by Irises are < 2 and > 4 (Vogel et al., 2014), we consider the results in terms of relative values rather than absolute values of DNA content. Nevertheless, the relative proportions for Low, Mid, and High bins in our pooled analysis roughly correlate with “Low” as late M phase (anaphase, telophase), G1, and early S; “Mid” as S; and “High” as late S/G2 and early M values. Using a combination of experimental methods, including EdU labeling for wild-type adult, Fox et al. (2011) report 2% M, 2% G1, 57% S and 39% G2 (Fox et al., 2011), whereas binned Irises values are 11% Low, 50% Mid and 38% High for the same age and genotype.
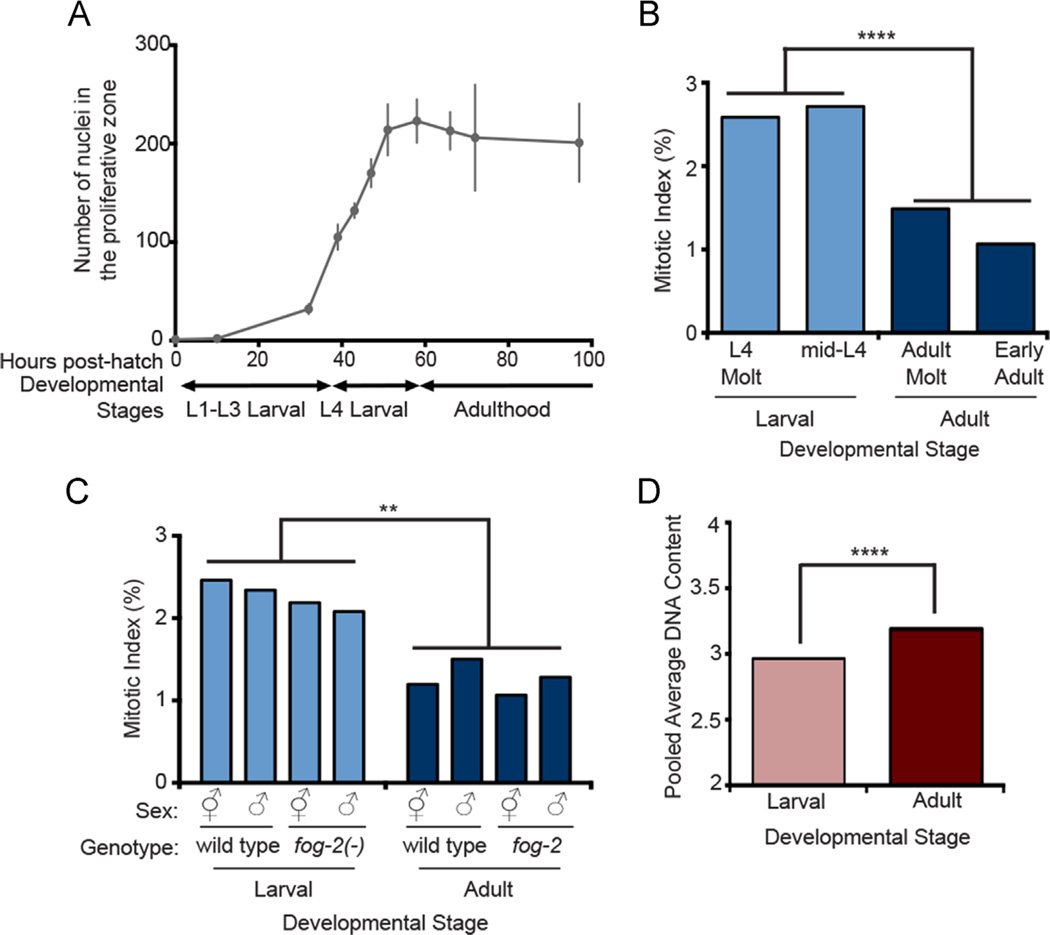
Fig. 1.
Mitotic index is higher and pooled average DNA content is lower in larval versus adult stages (A) The number of proliferative zone nuclei in wild-type animals under continuous feeding conditions at 20 °C from hatching to 96 h. Error bars indicate 1 standard deviation at each time point. Germ cells begin proliferating at ~ 10 h; N (numbers of gonad arms) are as follows for subsequent time points: 32 hr, N = 12; 39 hr, N = 10; 43 h, N = 10; 47 h, N = 10; 51 h, N = 10; 58 h, N = 10; 66 h, N = 10; 72 h, N = 10; 97 h, N = 9. (B) Mitotic Index (MI) at two larval and two adult stages: L3 to L4 Molt, mid-L4, L4 to Adult Molt and Early Adult (12-hours post-mid-L4) stages. Number of gonad arms (N) and nuclei (n): L4 Molt, N = 31, n = 1591; mid-L4, N = 68, n = 9466; Adult Molt, N = 27, n = 4300; Early Adult, N = 44, n = 9340. (C) MI in larval and adult wild-type and fog-2(oz40) mutant hermaphrodites and males. For larval (mid-L4): wild-type hermaphrodite, N = 22, n = 2939; wild-type male N = 22, n = 2519; fog-2 hermaphrodite N = 21, n = 3003; fog-2 male N = 21, n = 253. Adult: wild-type hermaphrodite N = 21, n = 4014; wild-type male N = 21, n = 3000; fog-2 hermaphrodite N = 21, n = 4052; fog-2 male N = 23, n = 3066. (D) Pooled average DNA content in larval and adult stages (see Materials and methods and Results). For larval, N = 27, n = 1524. For adult: N = 26, n = 1997. Statistics: two-tailed Mann–Whitney U test, **0.01 > p > 0.001, ****0.0001 > p > 0.00001.
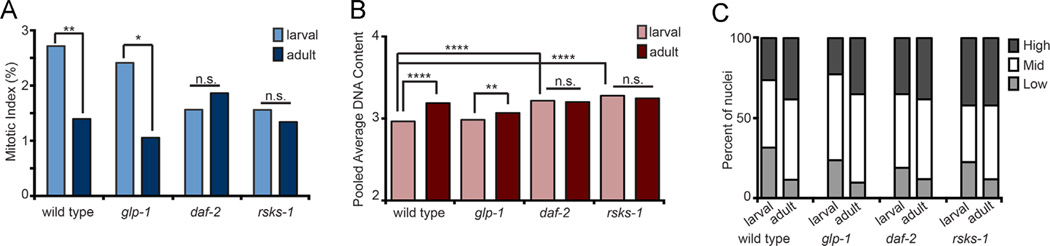
Fig. 2.
Mitotic index and DNA content in wild type, glp-1, daf-2, and rsks-1 larval and adult stages (A): Mitotic Index of wild type, glp-1(e2141), daf-2(e1370) and rsks-1(sv31) mutants at larval (mid-L4) and adult stages at 20 °C. Number of gonad arms (N) and nuclei (n) for larval: wild type, N = 22, n = 2989; glp-1, N = 69, n = 4610; daf-2, N = 15, n = 2279; rsks-1, N = 15, n = 1121. For adult: wild type, N = 21, n = 4014; glp-1, N = 32, n = 3193; daf-2, N = 22, n = 3047; rsks-1, N = 20, n = 2141. (B): Pooled average DNA content of wild type, glp-1(e2141), daf-2(e1370) and rsks-1(sv31) mutants, in larval and adult stages. Statistics: two-tailed Mann–Whitney U test, *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001 and n.s. indicates not significant. Note, pooled average DNA content of daf-2(e1370) and rsks-1(sv31) adults is not significantly different from wild type adult (p > 0.05). (C): Pooled average DNA content sub-divided into Low, Mid and High DNA content bins (see Materials and methods). Y-axis is the proportion of nuclei measured within the different DNA content bins. For larval, wild type, N = 27, n = 1524; glp-1(e2141), N = 56, n = 2090; daf-2(e1370), N = 44, n = 2487; rsks-1(sv31), N = 33, n = 1667. For adult: wild type, N = 26, n = 1940; glp-1(e2141), N = 34, n = 1899; daf-2(e1370), N = 33, n = 2530; rsks-1(sv31), N = 35, n = 3157.
“Spatial” or “positional” DNA content was first visualized by pooling all nuclei in a given CD position in a given age and genotype group without regard to number of animals in each group or the number of nuclei scored per animal. These data are displayed in Fig. S2 and Table S2.We then analyzed the positional DNA content (Fig. 3; Table S3) using methods that took into account all 288 animals of different ages and genotypes, and the position of the nuclei relative to the distal tip. We first calculated the average DNA content within each animal at each CD position because each animal contributed different numbers of nuclei to the total DNA content. Then, using regression models, we evaluated the effects of these components on DNA content (detailed further in the paragraph below). In each case, we binned the DNA content as described above in to Low, Mid, and High bins. The data were then categorized into CD regions: ≤ 5, 6 to 10, 11 to 15 and > 15 CD from the distal tip. Since averages were calculated per animal, per CD and then binned into Low, Mid and High for this analysis, the “Mid” bin appears expanded relative to a pooled average and binned proportions calculated on a per CD basis pooled across all animals (Table S2).
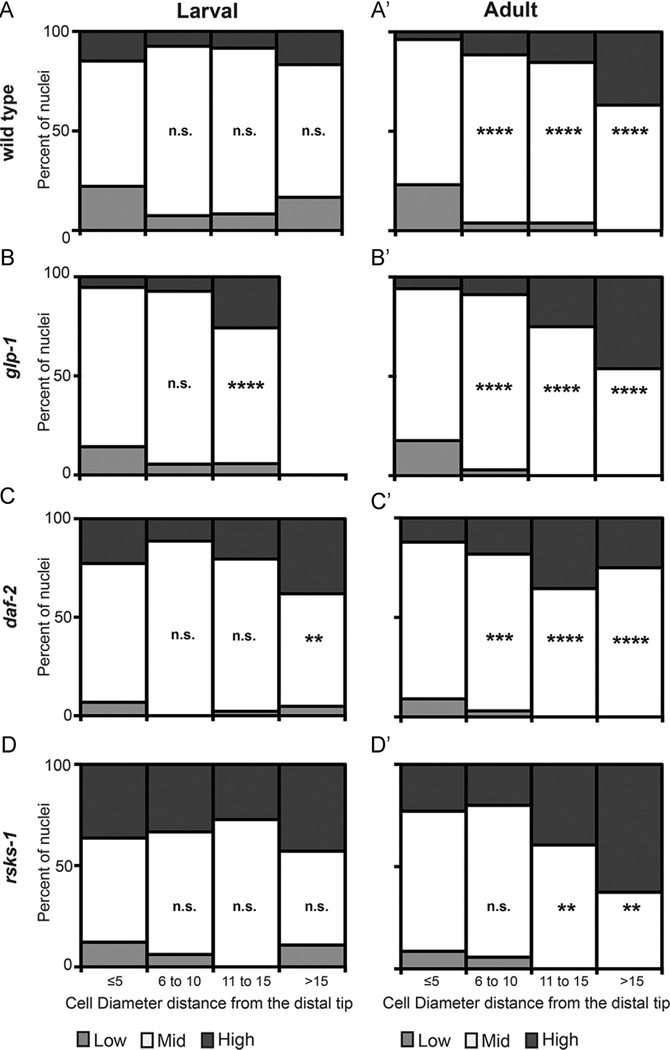
Fig. 3.
Summary of average DNA content across the proliferative zone. In all graphs, the Y-axis is the average percent of nuclei measured in Low, Mid and High DNA content bins (see Materials and methods) and the X-axis is the distance from the distal tip in cell diameters (grouped in increments of 5). Graphs on the left side (A–D) are larval and on the right side (A’-D’) are adult, both at 20 °C. Number of animals (N) and nuclei (n) are as follows: wild type: larval N = 27, n = 1524, adult N = 26, n = 1940; glp-1(e2141): larval N = 56, n = 2090, adult N = 34, n = 1899; daf-2(e1370): larval N = 44, n = 2487, adult N = 33, n = 2530; rsks-1(sv31): larval N = 33, n = 1667, adult N = 35, n = 3157 (see also Table S1). Statistics: mixed effect regression models (see Materials and methods) were used to determine effects of distance from the distal tip, age and genotype on DNA content. Note that the data are averaged and binned in this graphical representation, but the mixed effects regression model uses all observations. Region “CD ≤ 5” was used as reference to compute p-values for the remaining cell diameter regions in each genotype. **0.05 ≥ p ≤ 0.01, ***p ≤ 0.001 and ****p ≤ 0.0001 and n.s. is not significant.
Mixed effects regression models were used to incorporate the repeated observations within animals at each of the CD positions and the correlations within each animal across CD positions. These models also allow us to incorporate unequal numbers of animals in each of the age and genotype groups to evaluate the effects of age and genotype on DNA content (Fitzmaurice et al., 2011). To implement these models, genotype and age were fixed and CD position within animal was a random effect. We compared the results of these models that included the main effects of each of these variables, pairwise interactions, and 3 way interactions of all of these variables. These comparisons are based on the contributions of the individual and interaction components to the overall test statistic and the a priori hypotheses under study. We consider only hierarchical (nested) models in which the main effect of the individual variables must be present in order to consider interaction. We note that Fig. 3 represents a summary of the data (that is, binned and averaged). However, all observations were used in the mixed regression model.
We compared these nested models using Likelihood Ratio Tests. When there was a significant difference between models, we used the model with the greater number of components. Since all 3 way interactions were jointly statistically significant (likelihood ratio test p = 0.01 for all 3 way interactions compared to the model with only 2 way interactions and main effects), we stratified the data by genotype and age group and report the results of the analyses using mixed effect logit models within each of these 8 strata to elucidate the relationship between CD position and DNA content within each genotype and age combination.
The data used in the post-ARD regrowth analysis is shown in Table S4. Distribution of average DNA content at each CD position category across animals is shown in Table S5 for each time point. We developed mixed effect logit models using time point as the fixed effect and CD position within animal as the random effect.
3. Results
3.1. Mitotic index is elevated in larval compared to adult stages, regardless of germ cell sexual fate
Consistent with previous observations (Fox et al., 2011; Korta et al., 2012), we found that mitotic index – and therefore, by proxy, the cell cycle rate for the entire pool of cells – was elevated in larval stages relative to adult (Fig. 1A, B). We define mitotic index as the number of germ nuclei in the proliferative zone that are in metaphase or anaphase over the total number of nuclei in the zone as visualized by DAPI staining. We note that this DAPI-based method is applicable here since in all cases examined, the overall germline mitosis/meiosis pattern and normal nuclear morphology is maintained. Therefore, to the trained eye, the boundary of the proliferative zone is evident and mitotic figures are clearly distinguishable from crescent-shaped nuclei that indicate early stages of meiotic prophase. We also note that alternative markers for M phase, such as anti-phospho-histone H3 (pH3), give a higher average mitotic index since pH3 labels cells from pro-metaphase into early metaphase. However, pH3 may yield a somewhat less reliable measure than mitotic figures since the duration of pro-metaphase to fully congressed metaphase is variable (Gerhold et al., 2015). Regardless, even by pH3-positive mitotic prophase proxy, M phase occupies ~ 10% or less of the total cell cycle time (Gerhold et al., 2015) and mitotic figures appear on the order of 1–3% of the total proliferative zone nuclei. Therefore, the vast majority of proliferative germ cells are in interphase (G1, S and G2) at any one time, and various measures of mitotic index can therefore serve as proxies for cell cycle rate. We found that while mitotic index was significantly different between the larval and adult stages, it did not differ within the two larval stages (L3/L4 molt and mid-L4) or the two adult stages (L4/adult molt and young adult).
One plausible explanation for the difference between larval and adult mitotic index is the sexual identity of the proliferating cells. In C. elegans, the majority of cycling germ cells in early larvae likely become sperm whereas those in the adult become oocytes exclusively. To test the hypothesis that sperm-fated germ cells cycle faster than oocyte-fated germ cells, we measured mitotic index in the larval and adult stages in wild-type hermaphrodites that produce sperm and oocytes, fog-2 mutant hermaphrodites that produce only oocytes, wild-type males that produce only sperm and fog-2 males that produce only sperm. Similar to the wild type, we observed a significant difference in mitotic index between larval and adult stages regardless of the sperm or oocyte fate of the germ cells (Fig. 1C). Therefore, the elevated rate of cell cycle in larval relative to adult stages cannot be attributed to differences in the sexual fate of the germ cells.
3.2. Pooled average DNA content is lower in larval versus adult stages in the wild type
To further investigate cell cycle behavior we estimated DNA content of germ cells under a variety of conditions. To obtain DNA content information, we utilized Irises, a tool that allows semi-automated quantification of nuclear fluorescence to estimate DNA ploidy (Vogel et al., 2014). We found that the pooled average DNA content (see Materials and methods) in the proliferative zone within the larval (L3/L4 molt, mid-L4) and within adult (L4/adult molt, young adult) stages did not differ significantly (Fig. S1). However, the pooled average DNA content was significantly different between the collective larval versus adult stages: larval germ cells display lower overall average DNA content compared to adult (Fig. 1D).
3.3. Mutants that affect cell fate versus cell cycle show distinct larval versus adult cell cycle features
Our previous results (Killian and Hubbard, 2004, 2005; Korta et al., 2012; Voutev et al., 2006) and those of others (Ariz et al., 2009; Beanan and Strome, 1992; Fox et al., 2011) indicate that signals promoting the proliferative cell fate can be separated from those that promote robust mitotic cell cycle progression during larval stages. For example, GLP-1/Notch signaling maintains the undifferentiated proliferative-competent fate of germ cells and/or inhibits meiotic entry (Hansen and Schedl, 2013). However reducing glp-1 activity (e.g. by glp-1(2141) at the semi-permissive temperature; see Materials and methods) does not alter the larval mitotic index even though only half the normal number of proliferative cells accumulate by the early adult stage (Michaelson et al., 2010). By contrast, insulin-IGF-like signaling (mediated by the DAF-2/Insulin receptor) does not appear to influence cell fate but, rather, promotes robust larval cell cycle progression (Michaelson et al., 2010). Therefore, while reducing either glp-1 or daf-2 gene activity reduces the total number of proliferative zone nuclei that accumulate during larval stages, they do so by different cellular mechanisms. Previously, we also found that signaling by the S6 Kinase (rsks-1/S6K), promotes both the proliferative fate as well as robust larval cell cycle progression (Korta et al., 2012). Given that glp-1, daf-2 and rsks-1 have different effects on the larval cell cycle, we compared mitotic index at larval and adult stages (Fig. 2A). We found that, as in the wild type, mitotic index is higher in glp-1 mutant larvae relative to adult. However, neither daf-2 nor rsks-1 mutants display a significant difference between larval and adult mitotic index.
We further investigated the pooled average DNA content in these mutants using Irises (Table S1; see Materials and methods). We found that, similar to the wild type, mutants with reduced glp-1 activity display a modest but significant difference in the pooled average DNA content of germ nuclei in the proliferative zone between larval and adult animals (Fig. 2B, Fig. S1). In contrast, neither the daf-2 nor the rsks-1 mutants showed a significant difference in the pooled average DNA content of the larval versus adult germ nuclei (Fig. 2B, Fig. S1). When the data are separated into Low, Mid and High DNA content bins (Fig. 2C; see Materials and methods), adults contain a lower proportion of nuclei in the Low DNA content bin regardless of the genotype.
3.4. Average DNA content is non-uniform across the adult proliferative zone
The Irises tool collects spatial information together with fluorescence intensities for individual stem/progenitor germ nuclei. Therefore we extended the tool (Irises extension “Spatial_Analysis_of_DNA_Content”) to allow for facile calculation of DNA content as a function of distance from the distal tip. We sought to examine whether differences in DNA content correlate with distance from the distal tip in larval and adult stages. We first visualized pooled average DNA content on a CD-by-CD basis. Consistent with published analyses suggesting that the vast majority germ cells in the adult proliferative zone are in S and G2, we note a vast preponderance of DNA content values in the Mid and High ranges (Table S2; Fig. S2). These data were then subjected to per animal averaging and analyzed using an unbiased regression model to enable statistical comparisons (Table S3, Fig. 3). See Materials and methods for details.
Previous analyses suggested that the distal-most region of the proliferative zone has different properties compared to more proximal regions (Cinquin et al., 2010; Maciejowski et al., 2006). The DTC cap, which is the primary source of GLP-1/Notch ligands and cell fate regulation, is more closely associated with the distal-most 2–4 CD (Byrd et al., 2014), a region of lower mitotic index (Crittenden et al., 2006; Maciejowski et al., 2006) and of gap junctions between the DTC and germ cells (Starich et al., 2014). Therefore, for our analyses of the spatial distribution of DNA content, we divided the proliferative zone into regions of 5 CD (Table S6) and used CD ≤5 as a reference. We then determined whether there were statistical differences between this distal-most (≤ 5 CD) region and the remaining regions of the proliferative zone (Table S7).
First, we compared the spatial distribution of DNA content in wild-type larvae and adults (Fig. 3A, A’). We found that in wild-type larvae, the fraction of cells in each of the three DNA content bins (Low, Mid, High) did not differ statistically across the proliferative zone. However, in the adult stages, the 3 regions > 5 CD (that is, 6 to 10, 11 to 15, and > 15) differ significantly from the ≤ 5 CD region; each of the 3 regions contain a higher DNA content (as averaged per animal per CD) than the distal-most region. These results reconcile a previous contradiction in the literature (Feng et al., 1999; Fox et al., 2011) regarding DNA content measurements (see Discussion). Interestingly, the distal-most adult nuclei exhibit a modestly lower mitotic index (Maciejowski et al., 2006), suggesting that a simple inverse correlation between mitotic index and DNA content does not hold in the distal-most region.
The DTC is a source of ligands for the GLP-1/Notch receptor present on the germ cells, and the activity of this receptor maintains distal germ cells in an undifferentiated fate (Henderson et al., 1994; Nadarajan et al., 2009). We investigated whether DNA content is uniformly distributed in germ cells in mutants with reduced glp-1 activity (Fig. 3B, B’), and found that it is not. The general pattern of DNA content differences in the adult glp-1(rf) is similar to the wild-type adult in that there is a statistically significant elevation in the percentage of nuclei in late interphase further from the DTC. However, in glp-1 mutant larvae, unlike the wild type, a higher DNA content is also seen with increasing distance from the distal tip. Therefore, although the pooled average DNA content did not differ greatly between glp-1 and wild-type larvae (or adults) (Fig. 2B), differences become more apparent once the distance from the DTC is taken into account.
Next, we assessed the spatial pattern of DNA content in mutants with reduced daf-2 signaling (Fig. 3C, C’). These mutants primarily affect larval cell cycle progression (Michaelson et al., 2010). We found that, similar to the wild type, the DNA content in adult daf-2 germ cells increases with distance from the distal tip. However, unlike the wild type and consistent with an effect of daf-2 on larval germline development, the region > 15 CD in the daf-2 larval germ line displays a greater proportion of nuclei with higher DNA content.
Last, we investigated the DNA content in rsks-1 null animals. These mutants have defects in both cell cycle progression and cell fate regulation (Korta et al., 2012). Similar to the wild type, the rsks-1 mutant larvae show no effect of distance from the distal tip (Fig. 3D, D’). In adults, the effect of distance from the distal tip on DNA content is significant only in the two proximal-most regions, rather than all three regions, as in the wild type.
In summary, our results indicate that in adults, distance from the distal tip correlates with an elevation in DNA content in wild type and in all of the mutants tested (Table S7). In larvae, this effect is either absent (wild type and rsks-1) or present only in areas farthest from the distal tip (glp-1 and daf-2).
3.5. The regenerating wild-type adult stem/progenitor pool can accumulate at a rate similar to the larval germ line
The results presented thus far indicate that germ cells in wild-type larvae display a higher mitotic index (i.e., cycle faster) and a lower average DNA content compared to adults (Fig. 2). One possibility is that a “larval versus adult” developmental program underlies these differences. To test this possibility, we took advantage of germline regeneration that occurs following Adult Reproductive Diapause (ARD) in response to feeding. ARD occurs when early L4 animals are subject to complete starvation, and it is characterized by a dramatic loss of germ nuclei (including a severe reduction in the proliferative zone) while the animal continues to develop into adulthood. Consistent with previously published results (Angelo and Van Gilst, 2009; Seidel and Kimble, 2011), we see three distinct phenotypic classes after sudden and complete starvation at the early L4 stage: (1) L4 arrest, (2) adults undergoing matricide (“bagging”) and (3) ARD (Fig. 4A). After 5 days of starvation, we observed ~ 30–40% of the population exhibiting the previously-described shrunken and distinctive ARD germline morphology together with 1–2 oocytes or embryos. To examine the regeneration of the germ line, we moved these ARD animals (after 5 days of starvation; see Materials and methods) back to their original pre-starvation food source. We then studied the regrowth of the germline proliferative zone.
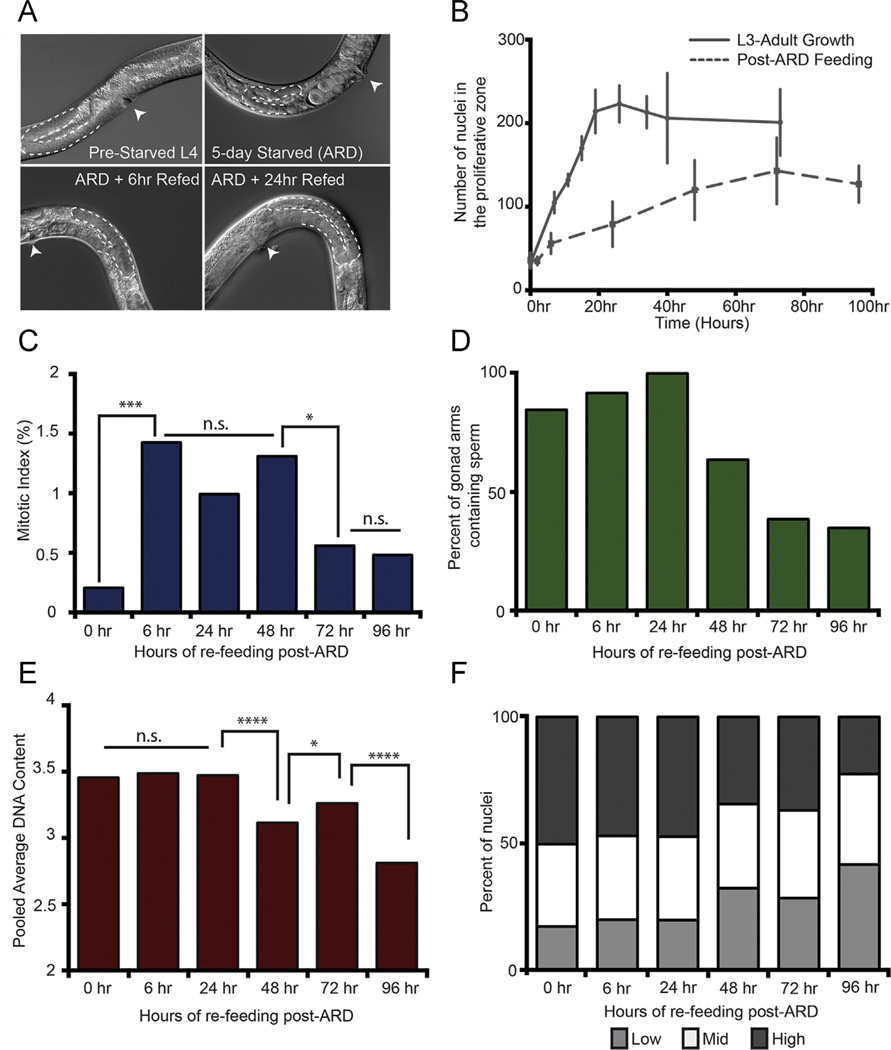
Fig. 4.
The post-ARD regenerating proliferative germ line recapitulates a subset of features of larval germ line development (A) Differential Interference Contrast (DIC) images of ARD and recovery upon re-feeding. The 5-day starved panel represents “0 hour” time point in subsequent panels. (B) The number of nuclei in the proliferative zone (Y-axis) of adult wild-type animals over time upon re-feeding. The curve for “L3-Adult Growth” is adapted from Fig. 1A to compare with post-ARD regenerative growth such that both curves start at approximately the same average number of proliferative zone nuclei: 36 for ARD and 32 for well-fed larvae. X-axis is time in hours as well as normal larval growth. The ‘0 hour’ time point for post-ARD re-feeding is therefore matched to a ‘32 hour post-hatching’ time point. Error bars are 1 standard deviation at each time point. N gonad arms for each time point are as follows: 0 h N = 16; 2 h N = 7, 6 h N = 20, 24 h N = 23, 48 h N = 11, 72 h N = 22, and 96 h N = 21. Panels C–F are time course analyses of (C) mitotic index, (D) penetrance of gonad arms containing sperm, (E) pooled average DNA content and (F) overall DNA content separated into Low, Mid and High DNA content bins (See Methods and materials). N gonad arms and n nuclei for each time point are as follows. For (C), 0 h, N = 16, n = 580; 6 h N = 20, n = 1138, 24 h N = 23, n = 1821, 48 h N = 11, n = 1319, 72 h N = 22, n = 2795, and 96 h, N = 21, n = 2679. For (D), 0 h N = 13, 6 h N = 12, 24 h N = 8, 48 h N = 10, 72 h N = 13, and 96 h N = 23. For (E) and (F), 0hr, N = 12, n = 408; 6 h N = 11, n = 558, 24 h N = 8, n = 427, 48 h N = 11, n = 967, 72 h N = 10, n = 1021, and 96 h, N = 7, n = 972. Statistics: two-tailed Mann–Whitney U test was used to compare between time points in panels C and E. *0.05≥p ≥ 0.01, ****0.00001 ≥ p ≥ 0.0001.
If developmental stage (larval versus adult) were the most important factor in determining cell cycle features, then adult regenerating proliferative germ cells may exhibit properties similar to otherwise continuously fed adults. Alternatively, if factors other than developmental stage (such as nutrition or expansion space) are important, then adult regenerating proliferative germ cells may display features more similar to larvae undergoing developmental expansion of the proliferative pool.
Since germline regeneration after ARD has not been characterized, we first conducted a time-course analysis to determine the rate of accumulation of the proliferative zone in terms of cell number at 0, 2, 6, 24, 48, 72 and 96 h post re-feeding (Fig. 4B). We reassessed proliferative germ cell accumulation in larval stages in parallel (using the same wild-type worm stock, same food and same growth conditions). From a starting point in the L3 with approximately 30 nuclei, larval proliferative germ cell accumulation was linear with respect to time over ~ 25 h reaching ~ 200–250 proliferative nuclei. These results suggest an approximate doubling of the pool every ~ 9 h on average. This is an underestimate, however, since it does not include cells donated to the meiotic pathway during this time. The linear expansion phase is followed by a plateau with little further accumulation over the next 7 h (consistent with Killian and Hubbard, 2005). Our time course analysis of proliferative germ cell accumulation during regeneration revealed several distinct stages of regrowth followed by homeostasis. First, we saw a lag of about 2 h before post-ARD proliferation-competent cells begin to accumulate rapidly (see Fig. 4B and Fig. S3). We observed a remarkable recovery phase during the first 6 h after re-feeding in which the rate of accumulation of proliferative germ cells was comparable to that of larval proliferative nuclei from a similar starting point of ~ 30–35 nuclei (Fig. 4B and Fig. S3). The rate of accumulation then slowed and eventually plateaued by the time ~ 140 nuclei had accumulated, 72 h post-ARD feeding. We conclude that in early stages of post-ARD recovery, the regenerating adult germ line is capable of expanding at a rate comparable to that of the well-fed larval germ line, indicating that the adult stage per se does not constrain the proliferative zone to a slower accumulation rate.
3.6. Certain features of the wild-type regenerating adult proliferative pool are reminiscent of larval development, while others are not
We next estimated the mitotic index and DNA content of the regenerating proliferative germ line in adults as they recovered from ARD. We found that, similar to larval growth, the mitotic index changed during the time course of re-establishment of the proliferative zone (Fig. 4C). After 5 days of starvation, the mitotic index of germ cells in animals that exhibit ARD characteristics was very low (~ 0.2%, corresponding to one mitotic figure seen in 17 gonad arms examined). During the early regeneration period, the mitotic index was relatively high (~ 1.5%) and remained at a high level before dropping by the 72 h time point. This trend is reminiscent of the changes in mitotic index during the normal development of well-fed animals: a higher mitotic index in larvae while the germ line is expanding and a lower mitotic index in adults in germline homeostasis.
We further note that oogenesis is taking place in post-ARD re-fed animals by the 24 h time point, and that the decrease in mitotic index at 72 h correlates with a drop in the percentage of recovering animals that contain sperm (Fig. 4D). Of the animals that do contain sperm at 72 and 96 h time points, the number of sperm is low (Fig. S3). Additionally, although the trend of mitotic index (higher during expansion and lower during homeostasis) was similar, the absolute value of the mitotic index in the developing larvae was greater than that of the regenerating adult germ line. The upper limit on the mitotic index of adult germ cells in this assay might reflect a developmental regulation or other aspects of the post-ARD adult germ line. However, collectively, these results show that similar to the larval expansion phase, the regenerating germ line mitotic index responds to feeding and then declines once the germ line is regrown.
Finally, we examined DNA content at the 0, 6, 24, 48, 72, and 96 h post-ARD re-feeding time points. We observed a relatively high and similar pooled average DNA content among nuclei in the 0, 6, and 24 h time points (Fig. 4E, F). Surprisingly, at the 48 h time point, we observed a significant reduction in average DNA content, which persisted 72 and 96 h time points.
When analyzed using a regression model (Table S8; Materials and methods), the interaction of CD position and time point during post-ARD recovery was not significant based on the contribution of the interaction to the model (likelihood ratio test Chi-Square = 15.39, p-value = 0.35). In addition, similar to the larval wild-type, comparing the DNA content by CD position showed that for lower CD positions (≤ 15 CD), DNA content did not deviate significantly from the ≤ 5 CD range. However, more similar to the adult, the region > 15 CD significantly predicted a higher DNA content category (odds ratio, OR: 2.39, 95% CI: 1.69–3.82). Surprisingly, later time points predict lower DNA content: DNA content in the 48 h and the 96 h groups were significantly lower relative to the 0 time point with odds ratios of 0.45 (95% CI: 0.24–0.85) and 0.36 (95% CI:0.18–0.74), respectively.
In summary, while cell accumulation and mitotic index among post-ARD re-fed germ cells are reminiscent of larval expansion, average DNA content and the spatial effect of distance from the distal tip were not re-capitulated in the regenerating adult germ line. Further, despite access to the same food source, the proliferative zone of the regenerating adult germ line did not accumulate to the same cell numbers nor sustain the same mitotic index as it would have done under continuously fed conditions.
4. Discussion
Here, we report mitotic index, proliferative cell numbers and DNA content over a variety of age, genetic and physiological conditions. We found that germ cells in late larval stages display a higher mitotic index than in the adult and that this difference cannot be attributed to sperm versus oocyte germ cell fate. In addition, we found that the average DNA content of proliferative nuclei is affected by age and genotype. Extending the Irises tool, we examined DNA content as a function of distance from the distal tip, and found that in the wild type, larvae display a uniform distribution while adults display a lower average DNA content distally. This spatial pattern was also influenced by genotype. Finally, we characterized growth of the adult proliferative zone following post-ARD re-feeding. We found that while accumulation of proliferative nuclei and mitotic index followed trends similar to larval into adult growth, the overall DNA content did not recapitulate the developmental scenario.
Our analysis of the temporal and spatial patterns of DNA content resolves several discrepancies in the literature. First, our comparison of larval and adult stages in this study suggest that much (though not all) of the difference in proportions of nuclei in early interphase reported by Michaelson et al. (2010) (Michaelson et al., 2010) versus Fox et al. (2011) (Fox et al., 2011) can be attributed to the different stages at which animals were examined (larval versus adult) in the two studies. Second, an apparent inconsistency between previously published results within the adult stage is resolved by comparing DNA content over the distal-to-proximal axis. Feng et al. (1999) (Feng et al., 1999) reported a greater percentage of early interphase in the young adult than did Fox et al. (2011) (Fox et al., 2011). However, Feng et al. (1999) measured only the distal-most 5 CD while Fox et al. (2011) measured the entire pool of REC-8-positive proliferative zone cells. Therefore, the latter data set includes proximal cells that, we find, have a higher average DNA content. In addition, the results from Fox et al. (2011) include the proximal-most REC-8-positive cells (in pre-meiotic S) that uniformly display high DNA content. Although we did not include the proximal-most 2 rows of cells in our study (see Materials and methods), we still observe a higher DNA content in proximal regions.
The combination of total proliferative cell number (accumulation), mitotic index, and DNA content among different mutants relative to wild type may help define phenotypic signatures for different functional categories. With respect to cell fate, reducing glp-1 activity causes germ cells to enter meiosis at the expense of maintaining undifferentiated cells in the proliferative zone (Kershner et al., 2013). Consistent with a more prominent role in cell fate than cell cycle control, at semi-permissive temperatures, reduced glp-1 activity does not alter larval mitotic index but the number of proliferative cells in the adult is reduced relative to wild type (Fox and Schedl, 2015; Korta et al., 2012; Michaelson et al., 2010). Here we show that in the glp-1 mutant, neither larval nor adult mitotic index differs from the wild type (p > 0.05, see Fig. 2), and that the adult pooled average DNA content is significantly lower in the mutant than in the wild type (p < 0.0001, see Fig. 2). While no one measure is sufficient to suggest a role in cell fate versus cell cycle, we speculate that a phenotypic combination of reduced proliferative zone cell number, unchanged mitotic index, and reduced average adult DNA content (all with respect to the wild type) may be a signature for mutants that primarily affect cell fate. Another aspect of this signature may be the influence of distance from the distal tip on DNA content in larval stages. We found that unlike the wild type, the DNA content of glp-1 larval germ cells is elevated with distance from the distal end. This result is consistent with the idea that lower levels of glp-1 effectively force a meiotic entry threshold at a shorter distance from the distal tip, which then displays features more similar to the wild-type adult at a further distance from the distal end (see Fox and Schedl, 2015).
With respect to cell cycle, reducing daf-2 or rsks-1 impairs the accumulation of proliferative cells through larval stages and causes a reduced mitotic index in larvae, but not in adults, relative to wild type. These mutations also elevated the average DNA content in larvae, suggesting a normal role for daf-2 and rsks-1 in promoting progression through S or G2 and/or in slowing progression through M or G1. Here we speculate that a phenotypic combination of reduced proliferative zone cell number, reduced larval mitotic index, and unchanged average adult DNA content (all with respect to the wild type) may be a signature for mutants that primarily affect cell cycle.
How the relative DNA content relates to mitotic index is not simple. Our results and those of others indicate that, on average, larval germ cells cycle faster than adult (Gerhold et al., 2015; Korta et al., 2012). In addition, here we show that on average, larval germ cells contain lower DNA content. One simple idea is that the faster-cycling cells spend more time in the G1/early S. However, this simple relationship does not hold once the DNA content is examined as a function of distance from the distal tip. The nuclei in the distal-most 1–3 CD of the adult display a reduced mitotic index (Crittenden et al., 2006; Fox et al., 2011; Jaramillo-Lambert et al., 2007; Maciejowski et al., 2006) and, as shown here, a lower DNA content. This may imply additional input close to the DTC body (see below).
Two very recent studies address proliferating germ cell dynamics in the adult under continuous feeding conditions: Fox and Schedl (2015) and Chiang et al. (2015) (Chiang et al., 2015; Fox and Schedl, 2015). Building on previous results (Fox et al., 2011) and on extensive temperature shift and S-phase labeling experiments Fox and Schedl (2015) propose a model for adult proliferative cell behavior in which cells in the first 10 CD experience a high (over-threshold) level of GLP-1 activity. Once falling below threshold, they propose that cells complete the ongoing mitotic cell cycle and enter the meiotic entry pathway. Similar to our study, but using different methods, Chiang et al. (2015) investigate the spatial pattern of DNA content. Their study corroborates ours in that they find higher DNA content furthest from the DTC. Similar to our studies, they also observe a difference in DNA content starting at CD 6. Therefore, both our study and that of Chiang et al. (2015) suggest that cells within the first (distal-most) 5 CD display differences in average DNA content compared to the next 5 CD. The distal-most cells (1–5 CD) experience extensive physical contact with the DTC body, including gap junctions that are critical for maintenance of the proliferative zone (Starich et al., 2014). One possibility is that in addition to signals from the DTC that activate GLP-1, extensive DTC-germ cell contact confers additional regulation. However, this difference in average DNA content may not appreciably affect the ability of individual distal-most cells to enter meiosis when glp-1 activity is withdrawn, as shown by Fox and Schedl (2015). Simultaneous evaluation of DNA content and differentiation status on a nucleus-by-nucleus basis would be required to resolve this apparent contradiction.
Finally, we analyzed adult germline regeneration. First we note that the regenerating germline is fragile, possibly limiting certain forms of data collection. Our time-course analysis of proliferative zone accumulation revealed a short lag, followed by a two-phase accumulation, the first short and robust and the second longer and slower. The lag indicates that the first cell cycle out of ARD may be slower than subsequent cell cycles or perhaps that time may be required for asynchronously arrested cells to all enter the cell cycle. Nevertheless, the proliferative zone cell count never reaches that of the continuously-fed adult. Our analysis of mitotic index during germline regeneration suggests that availability of space and food may also influence germ cell proliferation dynamics. We found that, similar to the larval expansion phase, cells in early stages of post-feeding regeneration exhibit a relatively high mitotic index that declines upon reestablishment of the mature germ line. The possible influence of a spatial constraint is implied by a recent modeling study (Atwell et al., 2015). Our DNA content analysis was surprising. If, as we hypothesized, larval expansion followed by adult maintenance were recapitulated in the post-ARD re-feeding scenario, we would have expected that the overall average DNA content would be higher over time. However, this is not what was observed. Instead we saw a greater proportion of nuclei with a higher overall average DNA content in the early stages of ARD recovery compared to the later maintenance phase. These differences may reflect the effects of one or more factors that are altered during ARD and recovery compared to development under continuously fed conditions. These include starvation (and consequent stress responses and changes in metabolism), age, persistence of sperm, and cell death. For example, post-ARD re-fed worms are already 5-day old adults and the majority still contain sperm. This combination is not observed in continuously fed adults. Taken together, our results suggest that while cell cycle dynamics during post-ARD regeneration share certain features with expansion and maturation of the well-fed germ line, the dynamics are also influenced by developmental stage-specific cues and altered life-history.
Supplementary Material
Acknowledgments
The authors acknowledge support from NIH R01 GM061706 to EJAH, NYSTEM Training Grant contract #C026880, GM097576 and HD075602 to ZB, and the NYU CTSA grant UL1 TR000038 from the NIH National Center for Advancing Translational Sciences for statistical support and NCI Cancer Center Support Grant P30 CA016087-32 (JDG). Some strains were obtained from the Caenorhabditis Genetics Center that is supported by the NIH Office of Research Infrastructure Programs P40 OD010440.
Footnotes
Appendix A. Supplementary material
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.ydbio.2015.10.031.
References
- Ambros V. Cell cycle-dependent sequencing of cell fate decisions in Caenorhabditis elegans vulva precursor cells. Development. 1999;126:1947–1956. doi: 10.1242/dev.126.9.1947. [DOI] [PubMed] [Google Scholar]
- Angelo G, Van Gilst MR. Starvation protects germline stem cells and extends reproductive longevity in C. elegans. Science. 2009;326:954–958. doi: 10.1126/science.1178343. [DOI] [PubMed] [Google Scholar]
- Atwell K, Qin Z, Gavaghan D, Kugler H, Hubbard EJA, Osborne JM. Mechano-logical model of C. elegans germ line suggests feedback on the cell cycle. Development. 2015 doi: 10.1242/dev.126359. http://dx.doi.org/10.1242/dev.126359 (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariz M, Mainpal R, Subramaniam K. C. elegans RNA-binding proteins PUF-8 and MEX-3 function redundantly to promote germline stem cell mitosis. Dev. Biol. 2009;326:295–304. doi: 10.1016/j.ydbio.2008.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beanan MJ, Strome S. Characterization of a germ-line proliferation mutation in C. elegans. Development. 1992;116:755–766. doi: 10.1242/dev.116.3.755. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd DT, Knobel K, Affeldt K, Crittenden SL, Kimble J. A DTC Niche Plexus Surrounds the Germline Stem Cell Pool in Caenorhabditis elegans. Plos ONE. 2014;9:e88372. doi: 10.1371/journal.pone.0088372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang M, Cinquin A, Paz A, Meeds E, Price CA, Welling M, Cinquin O. Control of C. elegans germline stem cell cycling speed meets requirements of design to minimize mutation accumulation. BMC Biol. 2015;13:51. doi: 10.1186/s12915-015-0148-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinquin O, Crittenden SL, Morgan DE, Kimble J. Progression from a stem cell-like state to early differentiation in the C. elegans germ line. Proc. Natl. Acad. Sci. 2010;107:2048–2053. doi: 10.1073/pnas.0912704107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crittenden SL, Leonhard KA, Byrd DT, Kimble J. Cellular analyses of the mitotic region in the Caenorhabditis elegans adult germ line. Mol. Biol. Cell. 2006;17:3051–3061. doi: 10.1091/mbc.E06-03-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H, Zhong W, Punkosdy G, Gu S, Zhou L, Seabolt EK, Kipreos ET. CUL-2 is required for the G1-to-S-phase transition and mitotic chromosome condensation in Caenorhabditis elegans. Nat. Cell. Biol. 1999;1:486–492. doi: 10.1038/70272. [DOI] [PubMed] [Google Scholar]
- Fitzmaurice GM, Laird NM, Ware JH. Applied Longitudinal Analysis. 2nd. Hoboken, NJ: John Wiley & Sons; 2011. [Google Scholar]
- Fox PM, Schedl T. Analysis of Germline Stem Cell Differentiation Following Loss of GLP-1 Notch Activity in Caenorhabditis elegans. Genetics. 2015;201:167–184. doi: 10.1534/genetics.115.178061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox PM, Vought VE, Hanazawa M, Lee M-H, Maine EM, Schedl T. Cyclin E and CDK-2 regulate proliferative cell fate and cell cycle progression in the C. elegans germline. Development. 2011;138:2223–2234. doi: 10.1242/dev.059535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhold AR, Ryan J, Vallée-Trudeau J-N, Dorn JF, Labbé J-C, Maddox PS. Investigating the Regulation of Stem and Progenitor Cell Mitotic Progression by In Situ Imaging. Curr. Biol.: CB. 2015:1–13. doi: 10.1016/j.cub.2015.02.054. [DOI] [PubMed] [Google Scholar]
- Hansen D, Hubbard EJA, Schedl T. Multi-pathway control of the proliferation versus meiotic development decision in the Caenorhabditis elegans germline. Dev. Biol. 2004;268:342–357. doi: 10.1016/j.ydbio.2003.12.023. [DOI] [PubMed] [Google Scholar]
- Hansen D, Pilgrim D. Sex and the single worm: sex determination in the nematode C. elegans. Mech. Dev. 1999;83:3–15. doi: 10.1016/s0925-4773(99)00024-6. [DOI] [PubMed] [Google Scholar]
- Hansen D, Schedl T. Stem cell proliferation versus meiotic fate decision in Caenorhabditis elegans. Adv. Exp. Med. Biol. 2013;757:71–99. doi: 10.1007/978-1-4614-4015-4_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson ST, Gao D, Lambie EJ, Kimble J. lag-2 may encode a signaling ligand for the GLP-1 and LIN-12 receptors of C. elegans. Development. 1994;120:2913–2924. doi: 10.1242/dev.120.10.2913. [DOI] [PubMed] [Google Scholar]
- Hirsh D, Oppenheim D, Klass M. Development of the reproductive system of Caenorhabditis elegans. Dev. Biol. 1976;49:200–219. doi: 10.1016/0012-1606(76)90267-0. [DOI] [PubMed] [Google Scholar]
- Hubbard EJA, Korta DZ, Dalfó D. Physiological Control of Germline Development. New York, New York, NY: Springer; 2012. pp. 101–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaramillo-Lambert A, Ellefson M, Villeneuve AM, Engebrecht J. Differential timing of S phases, X chromosome replication, and meiotic prophase in the C. elegans germ line. Dev. Biol. 2007;308:206–221. doi: 10.1016/j.ydbio.2007.05.019. [DOI] [PubMed] [Google Scholar]
- Kadyk LC, Lambie EJ, Kimble J. glp-3 is required for mitosis and meiosis in the Caenorhabditis elegans germ line. Genetics. 1997;145:111–121. doi: 10.1093/genetics/145.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kershner A, Crittenden SL, Friend K, Sorensen EB, Porter DF, Kimble J. Germline Stem Cells and Their Regulation in the Nematode Caenorhabditis elegans. Springer Netherlands: Dordrecht; 2013. pp. 29–46. [DOI] [PubMed] [Google Scholar]
- Killian DJ, Hubbard EJA. C. elegans pro-1 activity is required for soma/germline interactions that influence proliferation and differentiation in the germ line. Development. 2004;131:1267–1278. doi: 10.1242/dev.01002. [DOI] [PubMed] [Google Scholar]
- Killian DJ, Hubbard EJA. Caenorhabditis elegans germline patterning requires coordinated development of the somatic gonadal sheath and the germ line. Dev. Biol. 2005;279:322–335. doi: 10.1016/j.ydbio.2004.12.021. [DOI] [PubMed] [Google Scholar]
- Kimble JE, White JG. On the control of germ cell development in Caenorhabditis elegans. Dev. Biol. 1981;81(2):208–219. doi: 10.1016/0012-1606(81)90284-0. [DOI] [PubMed] [Google Scholar]
- Korta DZ, Tuck S, Hubbard EJA. S6K links cell fate, cell cycle and nutrient response in C. elegans germline stem/progenitor cells. Development. 2012;139:859–870. doi: 10.1242/dev.074047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudron MM, Reinke V. C. elegans nucleostemin is required for larval growth and germline stem cell division. Plos Genet. 2008;4:e1000181. doi: 10.1371/journal.pgen.1000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciejowski J, Ahn JH, Cipriani PG, Killian DJ, Chaudhary AL, Lee JI, Voutev R, Johnsen RC, Baillie DL, Gunsalus KC, Fitch DHA, Hubbard EJA. Autosomal genes of autosomal/X-linked duplicated gene pairs and germ-line proliferation in Caenorhabditis elegans. Genetics. 2005;169:1997–2011. doi: 10.1534/genetics.104.040121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciejowski J, Ugel N, Mishra B, Isopi M, Hubbard EJA. Quantitative analysis of germline mitosis in adult C. elegans. Dev. Biol. 2006;292:142–151. doi: 10.1016/j.ydbio.2005.12.046. [DOI] [PubMed] [Google Scholar]
- Michaelson D, Korta DZ, Capua Y, Hubbard EJA. Insulin signaling promotes germline proliferation in C. elegans. Development. 2010;137:671–680. doi: 10.1242/dev.042523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadarajan S, Govindan JA, McGovern M, Hubbard EJA, Greenstein D. MSP and GLP-1/Notch signaling coordinately regulate actomyosin-dependent cytoplasmic streaming and oocyte growth in C. elegans. Development. 2009;136:2223–2234. doi: 10.1242/dev.034603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusser-Stein S, Beyer A, Rimann I, Adamczyk M, Piterman N, Hajnal A, Fisher J. Cell-cycle regulation of NOTCH signaling during C. elegans vulval development. Mol. Syst. Biol. 2012;8:1–14. doi: 10.1038/msb.2012.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepper AS-R, Killian DJ, Hubbard EJA. Genetic analysis of Caenorhabditis elegans glp-1 mutants suggests receptor interaction or competition. Genetics. 2003;163:115–132. doi: 10.1093/genetics/163.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel HS, Kimble J. The oogenic germline starvation response in C. elegans. Plos ONE. 2011;6:e28074. doi: 10.1371/journal.pone.0028074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydoux G, Savage C, Greenwald I. Isolation and characterization of mutations causing abnormal eversion of the vulva in Caenorhabditis elegans. Dev. Biol. 1993;157:423–436. doi: 10.1006/dbio.1993.1146. [DOI] [PubMed] [Google Scholar]
- Starich TA, Hall DH, Greenstein D. Two classes of gap junction channels mediate soma-germline interactions essential for germline proliferation and gametogenesis in Caenorhabditis elegans. Genetics. 2014;198:1127–1153. doi: 10.1534/genetics.114.168815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiernagle T. Maintenance of C. elegans. In: WormBook, editor. The C. elegans Research Community. WormBook; 2006. Feb 11, http://dx.doi.org/10.1895/wormbook.1.101.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JLM, Michaelson D, Santella A, Hubbard EJA, Bao Z. Irises: A practical tool for image-based analysis of cellular DNA content. Worm. 2014;3:e29041. doi: 10.4161/worm.29041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voutev R, Killian DJ, Ahn JH, Hubbard EJA. Alterations in ribosome biogenesis cause specific defects in C. elegans hermaphrodite gonadogenesis. Dev. Biol. 2006;298:45–58. doi: 10.1016/j.ydbio.2006.06.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.