Abstract
Homeostatic adaptations to stress are regulated by interactions between the brainstem and regions of the forebrain, including limbic sites related to respiratory, autonomic, affective, and cognitive processing. Neuroanatomic connections between these homeostatic regions, however, have not been thoroughly identified in the human brain. In this study, we perform diffusion spectrum imaging tractography using the MGH-USC Connectome MRI scanner to visualize structural connections in the human brain linking autonomic and cardiorespiratory nuclei in the midbrain, pons, and medulla oblongata with forebrain sites critical to homeostatic control. Probabilistic tractography analyses in six healthy adults revealed connections between six brainstem nuclei and seven forebrain regions, several over long distances between the caudal medulla and cerebral cortex. The strongest evidence for brainstem-homeostatic forebrain connectivity in this study was between the brainstem midline raphe and the medial temporal lobe. The subiculum and amygdala were the sampled forebrain nodes with the most extensive brainstem connections. Within the human brainstem-homeostatic forebrain connectome, we observed that a lateral forebrain bundle, whose connectivity is distinct from that of rodents and nonhuman primates, is the primary conduit for connections between the brainstem and medial temporal lobe. This study supports the concept that interconnected brainstem and forebrain nodes form an integrated central homeostatic network (CHN) in the human brain. Our findings provide an initial foundation for elucidating the neuroanatomic basis of homeostasis in the normal human brain, as well as for mapping CHN disconnections in patients with disorders of homeostasis, including sudden and unexpected death, and epilepsy.
Key words: : brainstem, diffusion MRI, limbic system, medial forebrain bundle, sudden unexplained death in epilepsy (SUDEP), tractography
Introduction
Deep within the human forebrain lies a group of structures that play major roles in autonomic, respiratory, neuroendocrine, emotional, immune, and cognitive adaptations to stress. Collectively, these forebrain structures have been known, in part, as the limbic system, a term that retains usefulness today due to their anatomic proximity to the hypothalamus, robust mono- and/or oligo-synaptic connectivity to one another, and shared participation in homeostasis. The definition of the limbic system, as apart from the limbic lobe, has evolved since the classical descriptions by Broca (1878), Papez (1937), and Maclean (1952), reflecting different perspectives as to the overarching function(s) of the system.
In this study, we consider an expanded limbic network in which inclusion of neuroanatomic nodes is based upon human and experimental data indicating nodal participation in homeostasis, that is, adaption to stress. Homeostatic forebrain nodes within this network receive sensory information concerning extrinsic threats and intrinsic metabolic derangements from the brainstem, resulting in arousal from sleep, heightened attention and vigilance during waking, and visceral and somatic motor defenses (Ulrich-Lai and Herman, 2009). Yet despite emerging evidence for brainstem–forebrain interactions in regulating homeostasis, little direct information about the neuroanatomic connections between homeostatic regions of the brainstem and forebrain is available in the human brain. Current knowledge is based almost solely upon extrapolations from animal studies (Barger et al., 2014; Kaas, 2013), which are inherently limited due to major species differences in limbic anatomy. Temporal lobes, for example, occur only in primates and are most fully developed in humans (Barger et al., 2014; Kaas, 2013).
In this study, we performed ultra-high resolution diffusion spectrum imaging tractography in the brains of six healthy human adults using the MGH-USC Connectome MRI scanner to elucidate the structural connectome of selected brainstem and forebrain regions related to homeostasis. We tested the hypothesis that brainstem nuclei known to mediate autonomic and cardiorespiratory function are structurally connected with functionally related forebrain regions relevant to autonomic control, respiration, arousal, and affective and cognitive responses to homeostatic challenges in the adult human brain, including traditionally considered limbic regions. Based upon our connectivity findings, we propose the concept of a “central homeostatic network” (CHN) that expands upon prior models of the limbic system by integrating forebrain and brainstem structures involved in human homeostasis. Furthermore, the concept of a CHN is based upon the recognition that the network regulates not only autonomic functions (i.e., cardiac muscle, smooth muscle, and sweat glands) but also respiratory functions, which are mediated by skeletal muscle. The CHN connectome identified in this study represents an initial step toward elucidating the neuroanatomy of human homeostasis and defining altered connectivity in future studies of patients with disorders of homeostasis, such as sudden infant death syndrome (SIDS) (Kinney et al., 2015), sudden unexplained death in childhood (SUDC) (Kinney et al., 2007, 2009), and sudden unexplained death in epilepsy (SUDEP) (Massey et al., 2014).
Materials and Methods
Clinical dataset
We analyzed the brains of six healthy human subjects without a history of neurological, psychiatric, or medical disease. All studies were performed with written informed consent and the approval of the Institutional Review Board at the Massachusetts General Hospital (Boston, MA). The three men and women ranged in age from 23 to 37 (median 27.5) years.
Diffusion data acquisition and processing
Data were acquired on the MGH-USC Connectome 3 Tesla MRI scanner (McNab et al., 2013; Setsompop et al., 2013) equipped with 300 mT/m maximum gradient strengths. A custom-made 64-channel phased array coil was used for signal reception (Keil et al., 2013). We utilized probabilistic tractography for quantitative connectivity measurements and deterministic tractography for qualitative visualization of the pathways by which streamlines travel between seed and target nodes (Edlow et al., 2012; McNab et al., 2013). We have previously described our methods in detail (McNab et al., 2013) applying a 55-min diffusion spectrum imaging protocol with a 515 q-space lattice, bmax = 10,000 sec/mm2, and 2-mm isotropic voxels.
Brainstem seeds and forebrain targets
We focused on connectivity between six brainstem seed nuclei and seven limbic forebrain target regions (Fig. 1). Four of the six brainstem nuclei were selected because they are key components of the raphe/extra-raphe system and contain neuronal cell bodies that produce serotonin (5-HT), which is critical for homeostatic regulation: (1) median raphe in rostral pons; (2) dorsal raphe in rostral pons and caudal midbrain; (3) caudal raphe (mainly the raphe obscurus) throughout the rostrocaudal length of the medulla oblongata; and (4) paragigantocellularis lateralis (PGCL) in the rostral medulla (Azmitia and Gannon, 1986; Brust et al., 2014; Ray et al., 2011; Severson et al., 2003). In addition, the PGCL is thought to contain the human homologue of the preBotzinger complex, the central rhythm generator of respiration (Feldman et al., 2013; Schwarzacher et al., 2011). The locus coeruleus of the rostral pons was analyzed because it contains neuronal cell bodies that produce norepinephrine (NE), likewise important for homeostatic regulation and arousal (Aston-Jones and Cohen, 2005; Gompf et al., 2010; Li and Nattie, 2006). With diffusion tractography, we were not able to delineate 5-HT- or NE-specific fiber pathways within the raphe/extra-raphe and locus coeruleus pathways, respectively; nevertheless, the connectivity of these brainstem regions of interest (ROIs) as a whole was a surrogate for the transmitter-specific subsets. Two vagal nuclei in the medulla, the dorsal motor nucleus of the vagus, and nucleus of the solitary tract were selected for analysis because, in addition to their roles in sensorimotor aspects of autonomic control, they are involved in the treatment of epilepsy with vagal nerve stimulation (Ogbonnaya and Kaliaperumal, 2013). Due to the very small size of these two nuclei (largest combined diameter in adult human brainstem ∼5 mm), we placed the ROI around these two nuclei together, combined as the vagal complex (VC).
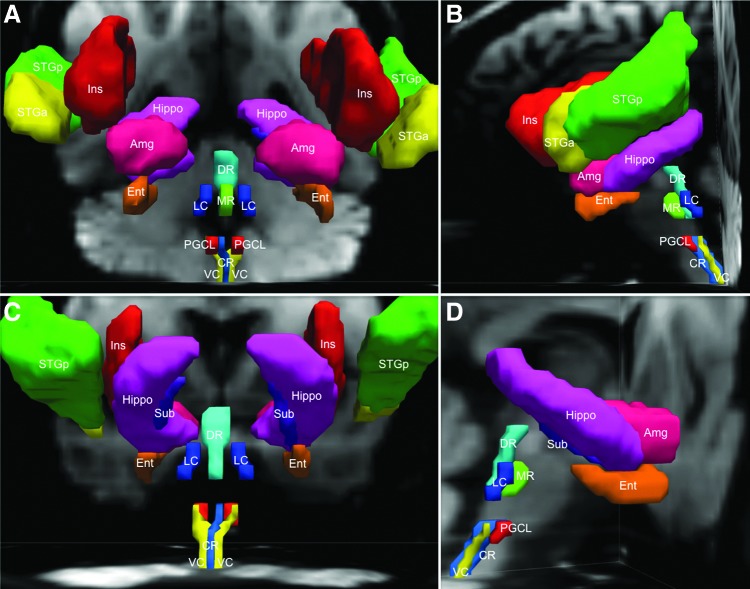
FIG. 1.
Brainstem seed regions and homeostatic forebrain regions are demonstrated with three-dimensional reconstructions in native diffusion space from an anterior (A), left lateral (B), posterior (C), and right lateral (D) perspective. Amg, amygdala; CR, caudal raphe; DR, dorsal raphe; Ent, entorhinal cortex; Hippo, hippocampus; Ins, insula; LC, locus coeruleus; MR, median raphe; PGCL, paragigantocellularis lateralis; STGa, superior temporal gyrus (anterior); STGp, superior temporal gyrus (posterior); Sub, subiculum; VC, vagal complex.
The forebrain target ROIs were the hippocampus proper, subiculum, entorhinal cortex, and amygdala in the medial temporal lobe, the superior temporal gyrus (anterior and posterior) in the lateral temporal lobe, and the insula (Frysinger and Harper, 1990; Koseoglu et al., 2009; Oppenheimer et al., 1992; Ter Horst and Postema, 1997; Ulrich-Lai and Herman, 2009; Vertes et al., 1999). The two lateral sites have been shown to participate in modulation of autonomic and/or respiratory processes in humans and experimental animals (Augustine, 1996; Harper et al., 2013). We did not examine the hypothalamus, which is regarded historically as the head ganglion of the autonomic nervous system (Saper and Lowell, 2014), or the cingulate cortex, which was included in early descriptions of the limbic system by Broca and Papez. Rather, because of our group's focus, we concentrated upon those sites known to be responsible for the greatest burden of homeostasis-related disorders, seizures, and sudden death in clinical practice, that is, hippocampus, amygdala, superior temporal lobe, and insula.
Brainstem seed ROIs were traced manually on the diffusion dataset of each subject by a neurologist (B.L.E.) and neuropathologist (H.C.K.). We determined the neuroanatomic boundaries of the seed ROIs by precise correlation of the radiologic data with three reference templates: (1) an ex vivo human brainstem template of ROIs generated by direct correlation with serially sectioned cytoarchitectural data in the same specimen (stained with hematoxylin and eosin and Luxol-fast-blue) (Edlow et al., 2012); (2) a human brainstem atlas (Paxinos et al., 2011); and (3) a novel template of brainstem ROIs in Montreal Neurologic Institute space that is being made available to the academic community as the Harvard Ascending Arousal Network Atlas (www.martinos.org/resources/aan-atlas). Target forebrain regions were generated using atlases distributed with the FMRIB Software Library (FSL; www.fmrib.ox.ac.uk/fsl) (Jenkinson et al., 2011) (Table 1). These target ROIs were transformed from Montreal Neurologic Institute space to native diffusion space using FSL's nonlinear image registration tool (FNIRT). Of note, diffusion tractography does not provide information about the direction of electrical signaling (i.e., anterograde versus retrograde) along a streamline. Thus, we refer to “brainstem–forebrain connections” based upon our probabilistic tractography methodology, in which brainstem nuclei were used as seeds and forebrain regions as targets. This nomenclature convention is not intended to suggest that homeostasis is mediated solely by ascending pathways from the brainstem to forebrain. Rather, prior studies suggest that homeostatic functions are modulated by bidirectional “bottom-up” and “top-down” signaling between the brainstem and forebrain (Saper, 2002; Ulrich-Lai and Herman, 2009).
Table 1.
Definition of Target ROIs
| Target ROI | Atlas | Atlas ROI(s) used to create target ROI | Initial thresholding before coregistrationa |
|---|---|---|---|
| Insula | Harvard-Oxford Cortical Structural Atlas | Insular cortex | 10–100 |
| STG anterior | Harvard-Oxford Cortical Structural Atlas | STG anterior division | 10–100 |
| STG posterior | Harvard-Oxford Cortical Structural Atlas | STG posterior division | 10–100 |
| Amygdala | Harvard-Oxford Subcortical Structural Atlas | Left amygdala + right amygdala | 10–100 |
| Hippocampus proper | Harvard-Oxford Subcortical Structural Atlas | Left hippocampus + right hippocampus | 10–100 |
| Subiculum | Juelich Histological Atlas | GM hippocampus subiculum L + GM hippocampus subiculum R | 75–100 |
| Entorhinal cortex | Juelich Histological Atlas | GM hippocampus entorrhinal cortex L + GM hippocampus entorrhinal cortex R | 75–100 |
Atlas ROIs were acquired from the FMRIB Software Library (www.fmrib.ox.ac.uk/fsl).
Initial thresholding was individualized for each ROI. After nonlinear coregistration of the target ROIs to native subject space, additional thresholding was performed (80–100).
GM, grey matter; L, left; R, right; ROIs, regions of interest; STG, superior temporal gyrus.
Connectivity analyses
Probabilistic tractography was performed using FMRIB's Diffusion Toolbox with the ball and two stick model (Behrens et al., 2003, 2007). Five thousand streamlines were propagated from each voxel within the seed ROIs, and target ROIs were used as termination masks. To quantitatively compare the probabilistic tractography results for seed–target pairs, we report a “streamline probability” (SP):
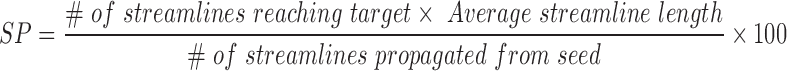 |
The SP is a measure of the probability of a streamline connecting a seed ROI and target ROI, and does not reflect the strength of the neuroanatomic connection (Jbabdi and Johansen-Berg, 2011). Since probabilistic tractography yields a flare of high SP near the seed ROI compared to more distant locations, we used the distance correction option (−pd) in FSL (probtrackx). With this option, the SP at each target voxel equals the number of streamline samples that cross the target voxel multiplied by the average length of those streamlines. The distance correction was applied because the distances between the seed and target ROIs are different, and we did not want the SP measurements to be biased by distance. Quantitative SP-based connectivity of the brainstem–forebrain network was visually displayed using an adaptation of the connectogram technique (Irimia et al., 2012) developed by our laboratory (Edlow et al., 2013).
SP may be influenced by a variety of methodological factors pertaining to diffusion data acquisition and postprocessing. Furthermore, the SP value between node A and node B will be lower if node A has widely distributed connectivity because the probability of a fiber tract connecting with node B goes down as the probability of that fiber tract connecting with other nodes goes up. Nevertheless, quantitative measures, such as SP, obtained from probabilistic tractography provide valuable information about the validity of the reconstructed streamline trajectories. In accordance with recently published recommendations for reporting quantitative imaging biomarker results (Kessler et al., 2015), we calculated the between-subject coefficient of variation (standard deviation/mean) for the SP values of each seed–target pair.
Deterministic tractography was also performed for visualization purposes using the Diffusion Toolkit and TrackVis (www.trackvis.org), as previously described (McNab et al., 2013). To reduce the likelihood of identifying spurious streamlines at sites of white matter crossing (Jones et al., 2013), we rigorously excluded nearby, nonlimbic white matter pathways by manually tracing these nonlimbic ROIs and performing a “NOT” function in TrackVis.
Results
Connectivity overview
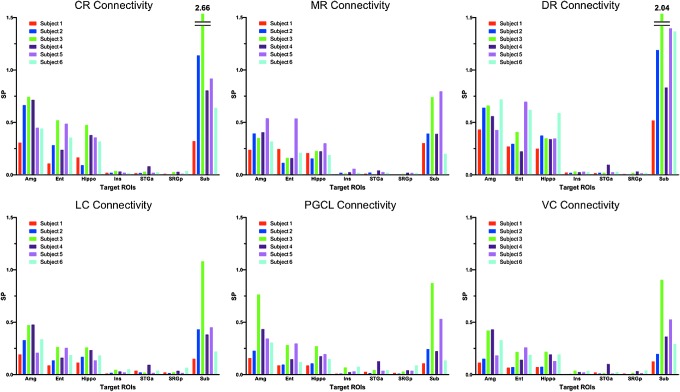
Probabilistic tractography utilizing the MGH-USC Connectome scanner demonstrated reproducible streamlines in all six subjects that connected the selected seed brainstem nuclei (dorsal raphe, median raphe, locus coeruleus, caudal raphe, PGCL, and VC) with target limbic forebrain regions (hippocampus, amygdala, subiculum, entorhinal cortex, insula, anterior superior temporal gyrus, and posterior superior temporal gyrus) (Figs. 2 and 3). The brainstem nucleus with the highest SP with the sampled forebrain regions overall was the dorsal raphe, and with the lowest SP, the VC. While the trajectories of streamlines between the brainstem and limbic forebrain were consistent across the brains of the six subjects studied, the SP measurements for each pathway among the subjects were variable (Fig. 4). The seed-target pairs with the highest between-subject SP coefficients of variation were the VC-superior temporal gyrus anterior (1.10), dorsal raphe-superior temporal gyrus anterior (0.83), and PGCL-subiculum (0.77). The seed-target pairs with the lowest between-subject SP coefficients of variation were the dorsal raphe-insula (0.19), dorsal raphe-amygdala (0.20), and median raphe-hippocampus (0.20). Overall, the between-subject SP coefficients of variation for the six brainstem seeds were within the following ranges for each forebrain target: amygdala (0.20–0.53), entorhinal cortex (0.34–0.59), hippocampus (0.20–0.44), insula (0.19–0.74), superior temporal gyrus anterior (0.55–1.10), superior temporal gyrus posterior (0.53–0.70), and subiculum (0.39–0.77).
FIG. 2.
Streamline probability (SP) measurements and connectogram. (A) SP measurements (mean ± SD) are provided for each seed–target pair. (B) The connectogram of the human central homeostatic network (CHN). Brainstem seed nodes are displayed on the outside of the connectogram and limbic forebrain target nodes at its center. Connectivity is represented quantitatively, with line thickness being proportional to the SP for each dyad. Connectogram lines are color coded according to the brainstem nucleus of origin: turquoise, DR; green, MR; dark blue, LC; red, PGCL; light blue, CR; yellow, VC.
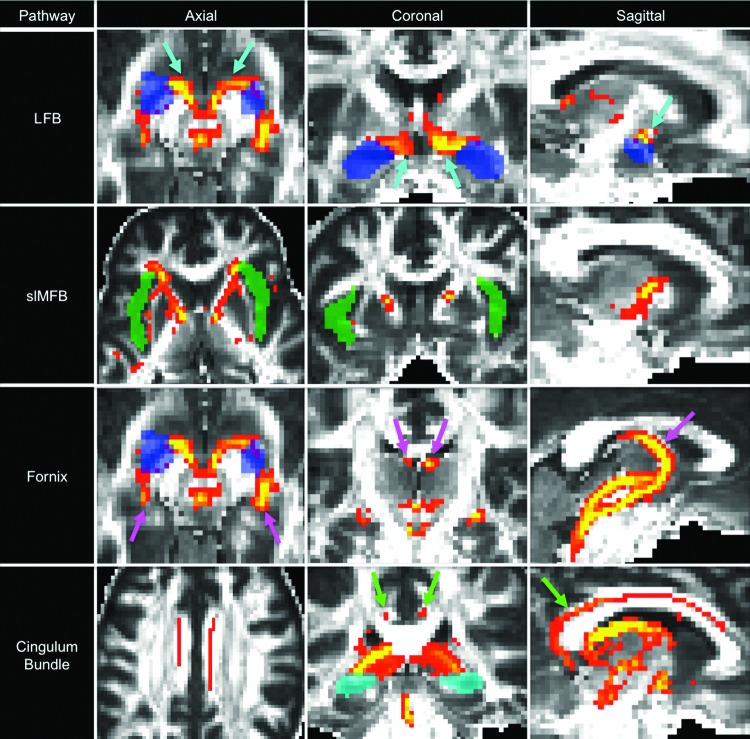
FIG. 3.
Montage of probabilistic tractography data from representative cases. Probabilistic streamlines are shown for the lateral forebrain bundle (LFB), superolateral medial forebrain bundle (slMFB), fornix, and cingulum bundle in the axial, coronal, and sagittal planes. Voxels are color coded according to the number of streamlines passing through, with yellow indicating higher numbers and red indicating fewer. Streamline thresholding (i.e., minimum and maximum number displayed) was adjusted to optimize visualization of the pathways of interest. In the LFB analysis, the turquoise arrows indicate LFB streamlines connecting with the amygdala (semitransparent blue target region of interest [ROI]). In the slMFB analysis, streamlines connect with the insula (semitransparent green ROI). In the fornix analysis, the pink arrows indicate fornix streamlines connecting with the amygdala (axial and coronal images) and hippocampus (sagittal image). In the cingulum bundle analysis, the green arrows indicate cingulum streamlines connecting with the hippocampus (semitransparent turquoise, axial and coronal images) and the entorhinal cortex (sagittal image). All streamlines are propagated from the caudal raphe seed ROI to demonstrate long-range connectivity between the caudal medulla and forebrain targets.
FIG. 4.
SP bar graphs for all seed–target analyses and all subjects (n = 6).
Despite different streamline trajectories between the seed and target ROIs, the subiculum was the site that shared the highest SP with all but one of the brainstem nuclei sampled. The exception was the PGCL, whose highest SP was with the amygdala, not the subiculum. The medial temporal lobe structures (hippocampus, entorhinal cortex, amygdala, and subiculum) demonstrated higher SPs with all brainstem nuclei sampled compared to the lateral temporal lobe and insula (Figs. 2 and 4). The dorsal raphe was the brainstem monoaminergic seed with the highest SP with the medial forebrain targets, followed in decreasing order by the caudal raphe, median raphe, and locus coeruleus.
Pathways within the brainstem
Streamlines from the three medullary seed nuclei ran in three separate pathways within the medullary and pontine tegmentum (Fig. 5). Streamlines of the caudal raphe traversed the midline medullary and pontine tegmentum, the latter level where they passed through the median and dorsal raphe in the rostral pons (Fig. 6). Streamlines of the VC traversed both sides of the dorsolateral tegmentum of the medulla and pons. Streamlines of the PGCL likewise traveled bilaterally, but in the ventrolateral tegmentum of the medulla and pons, overlapping partially with the central tegmental tract on each side. Streamlines of the locus coeruleus traveled bilaterally in the dorsolateral tegmentum of the pons, and those of the median and dorsal raphe in the midline tegmentum of the pons (Fig. 5). Notably, the vast majority of streamlines from bilateral nuclei (e.g., left- and right-sided PGCL, VC, and LC) were visualized ipsilateral to their respective seed nuclei. Thus the term “bilateral” refers to streamlines traversing both sides of the brainstem tegmentum and is not intended to imply that streamlines crossed the midline.
FIG. 5.
Pathways of the human CHN. Connections between brainstem seeds and forebrain targets occur by multiple pathways traveling through the medulla, pons, mesencephalon, and forebrain. Each brainstem seed generates streamlines that travel a unique pathway within the medulla and/or pons. All CHN streamlines converge at the pontomesencephalic junction to travel through the rostral and caudal mesencephalic homeostatic bundles (MHBR and MHBC). At the mesencephalic–diencephalic junction, streamlines from the MHBR and MHBC converge and then distribute to five different forebrain pathways: LFB, slMFB, inferomedial medial forebrain bundle (imMFB), fornix, and cingulum bundle. CTT, central tegmental tract; Hypothal, hypothalamus; N/A, not applicable; PBC, parabrachial complex; VTTC, caudal ventral tegmental tract.
FIG. 6.
Shared raphe connectivity with limbic medial temporal lobe sites. Streamlines between raphe nuclei and the Hippo, Sub, and Amg are shown from a right lateral view, superimposed on axial and sagittal T1-weighted images in a representative subject. Streamlines passing through the CR (blue) and DR (turquoise) are color coded turquoise, while fiber tracts passing through CR and MR (green) are green. Though not measured with tractography, mono-and/or oligo-synaptic connections may be present between the caudal and rostral (median and dorsal) raphe. All network nodes are semitransparent so that streamlines are seen within the nodes, except for Sub, which is opaque and visualized along the inferomedial margin of Hippo. Fiber tracts from the raphe system connect with Hippo, Sub, and Amg through the fornix (Fx) and cingulum bundle (CB), as well as through the LFB, which is located medial to Hippo, Sub, and Amg in this figure. The slMFB and imMFB also contain streamlines that originate in CR and pass through DR and MR.
At the junction of the pons and midbrain, all streamlines from the six brainstem seed nuclei joined two bundles, which we identified and named the rostral and caudal mesencephalic homeostatic bundles (MHBR and MHBC) (Fig. 7). The MHBR continued along the dorsal tegmentum of the midbrain bilaterally, overlapping partially with the central tegmental tract, and then turned ventrally at the rostral midbrain to enter the posterior hypothalamus (level of the mammillary bodies). The MHBC turned ventrally at the pontomesencephalic junction and traveled within the raphe (midline) toward the ventral tegmental area. Upon reaching the posterior aspect of the ventral tegmental area, the MHBC diverged into two distinct bundles, which traveled between the ventral tegmental area on their medial border and the cerebral peduncles on their lateral border, before entering the posterior hypothalamus. Both the MHBR and MHBC contained streamlines from all six brainstem seed nuclei, and streamlines from both bundles became intermingled as they entered the posterior hypothalamus (Fig. 7).
FIG. 7.
The MHBR and MHBC. Streamlines generated from (A) CR (light blue), (B) MR (green), (C) DR (turquoise), (D) PGCL (red), (E) VC (yellow), and (F) LC (blue), all connect with limbic forebrain sites by two discreet bundles in the midbrain: the MHBR and MHBC. All streamlines are shown from a right lateral view, superimposed on axial and sagittal T1-weighted images in a representative subject. Cerebellar, basis pontis, and superior cerebellar peduncle tracts are excluded for clarity.
Pathways within the forebrain
Upon converging at the level of the posterior hypothalamus, streamlines from the MHBR and MHBC yet again diverged, forming four distinct bundles bilaterally (Figs. 8 and 9). Three of these bundles connected with target forebrain homeostatic-related sites; the fourth was the rostral ventral tegmental tract (VTTR), recently described as the primary conduit for connections between brainstem arousal-related nuclei (e.g., locus coeruleus and median and dorsal raphe) and the paraventricular nuclei of the thalamus (Edlow et al., 2012). Of the three bundles that connected with the sampled forebrain sites, we considered two to be branches of the medial forebrain bundle (MFB). The other was the newly identified lateral forebrain bundle (LFB).
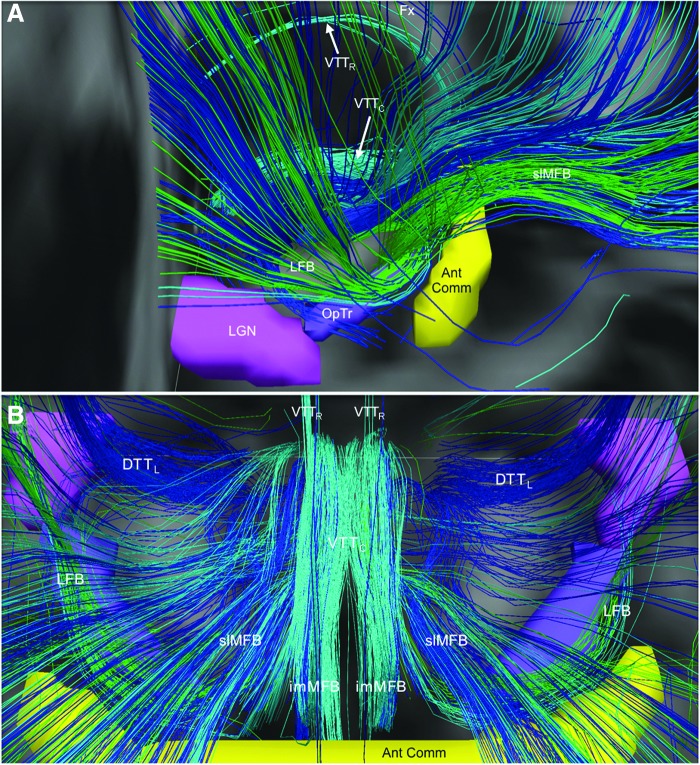
FIG. 8.
Neuroanatomic trajectory of the human LFB. The LFB is shown from right lateral (A) and superior (B) perspectives. LFB streamlines diverge from the mesencephalic homeostatic bundles in the posterior hypothalamus, then travel laterally around the posterior limbs of the internal capsules just posterior to the anterior commissure (Ant Comm, yellow), and superior to the optic tracts (OpTr, purple) and lateral geniculate nuclei (LGN, pink). LFB streamlines enter the external capsule, from which they branch out to limbic targets in the medial temporal lobes. All streamlines are color-coded according to their brainstem nucleus of origin: turquoise, dorsal raphe; green, medial raphe; and blue, locus coeruleus. Streamlines running in close proximity to the LFB are shown: lateral dorsal tegmental tract (DTTL), Fx, imMFB, slMFB, VTTC, and rostral ventral tegmental tract (VTTR).
FIG. 9.
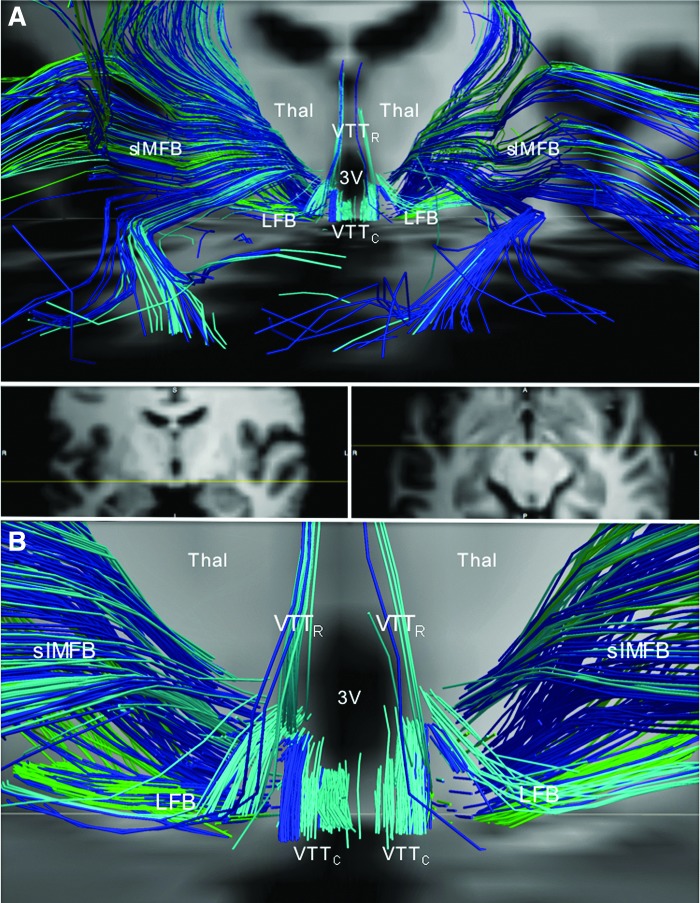
Divergence of the medial forebrain bundle, LFB, and ventral tegmental tracts in the posterior hypothalamus. (A) Anterior view of streamlines generated from the locus coeruleus (dark blue), dorsal raphe (turquoise), and median raphe (green) superimposed upon axial and coronal T1-weighted images (center inset) for a representative subject. In the posterior hypothalamus, streamlines from the locus coeruleus, dorsal raphe, and median raphe diverge as the follows: (1) slMFB, connecting to the prefrontal cortex; (2) VTTR, connecting to the paraventricular nuclei of the thalamus (Thal); (3) the VTTC, connecting to the anterior hypothalamus and basal forebrain, running alongside the imMFB; and (4) LFB, connecting to temporal limbic sites. (B) Zoomed view of the image in (A) demonstrates the divergence of the slMFB, VTTR, VTTC, and LFB in the posterior hypothalamus. Anatomic landmark; third ventricle (3V).
Medial forebrain bundle
The two branches of the MFB were the inferomedial medial forebrain bundle (imMFB) and superolateral medial forebrain bundle (slMFB), consistent with nomenclature recently proposed (Coenen et al., 2012). The main site of imMFB connectivity was the orbitofrontal cortex, a region that was not a prespecified target in this study, but that was readily identified as a site that connected with all of the brainstem nuclei analyzed (Fig. 9). Within the medial hypothalamus and basal forebrain, the imMFB traveled alongside fiber tracts of the caudal ventral tegmental tract (VTTC), recently described as the primary pathway connecting brainstem arousal nuclei to the human hypothalamus and basal forebrain (Edlow et al., 2012).
The slMFB diverged from the imMFB in the posterior hypothalamus and entered the anterior limb of the internal capsule (Fig. 3), as previously described in human adults (Coenen et al., 2012). From there, slMFB streamlines connected with the prefrontal lobe. The slMFB was also the primary conduit of streamlines connecting with the insula and superior temporal gyrus. These latter connections occurred by streamlines that traveled through the anterior limb of the internal capsule and then curved posterolaterally to enter the external capsule before reaching their insular and lateral temporal lobe targets (Fig. 3).
Lateral forebrain bundle
The fourth distinct pathway diverging at the level of the posterior hypothalamus was named by us as the LFB. The LFB was identified bilaterally in probabilistic and deterministic analyses of the brains of all six adult subjects, traveling laterally and circumferentially around the posterior limb of the internal capsule, posterior to the anterior commissure and superior to the optic tracts (Figs. 8 and 9). After this initial course, LFB streamlines passed laterally to the medial temporal lobe targets, that is, hippocampus, subiculum, entorhinal cortex, and amygdala and posteriorly through the external capsule to widely distributed frontal, parietal, and occipital targets.
Fornix and cingulum bundle
In addition to the imMFB, slMFB, and LFB, two additional fiber bundles included streamlines that connected brainstem seed nuclei to forebrain limbic targets: the fornix and cingulum bundle (Figs. 3 and 5–8). Streamlines from all six brainstem nuclei sampled entered the fornix through the hypothalamus and traveled within it to differentially branch to the hippocampus proper, subiculum, entorhinal cortex, and amygdala. Streamlines entered the cingulum bundle after traveling from brainstem nuclei to the basal forebrain through the VTTC; they traversed the cingulum bundle to connect with the hippocampus proper, subiculum, entorhinal cortex, and amygdala.
The human CHN connectogram
The CHN connectogram conveys that all six brainstem seed nuclei were interconnected with all seven limbic forebrain target sites, but with markedly different SPs (Fig. 2B). While the SP results appeared to correlate with the size of the target regions, this effect is not easily corrected for because the probability of streamline connections at each voxel in the target is not expected to be equal. Therefore, the size of the target ROI remains a potential confound in these SP measurements. To ensure that the target ROI size was not the only factor contributing to the SP, we verified that the SP measurements were derived from anatomically plausible pathways for which information was available from animal models (Azmitia and Gannon, 1986; Vertes, 1984a,b) or other diffusion tractography studies of subcortical pathways in the human brain (Coenen et al., 2012; Edlow et al., 2012).
Discussion
The conceptual framework for forebrain modulation of subcortical homeostatic functions has been in place since at least the last century with the canonical observations of Cannon (1929) on the relationship between emotional states and autonomic responses. Mounting evidence from experimental animals supports the idea that homeostasis is mediated by ascending and descending interconnections between brainstem nuclei and forebrain regions, which together regulate autonomic, respiratory, and arousal responses to stress (Azmitia and Gannon, 1986; Feldman et al., 2013; Harper, 1986; Vertes, 1984a,b). While the limbic lobe/system was historically regarded as the neuroanatomic substrate of emotion (Barger et al., 2014; Kaas, 2013), its role in the regulation of homeostasis has been increasingly recognized, and the originally defined sites have been encompassed in the central autonomic network (Beissner et al., 2013; Benarroch, 1993; Mraovitch and Calando, 1999; Saper, 2002), or “flight or fight” system (Nicolaides et al., 2015; Ulrich-Lai and Herman, 2009). In this study, we provide initial evidence for connectivity between forebrain and caudal brainstem regions that participate in the regulation of homeostasis in the human brain. These nodes and connections form, we propose, a CHN because its nodes not only regulate autonomic functions such as “fight or flight” and arousal (e.g., median and dorsal raphe, and locus coeruleus) but also nonautonomic homeostatic functions such as respiration (i.e., PGCL) and regulation of emotion/affect (e.g., amygdala). Within the human CHN, forebrain nodes are interconnected with multiple brainstem nodes over long distances between the cerebral cortex (e.g., insula), pons and medulla, suggesting a “limbic pons” and “limbic medulla” to add to Nauta's (1958) classic recognition of the “limbic midbrain.”
The highest probability of connectivity (SP value) for five of the six brainstem nuclei analyzed was with the subiculum. In animals, the subiculum by inhibitory hypothalamic projections limits or terminates the “fight or flight” stress response mediated by the hypothalamic–pituitary–adrenal (HPA) axis (Lowry, 2002). We found that the human subiculum is connected with brainstem nuclei known to be involved with homeostatic responses, including the median and dorsal raphe, locus coeruleus, VC, caudal raphe, and PGCL. We therefore speculate that these subicular-brainstem interconnections underlie, at least in part, the coordinated HPA axis response in human “fight or flight.”
The MFB is a major white matter conduit for forebrain–brainstem interconnections within the human CHN, consistent with animal studies (Vertes, 1984a,b). We observed that brainstem pathways in the tegmentum of the medulla and pons sampled in this study converge into newly described caudal and rostral divisions of an MHB in the midbrain, and in turn, into the MFB on either side of the third ventricle. Recently, the MFB was visualized for the first time in the human brain with diffusion tractography (Coenen et al., 2009, 2012) and was shown to traverse the anterior limb of the internal capsule on its way to the prefrontal cortex. We confirmed the capsular trajectory of the slMFB and its prefrontal connections, but we also visualized connections with the insula and superior temporal gyrus, two forebrain targets that are known to contribute to homeostatic regulation.
The identification of an LFB in the human brain sheds new light on the white matter pathways by which the medial temporal lobe connects with the brainstem. We observed that the LFB, but not the MFB, connects brainstem homeostatic nuclei with the hippocampus, amygdala, subiculum, and entorhinal cortex. The human LFB shares homologous connectivity properties with the rodent and primate MFB and dorsal raphe cortical tract (Azmitia and Gannon, 1986), but the pattern of brainstem–temporal lobe connectivity visualized in this study by the human LFB has not been previously reported in rodent, primate, or human studies. These human LFB connectivity findings have significant implications for future studies of homeostatic dysfunction in neurologic diseases. Temporal lobe epilepsy, for example, is postulated to be a disorder of the limbic system (Bartolomei et al., 2001; Bonilha et al., 2012; Spencer, 2002), and seizure disorders are increasingly conceptualized as network disorders (Blumenfeld, 2014; Stefan and Lopes da Silva, 2013). Seizures originating in temporal sites have been shown to spread along established axonal pathways (Mraovitch and Calando, 1999; Yoo et al., 2014), allowing the seizure activity to propagate to subcortical arousal nuclei necessary for cortical activation. Our identification of a human LFB thus provides a putative pathway by which temporal lobe seizures may cause dysfunction within brainstem arousal nuclei, with subsequent inhibition of the cerebral cortex and impaired consciousness (Blumenfeld, 2012).
Medullary–forebrain connectivity through the human LFB also provides a potential neuroanatomic basis for CHN disorders such as SUDEP (Engel et al., 2013; Richardson, 2012; Sowers et al., 2013), which is associated with temporal lobe epilepsy (Mueller et al., 2014; Schuele et al., 2011), as well as SIDS and SUDC, which are associated with hippocampal anomalies in infants and young children, respectively (Kinney et al., 2007, 2009, 2015). A paradoxical feature of the CHN is its marked susceptibility to generate and propagate seizures (Harper, 1986; Oliveira et al., 2011)—paradoxical in that a network so vital to survival is so prone to seizures that are inherently dangerous and potentially lethal. We found that the hippocampus, amygdala, and insula—seizure-prone regions of the CHN—are connected to medullary nuclei involved in heart rate and rhythm (VC), respiratory rhythm genesis (PGCL), and cardiorespiratory integration (caudal raphe). We speculate that seizure discharges originating in a hippocampus with a structural or molecular defect may propagate along axonal pathways of the LFB, fornix, and/or cingulum bundle to medullary nuclei, resulting in a dysfunction in brainstem nodes that manifests in lethal cardiac dysrhythmias, respiratory arrest, and/or blood pressure changes.
Notably, recent animal data suggest that SUDEP is linked to cardiorespiratory collapse after a seizure that is associated with spreading depolarization, defined as a self-propagating depolarizing wave that silences neuronal networks (Aiba and Noebels, 2015). Since depolarization waves do not travel along axons, but rather as large gradients of excess extracellular glutamate and potassium, it may be that seizure discharges from a forebrain node of the CHN travel rapidly along axonal pathways to medullary nuclei, where they contribute to the generation of local spreading depolarization. These depolarizations may silence medullary nodes of the CHN, leading to cardiorespiratory arrest and sudden death. The fornix and cingulum bundle were also observed to connect the brainstem to the medial temporal lobe, but these pathways are known to be oligosynaptic, with synapses in the hypothalamus and basal forebrain, respectively. Although diffusion tractography cannot provide information about the number of synapses in a pathway or the direction of electrical signaling (i.e., anterograde or retrograde), we speculate that the LFB may be the most direct route (i.e., containing the fewest synapses) by which electrical signals travel from the medial temporal lobe to the brainstem within the human CHN.
An important consideration in interpreting the findings of this study, as in all studies utilizing diffusion tractography to map brain networks, is the potential for false positive and negative connections (Jbabdi and Johansen-Berg, 2011; Jones et al., 2013; Thomas et al., 2014). The SP measurement is influenced by the structure of neuroanatomic connections (e.g., myelinated versus unmyelinated axons), the trajectories of these connections (e.g., tight bundles versus diffuse projections or crossing fibers), and multiple methodological parameters, including spatial resolution, diffusion weighting (i.e., b value), and the signal-to-noise ratio. For these reasons, the SP cannot be interpreted as a direct measure of connection strength, and we are careful to avoid this terminology. Nonetheless, diffusion imaging techniques detect significant anisotropic water diffusion even within unmyelinated axons (Beaulieu, 2002), which is particularly relevant to imaging diffuse ascending arousal pathways that contain widespread projections of unmyelinated fibers (Azmitia and Gannon, 1986). Furthermore, the dominant diffusion orientations in a voxel are known to correspond to the white matter fiber orientations within that voxel (Basser et al., 1994), and prior tractography studies of complex pathways in the human brainstem show associations with correlative histopathological data (Edlow et al., 2013). Even if diffusely branching axonal projections within the CHN may exceed detection by diffusion tractography, the human CHN's large white matter bundles—the MFB and LFB—share homologous properties with bundles described in rodents and primates (Azmitia and Gannon, 1986; Vertes, 1984a,b) and therefore provide novel targets for in vivo mapping of altered CHN connectivity in human disease. It should be emphasized that human and animal data further indicate that the projections of the ascending arousal systems are not homogeneous across the cerebral cortex, but rather, preferentially innervate different cortical regions (e.g., motor versus sensory sites), as well as different lamina of cerebral cortex.
Perhaps the most challenging methodological limitation, which may not be fully surmountable even with the highly optimized Connectome MRI scanner utilized here, is that axonal pathways that travel closely to one another can be difficult to map independently, as streamlines are prone to “jump” from one pathway to another, creating false positive connections between seed–target pairs. In this study, the convergence of medullary and pontine streamlines into two mesencephalic homeostatic bundles raises the possibility of a “highway effect,” whereby caudal streamlines all enter the same two highways at the pontomesencephalic junction and thereafter cease to demonstrate unique patterns of connectivity. Yet, arguing against the possibility of a “highway effect” confounding our CHN structural connectome are the following results: (1) the qualitative observation that deterministic streamlines emerge from the mesencephalic homeostatic bundles (i.e., exit the highways) in unique neuroanatomic patterns at the mesencephalic–diencephalic junction and (2) the quantitative observation that brainstem probabilistic streamlines demonstrate variable SP measurements with forebrain targets. If all streamlines from the caudal brainstem lost their target specificity once they intermingled within the mesencephalic bundles, one would expect random equally distributed connections with forebrain targets, not the highly specific and reproducible patterns of target connectivity observed here.
Furthermore, although manual segmentation of brainstem homeostatic nuclei in native diffusion space may introduce variability with respect to the neuroanatomic localization of streamline seeds, we observed that caudal streamlines entered the rostral “highways” (i.e., mesencephalic forebrain bundles) at consistent neuroanatomic locations in all subjects. Nevertheless, in recognition of the inherent challenges associated with precise localization of brainstem nuclei, we are distributing to the academic community the Harvard Ascending Arousal Network Atlas (www.martinos.org/resources/aan-atlas)—a resource that we hope will enable replication of these connectivity results and facilitate the ongoing development of automated algorithms for segmentation of homeostatic brainstem nuclei (Bianciardi et al., 2015).
Ultimately, while tractography-based connectivity measurements will always be vulnerable to errors, we believe that the potential discovery and validity of new connectivity patterns in the human brain, as suggested in this study, are grounded in the following: (1) the technical soundness of the MGH-USC Connectome MRI scanner (McNab et al., 2013; Setsompop et al., 2013), which has been optimized for diffusion imaging with unique 300 mT/m gradients; (2) the high angular resolution of the 55-min diffusion spectrum imaging sequence, which has been optimized for mapping complex white matter microstructure with bmax = 10,000 sec/mm2 and a 515 q-space lattice (McNab et al., 2013; Wedeen et al., 2008); (3) the quantitative SP measures provided by the probabilistic tractography analysis (Behrens et al., 2007); and (4) the anatomic congruence in evolutionarily sustained fiber bundles between the brains of humans and those of experimental animals, particularly primates (Azmitia and Gannon, 1986).
Our tractography findings provide novel evidence for the integration of forebrain and brainstem nodes into a single network by long-range fiber bundles to subserve the complex and multifaceted function of homeostasis. Although the well-recognized methodological limitations of diffusion tractography (Jbabdi and Johansen-Berg, 2011; Jones et al., 2013; Thomas et al., 2014) necessitate further validation and refinement of the CHN connectome proposed here, our in vivo tractography analyses provide a structural connectivity map that until now has not been achievable with postmortem labeling or other in vivo imaging methods in the human brain. These findings thus enable future hypothesis testing in experimental models and clinical studies of myriad homeostatic disorders, such as those associated with sudden death (e.g., SIDS, SUDEP, SUDC), psychiatric diseases (e.g., depression, addiction), and severe brain injuries (e.g., paroxysmal sympathetic hyperactivity).
We envision that a connectogram can be built for each disorder of the CHN based upon cohort analysis, enabling clinicians to define which nodes and/or connections are pathogenic and warrant targeted therapies. It may be possible to use structural connectivity maps to pinpoint specific fiber bundles involved in different homeostatic disorders. Of major interest is the confirmation in future large cohorts of intersubject variability in connectivity, which could make an individual more vulnerable to a disease process. Of additional interest is the development of the CHN connectogram from fetal life through adulthood to determine maturational differences in connectivity that may underlie age-related risk. Ultimately, building CHN connectograms for multiple patients with different homeostatic disorders associated with sudden unexpected death may lead to the discovery of a single, final common axonal pathway shared by all entities, or alternatively, multiple pathways with a common lethal outcome.
Acknowledgments
This work was supported by the Cooper Trewin Brighter Days Neuroimaging Study (H.C.K.), Robert's Program on Sudden Unexpected Death in Pediatrics (H.C.K.), CJ Foundation, Barrett Edward Tallman Memorial Fund (H.C.K.), Center for Integration of Medicine and Innovative Technology (B.L.E.), American Academy of Neurology/American Brain Foundation (B.L.E.), National Institutes of Health (K23NS094538, B.L.E.), Oxford-Stanford Big Data in Biomedicine Seed Grant (J.A.M.), and Stanford Department of Radiology Angel Funds (J.A.M.). This research utilized resources provided by National Institutes of Health (P41RR014075) and shared instrumentation grants (1S10RR023401, 1S10RR019307, and 1S10RR023043), as well as by the Massachusetts General Hospital Department of Neurology and Division of Neurocritical Care and Emergency Neurology. The authors thank Drs. Joseph J. Volpe, Eugene E. Nattie, and Bruce R. Rosen for critical reading of the article and Drs. Lawrence L. Wald and Boris Keil for their support in developing, maintaining, and optimizing the hardware components of the MGH-USC Connectome scanner.
Author Disclosure Statement
No competing financial interests exist.
References
- Aiba I, Noebels JL. 2015. Spreading depolarization in the brainstem mediates sudden cardiorespiratory arrest in mouse SUDEP models. Sci Transl Med 7:282ra246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aston-Jones G, Cohen JD. 2005. An integrative theory of locus coeruleus-norepinephrine function: adaptive gain and optimal performance. Annu Rev Neurosci 28:403–450 [DOI] [PubMed] [Google Scholar]
- Augustine JR. 1996. Circuitry and functional aspects of the insular lobe in primates including humans. Brain Res Brain Res Rev 22:229–244 [DOI] [PubMed] [Google Scholar]
- Azmitia EC, Gannon PJ. 1986. The primate serotonergic system: a review of human and animal studies and a report on Macaca fascicularis. Adv Neurol 43:407–468 [PubMed] [Google Scholar]
- Barger N, Hanson KL, Teffer K, Schenker-Ahmed NM, Semendeferi K. 2014. Evidence for evolutionary specialization in human limbic structures. Front Hum Neurosci 8:277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolomei F, Wendling F, Bellanger JJ, Regis J, Chauvel P. 2001. Neural networks involving the medial temporal structures in temporal lobe epilepsy. Clin Neurophysiol 112:1746–1760 [DOI] [PubMed] [Google Scholar]
- Basser PJ, Mattiello J, LeBihan D. 1994. Estimation of the effective self-diffusion tensor from the NMR spin echo. J Magn Reson B 103:247–254 [DOI] [PubMed] [Google Scholar]
- Beaulieu C. 2002. The basis of anisotropic water diffusion in the nervous system – a technical review. NMR Biomed 15:435–455 [DOI] [PubMed] [Google Scholar]
- Behrens TE, Berg HJ, Jbabdi S, Rushworth MF, Woolrich MW. 2007. Probabilistic diffusion tractography with multiple fibre orientations: What can we gain? Neuroimage 34:144–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens TE, Woolrich MW, Jenkinson M, Johansen-Berg H, Nunes RG, Clare S, et al. 2003. Characterization and propagation of uncertainty in diffusion-weighted MR imaging. Magn Reson Med 50:1077–1088 [DOI] [PubMed] [Google Scholar]
- Beissner F, Meissner K, Bar KJ, Napadow V. 2013. The autonomic brain: an activation likelihood estimation meta-analysis for central processing of autonomic function. J Neurosci 33:10503–10511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benarroch EE. 1993. The central autonomic network: functional organization, dysfunction, and perspective. Mayo Clin Proc 68:988–1001 [DOI] [PubMed] [Google Scholar]
- Bianciardi M, Toschi N, Edlow BL, Eichner C, Setsompop K, Polimeni JR, et al. 2015. Toward an in vivo neuroimaging template of human brainstem nuclei of the ascending arousal, autonomic, and motor systems. Brain Connect. [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenfeld H. 2012. Impaired consciousness in epilepsy. Lancet Neurol 11:814–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenfeld H. 2014. What is a seizure network? Long-range network consequences of focal seizures. Adv Exp Med Biol 813:63–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonilha L, Nesland T, Martz GU, Joseph JE, Spampinato MV, Edwards JC, et al. 2012. Medial temporal lobe epilepsy is associated with neuronal fibre loss and paradoxical increase in structural connectivity of limbic structures. J Neurol Neurosurg Psychiatry 83:903–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broca P. 1878. Anatomie comparee des circonvolutions cerebrales: Le grand lobe limbique et la scissure limbique dans la serie des mammifères. Revue d'Anthropologie 1:385–498 [Google Scholar]
- Brust RD, Corcoran AE, Richerson GB, Nattie E, Dymecki SM. 2014. Functional and developmental identification of a molecular subtype of brain serotonergic neuron specialized to regulate breathing dynamics. Cell Rep 9:2152–2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon WB. 1929. Bodily Changes in Pain, Hunger, Fear, and Rage. New York: Appleton-Century-Crofts [Google Scholar]
- Coenen VA, Honey CR, Hurwitz T, Rahman AA, McMaster J, Burgel U, et al. 2009. Medial forebrain bundle stimulation as a pathophysiological mechanism for hypomania in subthalamic nucleus deep brain stimulation for Parkinson's disease. Neurosurgery 64:1106–1114; discussion 1114–1105 [DOI] [PubMed] [Google Scholar]
- Coenen VA, Panksepp J, Hurwitz TA, Urbach H, Madler B. 2012. Human medial forebrain bundle (MFB) and anterior thalamic radiation (ATR): imaging of two major subcortical pathways and the dynamic balance of opposite affects in understanding depression. J Neuropsychiatry Clin Neurosci 24:223–236 [DOI] [PubMed] [Google Scholar]
- Edlow BL, Haynes RL, Takahashi E, Klein JP, Cummings P, Benner T, et al. 2013. Disconnection of the ascending arousal system in traumatic coma. J Neuropathol Exp Neurol 72:505–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlow BL, Takahashi E, Wu O, Benner T, Dai G, Bu L, et al. 2012. Neuroanatomic connectivity of the human ascending arousal system critical to consciousness and its disorders. J Neuropathol Exp Neurol 71:531–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel J, Jr., Thompson PM, Stern JM, Staba RJ, Bragin A, Mody I. 2013. Connectomics and epilepsy. Curr Opin Neurol 26:186–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman JL, Del Negro CA, Gray PA. 2013. Understanding the rhythm of breathing: so near, yet so far. Annu Rev Physiol 75:423–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frysinger RC, Harper RM. 1990. Cardiac and respiratory correlations with unit discharge in epileptic human temporal lobe. Epilepsia 31:162–171 [DOI] [PubMed] [Google Scholar]
- Gompf HS, Mathai C, Fuller PM, Wood DA, Pedersen NP, Saper CB, et al. 2010. Locus ceruleus and anterior cingulate cortex sustain wakefulness in a novel environment. J Neurosci 30:14543–14551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper RM. 1986. State-related physiological changes and risk for the sudden infant death syndrome. Aust Paediatr J 22 Suppl 1:55–58 [PubMed] [Google Scholar]
- Harper RM, Kumar R, Ogren JA, Macey PM. 2013. Sleep-disordered breathing: effects on brain structure and function. Respir Physiol Neurobiol 188:383–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia A, Chambers MC, Torgerson CM, Van Horn JD. 2012. Circular representation of human cortical networks for subject and population-level connectomic visualization. Neuroimage 60:1340–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jbabdi S, Johansen-Berg H. 2011. Tractography: where do we go from here? Brain Connect 1:169–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM. 2011. FSL. Neuroimage 62:782–790 [DOI] [PubMed] [Google Scholar]
- Jones DK, Knosche TR, Turner R. 2013. White matter integrity, fiber count, and other fallacies: the do's and don'ts of diffusion MRI. Neuroimage 73:239–254 [DOI] [PubMed] [Google Scholar]
- Kaas JH. 2013. The evolution of brains from early mammals to humans. Wiley Interdiscip Rev Cogn Sci 4:33–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keil B, Blau JN, Biber S, Hoecht P, Tountcheva V, Setsompop K, et al. 2013. A 64-channel 3T array coil for accelerated brain MRI. Magn Reson Med 70:248–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler LG, Barnhart HX, Buckler AJ, Choudhury KR, Kondratovich MV, Toledano A, et al. 2015. The emerging science of quantitative imaging biomarkers terminology and definitions for scientific studies and regulatory submissions. Stat Methods Med Res 24:9–26 [DOI] [PubMed] [Google Scholar]
- Kinney HC, Armstrong DL, Chadwick AE, Crandall LA, Hilbert C, Belliveau RA, et al. 2007. Sudden death in toddlers associated with developmental abnormalities of the hippocampus: a report of five cases. Pediatr Dev Pathol 10:208–223 [DOI] [PubMed] [Google Scholar]
- Kinney HC, Chadwick AE, Crandall LA, Grafe M, Armstrong DL, Kupsky WJ, et al. 2009. Sudden death, febrile seizures, and hippocampal and temporal lobe maldevelopment in toddlers: a new entity. Pediatr Dev Pathol 12:455–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinney HC, Cryan JB, Haynes RL, Paterson DS, Haas EA, Mena OJ, et al. 2015. Dentate gyrus abnormalities in sudden unexplained death in infants: morphological marker of underlying brain vulnerability. Acta Neuropathol 129:65–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koseoglu E, Kucuk S, Arman F, Ersoy AO. 2009. Factors that affect interictal cardiovascular autonomic dysfunction in temporal lobe epilepsy: role of hippocampal sclerosis. Epilepsy Behav 16:617–621 [DOI] [PubMed] [Google Scholar]
- Li A, Nattie E. 2006. Catecholamine neurones in rats modulate sleep, breathing, central chemoreception and breathing variability. J Physiol 570:385–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry CA. 2002. Functional subsets of serotonergic neurones: implications for control of the hypothalamic-pituitary-adrenal axis. J Neuroendocrinol 14:911–923 [DOI] [PubMed] [Google Scholar]
- Maclean PD. 1952. Some psychiatric implications of physiological studies on frontotemporal portion of limbic system (visceral brain). Electroencephalogr Clin Neurophysiol 4:407–418 [DOI] [PubMed] [Google Scholar]
- Massey CA, Sowers LP, Dlouhy BJ, Richerson GB. 2014. Mechanisms of sudden unexpected death in epilepsy: the pathway to prevention. Nat Rev Neurol 10:271–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNab JA, Edlow BL, Witzel T, Huang SY, Bhat H, Heberlein K, et al. 2013. The human connectome project and beyond: initial applications of 300 mT/m gradients. Neuroimage 80:234–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mraovitch S, Calando Y. 1999. Interactions between limbic, thalamo-striatal-cortical, and central autonomic pathways during epileptic seizure progression. J Comp Neurol 411:145–161 [PubMed] [Google Scholar]
- Mueller SG, Bateman LM, Laxer KD. 2014. Evidence for brainstem network disruption in temporal lobe epilepsy and sudden unexplained death in epilepsy. Neuroimage Clin 5:208–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nauta WJ. 1958. Hippocampal projections and related neural pathways to the midbrain in the cat. Brain 81:319–340 [DOI] [PubMed] [Google Scholar]
- Nicolaides NC, Kyratzi E, Lamprokostopoulou A, Chrousos GP, Charmandari E. 2015. Stress, the stress system and the role of glucocorticoids. Neuroimmunomodulation 22:6–19 [DOI] [PubMed] [Google Scholar]
- Ogbonnaya S, Kaliaperumal C. 2013. Vagal nerve stimulator: evolving trends. J Nat Sci Biol Med 4:8–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira MS, Pacheco LF, Mello CF, Cavalheiro EA, Garrido-Sanabria ER. 2011. Epileptiform activity in the limbic system. Front Biosci (Schol Ed) 3:565–593 [DOI] [PubMed] [Google Scholar]
- Oppenheimer SM, Gelb A, Girvin JP, Hachinski VC. 1992. Cardiovascular effects of human insular cortex stimulation. Neurology 42:1727–1732 [DOI] [PubMed] [Google Scholar]
- Papez JW. 1937. A proposed mechanism of emotion. Arch Neur Psych 38:725–743 [DOI] [PubMed] [Google Scholar]
- Paxinos G, Xu-Feng H, Sengul G, Watson C, 2011. Organization of brainstem nuclei. In: Mai JK, Paxinos G. (eds.) The Human Nervous System. Amsterdam: Elsevier, pp. 260–327 [Google Scholar]
- Ray RS, Corcoran AE, Brust RD, Kim JC, Richerson GB, Nattie E, et al. 2011. Impaired respiratory and body temperature control upon acute serotonergic neuron inhibition. Science 333:637–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson MP. 2012. Large scale brain models of epilepsy: dynamics meets connectomics. J Neurol Neurosurg Psychiatry 83:1238–1248 [DOI] [PubMed] [Google Scholar]
- Saper CB. 2002. The central autonomic nervous system: conscious visceral perception and autonomic pattern generation. Annu Rev Neurosci 25:433–469 [DOI] [PubMed] [Google Scholar]
- Saper CB, Lowell BB. 2014. The hypothalamus. Curr Biol 24:R1111–R1116 [DOI] [PubMed] [Google Scholar]
- Schuele SU, Afshari M, Afshari ZS, Macken MP, Asconape J, Wolfe L, et al. 2011. Ictal central apnea as a predictor for sudden unexpected death in epilepsy. Epilepsy Behav 22:401–403 [DOI] [PubMed] [Google Scholar]
- Schwarzacher SW, Rub U, Deller T. 2011. Neuroanatomical characteristics of the human pre-Botzinger complex and its involvement in neurodegenerative brainstem diseases. Brain 134:24–35 [DOI] [PubMed] [Google Scholar]
- Setsompop K, Kimmlingen R, Eberlein E, Witzel T, Cohen-Adad J, McNab JA, et al. 2013. Pushing the limits of in vivo diffusion MRI for the Human Connectome Project. Neuroimage 80:220–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severson CA, Wang W, Pieribone VA, Dohle CI, Richerson GB. 2003. Midbrain serotonergic neurons are central pH chemoreceptors. Nat Neurosci 6:1139–1140 [DOI] [PubMed] [Google Scholar]
- Sowers LP, Massey CA, Gehlbach BK, Granner MA, Richerson GB. 2013. Sudden unexpected death in epilepsy: fatal post-ictal respiratory and arousal mechanisms. Respir Physiol Neurobiol 189:315–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer SS. 2002. Neural networks in human epilepsy: evidence of and implications for treatment. Epilepsia 43:219–227 [DOI] [PubMed] [Google Scholar]
- Stefan H, Lopes da Silva FH. 2013. Epileptic neuronal networks: methods of identification and clinical relevance. Front Neurol 4:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ter Horst GJ, Postema F. 1997. Forebrain parasympathetic control of heart activity: retrograde transneuronal viral labeling in rats. Am J Physiol 273:H2926–H2930 [DOI] [PubMed] [Google Scholar]
- Thomas C, Ye FQ, Irfanoglu MO, Modi P, Saleem KS, Leopold DA, et al. 2014. Anatomical accuracy of brain connections derived from diffusion MRI tractography is inherently limited. Proc Natl Acad Sci U S A 111:16574–16579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich-Lai YM, Herman JP. 2009. Neural regulation of endocrine and autonomic stress responses. Nat Rev Neurosci 10:397–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vertes RP. 1984a. A lectin horseradish peroxidase study of the origin of ascending fibers in the medial forebrain bundle of the rat. The lower brainstem. Neuroscience 11:651–668 [DOI] [PubMed] [Google Scholar]
- Vertes RP. 1984b. A lectin horseradish peroxidase study of the origin of ascending fibers in the medial forebrain bundle of the rat. The upper brainstem. Neuroscience 11:669–690 [DOI] [PubMed] [Google Scholar]
- Vertes RP, Fortin WJ, Crane AM. 1999. Projections of the median raphe nucleus in the rat. J Comp Neurol 407:555–582 [PubMed] [Google Scholar]
- Wedeen VJ, Wang RP, Schmahmann JD, Benner T, Tseng WY, Dai G, et al. 2008. Diffusion spectrum magnetic resonance imaging (DSI) tractography of crossing fibers. Neuroimage 41:1267–1277 [DOI] [PubMed] [Google Scholar]
- Yoo JY, Farooque P, Chen WC, Youngblood MW, Zaveri HP, Gerrard JL, et al. 2014. Ictal spread of medial temporal lobe seizures with and without secondary generalization: an intracranial electroencephalography analysis. Epilepsia 55:289–295 [DOI] [PMC free article] [PubMed] [Google Scholar]