Abstract
The inability to coordinate cellular metabolic processes with the cellular and organismal nutrient environment leads to a variety of disorders, including diabetes and obesity. Nutrient-sensing protein kinases, such as AMPK and mTOR, play a pivotal role in metabolic regulation and are promising therapeutic targets for the treatment of disease. In this Extra View, we describe another member of the nutrient-sensing protein kinase group, PAS kinase, which plays a role in the regulation of glucose utilization in both mammals and yeast. PAS kinase deficient mice are resistant to high fat diet-induced weight gain, insulin resistance and hepatic triglyceride hyperaccumulation, suggesting a role for PAS kinase in the regulation of glucose and lipid metabolism in mammals. Likewise, PAS kinase deficient yeast display altered glucose partitioning, favoring glycogen biosynthesis at the expense of cell wall biosynthesis. As a result, PAS kinase deficient yeast are sensitive to cell wall perturbing agents. This partitioning of glucose in response to PAS kinase activation is due to phosphorylation of Ugp1, the enzyme primarily responsible for UDP-glucose production. The two yeast PAS kinase homologs, Psk1 and Psk2, are activated by two stimuli, cell integrity stress and nonfermentative carbon sources. We review what is known about yeast PAS kinase and describe a genetic screen that may help elucidate pathways involved in PAS kinase activation and function.
Keywords: PSK, PAS kinase, Ugp1, cell integrity, cell wall, metabolic regulation, sensory protein kinase
Cells have evolved complex mechanisms to sense their nutritional environment and regulate growth and proliferation accordingly. Failure to properly coordinate cellular metabolism results in a variety of human diseases.1 The well-known 5′-AMP Activated Protein Kinase (AMPK) and Target of Rapamycin (mTOR) nutrient-sensing protein kinases play a critical role in the coordination of cellular functions with nutritional status and have been implicated in the pathogenesis of human disease.2,3 PAS kinase, another nutrient-sensing protein kinase, has been shown to be involved in glucose sensing and metabolic regulation. Specifically, PAS kinase deficient mice have an increased metabolic rate and are resistant to high fat diet-induced obesity, liver triglyceride accumulation and insulin resistance.4 The resistance to diet-induced obesity has also recently been reported by another group.5 In addition, PAS kinase is activated both transcriptionally and post-translationally in pancreatic islet beta-cells in response to high glucose.6 Taken together, these results suggest that mammalian PAS kinase participates in the coordination of metabolism with nutrient status and may contribute to the development of diabetes and obesity (reviewed in ref. 7). However, the molecular mechanisms of PAS kinase activation and function, including mammalian substrates or interacting partners, remain largely unknown.
PAS kinase is highly conserved from yeast to man. We are currently engaged in efforts to identify PAS kinase regulators and substrates in the budding yeast S. cerevisiae. We recently showed that yeast PAS kinase is regulated in response to nutrient conditions,8 providing powerful genetic and biochemical tools to study the regulation and function of PAS kinase in a simpler eukaryotic model organism. The identification of activation and function mechanisms in S. cerevisiae may contribute to understanding the role of human PAS kinase in metabolic regulation and the pathogenesis of metabolic disease.
PAS Kinase Structure
PAS kinase contains both a canonical serine/threonine kinase catalytic domain and a regulatory PAS (Per-Arnt-Sim) domain. PAS domains are sensory modules that regulate an attached functional domain in cis, such as histidine kinases (i.e., bacterial FixL, DosT), bHLH DNA binding domains (i.e., AhR, HIF-1α and Clock), and potassium channels (i.e., hERG). PAS domains have been identified in over 1,100 proteins from all phylogenetic kingdoms.9–11 The structure of the PAS domain is malleable, allowing adaptation of this domain to a variety of functions by varying the ligand-binding capacity to favor protein-protein interactions or binding of discrete small molecules. Many PAS domains have been shown to sense the intracellular environment by reversibly binding small molecules (e.g., ATP or citrate) 11,12 or sensing environmental changes through bound cofactors (e.g., heme to sense oxygen or FMN to sense blue light).13–15 Thus, PAS domains respond to a diverse array of nutrients and metabolites. The coupling of a sensory PAS domain with a protein kinase domain is consistent with a role for PAS kinase in sensory-coupled signal transduction.
The N-terminal human PAS kinase domain (hPASK) PAS domain specifically interacts with and inactivates the C-terminal kinase catalytic domain both in cis and in trans.16,17 The three-dimensional NMR structure of the hPASK PAS domain has been determined by the laboratory of Dr. Kevin Gardner.16,17 Although the biological ligand for PAS kinase is unknown, Gardner’s group demonstrated the ability of the hPASK PAS domain to bind small organic molecules from a chemical library. The hPASK PAS domain adopts the standard mixed α/β PAS fold that consists of a five-stranded antiparallel β-sheet flanked by several α helices (from N to C terminus Aβ, Bβ, Cα, Dα, Eα, Fα, Gβ, Hβ, Iβ); however, it contains an unusual and dynamic Fα helix and FG loop. In other PAS domain containing proteins, these two regions interact with the hydrophobic core or with bound cofactors. In hPASK, they were shown to be involved in two important interactions, the Fα helix and Gβ strands were shown to be involved in binding small molecules while the FG loop participates in direct interaction with the kinase domain. The three-dimensional NMR structure places these two flexible PAS domain regions in close proximity and supports a model in which PAS domain ligand binding regulates the interaction of the kinase and PAS domain. We propose that an endogenous small molecule binds to the PAS domain and disrupts the PAS and kinase domain interaction, thereby activating PAS kinase in vivo.
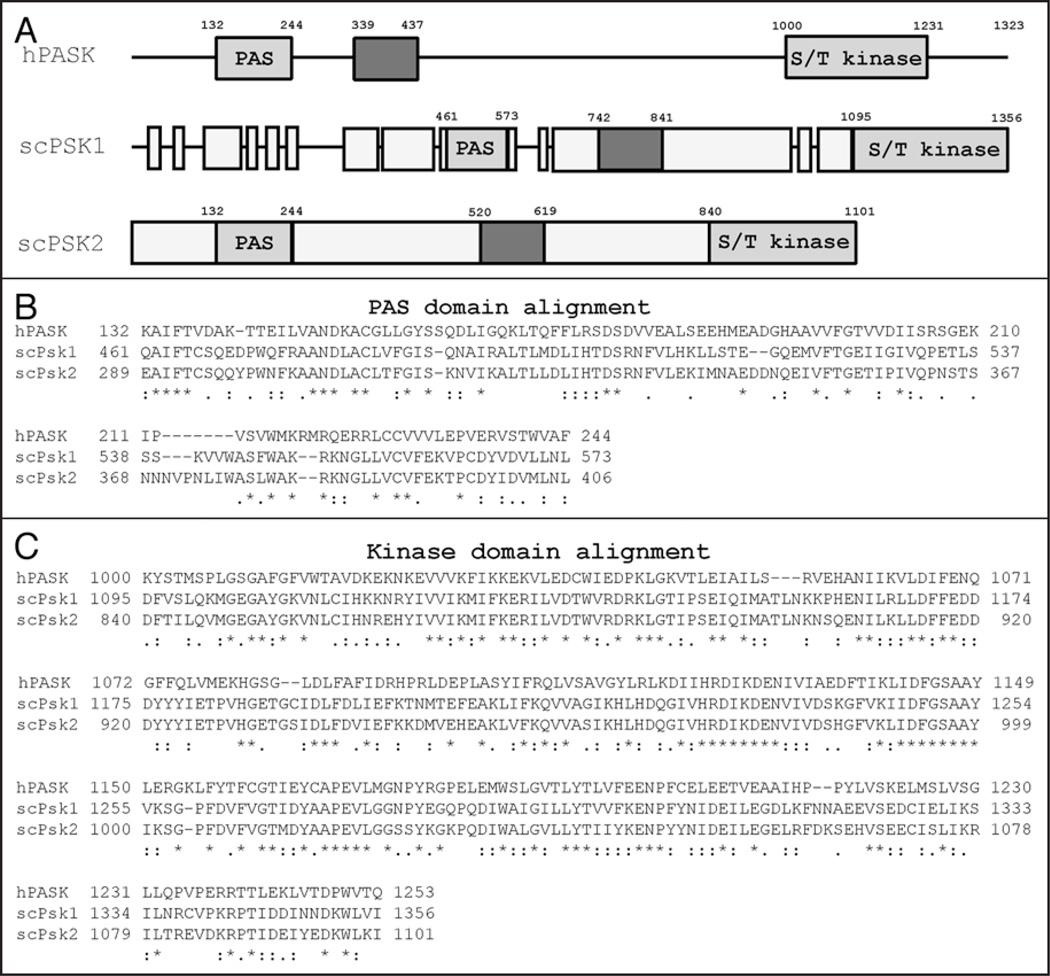
The S. cerevisiae genome contains two well-conserved homologs of hPASK, PSK1 and PSK2, which are highly similar to one another. An alignment of the most highly conserved regions of these proteins, the PAS and kinase domains, with hPASK is shown in Figure 1. The conservation of the PAS and kinase domains is consistent among the PASK orthologs identified from other organisms suggesting a prominent role for these two domains. Although no structural data from the yeast PAS kinase proteins are available, the Psk1 and Psk2 kinase domains are well conserved with 61% and 62% homology to hPASK, respectively. Yeast Psk1 and Psk2, share an overall homology to one another of 71%, but have high homology in the kinase (90%) and PAS (81%) domain regions. The major differences between the yeast proteins are the addition of 140 residues at the N-terminus of Psk1, another 70 immediately following the PAS domain, and 35 just prior to the kinase domain (see Fig. 1). The existence of two yeast PAS kinases is most likely the result of a whole-genome duplication that is believed to have occurred in an ancestor of S. cerevisiae18. Following this widespread duplication most redundant genes were lost while others evolved specialized roles that allowed for selection. These paralogous genes, which evolved following genome duplication, are known as ohnologous genes.19–21 The ohnologous PAS kinase genes have most likely evolved related but separate functions, allowing for evolutionary selection of both genes. One well-known example of gene duplication and divergence in protein function are the yeast Target of Rapamycin (Tor1/2) proteins. Tor1 and Tor2 have shared functions as well as unique regulatory functions that control cellular processes such as translation, transcription, autophagy, meiosis and actin/ cytoskeleton organization.22 As ohnologous genes, PSK1 and PSK2 may also have evolved different, but related, roles in yeast.
Figure 1.
Alignment of the human and yeast PAS kinase PAS and kinase domains. A schematic of hPASK and S. cerevisiae PAS kinase homologs (A) and alignment of the PAS (N-terminus) and kinase (C-terminus) domains (B) and (C). Grey boxes specify regions of similarity between the PAS kinase proteins and varying shades indicate discrete regions of homology (the PAS domains and kinase domains are indicated). Alignments of human PASK (hPASK) and S. cerevisiae Psk1 (scPsk1) and Psk2 (scPsk2) were produced using ClustalW.57 The degree of amino acid conservation is denoted by “*” (identical residues in all sequences), “:” (highly conserved amino acids) and “.” (weakly conserved amino acids).
Regulation and Function of Yeast PAS Kinase
A deletion of both yeast PAS kinase genes, PSK1 and PSK2, renders haploid yeast unable to grow on galactose at high temperatures (galts), while a psk2 deletion causes a similar growth defect and a deletion of psk1 causes only a minor defect. The phenotype of the double deletion may be rescued by a variety of high copy suppressors, most of which are involved in carbohydrate metabolism, protein translation or cell wall biosynthesis.23 In addition, psk1 psk2 mutant strains are sensitive to growth on cell wall perturbing agents, such as calcofluor white and congo red, supporting a role for PAS kinase in the maintenance of cell wall integrity.24
Although there are no confirmed mammalian PAS kinase substrates, yeast substrates have been identified through an in vitro biochemical screen.23 Purified Psk2 protein was combined with 32P-ATP and multi-dimensionally fractionated crude yeast protein lysates. Four proteins were identified as Psk2 substrates; three involved in protein translation (Caf20, Tif11, Sro9) and one involved in glucose metabolism (Ugp1). The Caf20 (Cap-Associated Factor 20) protein is the yeast homolog of the 4E binding protein (4E–BP) and negatively regulates translation by blocking the association of eIF4E and eIF4G.25,26 Interestingly, efficient phosphorylation of Caf20 by PAS kinase only occurred in the presence of eIF4E. Tif11 is the eukaryotic translation initiation factor 1A (eIF1A), which mediates the transfer of Met-tRNA to the 40S ribosomal subunit.27 Although there is no precise function for the Sro9 protein to date, it appears to function in translation since it binds RNA in vitro, it interacts with translating ribosomes and it’s deletion causes resistance to specific chemical translation inhibitors.28–30 In addition to the identification of these putative substrates, the role of PAS kinase in translation was further supported by the many high-copy suppressors of the PAS kinase double mutant phenotype that are translation factors or components of the translation apparatus (such as DED1, DBP1, EDC1, POP4, RPR1, UBA2, REF2 and RDN58).23 Although further studies are needed to elucidate the role of PAS kinase in translational regulation, overexpression of PSK2 rescues the temperature sensitive phenotype of a strain lacking the STM1 (TIF3) gene, which encodes the yeast eIF4B translation initiation factor.23 Of all of the putative PAS kinase substrates, phosphorylation of the enzyme UDP-glucose pyrophosphorylase (Ugp1), the source of intracellular UDP-glucose, has been the best characterized due to it’s pronounced physiological effect.
The Role of Phospho-Ugp1 in Cell Integrity Maintenance
The enzyme Ugp1 produces UDP-glucose, the immediate glucose donor for both glycogen and cell wall glucan biosynthesis as well as myriad other cellular processes (including galactose entry into glycolysis, biosynthesis of trehalose and golgi/ER protein modification).31 The predominant PAS kinase phosphorylation site has been mapped to serine 11 in a likely unstructured region at the N-terminus of Ugp1.23 When yeast cells harbor an unphosphorylatable form of Ugp1 (S11A) they display a phenotype identical to the PAS kinase double mutant (galts and sensitivity to cell wall perturbing agents), suggesting that Ugp1 phosphorylation is THE critical function of PAS kinase in yeast under these conditions.23,24 Surprisingly, phosphorylation of Ugp1 by PAS kinase does not change the catalytic activity of Ugp1 but instead alters the destination of its product UDP-glucose, favoring cell wall biosynthesis at the expense of glycogen synthesis.24 The inability to phosphorylate Ugp1 at Ser11, either through deletion of PSK1 and PSK2 or by the UGP1-S11A mutation, causes an elevation in glycogen levels and a decrease in β-1,6 glucan levels. This decrease in cell wall constituents is most likely responsible for the hypersensitivity of the PAS kinase double mutant strain to cell wall perturbing agents. The PAS kinase-dependent regulation of glucose partitioning is probably accomplished by enriching Ugp1 at the plasma membrane, where it delivers UDP-glucose to the cell wall biosynthetic machinery, thus altering the destination of UDP-glucose rather than the rate of production. This may occur via direct phospho-Ugp1 interaction with cell wall biosynthetic enzymes, allowing direct donation of UDP-glucose to cell wall biosynthetic enzymes. It is also possible that translocation of Ugp1 to the cell periphery simply causes the production of UDP-glucose in proximity to the cell wall synthesis machinery and this is sufficient to cause increased glucan synthesis. Finally, the translocation of Ugp1 to the cell periphery may increase cell wall biosynthesis through as yet unidentified interactions and pathways. In support of the Ugp1 transloction hypothesis, the psk1 psk2 mutant phenotype is suppressed by expression of three different Ugp1 fusion proteins that target the Ugp1 protein to the membrane.24
Cellular Conditions of PSK Activation in S. cerevisiae
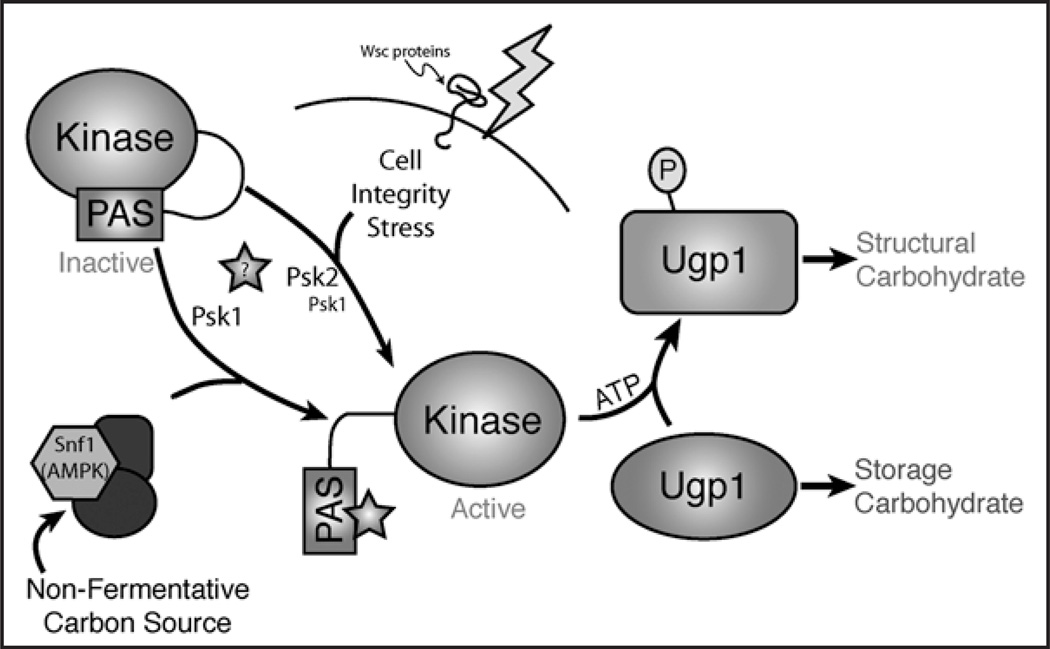
When Ugp1 is phosphorylated it adopts a distinct conformation that can be detected by protease digestion as well as ion-exchange chromatography.24 Thus, monitoring endogenous Ugp1 phosphorylation chromatographically has become a valuable tool in the study of yeast PAS kinase regulation and has facilitated the identification of growth conditions that stimulate PAS kinase activity.8 Two stimuli have been found to activate PAS kinase in S. cerevisiae: cell integrity stress and nutrient status, specifically, growth on nonfermentative carbon sources. A model for the regulation and function of yeast PAS kinase is shown in Figure 2.
Figure 2.
A model PAS kinase activation and function in S. cerevisiae. Autoinhibited PAS kinase can be activated by either growth on nonfermentative carbon sources or by cell integrity stress. The activation of PAS kinase by nonfermenative carbon source requires the Snf1 protein and may involve the production of a regulatory metabolite (star), while the cell integrity stress pathway can be activated by a range of cell wall or cell membrane perturbing agents as well as by overexpression of Wsc1. Activated PAS kinase then phosphorylates Ugp1, which leads to increased cell wall glucan biosynthesis at the expense of glycogen biosynthesis.8
PAS kinase activation occurs in response to perturbation of either the cell wall or cell membrane of S. cerevisiae. Specifically, we observed increased Ugp1 phosphorylation upon treatment with cell membrane perturbing agents (such as chlorpromazine), cell wall perturbing agents (such as calcofluor white) or nonspecific cell integrity perturbing agents (such as heat or SDS). The yeast cell wall is ~30% of the S. cerevisiae total dry weight and is composed of β-1,3-glucans (~85%), β-1,6-glucans (12%), chitin (~3%) as well as an outer layer of glycoproteins.32 The β-1,6-glucans connect the outer glycoproteins with the rigid β-1,3-glucan network and are essential under conditions of cell integrity stress.33 The cell wall is vital for cellular integrity and is thus constantly monitored for stability through plasma membrane-bound cell integrity sensing proteins known as the Wsc family of proteins.
The canonical Cell Integrity Pathway consists of these Wsc sensory proteins that activate the Rho1 guanine nucleotide exchange factors (GEF’s), Rom2 and Tus1, in response to membrane perturbation.34,35 Activated Rho1 then affects pathways that increase cell wall biosynthesis, including the PKC1-associated MAP kinase cascade and, through direct interaction with Fks1, the enzyme responsible for β-1,3-glucan synthesis.36 This canonical Cell Integrity Pathway appears to play a role in the activation of PAS kinase by cell integrity stress. Overexpression of WSC1, which encodes the major upstream sensory protein, increases phospho-Ugp1. PAS kinase may be a previously undescribed component of this canonical Cell Integrity Pathway or it may lie in a parallel pathway for maintaining cell integrity by promoting cell wall biosynthesis simply through the phosphorylation of Ugp1 and increased glucan synthesis. However it integrates with the known mediators of cell integrity signaling, genetic and biochemical experiments clearly demonstrate that it plays a role in the physiological response to cell integrity stress.
As mentioned above, PAS kinase is activated by a second, nutrient-dependent stimulus in addition to cell wall stress.8 While the PAS kinase substrate Ugp1 is found almost entirely in the unphosphorylated state in yeast grown on glucose (a carbon source that yeast preferentially ferment), it is almost completely phosphorylated when yeast are grown on nonfermentative carbon sources (carbon sources which they preferentially respire). Yeast repress a wide array of genes when growing on glucose, including those necessary for mitochondrial biogenesis and respiration.37 De-repression of these genes is dependent on the yeast AMPK ortholog Snf1, a key mediator of the regulatory switch between fermentative growth on glucose and respiratory growth on nonfermentative carbon sources. In contrast to the wild type parental strain, when yeast harboring constitutively active Snf1 (via deletion of the REG1 gene) are grown on glucose, Ugp1 is almost completely in the phosphorylated state. This increase in phospho-Ugp1 is prevented in the double reg1snf1 mutant. Thus, PAS kinase activation in response to nonfermentative growth is dependent on the Snf1 kinase. Yeast normally accumulate glycogen on nonfermentative carbon sources, however, PAS kinase activation appears to provide the necessary cell wall constituents in order to maintain cell integrity during rapid growth under respiratory conditions. Although Snf1 is known to respond to conditions of cellular stress, the activation of PAS kinase by nonfermentative carbon sources and cell integrity stress may involve two separate pathways because cell integrity stress activation occurred normally in the absence of SNF1.8 PAS kinase activation by nutrient status in S. cerevisiae is of particular interest because mammalian PAS kinase also displays nutrient-dependent regulation under conditions of high mitochondrial activity.6
Interestingly, the PAS kinase ohnologs display differential regulation in response to cell integrity stress and nonfermentative carbon source. When monitoring in vivo phosphorylation of Ugp1, only Psk1 is able to support Ugp1 phosphorylation in response to growth on a nonfermentative carbon source, while both Psk1 and Psk2 are capable of increasing phospho-Ugp1 in response to cell integrity stress.8 That is, deletion of PSK1 blocked the generation of phospho-Ugp1 during growth on nonfermentative carbon sources. As will be discussed below, this inability of Psk2 to support Ugp1 phosphorylation in response to nonfermentative carbon sources is almost certainly due to transcriptional repression of the PSK2 gene under these conditions and is consistent with the evolution of overlapping but distinct roles for the ohnologous yeast PAS kinase proteins.
In addition to assessing PAS kinase activity by monitoring endogenous phosphorylation of Ugp1 (as described above), we have measured PAS kinase activity directly through assay of purified TAP-tagged PAS kinase. This demonstrated that PAS kinase is indeed post-translationally activated and other possible mechanisms, such as induction of PAS kinase expression or alternate modes of phospho-Ugp1 regulation that are independent of PAS kinase, were not responsible for the observed increase in phospho-Ugp1 abundance.8 The PAS kinase proteins retain their activation state throughout TAP purification followed by an in vitro Ugp1 phosphorylation assay (i.e., cells with activated PAS kinase in vivo yield purified activated PAS kinase), providing a powerful system to biochemically dissect the modes of PAS kinase activation. Both Psk1 and Psk2 display increased activity if immunoprecipitated from cells grown under cell integrity stress conditions, however, only Psk1 is able to be efficiently immunoprecipitated from cells grown on nonfermentative carbon source. The decrease in Psk2 protein seen on nonfermentative carbon sources is accompanied by a decrease in PSK2 mRNA. In contrast, both Psk1 and Psk2 are activated by cell integrity stress and nonfermentative carbon sources when constitutive promoters are used to drive their expression.8 Thus transcriptional repression of PSK2 is responsible for the above-mentioned inability of Psk2 to phosphorylate Ugp1 during growth on nonfermentative carbon sources.
The activation of PAS kinase by cell integrity stress and nutrient status is physiologically relevant. That is, PAS kinase deficiency under conditions of cell integrity stress or growth on a nonfermentative carbon source leads to an increase in glycogen, presumably at the expense of cell wall structural carbohydrates.8 The activation of PAS kinase by cell integrity stress stimulates the biosynthesis of cell wall glucans necessary for repair through the phosphorylation of Ugp1 (the provider of cellular UDP-glucose). This altered glucose partitioning is necessary for cell survival under conditions of cell wall stress or damage.
The Role of PAS Kinase in Maintaining Cell Integrity: Support from a Genetic Screen
As mentioned above, the Ugp1-S11A mutant, which is unable to be phosphorylated by PAS kinase, has a phenotype that mimics the PAS kinase-deficient phenotype, namely hypersensitivity to conditions of cell integrity stress (growth in the presence of calcofluor white, congo red, or on galactose at 39°C). Sensitivity of Ugp1-S11A mutants to conditions of cell integrity stress is most likely due to their inability to produce the cell wall constituents necessary for proper cell wall maintenance. In addition to phospho-Ugp1 depletion (Ugp1-S11A), overexpression of UGP1 induced sensitivity to cell integrity stress conditions.23 Thus, overexpression of UGP1 is toxic to yeast grown on galactose (with an increased severity at high temperature) and on cell perturbing agents such as SDS. This toxicity was exacerbated by deletion of PAS kinase and can be suppressed by co-overexpression of PSK1 and/or PSK2 (Fig. 3). Overexpression of the UGP1-S11A mutant causes a growth defect that was not rescued by overexpression of PSK1 or PSK2 (data not shown) suggesting that the de-phospho form of Ugp1 is toxic when present at high concentrations under these conditions. These facts make UGP1 overexpression a powerful genetic tool allowing for the identification of genes that are able to suppress toxicity associated with hyperaccumulation of dephospho-Ugp1. These genes may suppress by hyperactivation of PAS kinase and/or by the expression of proteins that stimulate cell wall biosynthesis (including those that interact with phospho-Ugp1). Suppressors that are involved in PAS kinase activation or phospho-Ugp1 function may be delineated from PAS kinase-independent suppressors based on the ability to suppress in a psk1 psk2 mutant strain. In addition, those suppressors involved in PAS kinase activation may be separated from those augmenting phospho-Ugp1 function downstream by determining the phosphorylation state of Ugp1.
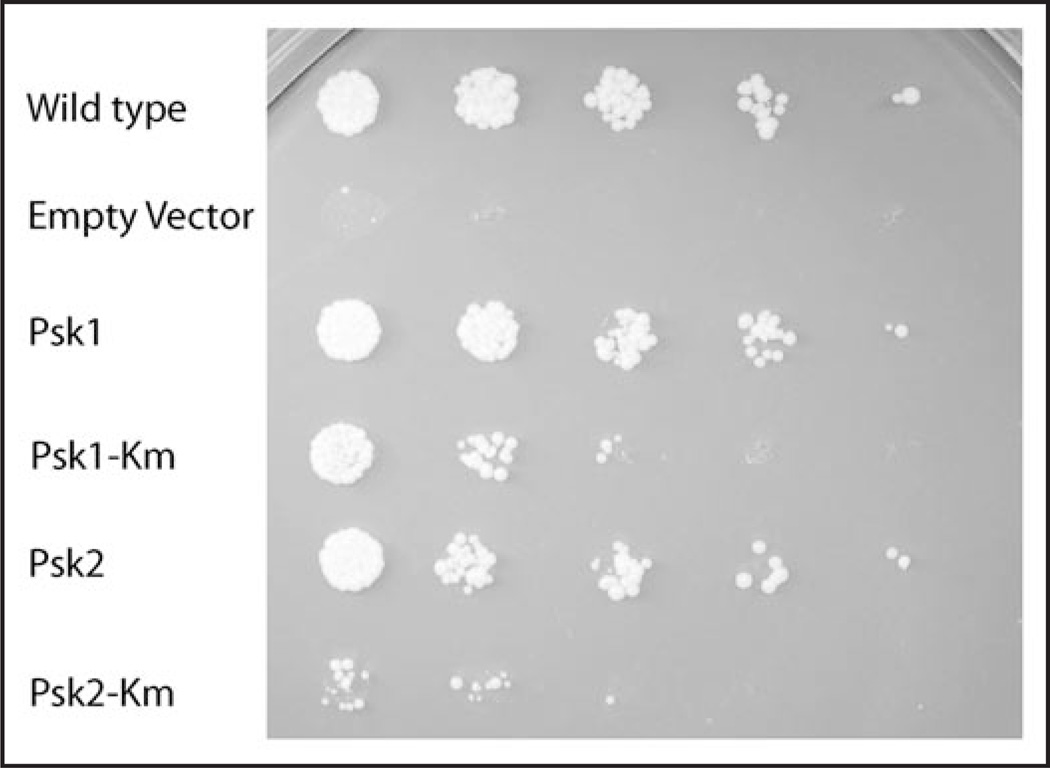
Figure 3.
Toxicity of Ugp1 overexpression is rescued by overexpression of PSK1 or PSK2. The PSK1psk2 (JRY277) strain overexpressing UGP1 (pJR3020b) displays a galts growth defect that may be rescued by over-expression of PSK1 or PSK2. A trp plasmid overexpressing UGP1 from the TetO regulatable promoter was constructed for this screen (pJR3020b) using the system of Gari et al.54 The JRY277 strain (PSK1psk2) was freshly transformed with either empty vector control (wild type, uppermost sample) or pJR3020b (remaining samples), and then one of five pRS426 constructs (empty vector, Psk1, Psk1 K1125R, Psk2, Psk2 K870R) and plated on SD-ura-trp + dox. The K1125R and K870R PAS kinase mutations (Km) are in the kinase domain and were constructed to decrease kinase activity.23 Liquid cultures were then grown for 48 hours in the same media and were used for serial dilution into water 1:20, 1:5, 1:5, 1:5 and 1:5. Each dilution was then spotted (5 uL) onto an SGal-ura-trp plate and incubated at 30°C. A control plate was made on SD-ura-trp + dox and the lack of phenotype for any of the five strains was confirmed (all dilutions were comparable). The galts phenotype of overexpressed UGP1 is not present in cells exposed to doxycycline. In contrast to the Psk2 Km mutation, the K1125R mutation in Psk1 appears to have little to no effect on its ability to suppress.
A high-copy plasmid suppressor (HCS) screen was performed using the galts phenotype due to overexpression of UGP1. This HCS screen was conducted in a psk2 deficient strain in order to exacerbate the growth phenotype while permitting isolation of PSK1-dependent suppressors. Plasmids were isolated from colonies able to grow on galactose at the non-permissive temperature and were retested for their ability to suppress by plasmid purification and retransformation. In addition, the Ugp1 activity of each suppressor strain was assayed and was shown to contain levels of activity that were similar to the control. Thus, no suppressors were isolated that simply diminished expression or activity of Ugp1. The screen was performed at two different temperatures (30 and 35°C) with the majority conducted at the higher temperature (35°C). The genes discovered thus far exhibited a range of suppression with the strongest suppressors displaying growth comparable to that observed with overexpression of PSK1 or PSK2 (Fig. 4).
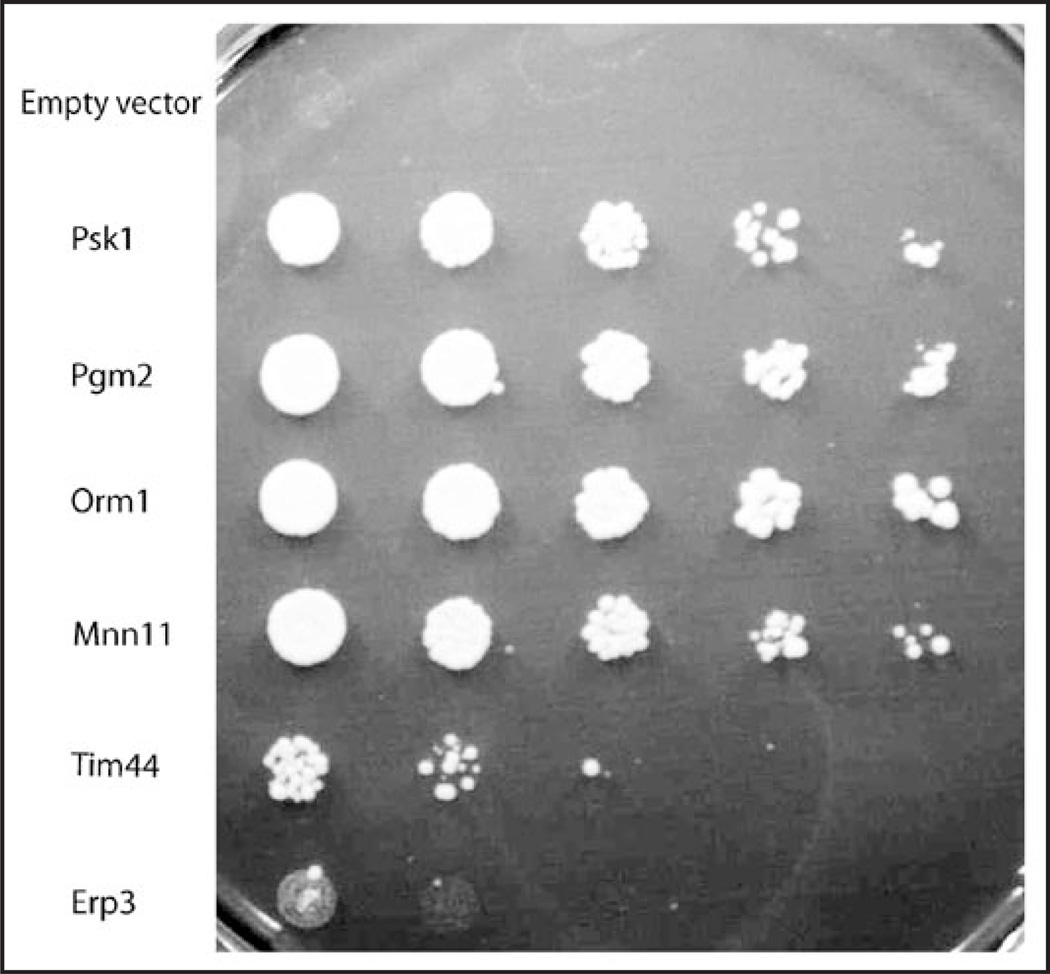
Figure 4.
Toxicity of Ugp1 overexpression is rescued by high copy suppressors from an S. cerevisiae pRS426-based library. The PSK1psk2 (JRY277) strain overexpressing UGP1 from a trp plasmid (pJR3020b) displays a galts growth defect that may be rescued by high copy suppressors (HCS) from a pRS426-based library. All samples have UGP1 overexpreessed and the suppressors are shown in comparison to the empty pRS426 vector control. Samples are, from top to bottom, pRS426 control, PSK1, PGM2, ORM1, MNN11, TIM44 and ERP3. Samples are shown in dilution series from left to right and were prepared as described in Figure 3 and plated on SGal-trp-ura at 30°C for 3 days.
A list of the genes encoded by the HCS plasmids obtained thus far, as well as their putative function, is in Table 1. Some suppressing plasmids contained multiple ORFs, thus the individual genes must be cloned in order to identify the responsible gene, however there are likely gene candidates identified based strictly on their previously characterized role in cell integrity (underlined). In most cases a gene was isolated several times in different plasmid constructs, thus the responsible suppressing gene was apparent. A majority of these genes appear to be involved in cell wall biosynthesis, the secretory pathway, nutrient partitioning, mitochondrial function, or translation. All HCS plasmids isolated function independently of PAS kinase, that is, they are capable of suppressing UGP1 over-expression toxicity in the absence of both PSK1 and PSK2 (data not shown). Although no PAS kinase-dependent suppressors have been isolated thus far, we believe that they may still be isolated from this screen. As mentioned above, a majority of the screen was conducted at 35°C. The high temperature combined with growth on galactose may produce a condition where most of the endogenous PAS kinase is hyperactivated since PAS kinase is activated by cell integrity stress and nonfermentative carbon source. Additional screens are being conducted at 30°C on galactose as well as on glucose with cell wall perturbing agents (such as calcofluor white). These may provide conditions were PAS kinase is only partially activated (by only the cell integrity or the nonfermentative carbon source pathway) and thus yield suppressors whose gene products are involved in PAS kinase activation pathways. In addition, it is likely that activation of PAS kinase may have a weak suppressing effect, thus attention to weak suppressors (small colonies) will be a focus in future rounds of suppressor isolation.
Table 1.
Genetic suppressors of the Ugp1 overexpression phenotype
| Gene(s) | # | Description | Efficacy | Temp |
|---|---|---|---|---|
| PSK1 | 8 | PAS kinase 1 | S | 35 |
| PSK2 | 3 | PAS kinase 2 | S | 30 |
| PGM2 | 6 | Phosphoglucomutase 2 | S | 30 |
| ORM1 | 1 | Required for resistance to agents that induce the unfolded protein response | S | 35 |
| MNN11 | 3 | Subunit of Golgi mannosyltransferase complex | S | 35 |
| ATG2 | 1 | Peripheral membrane protein required for vesicle formation during autophagy and for the CVT pathway |
M | 35 |
|
BET2, PRP4, HDA3 |
1 | Subunit of Type II geranylgeranyltransferase required for ER/Golgi vesicular transport; Splicing factor; Trichostatin A-sensitive class II histone deacetylase subunit |
M | 35 |
| ISD11 | 1 | Mitochondrial Fe-S cluster (FSC) biosynthesis | M | 35 |
| LEU1 | 3 | Isopropylmalate isomerase, catalyzes the second step in the leucine biosynthesis pathway | M | 35 |
| PRX1; KIP1 | 1 | Mitochondrial peroxiredoxin; Required for mitotic spindle assembly and chromosome segregation | M | 35 |
|
RDN58-2; ITS2-2; ITS1-2 |
5 | 5.8S ribosomal RNA, component of the 60S ribosomal subunit; Non-coding region between RDN58 and RDN25 |
M | 35 |
| STP22 | 2 | Component of the ESCRT-I complex | M | 35 |
| STT4 | 1 | Required for vacuole morphology, cell wall integrity, and actin cytoskeleton organization | M | 35 |
| TIM44 | 2 | Peripheral mitochondrial membrane protein involved in mitochondrial protein import, tethers essential chaperone Ssc1 to the mitochondrial membrane |
M | 35 |
| YRL125W | 4 | Putative protein of unknown function | M | 35 |
| OSH2; ERP3 | 1 | Involved in sterol metabolism; Member of the p24 family involved in ER to Golgi transport | W | 35 |
| ECM25 | 1 | Non-essential protein of unknown function; promoter contains a consensus binding sequence for factor Abf1p |
W | 35 |
|
HTB2; ECM15; NTH2 |
1 | Histone H2B; Non-essential protein of unknown function that may have a roll in cell wall biogeneiss; Putative neutral trehalase required for thermotolerance |
W | 35 |
The column labeled # indicates the number of independent clones recovered. Efficacy is the strength of suppression of each clone as compared to the PAS kinase suppression. S = strong, M = moderate and W = weak. Temperature indicates the temperature at which the screen yielding that suppressor was performed (30°C or 35°C). The strain JRY277(psk1 psk2) freshly transformed with pJR3020b (UGP1) was retransformed with a pRS426 library using the standard lithium acetate transformation protocol55 and screened for growth on SGal-trp-ura. Approximately 2 million transformants resulted in 401 initial candidate suppressors. These candidates were then patched to SD-trp containing dox (to inhibit UGP1 overexpression) and 5-FOA (to allow for the loss of the URA3-encoding pRS426 library plasmid) and then subsequently replica plated onto SGal-trp at the isolation temperature to test for plasmid-independent suppression. The high copy suppressor candidate (HCSC) library plasmids were then isolated and characterized by restriction endonuclease digestion with PvuII. The approximately 200 unique plasmids were then retransformed into JRY277 pJR3020B cells to confirm suppression on SGal-ura-trp (using empty pRS426 in place of the pRS426-library plasmid DNA as a control); only 71 were able to reproducibly rescue the Ugp1 galts phenotype. An UGPase enzymatic assay56 was used to eliminate regulation of Ugp1 activity as a possible mode of suppression. These candidate plasmids were then sequenced and the genes were identified using the Basic Local Alignment Search Tool for nucleotides (BLASTN) through the NIH National Center for Biotechnology Information website. The genes that are inferred to be suppressors by virtue of their rolls in cell integrity pathways are underlined.
Several HCS genes identified were expected outcomes of the screen, namely PSK1, PSK2, phosphoglucomutase 2 (PGM2) and RDN58-2. PSK1 was isolated seven times (all from screens performed at 35°C), while PSK2 was isolated only twice (both from screens at 30°C). In addition, only one of these PSK plasmids (a PSK1 plasmid) contained a full-length version of the protein, all others were N-terminal truncations that included the kinase domain (Fig. 5). This is consistent with biochemical data showing that the N-terminal PAS domain inhibits kinase activity.16,17 The recovery of PGM2 and RDN58-2 from this screen was expected as both were previously identified as high-copy suppressors of the double PAS kinase mutant phenotype.23 The enzyme Pgm2 is thought to suppress by increasing the flux of glucose-6-P to glucose-1-P, providing more glucose for conversion to UDP-glucose and for use in cell wall biosynthesis. In fact, PGM2 is transcriptionally upregulated in response to growth on galactose, a condition that is known to stress cells.38 The gene RDN58-2 encodes the 5.8S ribosomal RNA subunit and is therefore required for translation.39 As mentioned above, there were several other high copy suppressors of the double-PAS kinase mutant phenotype that are involved in translation and Psk2 has been shown to phosphorylate three proteins involved in translation in vitro.23 Therefore, RDN58-2 may suppress simply by enhancing translation under stressful conditions or it may directly compensate for loss of an unknown function of PAS kinase in translation. Interestingly, PGM2 and UGP1 contain stress response elements (STREs) in their promoter regions and are thus likely to be transcriptionally induced in response to stress.40 In contrast to the upregulation of PGM2 and UGP1, PSK1 is downregulated in response to cell integrity stress.8 This may account for the isolation of PSK1 dependent suppressor plasmids at higher temperatures where plasmid overexpression may compensate for decreased genomic expression.
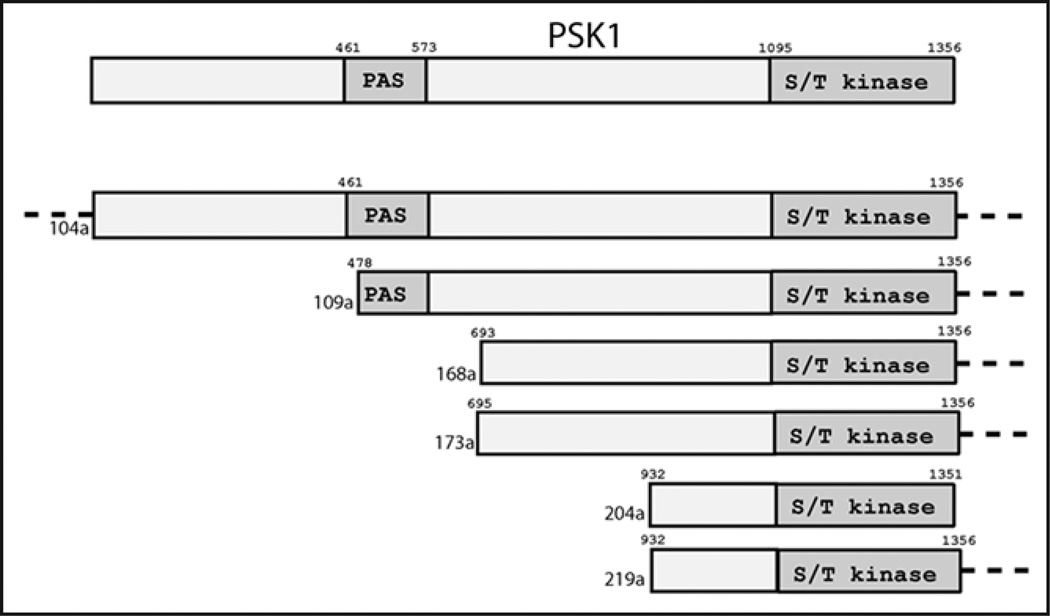
Figure 5.
Schematic alignment of pRS426-library clones encoding full-length PSK1 or PSK1 truncations. The uppermost Psk1 represents the full-length protein. For the clones listed below, dotted lines indicate additional DNA sequence to either side of the protein boundaries. Amino acid numbers are indicated above the Psk1 representations.
Based on the described role for Ugp1 phosphorylation, it is not surprising that the most robust suppressors participate in the maintenance of the cell wall, either through direct enzymatic participation in the biosynthesis of cell wall constituents, or in the delivery of these constituents from the ER to the cell wall through the secretory pathway. For example, the only genes that suppressed the growth defect as well as PSK1, PSK2 or PGM2 were ORM1 and MNN11. The Orm1 protein is one of two well-conserved yeast homologs of the human ORMDL proteins that localize to the ER membrane and are of unknown function. These yeast homologs (Orm1 and Orm2) have been shown to be required for resistance to ER stress and are thought to be involved in protein folding.41 Although the precise function of Orm1 is unknown, it is genetically linked to the secretory pathway as a phenotypic enhancer of mutations in nearly twelve secretory pathway genes (such as genes involved in sterol biosynthesis, ER stress, β-1,6-glucan biosynthesis, mannosylation and ER to golgi transport).42 The suppressor MNN11 is directly involved in cell integrity maintenance. As a subunit of the Golgi mannosyltransferase complex Mnn11 is directly involved in the formation of the polysaccharide mannan backbone core for extracellular mannosylated proteins (mannans), which make-up almost 40% of the dry cell wall weight.43 These extracellular mannans are essential for the S. cerevisiae response to cell-integrity stressors.32,44 The suppressing plasmid contained a truncation of MNN11. We cloned the full-length gene and found that it was a weaker suppressor than the truncated form, suggesting the full-length protein may be subject to autoinhibition (data not shown).
Many other suppressors are involved in secretory pathway trafficking and thus may be important for transmembrane and extracellular protein targeting, processes that are essential for maintenance of cellular integrity. For example, Erp3 and Bet2 are both involved in vesicular ER to Golgi transport.45,46 The precise function of Erp3 is unknown, however, it is a member of the p24 protein family and is involved in the early secretory pathway.42,47 The human p24 luminal protein has a cytoplasmic tail that facilitates vesicle formation.48 The Bet2 protein is a known geranylgeranyltransferase that modifies the Rab GTPases Ypt1 and Sec4 with a membrane-attachment moiety that allows them to bind and target secretory vesicles. Thus Erp3 and Bet2 are involved in proper functioning of the secretory pathway and may rescue the Ugp1 galts phenotype by increasing the ability of cells to respond to stress through the delivery of proteins and glucans to the cell membrane and cell wall. An additional suppressor, encoded by STP22, is a member of the ESCRT-I complex and is essential for normal membrane trafficking in the late endosome; an essential process for regulating signals transmitted from the extracellular environment to the cell.49 Many constituents necessary for a healthy cell wall are first synthesized and then delivered to the cell periphery through the secretory pathway, including the necessary enzymes for β-1,3-glucan and chitin synthesis.32,35,50 This may be true for β-1,6-glucan as well since recent findings have implicated ER-membrane bound proteins in their biosynthesis.51 Thus, the secretory pathway is critical for cell integrity maintenance, explaining why several genes involved in the secretory pathway were isolated as suppressors.
The secretory pathway may also be the link between the suppressors that have mitochondrial function and those involved in cell wall biosynthesis. The mitochondrial suppressors TIM44 and ISD11 are required for basic mitochondrial function while PRX1 encodes a mitochondrial peroxiredoxin that is activated under conditions of oxidative stress. Specifically, TIM44 is required for mitochondrial protein import and folding, while ISD11 is required for mitochondrial Fe-S cluster (FSC) biosynthesis. Interestingly, proper ER and Golgi trafficking is necessary for proper mitochondrial structure,52 thus cell integrity stress may trigger mitochondrial stress through changes in ER/Golgi trafficking.
Perhaps the most intriguing suppressors isolated are the genes encoding proteins of unknown function, YLR125W and ECM25. Interestingly, ECM25 mutants were previously identified in a screen for yeast with altered cell surface architecture.53 In addition, several of the genes discussed above (including ORM1 and ERP3) have roles in the cell wall integrity or secretory pathways; however, their precise function is unknown. Further study of these suppressors will provide a more detailed understanding of their roles through the analysis of specific pathways such as cell wall constituent biosynthesis, protein glycosylation, the unfolded protein response, or the efficiency of early and late secretory events. In addition, characterization of these as well as other PAS kinase-dependent suppressors may yield a more detailed understanding of the role of PAS kinase and phospho-Ugp1 in the maintenance of cell integrity as well as pathways responsible for the activation of PAS kinase.
References
- 1.Lindsley JE, Rutter J. Nutrient sensing and metabolic decisions. Comp Biochem Physiol B Biochem Mol Biol. 2004;139:543–559. doi: 10.1016/j.cbpc.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 2.Lage R, Dieguez C, Vidal-Puig A, Lopez M. AMPK: a metabolic gauge regulating whole-body energy homeostasis. Trends Mol Med. 2008;14:539–549. doi: 10.1016/j.molmed.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 3.Goberdhan DC, Boyd CA. mTOR: dissecting regulation and mechanism of action to understand human disease. Biochem Soc Trans. 2009;37:213–216. doi: 10.1042/BST0370213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hao HX, Cardon CM, Swiatek W, Cooksey RC, Smith TL, Wilde J, et al. PAS kinase is required for normal cellular energy balance. Proc Natl Acad Sci USA. 2007;104:15466–15471. doi: 10.1073/pnas.0705407104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schlafli P, Borter E, Spielmann P, Wenger RH. The PAS-domain kinase PASKIN: a new sensor in energy homeostasis. Cell Mol Life Sci. 2009;66:876–883. doi: 10.1007/s00018-009-8699-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.da Silva Xavier G, Rutter J, Rutter GA. Involvement of Per-Arnt-Sim (PAS) kinase in the stimulation of preproinsulin and pancreatic duodenum homeobox 1 gene expression by glucose. Proc Natl Acad Sci USA. 2004;101:8319–8324. doi: 10.1073/pnas.0307737101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hao HX, Rutter J. The role of PAS kinase in regulating energy metabolism. IUBMB Life. 2008;60:204–209. doi: 10.1002/iub.32. [DOI] [PubMed] [Google Scholar]
- 8.Grose JH, Smith TL, Sabic H, Rutter J. Yeast PAS kinase coordinates glucose partitioning in response to metabolic and cell integrity signaling. EMBO J. 2007;26:4824–4830. doi: 10.1038/sj.emboj.7601914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gu YZ, Hogenesch JB, Bradfield CA. The PAS superfamily: sensors of environmental and developmental signals. Annual review of pharmacology and toxicology. 2000;40:519–561. doi: 10.1146/annurev.pharmtox.40.1.519. [DOI] [PubMed] [Google Scholar]
- 10.Zhulin IB, Taylor BL, Dixon R. PAS domain S-boxes in Archaea, Bacteria and sensors for oxygen and redox. Trends Biochem Sci. 1997;22:331–333. doi: 10.1016/s0968-0004(97)01110-9. [DOI] [PubMed] [Google Scholar]
- 11.Kaspar S, Bott M. The sensor kinase CitA (DpiB) of Escherichia coli functions as a high-affinity citrate receptor. Arch Microbiol. 2002;177:313–321. doi: 10.1007/s00203-001-0393-z. [DOI] [PubMed] [Google Scholar]
- 12.Stephenson K, Hoch JA. PAS-A domain of phosphorelay sensor kinase A: a catalytic ATP-binding domain involved in the initiation of development in Bacillus subtilis. Proc Natl Acad Sci USA. 2001;98:15251–15256. doi: 10.1073/pnas.251408398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christie JM, Swartz TE, Bogomolni RA, Briggs WR. Phototropin LOV domains exhibit distinct roles in regulating photoreceptor function. Plant J. 2002;32:205–219. doi: 10.1046/j.1365-313x.2002.01415.x. [DOI] [PubMed] [Google Scholar]
- 14.Gilles-Gonzalez MA. Oxygen signal transduction. IUBMB Life. 2001;51:165–173. doi: 10.1080/152165401753544232. [DOI] [PubMed] [Google Scholar]
- 15.Kaspar S, Perozzo R, Reinelt S, Meyer M, Pfister K, Scapozza L, et al. The periplasmic domain of the histidine autokinase CitA functions as a highly specific citrate receptor. Mol Microbiol. 1999;33:858–872. doi: 10.1046/j.1365-2958.1999.01536.x. [DOI] [PubMed] [Google Scholar]
- 16.Amezcua CA, Harper SM, Rutter J, Gardner KH. Structure and interactions of PAS kinase N-terminal PAS domain: model for intramolecular kinase regulation. Structure. 2002;10:1349–1361. doi: 10.1016/s0969-2126(02)00857-2. [DOI] [PubMed] [Google Scholar]
- 17.Rutter J, Michnoff CH, Harper SM, Gardner KH, McKnight SL. PAS kinase: an evolutionarily conserved PAS domain-regulated serine/threonine kinase. Proc Natl Acad Sci USA. 2001;98:8991–8996. doi: 10.1073/pnas.161284798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kellis M, Birren BW, Lander ES. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature. 2004;428:617–624. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- 19.Wolfe K. Robustness—it’s not where you think it is. Nat Genet. 2000;25:3–4. doi: 10.1038/75560. [DOI] [PubMed] [Google Scholar]
- 20.Bergthorsson U, Andersson DI, Roth JR. Ohno’s dilemma: evolution of new genes under continuous selection. Proc Natl Acad Sci USA. 2007;104:17004–17009. doi: 10.1073/pnas.0707158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Conant GC, Wolfe KH. Turning a hobby into a job: How duplicated genes find new functions. Nat Rev Genet. 2008;9:938–950. doi: 10.1038/nrg2482. [DOI] [PubMed] [Google Scholar]
- 22.Rohde JR, Bastidas R, Puria R, Cardenas ME. Nutritional control via Tor signaling in Saccharomyces cerevisiae. Curr Opin Microbiol. 2008;11:153–160. doi: 10.1016/j.mib.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rutter J, Probst BL, McKnight SL. Coordinate regulation of sugar flux and translation by PAS kinase. Cell. 2002;111:17–28. doi: 10.1016/s0092-8674(02)00974-1. [DOI] [PubMed] [Google Scholar]
- 24.Smith TL, Rutter J. Regulation of glucose partitioning by PAS kinase and Ugp1 phosphorylation. Mol Cell. 2007;26:491–499. doi: 10.1016/j.molcel.2007.03.025. [DOI] [PubMed] [Google Scholar]
- 25.Altmann M, Krieger M, Trachsel H. Nucleotide sequence of the gene encoding a 20 kDa protein associated with the cap binding protein eIF-4E from Saccharomyces cerevisiae. Nucleic Acids Res. 1989;17:7520. doi: 10.1093/nar/17.18.7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altmann M, Trachsel H. Altered mRNA cap recognition activity of initiation factor 4E in the yeast cell cycle division mutant cdc33. Nucleic Acids Res. 1989;17:5923–5931. doi: 10.1093/nar/17.15.5923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wei CL, Kainuma M, Hershey JW. Characterization of yeast translation initiation factor 1A and cloning of its essential gene. J Biol Chem. 1995;270:22788–22794. doi: 10.1074/jbc.270.39.22788. [DOI] [PubMed] [Google Scholar]
- 28.Sobel SG, Wolin SL. Two yeast La motif-containing proteins are RNA-binding proteins that associate with polyribosomes. Mol Biol Cell. 1999;10:3849–3862. doi: 10.1091/mbc.10.11.3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hon T, Lee HC, Hach A, Johnson JL, Craig EA, Erdjument-Bromage H, et al. The Hsp70-Ydj1 molecular chaperone represses the activity of the heme activator protein Hap1 in the absence of heme. Mol Cell Biol. 2001;21:7923–7932. doi: 10.1128/MCB.21.23.7923-7932.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kagami M, Toh-e A, Matsui Y. SRO9, a multicopy suppressor of the bud growth defect in the Saccharomyces cerevisiae rho3-deficient cells, shows strong genetic interactions with tropomyosin genes, suggesting its role in organization of the actin cytoskeleton. Genetics. 1997;147:1003–1016. doi: 10.1093/genetics/147.3.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parodi AJ. Role of N-oligosaccharide endoplasmic reticulum processing reactions in glycoprotein folding and degradation. Biochem J. 2000;348:1–13. [PMC free article] [PubMed] [Google Scholar]
- 32.Levin DE. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2005;69:262–291. doi: 10.1128/MMBR.69.2.262-291.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shahinian S, Bussey H. beta-1,6-Glucan synthesis in Saccharomyces cerevisiae. Mol Microbiol. 2000;35:477–489. doi: 10.1046/j.1365-2958.2000.01713.x. [DOI] [PubMed] [Google Scholar]
- 34.Bickle M, Delley PA, Schmidt A, Hall MN. Cell wall integrity modulates RHO1 activity via the exchange factor ROM2. EMBO J. 1998;17:2235–2245. doi: 10.1093/emboj/17.8.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmelzle T, Helliwell SB, Hall MN. Yeast protein kinases and the RHO1 exchange factor TUS1 are novel components of the cell integrity pathway in yeast. Mol Cell Biol. 2002;22:1329–1339. doi: 10.1128/mcb.22.5.1329-1339.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Delley PA, Hall MN. Cell wall stress depolarizes cell growth via hyperactivation of RHO1. J Cell Biol. 1999;147:163–174. doi: 10.1083/jcb.147.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hedbacker K, Carlson M. SNF1/AMPK pathways in yeast. Front Biosci. 2008;13:2408–2420. doi: 10.2741/2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oh D, Hopper JE. Transcription of a yeast phosphoglucomutase isozyme gene is galactose inducible and glucose repressible. Mol Cell Biol. 1990;10:1415–1422. doi: 10.1128/mcb.10.4.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uesono Y, Ashe MP, Toh EA. Simultaneous yet independent regulation of actin cytoskeletal organization and translation initiation by glucose in Saccharomyces cerevisiae. Mol Biol Cell. 2004;15:1544–1556. doi: 10.1091/mbc.E03-12-0877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Francois J, Parrou JL. Reserve carbohydrates metabolism in the yeast Saccharomyces cerevisiae. FEMS Microbiol Rev. 2001;25:125–145. doi: 10.1111/j.1574-6976.2001.tb00574.x. [DOI] [PubMed] [Google Scholar]
- 41.Hjelmqvist L, Tuson M, Marfany G, Herrero E, Balcells S, Gonzalez-Duarte R. ORMDL proteins are a conserved new family of endoplasmic reticulum membrane proteins. Genome Biol. 2002:3. doi: 10.1186/gb-2002-3-6-research0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schuldiner M, Collins SR, Thompson NJ, Denic V, Bhamidipati A, Punna T, et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 43.Jungmann J, Rayner JC, Munro S. The Saccharomyces cerevisiae protein Mnn10p/Bed1p is a subunit of a Golgi mannosyltransferase complex. J Biol Chem. 1999;274:6579–6585. doi: 10.1074/jbc.274.10.6579. [DOI] [PubMed] [Google Scholar]
- 44.Jigami Y, Odani T. Mannosylphosphate transfer to yeast mannan. Biochim Biophys Acta. 1999;1426:335–345. doi: 10.1016/s0304-4165(98)00134-2. [DOI] [PubMed] [Google Scholar]
- 45.Jiang Y, Ferro-Novick S. Identification of yeast component A: reconstitution of the geranylgeranyltransferase that modifies Ypt1p and Sec4p. Proc Natl Acad Sci USA. 1994;91:4377–4381. doi: 10.1073/pnas.91.10.4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marzioch M, Henthorn DC, Herrmann JM, Wilson R, Thomas DY, Bergeron JJ, et al. Erp1p and Erp2p, partners for Emp24p and Erv25p in a yeast p24 complex. Mol Biol Cell. 1999;10:1923–1938. doi: 10.1091/mbc.10.6.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Springer S, Chen E, Duden R, Marzioch M, Rowley A, Hamamoto S, et al. The p24 proteins are not essential for vesicular transport in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2000;97:4034–4039. doi: 10.1073/pnas.070044097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fiedler K, Rothman JE. Sorting determinants in the transmembrane domain of p24 proteins. J Biol Chem. 1997;272:24739–24742. doi: 10.1074/jbc.272.40.24739. [DOI] [PubMed] [Google Scholar]
- 49.Babst M, Odorizzi G, Estepa EJ, Emr SD. Mammalian tumor susceptibility gene 101 (TSG101) and the yeast homologue, Vps23p, both function in late endosomal trafficking. Traffic. 2000;1:248–258. doi: 10.1034/j.1600-0854.2000.010307.x. [DOI] [PubMed] [Google Scholar]
- 50.Sekiya-Kawasaki M, Abe M, Saka A, Watanabe D, Kono K, Minemura-Asakawa M, et al. Dissection of upstream regulatory components of the Rho1p effector, 1,3-beta-glucan synthase, in Saccharomyces cerevisiae. Genetics. 2002;162:663–676. doi: 10.1093/genetics/162.2.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakamata K, Kurita T, Bhuiyan MS, Sato K, Noda Y, Yoda K. KEG1/YFR042w encodes a novel Kre6-binding endoplasmic reticulum membrane protein responsible for beta-1,6-glucan synthesis in Saccharomyces cerevisiae. J Biol Chem. 2007;282:34315–34324. doi: 10.1074/jbc.M706486200. [DOI] [PubMed] [Google Scholar]
- 52.Prinz WA, Grzyb L, Veenhuis M, Kahana JA, Silver PA, Rapoport TA. Mutants affecting the structure of the cortical endoplasmic reticulum in Saccharomyces cerevisiae. J Cell Biol. 2000;150:461–474. doi: 10.1083/jcb.150.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lussier M, White AM, Sheraton J, di Paolo T, Treadwell J, Southard SB, et al. Large scale identification of genes involved in cell surface biosynthesis and architecture in Saccharomyces cerevisiae. Genetics. 1997;147:435–450. doi: 10.1093/genetics/147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gari E, Piedrafita L, Aldea M, Herrero E. A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast. 1997;13:837–848. doi: 10.1002/(SICI)1097-0061(199707)13:9<837::AID-YEA145>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 55.Gietz RD, Schiestl RH, Willems AR, Woods RA. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- 56.Daran JM, Dallies N, Thines-Sempoux D, Paquet V, Francois J. Genetic and biochemical characterization of the UGP1 gene encoding the UDP-glucose pyrophosphorylase from Saccharomyces cerevisiae. Eur J Biochem. 1995;233:520–530. doi: 10.1111/j.1432-1033.1995.520_2.x. [DOI] [PubMed] [Google Scholar]
- 57.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, et al. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]