Abstract
Background: Studies have demonstrated an association of the BRAFV600E mutation and microRNA (miR) expression with aggressive clinicopathologic features in papillary thyroid cancer (PTC). Analysis of BRAFV600E mutations with miR expression data may improve perioperative decision making for patients with PTC, specifically in identifying patients harboring central lymph node metastases (CLNM).
Methods: Between January 2012 and June 2013, 237 consecutive patients underwent total thyroidectomy and prophylactic central lymph node dissection (CLND) at four endocrine surgery centers. All tumors were tested for the presence of the BRAFV600E mutation and miR-21, miR-146b-3p, miR-146b-5p, miR-204, miR-221, miR-222, and miR-375 expression. Bivariate and multivariable analyses were performed to examine associations between molecular markers and aggressive clinicopathologic features of PTC.
Results: Multivariable logistic regression analysis of all clinicopathologic features found miR-146b-3p and miR-146b-5p to be independent predictors of CLNM, while the presence of BRAFV600E almost reached significance. Multivariable logistic regression analysis limited to only predictors available preoperatively (molecular markers, age, sex, and tumor size) found miR-146b-3p, miR-146b-5p, miR-222, and BRAFV600E mutation to predict CLNM independently. While BRAFV600E was found to be associated with CLNM (48% mutated in node-positive cases vs. 28% mutated in node-negative cases), its positive and negative predictive values (48% and 72%, respectively) limit its clinical utility as a stand-alone marker. In the subgroup analysis focusing on only classical variant of PTC cases (CVPTC), undergoing prophylactic lymph node dissection, multivariable logistic regression analysis found only miR-146b-5p and miR-222 to be independent predictors of CLNM, while BRAFV600E was not significantly associated with CLNM.
Conclusion: In the patients undergoing prophylactic CLNDs, miR-146b-3p, miR-146b-5p, and miR-222 were found to be predictive of CLNM preoperatively. However, there was significant overlap in expression of these miRs in the two outcome groups. The BRAFV600E mutation, while being a marker of CLNM when considering only preoperative variables among all histological subtypes, is likely not a useful stand-alone marker clinically because the difference between node-positive and node-negative cases was small. Furthermore, it lost significance when examining only CVPTC. Overall, our results speak to the concept and interpretation of statistical significance versus actual applicability of molecular markers, raising questions about their clinical usefulness as individual prognostic markers.
Introduction
The incidence of thyroid cancer worldwide has risen significantly in the last decade and papillary thyroid cancer (PTC) is the most prevalent histological type, comprising approximately 80% of reported cases (1,2). Overall, the majority of patients have an excellent clinical outcome when treated with appropriate surgical and medical therapy (3). However, 5–10% experience a more aggressive clinical course, characterized by recurrent disease and early metastases, resistance to radioactive iodine, and increased mortality (3,4). Thyroidectomy and therapeutic neck dissection is recommended for patients with known metastatic lymph nodes identified either pre- or intraoperatively (3). However, the benefit of prophylactic central lymph node dissection (CLND) in patients who have no pre- or intraoperative evidence of nodal metastasis remains controversial (3,5,6). Some argue that CLND may reduce the recurrence rate of lymph node metastasis, while others claim that patients receive no benefit, yet experience increased risk of permanent hypoparathyroidism and recurrent laryngeal nerve injury (6–8). Furthermore, currently available preoperative features have not accurately identified PTCs that are associated with lymph node metastases (LNM) (9), making it difficult to tailor the extent of surgery (3) and specifically whether to perform a prophylactic CLND (10).
The BRAFV600E mutation is the most common genetic mutation in PTC (11,12). The p.BRAFV600E mutation is caused by a c.1799 T > A transversion that results in a constitutively activated mitogen-activated protein kinase (MAPK) pathway, leading to tumorigenesis (13) and distinct biological consequences (14). The prevalence of the BRAFV600E mutation varies between the different histological subtypes of PTC (12). The tall-cell variant of PTC (TCVPTC) has the highest prevalence at 93% (15), followed by classical variant of PTC (CVPTC), comprising up to 75.3% (11), and lastly, follicular variant of PTC (FVPTC) at only 25% (16). Some studies have shown that the BRAFV600E mutation is associated with aggressive clinicopathologic features, such as extrathyroidal invasion, multifocality, lymphovascular invasion, large tumor size, local lymph node metastasis, distant metastasis, and advanced disease stages (17–22), whereas others studies have demonstrated no association between the BRAFV600E mutation and aggressive features (23–30). In a previous retrospective study, the BRAFV600E mutation was found to be an independent predictor of CLNM in the overall cohort of patients with PTC (23). However, this relationship lost significance when only CVPTC was included in the analysis (23). Despite the discrepancies that exist in the literature, some studies have suggested the incorporation of BRAFV600E mutation into the management algorithm of PTC to improve perioperative decision making (9,17,18,20,22). However, the usefulness of BRAFV600E in predicting the presence of LNM remains questionable (23,31).
The recent expansion of knowledge and the ongoing effort to characterize PTCs genetically by The Cancer Genome Atlas (TCGA) research thyroid working group have also suggested that micro-RNAs may influence the PTC phenotype and thereby play an important role in its prognosis (12). MicroRNAs (miRs) are small non-protein coding RNA molecules that are 21–25 nucleotides in length. They regulate gene expression at the post-transcriptional level by binding to imperfectly complementary sequences within target mRNAs, often in the 3′-untranslated regions, thereby leading to degradation or translational suppression (32). Although definitive studies are currently lacking, miRs may nevertheless have potential as prognostic indicators in PTC, thereby optimizing the surgical management of patients with PTC (28,33–44).
The BRAFV600E mutation and miRs have the potential to guide surgical decision making. However, most previous studies were retrospective, and did not include patients who had undergone routine CLND, challenging the validity of the findings. This underscores the need for well-designed prospective studies involving routine CLND to investigate whether the BRAFV600E mutation and miRs are independent predictors of aggressive clinicopathologic features before it can be recommended that these molecular markers be incorporated into the management algorithm for patients with PTC.
Accordingly, a multi-institutional prospective study was carried out that included consecutive patients who underwent total thyroidectomy (TT) and routine CLND at four tertiary endocrine surgery centers between January 1, 2012, and June 30, 2013. The objective was to determine, by multivariable analysis, whether the BRAFV600E mutation and miR expression levels of seven selected miRs are associated with aggressive clinicopathologic features of PTC. Their association with CLNM was examined in the subset of patients undergoing prophylactic rather than therapeutic CLND, in order to explore their potential future value as factors to assist in guiding the treatment of patients with PTC, specifically whether to perform prophylactic CLND in the absence of clinically apparent LNM.
Material and Methods
Patients and data collection
Under Institutional Review Board approval and patient consent, a prospective multi-institutional study was conducted that included Johns Hopkins Hospital, Weill Cornell Medical Center, the University of Michigan Health Systems, and the Mayo Clinic. This study was designed as validation of an initial retrospective pilot study (23). Consecutive patients between January 1, 2012, and June 30, 2013, diagnosed with PTC preoperatively on cytopathology or intraoperatively on frozen section and with a tumor size ≥1 cm were prospectively enrolled and included. Patients were operated on by surgeons who routinely perform TT and prophylactic CLND for patients with PTC ≥1 cm. At all centers, a unilateral CLND was performed, as described by Carty et al. (45), unless there was evidence of positive disease midline or contra-laterally in which case bilateral dissection was performed. Patients with suspicious or cytologically positive lymph node metastasis detected on ultrasound or by intraoperative evaluation underwent therapeutic CLND and were excluded, whereas those patients that did not exhibit signs of lymphadenopathy either pre- or intraoperatively, by definition, underwent a prophylactic CLND.
Clinical variables examined included patient age, sex, history of radiation exposure, and family history of thyroid cancer. Surgical pathology reports were reviewed for the following: tumor size, PTC subtype, presence LNM, extrathyroidal extension (ETE), multifocality, lymphovascular invasion (LVI), involvement of surgical margins, and American Joint Committee on Cancer (AJCC) stage.
Nucleic acid extraction
All molecular analyses were performed at Johns Hopkins Hospital. DNA and total RNA were extracted from formalin-fixed paraffin-embedded (FFPE) tumor samples, according to the protocol outlined by Kotorashvili et al. (46). Final DNA and RNA quantitation was determined by absorbance and fluorescence spectroscopy–based nucleic acid quantification using NanoDrop 2000c UV-Vis Spectrophotometer (Thermo Scientific) and Qubit® Fluorometric Quantitation (Life Technologies). PTC subtype and follicular and tall-cell architecture percentage was reviewed and confirmed by a single head and neck/endocrine pathologist (J.A.B.). Tumors were classified as FVPTC if they were 99% follicular patterned, and as TCVPTC if they had ≥50% tall-cell features.
BRAFV600E mutation detection
Detection of the BRAFV600E mutation was performed in all cases by pyrosequencing. Polymerase chain reaction (PCR) amplification of a 124-base region of BRAF exon 15 surrounding c.1799T was performed using 20–200 ng genomic DNA and 0.2 M 5′-GAA GAC CTC ACA GTA AAA ATA G-3′ (forward) and 5′-biotin-ATA GCC TCA ATT CTT ACC ATC C-3′ (reverse) primers, along with HotStarTaq DNA polymerase (Qiagen), according to the manufacturer's instructions. Thermocycling parameters were as follows: 95°C for 15 min, followed by 42 cycles of 95°C for 20 sec, 53°C for 50 sec, 72°C for 20 sec, followed by 72°C for 5 min. Pyrosequencing was performed using the Pyromark-24 and sequencing reagents (Qiagen), according to the manufacturer's instructions, with the sequencing primer 5′-GAC CTC ACA GTA AAA ATA GGT GAT TTT G-3′. The pyrosequencing assay used was designed to detect all reported COSMIC (catalog of somatic mutations in cancer; http://cancer.sanger.ac.uk/cosmic/) mutations from 596 to 605, including K601E, K601I, K601L, K601N, K601Q, and K601R mutations. Samples with low pyrosequencing signals were classified as indeterminate and re-amplified using 52 cycles. Positive, negative, and no-template controls were run with each sample. BRAFV600E mutations (c.1799T>A, GTG>GAG) were diagnosed as positive based on the appearance of an A in the chromatogram and a decrease of the wild-type T peak with a mutant peak cutoff of 5%. This cutoff was based on testing of normal colon and normal thyroid FFPE samples, which displayed percentages in the range 0–3%. More complex results were resolved using a combination of Sanger sequencing and Pyromaker software (23,47). The CLIA-approved laboratory in which the pyrosequencing was performed has run large series of validation experiments against both Sanger sequencing and next-generation sequencing (NGS) protocols, with 100% agreement among all three methods. In addition, postoperative molecular analysis for BRAFV600E mutation is known to be accurate, with several studies documenting concordance of molecular analysis in FNA and corresponding tumors (48,49).
miR analysis
Promising miR candidates for predicting aggressiveness were selected based on results from the TCGA thyroid study (12) and a recent systematic review of the literature (50). MiRs-21, -146-3p, -146-5p, -204, -221, -222, and -375 were measured along with candidate reference miR-16 in technical triplicates. Reverse transcription for each specific miR was performed using 2 μL (10 ng/μL) of total RNA for a total RT-PCR reaction of 12 μL. For the qPCR step, cDNA was diluted with 20 μL of water, and 2 μL of diluted cDNA added to 10 μL of master mix for total volume of 12 μL per qPCR reaction (TaqMan® MicroRNA Reverse Transcription kit, TaqMan® Universal Master Mix II, not containing Uracil-N glycosylase [UNG], and TaqMan® MicroRNA assays; Life Technologies). MicroRNA TaqMan® assays and their part numbers from Life Technologies were: miR-21-5p (000397), miR-146b-5p (001097), miR-146b-3p (002361), miR-204 (000508), miR-221 (000524), miR-222 (002276), miR-375 (000564), and miR-16-5p (000391).
In order to minimize batch effects, samples from all contributing centers were randomized on the microtiter plates. Mean Ct values were calculated for each sample and normalized against miR-16 to obtain dCT values. MiR-16-5p was used as endogenous control to normalize miR expression levels based on prior publications (43,51–55) as well as available TCGA thyroid data, where miR-16 appears as relatively invariant within thyroid tumor samples (12). Mean dCt values relative to expression of miR-16 were normally distributed in the data set.
Data analysis
Power calculation
The study sample size was calculated based upon the proportional prevalence of BRAFV600E mutations and the prevalence of lymph node metastases in patients treated for PTC reported in previous studies. The BRAFV600E mutation has been reported in 29–83% of patients with PTC overall (56), with the smallest proportion (29%) reported by Namba et al. (57). To be conservative, this sample-size calculation assumes that the BRAFV600E mutation will be present in 25% of study subjects overall. Based upon the assumption that LNM will be present in 18% of BRAFV600E-negative individuals and 38% of those who are BRAFV600E positive (58), a sample size of 224 individuals was required to examine the relationship between BRAFV600E status and lymph node metastasis with 80% power at an alpha level of 0.05 (two sided).
Statistical analysis
In order to explore the relationships between molecular markers, BRAFV600E and miRs, and routine clinicopathologic features of PTC, a univariate analysis was performed on patient sex, age, tumor size, multifocality, LVI, involvement of surgical margins, ETE, CLNM, advanced AJCC stage, histological subtype, and follicular and tall-cell component percentage. In order to find significant molecular predictors of CLNM, a multivariable logistic regression analysis was performed, adjusting for established clinicopathologic predictors, including patient age ≥45 years, sex, tumor size >2 cm, multifocality, LVI, involvement of surgical margins, ETE, and follicular and tall-cell component. Since relative levels of expression of the studied miRs were highly correlated with each other, these analyses were performed examining one miR at a time. In addition, multivariable analysis was performed examining each molecular marker individually controlling for factors that potentially would have been available preoperatively, including patient age, sex, tumor size, BRAFV600E mutation, and miR expression. To explore the possibility that variability in miR expression might be associated with the likelihood of CLNM in patients without indication of CLN involvement, only those patients who underwent prophylactic CLND both across the entire cohort and within the subset of patients diagnosed with CVPTC alone were examined. Since PTCs classified as CVPTC may harbor up to 98% follicular and up to 49% tall-cell components, the possibility that the follicular and tall-cell component percentages presented an influence in the CVPTC subgroup was also explored, and the respective percentages were included in the CVPTC analysis. Finally, statistical analyses for the entire cohort, including those who underwent therapeutic CLND, were performed and are presented in the Supplementary Data (Supplementary Data are available online at www.liebertpub.com/thy). All statistical analyses were conducted using R v0.98.507 (http://cran.r-project.org). A p-value of <0.05 was defined as statistically significant.
Results
Demographics
A total of 237 patients underwent prophylactic CLND and were included in the study. The prophylactic cohort of 237 patients included 173 women (73%) and 64 men (27%) with a mean age of 47.7 ± 14.5 years and a mean tumor size of 2 ± 1.2 cm. Multifocality was observed in 95 (40%) cases, involvement of surgical margins in 34 (14%) cases, LVI in 30 (13%) cases, ETE in 55 (23%) cases, and CLNM in 101 (43%) cases. One hundred and seventy (72%) tumors were positive for the BRAFV600E mutation. In three cases (1.3%), the results were equivocal for at least one of the molecular analyses (BRAFV600E mutation or miR expression); these were therefore excluded from those specific analyses. One hundred and ninety-two cases had CVPTC (81%), 27 had FVPTC (11%), and 18 had TCVPTC (8%; Table 1).
Table 1.
Patient Characteristics
| Clinicopathologic features | n = 237 (%) |
|---|---|
| Male | 64 (27) |
| Female | 173 (73) |
| Age at diagnosis (years), mean ± SD | 48 ± 15 |
| Tumor size (cm), mean ± SD | 2 ± 1.2 |
| Lymph node metastasis | |
| Central | 101 (43) |
| Total number of CLN dissected | 7.9 ± 6.0 |
| Positive number of CLN dissecteda | 3.9 ± 3.3 |
| Extrathyroidal extension | 55 (23) |
| Multifocality | 95 (40) |
| Lymphovascular invasion | 30 (13) |
| Involvement of surgical margins | 34 (14) |
| AJCC | |
| Stage I | 147 (62) |
| Stage II | 10 (4) |
| Stage III | 77 (33) |
| Stage IV | 3 (1) |
| Subtype | |
| Classical | 192 (81) |
| Follicular | 27 (11) |
| Tall cell | 18 (8) |
| BRAF mutation positive, n (%) | 170 (72) |
| Classical | 149 (78) |
| Follicular | 6 (22) |
| Tall cell | 15 (83) |
| microRNAs,b mean (SD) | |
| 21 | −0.50 (1.97) |
| 146b-3p | −7.75 (1.97) |
| 146b-5p | −0.12 (1.71) |
| 204 | −7.91 (1.80) |
| 221 | −2.27 (1.78) |
| 222 | 0.09 (1.35) |
| 375 | −6.30 (2.21) |
Only includes those patients with positive disease.
Inverse dCt values relative to miR-16.
AJCC, American Joint Committee on Cancer; CLN, central lymph node; SD, standard deviation.
Predictors of CLNM in all PTC subtypes
Univariate logistic regression analyses in patients who underwent prophylactic neck dissection demonstrated significant positive association between the BRAFV600E mutation (p = 0.01), miR-146b-3p (p = 0.01), miR-146b-5p (p < 0.01), miR-221 (p = 0.04), miR-222 (p < 0.01), tumor size (p < 0.01), multifocality (p = 0.01), positive surgical margins (p < 0.01), LVI (p < 0.01), ETE (p < 0.01), and advanced AJCC stage (p < 0.01) with CLNM. Multivariable logistic regression analysis (Table 2), controlling for each molecular marker, sex, age, tumor size, multifocality, LVI, positive surgical margins, ETE, and histological subtypes, found only miR-146b-3p (p = 0.03), miR-146b-5p (p = 0.02), multifocality (p < 0.05), LVI (p < 0.01), and ETE (p < 0.01) to be independent predictors of CLNM. The BRAFV600E mutation (p = 0.05) was only found to have a borderline association with CLNM in this analysis (Table 2).
Table 2.
Multivariate Analysis of Individual Molecular Marker with Postoperative Clinicopathologic Variables in Prophylactic PTCs (n = 237)
| Statistic of molecular marker | Significance of clinicopathologic factors (p-value only) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular marker | RR | CI | p-Value | Sex | Age (≥45 years) | Size (>2 cm) | Multifocality | LVI | ETE | Margins | FVPTC | TCVPTC |
| BRAFV600E | 1.55 | 0.99–2.41 | 0.05 | 0.72 | 0.25 | 0.29 | 0.83 | 0.71 | 0.05 | <0.01* | <0.01* | 0.48 |
| miR-21 | 1.00 | 0.90–1.12 | 1.00 | 0.74 | 0.53 | 0.28 | 0.31 | 0.82 | 0.02* | <0.01* | <0.01* | 0.36 |
| miR-146-3p | 1.09 | 1.01–1.18 | 0.03* | 0.41 | 0.27 | 0.17 | 0.69 | 0.79 | 0.06 | <0.01* | <0.01* | 0.21 |
| miR-146-5p | 1.12 | 1.02–1.24 | 0.02* | 0.54 | 0.15 | 0.20 | 0.79 | 0.75 | 0.02* | <0.01* | <0.01* | 0.44 |
| miR-204 | 0.99 | 0.91–1.07 | 0.74 | 0.87 | 0.26 | 0.23 | 0.40 | 0.49 | 0.07 | <0.01* | <0.01* | 0.23 |
| miR-221 | 1.05 | 0.97–1.14 | 0.20 | 0.72 | 0.17 | 0.27 | 0.33 | 0.67 | 0.03* | <0.01* | <0.01* | 0.36 |
| miR-222 | 1.11 | 0.99–1.25 | 0.08 | 0.90 | 0.23 | 0.22 | 0.49 | 0.51 | 0.04* | <0.01* | <0.01* | 0.43 |
| miR-375 | 1.04 | 0.98–1.10 | 0.22 | 0.90 | 0.33 | 0.11 | 0.51 | 0.98 | 0.05 | <0.01* | <0.01* | 0.22 |
Formula: CLNM = molecular marker + sex + age (>45 years) + size (>2 cm) + FVPTC + TCVPTC + multifocality + LVI + ETE + involvement of margins.
Each row represents an independent postoperative predictor model performed for CLNM. In the prophylactic cohort, independent predictors for CLNM were miR-146b-3p, miR-146b −5p multifocality, LVI, and ETE.
Statistically significant.
CI, confidence interval; CLNM, central lymph node metastasis; ETE, extrathyroidal extension; FVPT, follicular variance of PTC; LVI, lymphovascular invasion; PTC, papillary thyroid cancer; RR, relative risk; TCVPTC, tall-cell variance of PTC.
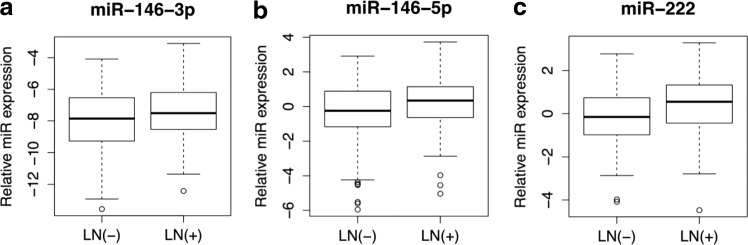
In a second multivariable logistic regression analysis limited to predictors available preoperatively only (molecular markers, age, sex, and tumor size) in prophylactic CLND patients, the BRAFV600E mutation (p = 0.01), miR-146b-3p (p = 0.01), miR-146b-5p (p = 0.01), miR-222 (p = 0.01), and tumor size >2 cm (p < 0.05) were found to predict CLNM independently (Table 3). The relative expression levels of miR-146b-3p, miR-146b-5p, and miR-222 were associated with a 10%, 15%, and 16% increase in the relative risk of CLNM, respectively (Table 3 and Fig. 1). Notably, in the prophylactic CLND cohort, patients who were BRAFV600E positive had a 48% chance of harboring CLNM, whereas those who were BRAFV600E negative had a 28% chance, resulting in a sensitivity of 65.9%, a specificity of 46.5%, a positive predictive value of 47.6%, and a negative predictive value of 71.8% for the BRAFV600E mutation.
Table 3.
Multivariate Analysis of Individual Molecular Marker with Preoperative Clinicopathologic Variables in Prophylactic PTCs (n = 237)
| Statistic of molecular marker | Clinicopathologic factors (p-value only) | |||||
|---|---|---|---|---|---|---|
| Molecular marker | RR | CI | p-Value | Sex | Age (≥45 years) | Size (>2 cm) |
| BRAFV600E | 1.69 | 1.11–2.57 | 0.01* | 0.52 | 0.85 | 0.01* |
| miR-21 | 1.04 | 0.92–1.16 | 0.56 | 0.67 | 0.72 | 0.01* |
| miR-146-3p | 1.10 | 1.02–1.19 | 0.01* | 0.96 | 0.98 | 0.01* |
| miR-146-5p | 1.15 | 1.04–1.26 | 0.01* | 0.75 | 0.69 | 0.01* |
| miR-204 | 0.96 | 0.88–1.04 | 0.30 | 0.54 | 0.80 | 0.01* |
| miR-221 | 1.07 | 0.99–1.16 | 0.07 | 0.66 | 0.64 | 0.01* |
| miR-222 | 1.16 | 1.03–1.30 | 0.01* | 0.47 | 0.76 | 0.01* |
| miR-375 | 1.05 | 0.98–1.11 | 0.15 | 0.51 | 0.87 | <0.01* |
Each row represents an independent preoperative predictor model performed for CLNM. In the prophylactic cohort, independent preoperative predictors for CLNM were BRAFV600E mutation, miR-146b-3p, miR-146-5p, miR-222, and tumor size.
Statistically significant.
FIG. 1.
Micro RNA (miRNA) expression levels of predictive miRs in the prophylactic CLND subgroup (n = 237). Tukey boxplots showing medians, interquartile range (IQR), and whiskers extending to 1.5 × IQR showing the inverse dCt values of (a) miR-146-3p, (b) miR-146-5p, and (c) miR-222 relative to miR-16. Outliers are represented by circles. CLND, central lymph node dissection; LN(+), lymph node–positive group; LN(–), lymph node–negative group.
Predictors of CLNM in the CVPTC histological subtype
In univariate logistic regression analyses focusing on the 192 patients with CVPTC who underwent prophylactic neck dissection, miR-146b-3p (p = 0.03), miR-146b-5p (p < 0.01), miR-221 (p = 0.02), miR-222 (p < 0.01), tumor size >2 cm (p = 0.01), multifocality (p = 0.04), positive surgical margins (p < 0.01), LVI (p < 0.01), ETE (p < 0.01), and advanced AJCC stage (p < 0.01) were found to be associated with CLNM. Furthermore, miR-204 (p = 0.02) was inversely associated, and the BRAFV600E mutation (p = 0.12) and follicular and tall-cell component percentages were not found to be associated with CLNM. Multivariable logistic regression analysis of only CVPTC cases undergoing prophylactic neck dissections (Table 4), while controlling for each molecular marker, sex, age, size, multifocality, LVI, positive surgical margins, ETE, and follicular and tall-cell component, found only miR-146b-5p (p = 0.01) and miR-222 (p = 0.02), LVI (p < 0.01), and ETE (p < 0.05) to be independent predictors of CLNM. BRAFV600E was not significantly associated with CLNM in this subset (p = 0.09). Multivariable logistic regression analysis in all CVPTC patients, including only predictors available preoperatively (molecular markers, age, sex, and tumor size), found miR-146b-5p (p = 0.01), miR-221 (p = 0.03), miR-222 (p < 0.01), and size >2 cm (p < 0.05) to be independent predictors of CLNM. BRAFV600E was again not associated with CLNM (p = 0.09; Table 5).
Table 4.
Multivariate Analysis of Individual Molecular Marker with Postoperative Clinicopathologic Variables in Prophylactic CVPTC (n = 192)
| Statistic of molecular marker | Significance of clinicopathologic factors (p-value only) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular marker | RR | CI | p-Value | Sex | Age (≥45 years) | Size (>2 cm) | Multifocality | Margins | Follicular % | Tall cell % | LVI | ETE |
| BRAFV600E | 1.49 | 0.94–2.37 | 0.09 | 0.96 | 0.33 | 0.15 | 0.15 | 0.47 | 0.99 | 0.97 | <0.01* | <0.01* |
| miR-21 | 0.97 | 0.86–1.11 | 0.69 | 0.96 | 0.55 | 0.23 | 0.07 | 0.36 | 0.74 | 0.91 | <0.01* | 0.01* |
| miR-146-3p | 1.09 | 0.99–1.18 | 0.07 | 0.67 | 0.29 | 0.16 | 0.17 | 0.30 | 0.59 | 0.75 | <0.01* | 0.01* |
| miR-146-5p | 1.15 | 1.03–1.29 | 0.01* | 0.74 | 0.15 | 0.30 | 0.05 | 0.64 | 0.75 | 0.97 | <0.01* | 0.02* |
| miR-204 | 0.94 | 0.86–1.02 | 0.15 | 0.99 | 0.24 | 0.41 | 0.12 | 0.43 | 0.89 | 0.80 | <0.01* | 0.03* |
| miR-221 | 1.07 | 0.98–1.17 | 0.11 | 0.97 | 0.16 | 0.33 | 0.07 | 0.48 | 0.72 | 0.90 | <0.01* | 0.02* |
| miR-222 | 1.16 | 1.02–1.32 | 0.02* | 0.76 | 0.22 | 0.28 | 0.05 | 0.67 | 0.74 | 0.85 | <0.01* | 0.02* |
| miR-375 | 1.05 | 0.98–1.13 | 0.14 | 0.85 | 0.28 | 0.12 | 0.12 | 0.33 | 0.85 | 0.89 | <0.01* | 0.03* |
Formula: CLNM = molecular marker + sex + age (≥45 years) + size (>2 cm) + FVPTC + TCVPTC + multifocality + LVI + ETE + involvement of margins.
Each row represents an independent postoperative predictor model performed for CLNM. In the prophylactic CVPTC subgroup, independent predictors for CLNM were miR-146b-5p, miR-222, LVI, and ETE.
Statistically significant.
CVPTC, classical variant of PTC.
Table 5.
Multivariate Analysis of Individual Molecular Marker with Preoperative Clinicopathologic Variables in Prophylactic CVPTCs (n = 192)
| Statistic of molecular marker | Clinicopathologic factors (p-value only) | |||||
|---|---|---|---|---|---|---|
| Molecular marker | RR | CI | p-Value | Sex | Age (≥45 years) | Size (>2 cm) |
| BRAFV600E | 1.50 | 0.93–2.40 | 0.09 | 0.32 | 0.61 | 0.02* |
| miR-21 | 1.02 | 0.90–1.15 | 0.80 | 0.42 | 0.95 | 0.01* |
| miR-146-3p | 1.09 | 1.00–1.19 | 0.06 | 0.69 | 0.70 | 0.01* |
| miR-146-5p | 1.17 | 1.05–1.32 | 0.01* | 0.56 | 0.41 | 0.02* |
| miR-204 | 0.92 | 0.84–1.01 | 0.08 | 0.42 | 0.61 | 0.04* |
| miR-221 | 1.10 | 1.01–1.19 | 0.03* | 0.44 | 0.36 | 0.03* |
| miR-222 | 1.20 | 1.06–1.36 | 0.00* | 0.28 | 0.50 | 0.02* |
| miR-375 | 1.06 | 0.99–1.14 | 0.09 | 0.33 | 0.76 | <0.01* |
Formula: CLNM = molecular marker + sex + age (>45 years) + size (>2 cm).
Each row represents an independent preoperative predictor model performed for CLNM. In the CVPTC prophylactic subgroup, independent preoperative predictors for CLNM were miR-146-5p, miR-221, miR-222, and tumor size.
Statistically significant.
Discussion
To the authors' knowledge, this is the largest prospective study to date examining the association of BRAFV600E and miRs with aggressive clinicopathologic variables of PTC, particularly the presence of CLNM. Due to current controversies in the surgical management of PTCs and prophylactic CLND as well as ambiguity in the literature, molecular markers that allow for risk stratification for the presence of CLNM would be invaluable in the clinical arena.
In the patients with no clinical indications of CLNM, where these molecular markers would be most valuable, miR-146b-3p, and miR-146b-5p were identified to be predictive of CLNM. Furthermore, because the clinical decision to perform a CLND is determined preoperatively, a multivariable analysis among prophylactic CLND patients, controlling for clinicopathologic variables that are available preoperatively (age, sex, and tumor size), is critical for the identification of molecular markers with clinical utility. This subgroup analysis of individual miRs identified elevated levels of miR-146b-3p, miR-146b-5p, and miR-222 as independent predictors of the presence of CLNM. More importantly, in this group, each doubling of expression of these markers was associated with a 10%, 15%, and 16%, increase in the risk of CLNM, respectively. These findings are consistent with previously reported studies that identified positive associations of these miRs with various features of PTC aggressiveness (28,33,34,37,39,40,42–44,59). Three of the miRs investigated were found to predict CLNM in the multivariate model. Inspection of the bar graphs in Figure 1, however, clearly shows significant overlap in expression of these miRs in the two outcome groups, raising legitimate questions about their clinical usefulness as individual prognostic markers. Each doubling of expression of these markers was associated with increased risk of CLNM. Unfortunately, this only segregated CLND cases at higher levels of expression that were only seen in a few cases, providing only poor sensitivity at acceptable levels of specificity. These results speak to the concept and interpretation of statistical significance versus actual applicability of molecular markers clinically.
The BRAFV600E mutation was significantly associated with CLNM in patients undergoing prophylactic central neck dissection when specifically controlling for preoperatively available clinicopathologic variables in all PTC subtypes. One explanation for this result, as exhibited in TCGA Thyroid Project data, is that the BRAFV600E mutation prevalence varies across PTC subtypes (12). Thus, the incorporation of the BRAFV600E mutation with subtypes of PTCs with low rates of the BRAFV600E mutation (e.g., FVPTC), along with a favorable phenotype, may confound the results to identify a positive association between BRAFV600E and aggressive behavior, rather than simply being a result of subtypes having different mutation rates. TCGA has reported that the BRAFV600E mutation status is strongly enriched in CVPTCs, which are more aggressive than their FVPTC counterparts are. Indeed, when a subgroup analysis was performed only in the CVPTCs of our cohort, the BRAFV600E mutation was no longer an independent predictor of CLNM. This finding is similar to a retrospective study previously performed with the same institutions (23). These findings are attributed to the fact that the entire cohort consisted of CVPTC as well as FVPTC and TCVPTC, the latter two having a lower and higher BRAF mutation rate with correspondingly lower and higher rates of LNM. Thus, it is believed that the FVPTC and TCVPTC may have contributed to finding a correlation in the entire series, but not in the CVPTC-only group.
In the present study, while BRAFV600E mutation status was identified as being statistically significantly associated with CLNM in the analysis of all histological subtypes that underwent prophylactic CLND, the ability to predict the presence of LNM with BRAFV600E analysis (48% for BRAFV600E positive vs. 28% for BRAFV600E negative) would likely not have any useful clinical applicability. In other words, its positive and negative predictive value (48% and 72%, respectively) limit its clinical utility as a stand-alone marker.
The TCGA molecular characterization of PTCs has revealed distinct molecular signatures between the different PTC subtypes. Furthermore, many of these molecular differences are associated with the percent follicular component in the tumor. Therefore, studying molecular markers while treating PTC as a homogenous disease may lead to inaccurate conclusions being drawn based on molecular markers and tumor phenotypes. To that end, a subgroup analysis was performed to consider the significance of molecular markers in the CVPTC subtype alone, excluding FVPTCs and TCVPTCs, but including a measure of the follicular and tall-cell component percentages present in the CVPTCs, a variable heretofore not incorporated in similar analyses. CVPTCs represented the largest cohort (192 cases) in the study, and a subgroup analysis, controlling for all clinicopathologic variables in the prophylactic group, identified miR-146b-5p and miR-222 to be predictive of CLNM; an analysis limited to preoperative clinicopathologic variables only found miR-146b-5p, miR-221, and miR-222 to be associated with CLNM. As discussed, BRAFV600E was not significantly associated with CLNM in CVPTCs, nor was the extent of the follicular and tall-cell components of the CVPTCs.
This prospective study examining a consecutive cohort of patients was adequately powered to examine relationships between potentially useful molecular markers and the prevalence of pathologically confirmed CLNM among patients with no clinical indication of CLNM. Our multi-institutional study is also the largest prospective study of patients with prophylactic CLND that examines the association of BRAFV600E and miRs with aggressive clinicopathologic variables of PTC. Most existing studies are retrospective and include non-consecutive, heterogeneous CLND cohorts and therapeutic CLND cases (50). Only one previous prospective study included patients who underwent prophylactic CLND, but was comprised of only 148 patients and from a single institution (18). While this prospective study overcomes major patient selection biases arising from retrospective non-consecutive and heterogeneous CLND cohorts that include therapeutic CLND cases, there are some limitations that require consideration. First, the study contains PTCs of various subtypes, namely CVPTC, FVPTC, and TCVPTC, which have different degrees of aggressiveness. The low number of patients with FVPTC and TCVPTC did not permit an adequately powered subgroup analysis within each disease subtype. Second, bias may be introduced by including patients with a clear diagnosis of PTC and excluding patients with indeterminate and suspicious Bethesda cytology categories, as the latter would have included a larger number of FVPTCs. However, in general, surgeons would not consider prophylactic CLND unless the diagnosis of PTC was clear preoperatively. Third, the pathologic spectrum of LNM based on size was not available. Therefore, a risk-stratification analysis based on these factors could not be performed. Lastly, variations in the exact surgical techniques and surgical practice patterns may have introduced undetected biases.
The TCGA experience characterized molecular phenotypes of PTC and highlighted the importance of considering molecular differences in treatment of the disease. It has provided a great resource to researchers. However, unfortunately, aggressive PTC cases are under-represented, and follow-up information is incomplete at best. Therefore, there is a need for a comprehensive characterization of the relationships between miRs and CLNM in studies that are adequately powered to permit examination across benign and aggressive PTCs with complete follow-up information, while avoiding the selection bias introduced by selective CLND.
While the current findings suggest the potential for developing miR-related predictive tools that may have clinical utility, this study was performed on FFPE tumor samples, and any future applicability would need to be applied to FNA samples. It is well known, however, that mutational analysis of the BRAF gene is reliable in FNA samples and correlates with corresponding tumors (48,49). Importantly, this exploratory study relied upon patient-specific relative values of miR expression rather than absolute values. In order to maximize the clinical usefulness of miR expression as a decision tool for determining the need for CLND, developing and validating techniques for measuring absolute levels of target miRs may be required. Along with others, this study has shown the ability to identify molecular alterations in PTCs, both at the DNA level and in terms of gene expression, from these sample types (23), and further consideration should be made to investigate their clinical utility.
Conclusions
In patients undergoing prophylactic CLNDs, miR-146b-3p, miR-146b-5p, and miR-222 were identified as potential markers of CLNM. BRAFV600E mutation was significantly associated with CLNM among patients with all PTC subtypes examined as a single group. When present, the BRAFV600E mutation was associated with a 48% chance of node positivity, while BRAFV600E negative patients had a 28% chance of node positivity. This proportional difference in the BRAF-associated likelihood of CLNM may not be useful clinically when deciding whether a patient should undergo prophylactic CLND. Furthermore, this statistical significance is likely due to the BRAFV600E mutation prevalence in the different PTC subtypes (FVPTC and TCVPTC). The results also demonstrate, however, that statistical significance does not necessarily equal clinical usefulness (48% vs. 28% LNM). The usefulness of BRAFV600E and other markers will depend strongly on the context and pretest probability of a positive test result. In cases with a high likelihood of BRAFV600E positivity, such as patients diagnosed with PTC on FNA, BRAFV600E testing is unlikely to be helpful. In more ambiguous cases, for example patients with indeterminate and suspicious Bethesda categories, the presence of a BRAFV600E mutation may increase the likelihood of being associated with LNM, but this has to be balanced with the lower overall risk of associated LNM in these categories. Before consideration can be given to creating a clinically useful prognostic prediction model for CLNM for patients with PTC, substantial additional work will be required to understand the observed relationships, paying particular attention to parameters that impact pretest probabilities, such as therapeutic versus prophylactic CLND intent, or the histological subtype composition of a given study cohort.
Supplementary Material
Acknowledgments
We would like to acknowledge Derek Anderson, Katie Beierl, Elizabeth Neylan, and Rouzbeh Mashayekhi for outstanding technical assistance.
Author Disclosure Statement
The authors have nothing to disclose.
References
- 1.Davies L, Welch HG. 2006. Increasing incidence of thyroid cancer in the United States, 1973–2002. JAMA 295:2164–2167 [DOI] [PubMed] [Google Scholar]
- 2.Horn-Ross PL, Lichtensztajn DY, Clarke CA, Dosiou C, Oakley-Girvan I, Reynolds P, Gomez SL, Nelson DO. 2014. Continued rapid increase in thyroid cancer incidence in california: trends by patient, tumor, and neighborhood characteristics. Cancer Epidemiol Biomarkers Prev 23:1067–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.American Thyroid Association Guidelines Taskforce on Thyroid Nodules and Differentiated Thyroid Cancer, Cooper DS, Doherty GM, Haugen BR, Kloos RT, Lee SL, Mandel SJ, Mazzaferri EL, McIver B, Pacini F, Schlumberger M, Sherman SI, Steward DL, Tuttle RM. 2009. Revised American Thyroid Association management guidelines for patients with thyroid nodules and differentiated thyroid cancer. Thyroid 19:1167–1214 [DOI] [PubMed] [Google Scholar]
- 4.Xing M, Haugen BR, Schlumberger M. 2013. Progress in molecular-based management of differentiated thyroid cancer. Lancet 381:1058–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calo PG, Pisano G, Medas F, Marcialis J, Gordini L, Erdas E, Nicolosi A. 2014. Total thyroidectomy without prophylactic central neck dissection in clinically node-negative papillary thyroid cancer: is it an adequate treatment? World J Surg Oncol 12:152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mazzaferri EL, Doherty GM, Steward DL. 2009. The pros and cons of prophylactic central compartment lymph node dissection for papillary thyroid carcinoma. Thyroid 19:683–689 [DOI] [PubMed] [Google Scholar]
- 7.Viola D, Materazzi G, Valerio L, Molinaro E, Agate L, Faviana P, Seccia V, Sensi E, Romei C, Piaggi P, Torregrossa L, Sellari-Franceschini S, Basolo F, Vitti P, Elisei R, Miccoli P. 2015. Prophylactic central compartment lymph node dissection in papillary thyroid carcinoma: clinical implications derived from the first prospective randomized controlled single institution study. J Clin Endocrinol Metab 100:1316–1324 [DOI] [PubMed] [Google Scholar]
- 8.McHenry CR, Stulberg JJ. 2014. Prophylactic central compartment neck dissection for papillary thyroid cancer. Surg Clin North Am 94:529–540 [DOI] [PubMed] [Google Scholar]
- 9.Baek SK, Jung KY, Kang SM, Kwon SY, Woo JS, Cho SH, Chung EJ. 2010. Clinical risk factors associated with cervical lymph node recurrence in papillary thyroid carcinoma. Thyroid 20:147–152 [DOI] [PubMed] [Google Scholar]
- 10.Mercante G, Frasoldati A, Pedroni C, Formisano D, Renna L, Piana S, Gardini G, Valcavi R, Barbieri V. 2009. Prognostic factors affecting neck lymph node recurrence and distant metastasis in papillary microcarcinoma of the thyroid: results of a study in 445 patients. Thyroid 19:707–716 [DOI] [PubMed] [Google Scholar]
- 11.Lim JY, Hong SW, Lee YS, Kim BW, Park CS, Chang HS, Cho JY. 2013. Clinicopathologic implications of the BRAF(V600E) mutation in papillary thyroid cancer: a subgroup analysis of 3130 cases in a single center. Thyroid 23:1423–1430 [DOI] [PubMed] [Google Scholar]
- 12.Agrawal N, Akbani R, Aksoy BA, Ally A, Arachchi H, Asa Sylvia L, Auman JT, Balasundaram M, Balu S, Baylin SB, Behera M, Bernard B, Beroukhim R, Bishop Justin A, Black AD, Bodenheimer T, Boice L, Bootwalla MS, Bowen J, Bowlby R, Bristow CA, Brookens R, Brooks D, Bryant R, Buda E, Butterfield YS, Carling T, Carlsen R, Carter SL, Carty SE, Chan TA, Chen AY, Cherniack AD, Cheung D, Chin L, Cho J, Chu A, Chuah E, Cibulskis K, Ciriello G, Clarke A, Clayman GL, Cope L, Copland JA, Covington K, Danilova L, Davidsen T, Demchok JA, DiCara D, Dhalla N, Dhir R, Dookran SS, Dresdner G, Eldridge J, Eley G, El-Naggar AK, Eng S, Fagin JA, Fennell T, Ferris RL, Fisher S, Frazer S, Frick J, Gabriel SB, Ganly I, Gao J, Garraway LA, Gastier-Foster JM, Getz G, Gehlenborg N, Ghossein R, Gibbs RA, Giordano TJ, Gomez-Hernandez K, Grimsby J, Gross B, Guin R, Hadjipanayis A, Harper HA, Hayes DN, Heiman DI, Herman JG, Hoadley KA, Hofree M, Holt RA, Hoyle AP, Huang FW, Huang M, Hutter CM, Ideker T, Iype L, Jacobsen A, Jefferys SR, Jones CD, Jones SJ, Kasaian K, Kebebew E, Khuri FR, Kim J, Kramer R, Kreisberg R, Kucherlapati R, Kwiatkowski DJ, Ladanyi M, Lai PH, Laird PW, Lander E, Lawrence MS, Lee D, Lee E, Lee S, Lee W, Leraas KM, Lichtenberg TM, Lichtenstein L, Lin P, Ling S, Liu J, Liu W, Liu Y, LiVolsi VA, Lu Y, Ma Y, Mahadeshwar HS, Marra MA, Mayo M, McFadden DG, Meng S, Meyerson M, Mieczkowski PA, Miller M, Mills G, Moore RA, Mose LE, Mungall AJ, Murray BA, Nikiforov YE, Noble MS, Ojesina AI, Owonikoko TK, Ozenberger BA, Pantazi A, Parfenov M, Park PJ, Parker JS, Paull EO, Pedamallu CS, Perou CM, Prins JF, Protopopov A, Ramalingam SS, Ramirez NC, Ramirez R, Raphael BJ, Rathmell WK, Ren X, Reynolds SM, Rheinbay E, Ringel MD, Rivera M, Roach J, Robertson AG, Rosenberg MW, Rosenthal M, Sadeghi S, Saksena G, Sander C, Santoso N, Schein JE, Schultz N, Schumacher SE, Seethala RR, Seidman J, Senbabaoglu Y, Seth S, Sharpe S, Shaw KR, Shen JP, Shen R, Sherman S, Sheth M, Shi Y, Shmulevich I, Sica GL, Simons JV, Sinha R, Sipahimalani P, Smallridge RC, Sofia HJ, Soloway MG, Song X, Sougnez C, Stewart C, Stojanov P, Stuart JM, Sumer SO, Sun Y, Tabak B, Tam A, Tan D, Tang J, Tarnuzzer R, Taylor BS, Thiessen N, Thorne L, Thorsson V, Tuttle RM, Umbricht CB, Van Den Berg DJ, Vandin F, Veluvolu U, Verhaak RG, Vinco M, Voet D, Walter V, Wang Z, Waring S, Weinberger PM, Weinhold N, Weinstein JN, Weisenberger DJ, Wheeler D, Wilkerson MD, Wilson J, Williams M, Winer DA, Wise L, Wu J, Xi L, Xu AW, Yang L, Yang L, Zack TI, Zeiger MA, Zeng D, Zenklusen JC, Zhao N, Zhang H, Zhang J, Zhang J, Zhang W, Zmuda E, Zou L. 2014. Integrated genomic characterization of papillary thyroid carcinoma. Cell 159:676–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, Davis N, Dicks E, Ewing R, Floyd Y, Gray K, Hall S, Hawes R, Hughes J, Kosmidou V, Menzies A, Mould C, Parker A, Stevens C, Watt S, Hooper S, Wilson R, Jayatilake H, Gusterson BA, Cooper C, Shipley J, Hargrave D, Pritchard-Jones K, Maitland N, Chenevix-Trench G, Riggins GJ, Bigner DD, Palmieri G, Cossu A, Flanagan A, Nicholson A, Ho JW, Leung SY, Yuen ST, Weber BL, Seigler HF, Darrow TL, Paterson H, Marais R, Marshall CJ, Wooster R, Stratton MR, Futreal PA. 2002. Mutations of the BRAF gene in human cancer. Nature 417:949–954 [DOI] [PubMed] [Google Scholar]
- 14.Giordano TJ, Kuick R, Thomas DG, Misek DE, Vinco M, Sanders D, Zhu Z, Ciampi R, Roh M, Shedden K, Gauger P, Doherty G, Thompson NW, Hanash S, Koenig RJ, Nikiforov YE. 2005. Molecular classification of papillary thyroid carcinoma: distinct BRAF, RAS, and RET/PTC mutation-specific gene expression profiles discovered by DNA microarray analysis. Oncogene 24:6646–6656 [DOI] [PubMed] [Google Scholar]
- 15.Bernstein J, Virk RK, Hui P, Prasad A, Westra WH, Tallini G, Adeniran AJ, Udelsman R, Sasaki CT, Roman SA, Sosa JA, Prasad ML. 2013. Tall cell variant of papillary thyroid microcarcinoma: clinicopathologic features with BRAF(V600E) mutational analysis. Thyroid 23:1525–1531 [DOI] [PubMed] [Google Scholar]
- 16.Nikiforov YE. 2011. Molecular analysis of thyroid tumors. Mod Pathol 24:S34–43 [DOI] [PubMed] [Google Scholar]
- 17.Howell GM, Nikiforova MN, Carty SE, Armstrong MJ, Hodak SP, Stang MT, McCoy KL, Nikiforov YE, Yip L. 2013. BRAF V600E mutation independently predicts central compartment lymph node metastasis in patients with papillary thyroid cancer. Ann Surg Oncol 20:47–52 [DOI] [PubMed] [Google Scholar]
- 18.Joo JY, Park JY, Yoon YH, Choi B, Kim JM, Jo YS, Shong M, Koo BS. 2012. Prediction of occult central lymph node metastasis in papillary thyroid carcinoma by preoperative BRAF analysis using fine-needle aspiration biopsy: a prospective study. J Clin Endocrinol Metab 97:3996–4003 [DOI] [PubMed] [Google Scholar]
- 19.Kim SJ, Lee KE, Myong JP, Park JH, Jeon YK, Min HS, Park SY, Jung KC, Koo do H, Youn YK. 2012. BRAF V600E mutation is associated with tumor aggressiveness in papillary thyroid cancer. World J Surg 36:310–317 [DOI] [PubMed] [Google Scholar]
- 20.Fernandez IJ, Piccin O, Sciascia S, Cavicchi O, Repaci A, Vicennati V, Fiorentino M. 2013. Clinical significance of BRAF mutation in thyroid papillary cancer. Otolaryngol Head Neck Surg 148:919–925 [DOI] [PubMed] [Google Scholar]
- 21.Chakraborty A, Narkar A, Mukhopadhyaya R, Kane S, D'Cruz A, Rajan MG. 2012. BRAF V600E mutation in papillary thyroid carcinoma: significant association with node metastases and extra thyroidal invasion. Endocr Pathol 23:83–93 [DOI] [PubMed] [Google Scholar]
- 22.Yang Y, Chen C, Chen Z, Jiang J, Chen Y, Jin L, Guo G, Zhang X, Ye T. 2014. Prediction of central compartment lymph node metastasis in papillary thyroid microcarcinoma. Clin Endocrinol 81:282–288 [DOI] [PubMed] [Google Scholar]
- 23.Li C, Aragon Han P, Lee KC, Lee LC, Fox AC, Beninato T, Thiess M, Dy BM, Sebo TJ, Thompson GB, Grant CS, Giordano TJ, Gauger PG, Doherty GM, Fahey TJ, 3rd, Bishop J, Eshleman JR, Umbricht CB, Schneider EB, Zeiger MA. 2013. Does BRAF V600E mutation predict aggressive features in papillary thyroid cancer? Results from four endocrine surgery centers. J Clin Endocrinol Metab 98:3702–3712 [DOI] [PubMed] [Google Scholar]
- 24.Ito Y, Yoshida H, Maruo R, Morita S, Takano T, Hirokawa M, Yabuta T, Fukushima M, Inoue H, Tomoda C, Kihara M, Uruno T, Higashiyama T, Takamura Y, Miya A, Kobayashi K, Matsuzuka F, Miyauchi A. 2009. BRAF mutation in papillary thyroid carcinoma in a Japanese population: its lack of correlation with high-risk clinicopathological features and disease-free survival of patients. Endocr J 56:89–97 [DOI] [PubMed] [Google Scholar]
- 25.Sancisi V, Nicoli D, Ragazzi M, Piana S, Ciarrocchi A. 2012. BRAFV600E mutation does not mean distant metastasis in thyroid papillary carcinomas. J Clin Endocrinol Metab 97:E1745–1749 [DOI] [PubMed] [Google Scholar]
- 26.Barbaro D, Incensati RM, Materazzi G, Boni G, Grosso M, Panicucci E, Lapi P, Pasquini C, Miccoli P. 2014. The BRAF V600E mutation in papillary thyroid cancer with positive or suspected pre-surgical cytological finding is not associated with advanced stages or worse prognosis. Endocrine 45:462–468 [DOI] [PubMed] [Google Scholar]
- 27.Nam JK, Jung CK, Song BJ, Lim DJ, Chae BJ, Lee NS, Park WC, Kim JS, Jung SS, Bae JS. 2012. Is the BRAF(V600E) mutation useful as a predictor of preoperative risk in papillary thyroid cancer? Am J Surg 203:436–441 [DOI] [PubMed] [Google Scholar]
- 28.Chou CK, Chen RF, Chou FF, Chang HW, Chen YJ, Lee YF, Yang KD, Cheng JT, Huang CC, Liu RT. 2010. miR-146b is highly expressed in adult papillary thyroid carcinomas with high risk features including extrathyroidal invasion and the BRAF(V600E) mutation. Thyroid 20:489–494 [DOI] [PubMed] [Google Scholar]
- 29.Danilovic DL, Lima EU, Domingues RB, Brandao LG, Hoff AO, Marui S. 2014. Pre-operative role of BRAF in the guidance of the surgical approach and prognosis of differentiated thyroid carcinoma. Eur J Endocrinol 170:619–625 [DOI] [PubMed] [Google Scholar]
- 30.Dutenhefner SE, Marui S, Santos AB, de Lima EU, Inoue M, Neto JS, Shiang C, Fukushima JT, Cernea CR, Friguglietti CU. 2013. BRAF: a tool in the decision to perform elective neck dissection? Thyroid 23:1541–1546 [DOI] [PubMed] [Google Scholar]
- 31.Aragon Han P, Olson MT, Fazeli R, Prescott JD, Pai SI, Schneider EB, Tufano RP, Zeiger MA. 2014. The impact of molecular testing on the surgical management of patients with thyroid nodules. Ann Surg Oncol 21:1862–1869 [DOI] [PubMed] [Google Scholar]
- 32.Esquela-Kerscher A, Slack FJ. 2006. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer 6:259–269 [DOI] [PubMed] [Google Scholar]
- 33.Chou CK, Yang KD, Chou FF, Huang CC, Lan YW, Lee YF, Kang HY, Liu RT. 2013. Prognostic implications of miR-146b expression and its functional role in papillary thyroid carcinoma. J Clin Endocrinol Metab 98:E196–205 [DOI] [PubMed] [Google Scholar]
- 34.Acibucu F, Dokmetas HS, Tutar Y, Elagoz S, Kilicli F. 2014. Correlations between the expression levels of micro-RNA146b, 221, 222 and p27Kip1 protein mRNA and the clinicopathologic parameters in papillary thyroid cancers. Exp Clin Endocrinol Diabetes 122:137–143 [DOI] [PubMed] [Google Scholar]
- 35.Benvenga S, Koch CA. 2014. Molecular pathways associated with aggressiveness of papillary thyroid cancer. Curr Genom 15:162–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dettmer M, Perren A, Moch H, Komminoth P, Nikiforov YE, Nikiforova MN. 2013. Comprehensive microRNA expression profiling identifies novel markers in follicular variant of papillary thyroid carcinoma. Thyroid 23:1383–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang YB, Liao DH, Pan LX, Ye RY, Li XX, Wang SM, Ye CS, Chen LH. 2013. Expressions of miRNAs in papillary thyroid carcinoma and their associations with the BRAF(V600E) mutation. Eur J Endocrinol 168:675–681 [DOI] [PubMed] [Google Scholar]
- 38.Peng Y, Li C, Luo DC, Ding JW, Zhang W, Pan G. 2014. Expression profile and clinical significance of microRNAs in papillary thyroid carcinoma. Molecules 19:11586–11599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sheu SY, Grabellus F, Schwertheim S, Handke S, Worm K, Schmid KW. 2009. Lack of correlation between BRAF V600E mutational status and the expression profile of a distinct set of miRNAs in papillary thyroid carcinoma. Horm Metab Res 41:482–487 [DOI] [PubMed] [Google Scholar]
- 40.Sun Y, Yu S, Liu Y, Wang F, Liu Y, Xiao H. 2013. Expression of miRNAs in papillary thyroid carcinomas is associated with BRAF mutation and clinicopathological features in Chinese patients. Int J Endocrinol 2013:128735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Z, Zhang H, He L, Dong W, Li J, Shan Z, Teng W. 2013. Association between the expression of four upregulated miRNAs and extrathyroidal invasion in papillary thyroid carcinoma. Onco Targets Ther 6:281–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yip L, Kelly L, Shuai Y, Armstrong MJ, Nikiforov YE, Carty SE, Nikiforova MN. 2011. MicroRNA signature distinguishes the degree of aggressiveness of papillary thyroid carcinoma. Ann Surg Oncol 18:2035–2041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu S, Liu Y, Wang J, Guo Z, Zhang Q, Yu F, Zhang Y, Huang K, Li Y, Song E, Zheng XL, Xiao H. 2012. Circulating microRNA profiles as potential biomarkers for diagnosis of papillary thyroid carcinoma. J Clin Endocrinol Metab 97:2084–2092 [DOI] [PubMed] [Google Scholar]
- 44.Zhou YL, Liu C, Dai XX, Zhang XH, Wang OC. 2012. Overexpression of miR-221 is associated with aggressive clinicopathologic characteristics and the BRAF mutation in papillary thyroid carcinomas. Med Oncol 29:3360–3366 [DOI] [PubMed] [Google Scholar]
- 45.Carty SE, Cooper DS, Doherty GM, Duh QY, Kloos RT, Mandel SJ, Randolph GW, Stack BC, Jr, Steward DL, Terris DJ, Thompson GB, Tufano RP, Tuttle RM, Udelsman R. 2009. Consensus statement on the terminology and classification of central neck dissection for thyroid cancer. Thyroid 19:1153–1158 [DOI] [PubMed] [Google Scholar]
- 46.Kotorashvili A, Ramnauth A, Liu C, Lin J, Ye K, Kim R, Hazan R, Rohan T, Fineberg S, Loudig O. 2012. Effective DNA/RNA co-extraction for analysis of microRNAs, mRNAs, and genomic DNA from formalin-fixed paraffin-embedded specimens. PloS One 7:e34683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen G, Olson MT, O'Neill A, Norris A, Beierl K, Harada S, Debeljak M, Rivera-Roman K, Finley S, Stafford A, Gocke CD, Lin MT, Eshleman JR. 2012. A virtual pyrogram generator to resolve complex pyrosequencing results. J Molec Diagn 14:149–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bentz BG, Miller BT, Holden JA, Rowe LR, Bentz JS. 2009. B-RAF V600E mutational analysis of fine needle aspirates correlates with diagnosis of thyroid nodules. Otolaryngol Head Neck Surg 140:709–714 [DOI] [PubMed] [Google Scholar]
- 49.Xing M, Tufano RP, Tufaro AP, Basaria S, Ewertz M, Rosenbaum E, Byrne PJ, Wang J, Sidransky D, Ladenson PW. 2004. Detection of BRAF mutation on fine needle aspiration biopsy specimens: a new diagnostic tool for papillary thyroid cancer. J Clin Endocrinol Metab 89:2867–2872 [DOI] [PubMed] [Google Scholar]
- 50.Aragon Han P, Weng CH, Khawaja HT, Nagarajan N, Schneider EB, Umbricht CB, Witwer KW, Zeiger MA. 2015. MicroRNA expression and association with clinicopathologic features in papillary thyroid cancer: a systematic review. Thyroid 25:1322–1329 [DOI] [PubMed] [Google Scholar]
- 51.Davoren PA, McNeill RE, Lowery AJ, Kerin MJ, Miller N. 2008. Identification of suitable endogenous control genes for microRNA gene expression analysis in human breast cancer. BMC Mol Biol 9:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xiang M, Zeng Y, Yang R, Xu H, Chen Z, Zhong J, Xie H, Xu Y, Zeng X. 2014. U6 is not a suitable endogenous control for the quantification of circulating microRNAs. Biochem Biophys Res Commun 454:210–214 [DOI] [PubMed] [Google Scholar]
- 53.Mattie MD, Benz CC, Bowers J, Sensinger K, Wong L, Scott GK, Fedele V, Ginzinger D, Getts R, Haqq C. 2006. Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Mol Cancer 5:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liang Y, Ridzon D, Wong L, Chen C. 2007. Characterization of microRNA expression profiles in normal human tissues. BMC Genom 8:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roberts TC, Blomberg KE, McClorey G, El Andaloussi S, Godfrey C, Betts C, Coursindel T, Gait MJ, Smith CI, Wood MJ. 2012. Expression analysis in multiple muscle groups and serum reveals complexity in the microRNA transcriptome of the mdx mouse with implications for therapy. Mol Ther Nucleic acids 1:e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xing M. 2005. BRAF mutation in thyroid cancer. Endocr Relat Cancer 12:245–262 [DOI] [PubMed] [Google Scholar]
- 57.Namba H, Nakashima M, Hayashi T, Hayashida N, Maeda S, Rogounovitch TI, Ohtsuru A, Saenko VA, Kanematsu T, Yamashita S. 2003. Clinical implication of hot spot BRAF mutation, V599E, in papillary thyroid cancers. J Clin Endocrinol Metab 88:4393–4397 [DOI] [PubMed] [Google Scholar]
- 58.Xing M, Clark D, Guan H, Ji M, Dackiw A, Carson KA, Kim M, Tufaro A, Ladenson P, Zeiger M, Tufano R. 2009. BRAF mutation testing of thyroid fine-needle aspiration biopsy specimens for preoperative risk stratification in papillary thyroid cancer. J Clin Oncol 27:2977–2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee JC, Zhao JT, Clifton-Bligh RJ, Gill A, Gundara JS, Ip JC, Glover A, Sywak MS, Delbridge LW, Robinson BG, Sidhu SB. 2013. MicroRNA-222 and microRNA-146b are tissue and circulating biomarkers of recurrent papillary thyroid cancer. Cancer 119:4358–4365 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.