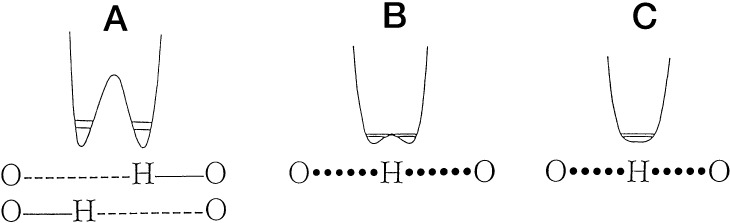
Hydrogen bonding represents one of the primary interactions controlling chemical structure and reactivity. The properties of H-bonds are predicted to vary with the distance between the H-donor and H-acceptor, Figure 1, from a canonical structure (>2.5 Å, Figure 1A), in which the proton resides within a double welled structure, to one in which the proton has become fully delocalized within a single well (≪2.5 Å, Figure 1C).
Figure 1.
Categorization of hydrogen bonds based on donor–acceptor distances, from >2.5 Å (A) to ≪2.5 Å (C). The horizontal lines within the wells represent zero point energies for protium and deuterium. The low barrier hydrogen bond, LBHB (B), has the unique property of a barrier height for H-transfer that has fallen below the ground state vibrational levels of the donor and acceptor. Reprinted with permission from ref (1). Copyright 1998 American Society for Biochemistry and Molecular Biology, Inc.
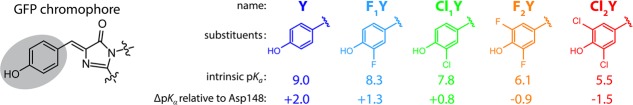
The low barrier hydrogen bond (LBHB), Figure 1B, lies between these extreme states, with a predicted donor–acceptor distance in the range of 2.5 Å and a hydrogen transfer just above the top of the reaction barrier. Analogous to Figure 1C, the hydrogen is equally shared between its donor and acceptor. Over a period of decades, the presence and role of such LBHBs has been a topic of ongoing controversy. At the heart of the controversy has been the question of whether enzymes are capable of accelerating rates for C–H activation by matching active site pKas for the hydrogen donor and acceptor, producing a greatly reduced reaction barrier height, and a resulting highly accelerated rate for bond cleavage.2 In a new investigation, Oltrogge and Boxer turn to the well-characterized green fluorescent protein (GFP) to pursue the properties of short hydrogen bonds within a biological context. The results from their elegant experiments, which depend on the incorporation of a series of unnatural amino acids to produce modified protein chromophores with pKas that vary by 3.5 units (Figure 2), indicate that the active site hydrogen bond within GFP is short but not delocalized.3
Figure 2.
Structure of the natural GFP chromophore and the modified tyrosines used to generate modified chromphores with altered pKa values. The referenced pKa values are those measured within the denatured protein variants. Reprinted with permission from ref (3). Copyright 2015 American Chemical Society.
Historically, evidence for LBHBs in proteins has come from a compelling and repeated observation of chemical shifts in 1H NMR spectra that are significantly downfield shifted (to ca. 17–21 ppm), together with inverse fractionation factors (of ca. 0.3) for the behavior of such H-bonds in D2O vs H2O.1 The latter is attributed to a decrease in bond order at the transferred hydrogen that increases discrimination against deuterium in favor of an accumulation of protium. The new studies by Oltrogge and Boxer are multifaceted. They start by solving the X-ray crystal structures for each of the proteins with modified chromophores, showing a consistently short donor–acceptor distance of ca. 2.45 Å. The experimental findings become especially intriguing when UV/vis spectra are interrogated at low pH where large differences in absorbance spectra emerge correlating with the pKa of the modified chromophore. (By contrast, high pH shows essentially identical properties among the variants.)
Oltrogge and Boxer proceed to simulate spectra for the low pH spectra, using methods developed by McKenzie4 for the analysis of the extent of coupling between H-bonded diabatic states. Focusing on the magnitude of the off-diagonal coupling factor, ΔDA, they are able to simulate the low pH trends in both spectral position and line width; although a first approximation fit of the data suggested a symmetrical H-bond, further adjustments to increase the goodness of fit produce a model in which the ground state vibrational levels of reactants clearly lie beneath the top of the barrier (closer to A in Figure 1). Further model discrimination was possible by collecting spectra in mixtures of H2O and D2O and analyzing their resulting isotopic spectral shifts and fractionation factors. Once again, comparison of experiments to computation shows behavior that deviates significantly from that expected for a symmetrical LBHB. Despite the short distances and apparent matching of pKa values within the modified forms of GFP, Oltrogge and Boxer conclude that the short H-bond in GFP is perturbed, but lacks the critical features diagnostic of an LBHB.3
While it is hoped that the authors will take the next step—to actually visualize the position of the proton within their available structures using neutron diffraction methods—the present analysis adds another brick to the edifice against truly symmetrical H-bond in protein structures. At the same time, these new data add support for the accumulated evidence in support of short, perturbed H-bonds within proteins.5 This raises important questions regarding the origin of such short bonds and their possible relevance to catalysis. In the area of enzymatic C–H cleavage, it has been shown repeatedly that hydrogen transfer occurs by tunneling mechanisms that require achievement of donor–acceptor distances that are reduced relative to initial van der Waals distances, typically attributed to protein conformational sampling that transiently achieves the requisite tunneling-ready distances.6
Studies of a methyl transfer reaction, catalyzed by the enzyme catechol O-methyltransferase (COMT), may offer findings more directly analogous to the cumulative evidence for short hydrogen bonded structures in ground-state structures of proteins. Extensive X-ray studies on COMT show distances between the sulfur atom of the methyl donor in S-adenosylmethionine and the oxygen anion of a catechol-like inhibitor in the range of 2.7 Å, significantly reduced from a van der Waals distance of ca. 3.2 Å (cf. ref (7)). QM/MM modeling of an enzyme–S-adenosylmethionine–substrate ground-state structure (using graphical processing units that enable interrogation of a greatly extended QM region) indicates that ca. 25 residues of COMT within the QM regime are required to reproduce the experimentally observed short methyl donor to acceptor distances.8 These experimental and computational findings indicate that an extensive region of protein participates in generating the tight/precise packing within enzyme active sites that is increasingly linked to highly efficient catalysis (cf. ref (6)). Such considerations suggest that the frequent detection of short H-bonded structures within proteins may be a consequence of the strategies used by proteins to achieve high turnover rates, as opposed to the direct origin of the catalytic rate acceleration itself.
References
- Cleland W. W.; Frey P. A.; Gerlt J. A. The Low Barrier Hydrogen Bond in Enzymatic Catalysis. J. Biol. Chem. 1998, 273, 25529–25532. [DOI] [PubMed] [Google Scholar]
- Gerlt J. A.; Gassman P. G. An Explanation for Rapid Enzyme Catalyzed Proton Abstraction from Carbon Acids: Importance of Late Transition States in Concerted Mechanisms. J. Am. Chem. Soc. 1993, 115, 11552–11568. [Google Scholar]
- Oltrogge L. M.; Boxer S. G. Short Hydrogen Bonds and Proton Delocalization in Green Fluorescent Protein (GFP). ACS Cent. Sci. 2015, 10.1021/acscentsci.5b00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie R. H. A Diabatic State Model for Donor-Acceptor Hydrogen Vibrational Frequency Shifts in Hydrogen Bonded Complexes. Chem. Phys. Lett. 2012, 535, 196–200. [Google Scholar]
- Jacob D. G.; Buytendyk A. M.; Wang D.; Bowen K. H.; Collins K. D. Strong, Low-Barrier Hydrogen Bonds May Be Available to Enzymes. Biochemistry 2014, 53, 344–349. [DOI] [PubMed] [Google Scholar]
- Klinman J. P.; Kohen A. Hydrogen Tunneling Links Protein Dynamics to Enyme Catalysis. Annu. Rev. Biochem. 2013, 82, 471–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford K.; Le Trong I.; Stenkamp R. E.; Parson W. W. Crystal Structures of Human 108V and 108M Catechol O-Methyltransferase. J. Mol. Biol. 2008, 380, 120–130. [DOI] [PubMed] [Google Scholar]
- Zhang J.; Kulik H. J.; Martinez T. J.; Klinman J. P.. Mediation of Donor Acceptor Distance in an Enzymatic Methyl Transfer Reaction. Proc. Natl. Acad. Sci. U.S.A., in press, DOI: 10.1073/pnas.1506792112. [DOI] [PMC free article] [PubMed] [Google Scholar]