Abstract
With the recent burst of technological developments in genomics, and the clinical implementation of genome-wide assays, our understanding of the molecular basis of genomic disorders, specifically the contribution of structural variation to disease burden, is evolving quickly. Ongoing studies have revealed a ubiquitous role for genome architecture in the formation of structural variants at a given locus, both in DNA recombination-based processes and in replication-based processes. These reports showcase the influence of repeat sequences on genomic stability and structural variant complexity and also highlight the tremendous plasticity and dynamic nature of our genome in evolution, health and disease susceptibility.
Genomic disorders are a group of diseases caused by rearrangements of the human genome due to inherent genomic instability that results in susceptibility to structural variation mutagenesis1. In the context of human disease, structural variants include deletions, duplications, triplications, added amplifications (for example, quadruplications) and other large-scale (that is, from ~50–200 bp, the size of an average exon, to megabases of DNA) copy number variants (CNVs) that are not resolved by chromosome karyotype studies (<5 Mb), as well as copy number-neutral inversions, insertions and trans-locations. Structural variants differ from the concept of single nucleotide polymorphisms (SNPs) or single nucleotide variants (SNVs), which only change a single base or a few bases2, as the former requires the disruption of the sugar–phosphate backbone of DNA and involve many base pairs. The definition of a structural variant can overlap with the concept of small insertions and deletions (indels), which were previously defined as variants of <10,000 bp in length3. However, in this Review we consider indels as <50–100 bp in size, that is, a variant size that can be detected within a single next-generation sequencing read.
Structural variants result from different mutational mechanisms, including DNA recombination-, replication- and repair-associated processes. Current approaches to model the formation of structural variants in the human genome include: the use of model organisms4; the use of in vitro human cells subjected to stress5; and the direct observation of human genomic alterations, or rearrangement end products, that convey a disease trait. The manifested trait or genomic disorder enables one to both ascertain the mutational event and distinguish the affected individual from the population.
Studying disease-causing structural variants that have been classified as either extremely rare or de novo provides a unique opportunity to glean insights into mutational mechanisms. Such studies have revealed complex exonic, genic and chromosomal rearrangements that can be generated in a single mutagenic event, for example, in disease-associated loci at 17p11.2 and 17p12 (REF. 6), in MECP2 duplication syndrome (OMIM 300260)7 and due to chromothripsis-like events in multiple congenital anomalies8,9. Other studies have shown that the formation of structural variants can be accompanied by additional genome modification that may result in a disease trait. For example, CNVs and SNVs can be generated concomitantly10, and SNVs created during mutagenic repair can potentially affect the function of genes that do not map within the CNV. Also, CNVs can be followed by extended regions of absence of heterozygosity (AOH)11,12. If the AOH that is generated from template switching between homologues versus sister chromatids occurs at an imprinted locus, disease can result. Alternatively, the AOH region may encompass a variant in a gene for a recessive disease and reduce it to homozygosity when only one parent is a carrier, thus distorting Mendelian expectations.
The accurate detection of the complete mutagenic event at the single-base-pair level requires either the use of Sanger sequencing, along with techniques that enable large-scale CNV identification, or the use of composite pipelines for CNV analysis of next-generation sequencing data13. Therefore, using combined molecular analytic tools is necessary to delineate the entire range of variation that is associated with a particular structural variant in an individual personal genome. Methods that can be combined include fluorescence in situ hybridization (FISH), array comparative genomic hybridization (aCGH), SNP arrays, next-generation sequencing and Sanger sequencing13,14. Potential implications for the evolution of genes and genomes remain to be further explored, but mutational signatures — that is, the ‘scars’ left in the genome by DNA repair and replication mechanisms after structural variant mutagenesis has occurred — can partially explain the complex pattern of polymorphic human structural variants that has been revealed by the 1000 Genomes Project15.
New mechanistic discoveries in humans are elucidating how the formation of structural variants can re-structure a specific region of the genome to change gene expression either locally or genome-wide16,17, including through the alteration of chromatin architecture, with pathological consequences for carriers18. These discoveries are also paving the way to understanding the molecular basis of human disease, including diseases that result from somatic mutagenesis19.
In this Review, we first discuss the types of structural variants that cause genomic disorders, before reviewing in detail the recombination and replication-based mechanisms (RBMs) by which these arise. Our focus is on the pervasive role of the genomic architecture, including DNA sequence repeat structure and orientation, in the mechanism of formation of structural variants that have been implicated in human genomic disorders. We also describe the most common types of complex rearrangements and review current knowledge underlying their formation. We do not review the phenotypic consequences of specific genomic disorders; for a recent review on this topic please see REF. 20.
Repeat sequences in the human genome
The genomic architecture — that is, the local organization of repeat and other sequences, including their relative orientation, size, density and distribution — is a key feature that helps to predict the type of structural variants, rearrangement susceptibility and potential major mechanism that underlie variation at a specific locus. Approximately 50% of the human genome consists of repeat sequences21, which include mobile elements such as Alu-processed pseudogenes, simple sequence repeats, tandemly repeated sequences (which feature at centromeres, telomeres, the short arm of acrocentric chromosomes and ribosomal gene clusters) and low-copy repeats (LCRs) such as segmental duplications (SDs). SDs have been computationally defined as segments of DNA that contain ≥90% of sequence identity and ≥1 kb in length in the reference haploid genome, and they constitute approximately 4–5% of the human genome22. One of the surprising results from comparative genomic studies has been the observation that the human genome sequence distinguishes itself from other species genomes, such as the fly or worm, by a greater genome-wide presence of SDs and other LCRs21, which indicates that these repeat sequences greatly contribute to individual human variation23,24.
Most large LCRs (>10 kb) consist of a cluster of paralogous sequences of diverse genomic origin. The molecular characterization of such repeated regions in the human genome has revealed a mosaic architecture25,26; that is, such repeats are organized in hierarchical groups of direct and inversely orientated sequences as opposed to simple interchromosomal and intrachromosomal SDs27–29. The genome-wide distribution patterns of large LCRs show that they overlap with regions that frequently undergo genomic rearrangements that are associated with disease22. This finding can be explained by the fact that the large LCRs that flank a unique genomic region negatively affect genome stability30, making the flanked sequences prone to DNA rearrangements via nonallelic homologous recombination (NAHR; also known as ectopic homologous recombination) or RBMs. These characteristic genomic sequence features can thus be used to identify novel genomic loci that are susceptible to structural variant formation. Indeed, the first definition of genomic disorders1 arose from the susceptibility of specific variant regions that are associated with LCRs to undergo rearrangement in certain human diseases; for example, a 1.4 Mb duplication of a 17p12 interval causes Charcot–Marie–Tooth disease type 1A (CMT1A; OMIM 118220), and a reciprocal recombination deletion product causes hereditary neuropathy with liability to pressures palsies (HNPP; OMIM 162500).
Recurrent versus nonrecurrent rearrangements
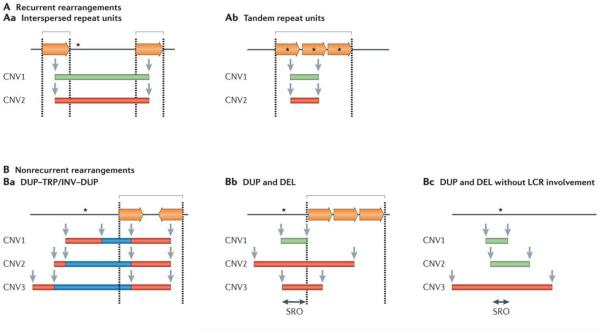
The formation of a CNV depends on the joining of two formerly separated DNA segments; these breakpoint junctions yield insights into the mechanisms that cause the chromosomal structural change. Recurrent rearrangements share the same size and genomic content in unrelated individuals. The breakpoints map within long, highly identical, flanking interspersed paralogous repeats (FIG. 1Aa), which mostly consist of LCRs28. One or more dosage-sensitive genes are present in-between the repeats and will be affected by the copy number change. Pathogenic CNVs are preferential rearrangement products at such loci31. Recurrent structural variants can also occur within tandem paralogous repeats (FIG. 1Ab) and alter the copy number of a dosage-sensitive gene that is present within32.
Figure 1. Schematic representation of recurrent and nonrecurrent genomic rearrangements observed in genomic disorders.
Black lines represent the genomic segments of a given locus. Dosage-sensitive genes that are involved in the rearrangement are represented by black asterisks. Paralogous repeats or low-copy repeats (LCRs) are represented by inverted and directly oriented horizontal orange arrows. Dashed lines indicate LCR regions. Individual genomic copy number variants (CNVs) are represented by colour-matched rectangles (deleted segments (green); duplicated segments (red); and triplicated segments (blue)) and identified as CNV1, CNV2 and CNV3. Locations of CNV breakpoints are indicated by grey vertical arrows. Square brackets denote regions with clustering or grouping of breakpoint junctions. A | Recurrent rearrangements share the same size and genomic content in unrelated individuals. More than 40 non-overlapping genomic disorders are caused by recurrent rearrangements49. Aa | In these types of structural variants the duplication or deletion breakpoints cluster within long, highly identical flanking interspersed paralogous repeats (represented by directly oriented horizontal orange arrows) that serve as substrates for nonallelic homologous recombination (NAHR). Ab | Alternatively, recurrent rearrangements can occur within tandem paralogous repeats and affect the copy number of a dosage-sensitive gene that is present within the repeat. For example, ectopic recombination of a tandem repeat consisting of opsin genes at Xq28 can cause red–green colour blindness (OMIM 303900, OMIM 303800) or it can lead to copy number polymorphism at this locus32. B | Nonrecurrent rearrangements present a unique size and genomic content at a given locus in unrelated individuals. At least 70 genomic disorders that are caused by nonrecurrent rearrangements have been described49. Ba | DUP–TRP/INV–DUP (duplication–inverted triplication–duplication) structures are nonrecurrent but, in some cases, have a limited genomic recurrence with two of four breakpoints mapping to an inverted repeat pair7,33–36. In these cases, the triplicated segments are inverted in relation to the duplications. Bb | Some genomic disorders are characterized by nonrecurrent rearrangements that show breakpoint grouping (rather than clustering as in the recurrent cases discussed above) within paralogous repeats38–42. Bc | By contrast, some genomic disorders are characterized by nonrecurrent rearrangements without any clustering or grouping of breakpoints43–45. Green rectangles represent deletions (DEL); red rectangles represent duplications (DUP); blue rectangles represent triplications (TRP). INV, inversion; SRO, smallest region of overlap.
Nonrecurrent rearrangements are rearrangements that have a unique size and genomic content at a given locus in unrelated individuals. A dosage-sensitive gene (or genes) can be identified within the structural variant smallest region of overlap (SRO) among individuals who share overlapping clinical phenotypes. Distinct types of nonrecurrent rearrangements are observed in genomic disorders, some of which are also delimited by LCRs. The formation of complex patterns of rearrangements can frequently be observed in some of these loci. For example, paralogous inverted repeats mediate the formation of a unique end-product pattern that consists of an inverted triplication that is interspersed with duplicated genomic segments. This structure is designated DUP–TRP/INV–DUP (duplication–inverted triplication–duplication)7,33–36 (FIG. 1Ba). In some cases, the inverted repeat pair consists of small repeats such as Alu elements37, but the majority of DUP–TRP/INV–DUP rearrangements described thus far have been LCR-mediated. Genomic disorders such as MECP2 duplication syndrome and Pelizaeus–Merzbacher disease (PMD; OMIM 312080) are characterized by nonrecurrent rearrangements, mostly duplications, that present with one breakpoint grouping within an LCR-laden region38–42 (FIG. 1Bb). By contrast, most of the nonrecurrent rearrangements in genomic disorders seem to be formed without LCR involvement (FIG. 1Bc), although many deletion and duplication variants are mediated by recombination between Alu elements43–45.
Thus, two general types of genomic rearrangement, recurrent and nonrecurrent, are observed and show intrinsically distinct features that reflect their underlying mechanisms of formation. They also have a distinct propensity for the formation of complex rearrangements such as interspersed duplications or triplications, as well as different meiotic versus mitotic risk (discussed below).
Recurrent structural variants: key mechanisms NAHR
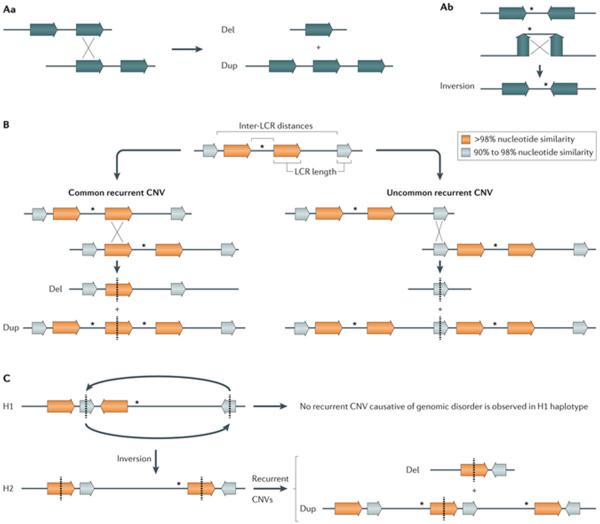
Recurrent structural variants often result from NAHR between directly-oriented or inverted LCRs that flank unique sequence genomic regions. According to the ectopic synapsis model, the recurrent size and genomic content generated by events that are mediated by NAHR results from the homologous recombination requirement for an ectopic synapsis preceding an ectopic crossing over that is dependent on the distance and the length of the homologous sequence46. By contrast, RBMs do not show such a requirement, therefore generating rearrangements that vary in size and genomic content. Ectopic homologous recombination47 that occurs during meiosis has been known to contribute to human disease for more than 40 years48 and is perhaps one of the best studied causative mechanisms for genomic disorders30. In fact, large LCR pairs located <5 Mb apart can lead to localized genomic instability via NAHR1,30 (FIG. 2A). Approximately 40 non-overlapping genomic loci are associated with meiotic NAHR that manifest as genomic disorders of microdeletion and microduplication syndromes49. Importantly, the duplication of a locus predisposes the region to undergo triplications in subsequent generations by more than 100-fold using the same molecular mechanism50. Most NAHR events that are causative of genomic disorders result from crossovers between LCRs located on the same chromosome, that is, intrachromosomal NAHR, or between non-allelic LCRs located in homologous chromosomes, that is, interchromosomal NAHR30. Rare cases of recurrent or de novo unbalanced translocations51,52 result from crossovers between repeats located on nonhomologous chromosomes.
Figure 2. Nonallelic homologous recombination mechanism.
A | Nonallelic homologous recombination (NAHR) between directly oriented repeats generates deletions and duplications (part Aa), whereas NAHR between inverted repeats generates the inversion of the segment in between the repeats (part Ab). B | The rate of NAHR may vary within a particular locus owing to the presence of low-copy repeat (LCR) pairs with distinct susceptibility risk. For example, NAHR rate positively correlates with LCR length and inversely correlates with inter-LCR distance, which can lead to common recurrent and uncommon recurrent copy number variants (CNVs) at a given locus identified in different patients with a genomic disorder30 (for example, in common recurrent and uncommon recurrent deletions and reciprocal duplications in Smith–Magenis syndrome and Potocki–Lupski syndrome, respectively)46. C | NAHR rate may vary among individuals with distinct genomic architecture in a given locus or a structural variant haplotype that results in directly oriented LCR. This is exemplified by inverted structural haplotypes that present with distinct risks of undergoing NAHR owing to structural variability in LCR content and orientation. For instance, two divergent structural variant haplotypes, H1 and H2, are observed in 17q21.31 region but deletions that cause Koolen-De Vries syndrome (OMIM 610443)150 occur on chromosomes carrying the H2 haplotype. Dosage-sensitive genes flanked by LCRs are represented by asterisks. LCRs are represented by horizontal colour-matched arrows.
The definition of the molecular features of NAHR has been instrumental to uncovering susceptible regions in the human genome28,53, as well as to discovering new genomic disorders31. The reciprocal nature of the NAHR model proposes that interchromatid or interchromosomal crossover will concomitantly generate a deletion and a duplication of the same interval; therefore, some genomic disorders that arise from a deletion CNV have also had a reciprocal duplication counterpart that is associated with a clinical phenotype, whereas other disorders remain to be clinically described1,30. Remarkably, reciprocal duplication and deletion syndromes can sometimes manifest mirror image traits for selected phenotypes, for example, weight (overweight and underweight in 17p11.2 microdeletion and microduplication syndromes, respectively), head size (microcephaly versus macrocephaly in 1q21.1 and 16p11.2), microdeletion and microduplication syndromes associated with schizophrenia (del1q21.1 and dup16p11.2) and autism (dup1q21.1 and del16p11.2) (reviewed in REF. 54).
Intrachromatidal NAHR is predicted to exclusively produce deletions, whereas interchromatidal and interchromosomal NAHR can mediate both deletions and duplications30. This hypothesis has been confirmed experimentally by analysing the rate of de novo NAHR-generated deletions in the germ line, which indicated that deletions occur approximately twice as frequently as duplications55, suggesting that NAHR has a preference for generating copy-number losses rather than copy-number gains.
NAHR frequency is driven by local genomic architecture
The complex structure of the LCR clusters suggests that more than one repeat pair within a certain cluster can be used as substrates for a de novo event. Nevertheless, the frequency of NAHR that occurs within and between LCRs mostly depends on distance, homology and size1. The distribution and relative frequencies of NAHR-mediated rearrangements positively correlate with LCR length, whereas the frequency of rearrangement is inversely influenced by inter-LCR distances46. Such correlation is even stronger when both LCR length and distance are considered (ln(LCR length/inter-LCR distance))28,46. Such coupled dependence suggests that, in meiotic NAHR, an ectopic synapsis occurs as a precursor to ectopic crossing-over46. Other LCR features such as DNA sequence identity, GC content and concentration of the PRDM9 homologous recombination hotspot motif 5′-CCNCCNTNNCCNC-3′ (REFS 56,57) are also positively correlated with the frequency of NAHR28. Experimentally, well-studied genomic disorders such as the 17p11.2 microdeletion Smith–Magenis syndrome (SMS; OMIM 182290) and its reciprocal duplication Potocki–Lupski syndrome (PTLS; OMIM 610883) indicate that preferential use of a particular LCR prevails, that is, common and uncommon recurrent CNVs46,58 (FIG. 2B). Evidence also supports the definition of active (hotspots) and inactive (coldspots) crossover regions or intervals within LCRs59.
Inter-individual variation in the rate of NAHR-mediated deletions and duplications was observed in sperm-based assay studies55,60. One of the potential causes is a personal or population locus-specific structural variation that predisposes individuals who carry particular structural variant haplotypes to present with distinct NAHR susceptibility (FIG. 2C). Nevertheless, the contribution of inter-individual structural variants to NAHR frequency is still not well understood. A recent study revealed that the rate of meiotic NAHR correlated between identical twins and was independent of age61, which indicates that other as yet unidentified genetic or environmental factors probably have an important role in the regulation of NAHR.
Inversions are particularly challenging to assess in an individual personal genome owing to the lack of high-throughput genome-wide screening techniques to assay such structures. Early evidence suggested that mitotic NAHR-mediated inversions frequently occur in somatic cells and potentially increase with age62. Meiotic inversions may also be frequent in the human genome. A study using fosmid paired-end sequencing data from eight human personal genomes from diverse populations identified 50–100 large genomic inversions that are not represented in the human reference genome63. Furthermore, genome-wide identification and mapping of inverted LCRs revealed that as much as ~12% of the genome may be susceptible to inversion that is mediated by NAHR53; inversions are therefore probably underestimated.
In summary, evidence suggests that paralogous sequence substrate size is important to ectopic synapsis28,46, and PRDM9 variation and the PRDM9 hotspot motif facilitate crossing over56,57.
Homologous versus homeologous recombination substrates
Strand exchange during intrachromosomal homologous recombination in mammalian cells preferentially occurs in regions of uninterrupted sequence identity64. A critical target size between consecutive nucleotide mismatches or minimum efficient processing segments (MEPSs) seems to be required. In mammalian genomes, MEPSs were experimentally defined as being between 134 and 232 bases in length; shortening of that segment of identity caused a sharp decrease in the spontaneous recombination rate in mammalian cells64. NAHR favours recombination between substrates that share perfect or near-perfect homology, as evidenced by the frequent use of LCRs ≥10 kb in length with ≥97% sequence identity30. Nonetheless, disease-causing rearrangements between repeat regions with a lower percentage of sequence identity occurs in some pathogenic CNVs (for example, HERV elements with 94–95% similarity65–67). HERV-mediated CNVs show enrichment for PRDM9 hotspot motifs as well as the property of recurrence65,67, which suggests NAHR as their underlying mechanism. Other examples of short NAHR substrates generating CNVs include neurofibromatosis type I (NF1; OMIM 162200), where strand exchange occurs between intervals as short as 114 bp (REF. 68), and α-thalassemia, where recombination between shorter intervals (34 bp in length) generates deletions of the human α-globin gene69.
By contrast, recombination between imperfectly matched substrates, that is, homeologous sequences 70, may be mediated by mechanisms other than NAHR. Homeologous sequences that underlie structural variant formation do not show enrichment for PRDM9 hotspot motifs and are shorter, with a lower percentage homology, than sequences used by NAHR. Structural variant formation events using homeologous sequences mostly generate nonrecurrent rearrangements that can be important contributors to genomic disorders43,44,71–73. For example, recombination between retroelements, such as short non-autonomous ~300 bp Alu elements, leading to CNVs is a predominant event in some genomic disorders74, such as autosomal-dominant spastic paraplegia 4 (SPG4; OMIM 182601)43,72 and alveolar capillary dysplasia with misalignment of pulmonary veins (ACDMPV; OMIM 265380)44. Alu-Alu-mediated rearrangement events have been historically attributed to NAHR; however, the low nucleotide sequence identity shared between the Alu family members, which can be as low as 75%, suggests that alternative recombination mechanisms also take place43. A recent report of complex genomic variants such as interspersed duplications and triplications mediated by multiple iterative template switches between Alu indicates that DNA synthesis can be involved in Alu-Alu-mediated events and supports a role for DNA replication in their formation37.
Nonrecurrent structural variants: key mechanisms
At least 70 genomic disorders are known to be caused by nonrecurrent structural variants49. This number is likely to be underestimated, because many of the genomic disorders that are associated with recurrent structural variants co-occur with nonrecurrent events in affected individuals (for selected examples see TABLE 1). Nonrecurrent rearrangements exhibit characteristic genomic features that are distinct from the recurrent structural variants that are mediated by NAHR (FIG. 1), for example, the scattered locations of their breakpoint junctions. Therefore, nonrecurrent rearrangements have a genomic content that is unique to the individual in which the rearrangement was generated (FIG. 1B), which makes the interpretation of potential clinical consequences challenging75. Moreover, predicting the occurrence of nonrecurrent events genome-wide is more difficult than that of recurrent structural variants.
Table 1.
Representative LCRs that mediate recurrent SVs as well as stimulate nonrecurrent genomic disorder associated
| LCR | Chromosome | Nonrecurrent SV | Size range (Mb) | Associated clinical phenotype (OMIM) | Refs |
|---|---|---|---|---|---|
| A, B, C | 3q29 |
|
|
|
151 |
|
| |||||
| SoS-PREP, SoS-DREP |
5q35 |
|
|
|
152,153 |
|
| |||||
| A, B, C | 7q11.23 |
|
|
|
154 – 156 |
|
| |||||
| BP3, BP4, BP5 | 15q13.1 |
|
|
|
36 |
|
| |||||
| 1-8 or A-H | 22q11.2 |
|
|
|
157 – 159 |
|
| |||||
| SMS-REPs | 17p11.2 |
|
|
|
6,46,160 |
|
| |||||
| CMT1A-REPs | 17p12 |
|
|
|
6,161 |
|
| |||||
| NF1-REP-A, B, C | 17q12 |
|
|
|
162 |
|
| |||||
| CRI-S232 | Xp22.31 |
|
|
|
105 |
|
| |||||
| PMDA-D | Xq22 |
|
|
|
7,33,40,42,77 |
|
| |||||
| JA, JB, JC K1, K2 |
Xq28 |
|
|
|
38,39,41,163 |
|
| |||||
| L1 (LCR1), L2 (LCR2) |
Xq28 |
|
|
|
164 |
DEL, deletion; DUP, duplication; INV, inversion; LCR, low-copy repeat; NML, normal (copy number neutral region); SV, structural variant; TRP, triplication. *Recurrent SVs mediated by LCRs PMDA-D, K1, K2, L1 and L2 are not causative of diseases. Distinct SV patterns of complex rearrangements, such as DUP–NML–INV/DUP, DUP–NML–DUP, DEL–NML–DEL, are observed at the loci as described.
The breakpoint junctions of nonrecurrent rearrangements can be characterized by simple blunt ends or microhomologies76, which is in sharp contrast to the extensive homology provided by LCRs that flank recurrent structural variants. In addition, small insertions have been identified in a number of junctions77, a characteristic that is not observed in NAHR-mediated events. Such features indicate that nonrecurrent structural variants are formed by molecular mechanisms that are distinct from NAHR, for example, non-homologous end joining (NHEJ) and RBMs such as break-induced replication (BIR), microhomology-mediated break-induced replication (MMBIR), serial replication slippage (SRS) and fork stalling and template switching (FoSTeS) (reviewed in REF. 78). NHEJ generates mostly simple, blunt CNV end points and, infrequently, short homology (1–3 nucleotides); however, small deletions or the insertion of random nucleotides can also be observed at breakpoint junctions79. By contrast, most of the nonrecurrent structural variants that are associated with genomic disorders present 2–33-bp-long microhomologies at breakpoint junctions80. In addition, the insertion of short segments (<100 bp) copied from nearby genomic regions is observed in up to 35% of nonrecurrent structural variant junctions10. These experimental observations support the notion that the underlying mechanism of formation of nonrecurrent structural variants involves DNA replication in which microhomology is used as a primer to assist replication initiation77,81.
RBMs
Evidence of DNA synthesis, which implicates RBMs in the formation of structural variants, has been documented in human diseases for almost a decade77,82. A key finding was the discovery that complex genomic rearrangements (CGRs), that is, rearrangements that consist of more than two breakpoint junctions80, can be formed in a single mutational event during DNA repair77,83. This contention is strongly supported by observations of a de novo nonrecurrent triplication that exclusively involves a single X-chromosome inherited from a non-carrier father7, and the finding that multiple, interspersed de novo copy number-amplified segments can all be stitched together in a single mutational event (chromoanasynthesis)8,9. Remarkably, copy number gains are more frequently associated with CGRs than are copy number losses46. This phenomenon may reflect the fact that the formation of nonrecurrent copy number gains is particularly likely to be generated by RBMs rather than by any other mechanism, although this observation is potentially biased by the fact that the signatures of DNA synthesis are more likely to remain in rearrangement structures that are associated with copy number gains46.
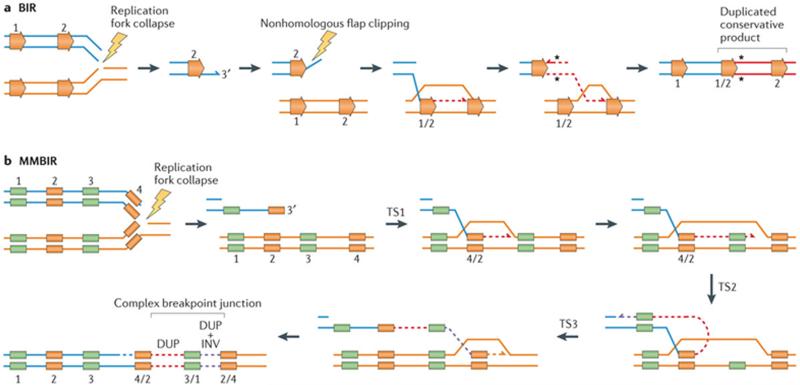
The replicative process BIR is a homologous recombination pathway that is conserved from phage to eukaryotes and serves to repair single-ended double-stranded breaks (seDSBs)84–86 (FIG. 3a). Spontaneous generation of such free DNA ends occurs, for example, during replication fork collapse at stalled forks or during the S phase of the cell cycle, in regions where there is no replication fork coming from the opposite direction (for example, subtelomeric regions). In BIR, the broken chromosome 3′ end invades a homologous template and initiates DNA replication that may extend hundreds of kilobases up to the telomere, in which case homozygosity of all markers distal to the double-stranded breaks (DSBs) can follow86. In yeast, BIR can also occur outside of the S phase, for example, in the G2 phase of the cell cycle. BIR requires almost all S phase replication proteins to be initiated, including all three major DNA polymerases (Polα–primase, Polδ and Polε) in addition to the replicative nonessential subunit of Polδ, Pol32 (REFS 87–89).
Figure 3. Replication-based mechanisms generate structural variants and single-nucleotide variants.
a | Break-induced replication (BIR) can be triggered by a nick on the template strand that may cause stalling or the collapse of the replication fork. Resection of the 5′ end of the broken single-ended, double-stranded DNA (seDNA) molecule exposes a 3′ tail that can invade an allelic (not shown) or a paralogous genomic segment with shared homology (ectopic recombination) to prime replication. Paralogous segments are represented by horizontal orange arrows. The use of ectopic homology to repair broken molecules by BIR can lead to structural variants (for example, duplication (shown here), triplications, deletions and inversions). A conservative mode of repair was recently proposed for BIR that can contribute to perpetuate repair indels or single-nucleotide variant (SNV) (black asterisks) mutations that are acquired during the replicative repair116,117. b | Microhomology-mediated break-induced replication (MMBIR) can be triggered by a nick on the template strand that may cause stalling or the collapse of the replication fork92. Alternatively, MMBIR can also occur by disrupted BIR111. Resection of the 5′ end of the broken seDNA molecule exposes a 3′ tail that can anneal to a single-strand DNA sharing microhomology (colour-matched boxes) to prime replication. The initial polymerase extension and replication is carried out by a low processivity polymerase, rendering this repair process prone to undergoing multiple rounds of disengagement and template switches until a fully processive replication fork is established. Short and/or long template switches during repair can generate complexity due to the insertion of templated segments at the rearrangement junction. DUP, duplication; INV, inversion; TS, template switches.
Recent in vitro studies in human cells under replication stress have implicated BIR in the formation of genomic rearrangements, including segmental amplifications5. Furthermore, nonrecurrent CNVs that are induced in mammalian cells through the use of replication inhibitors show many similarities to nonrecurrent structural variants that are formed spontaneously in genomic disorders. Similarities include breakpoint junctions that consist of microhomologies or blunt ends and contain small insertions, as well as the occurrence of complex rearrangements90. Importantly, in vitro inactivation of Xrcc4, which encodes proteins required for canonical NHEJ (c-NHEJ), did not alter the frequency or the features of the resulting structural variants in mouse embryonic stem cells, which suggests that c-NHEJ is unlikely to be the underlying mechanism90. Therefore, in vitro experiments support the argument that RBMs underlie the formation of nonrecurrent structural variants, a hypothesis that was first proposed almost 10 years ago from the study of rearrangements that are causative for genomic disorders77,82.
RAD51-dependent and RAD51-independent BIR (that is, MMBIR) seem to have different requirements for homology during DSB repair91,92. MMBIR uses microhomology to resume a stalled or collapsed replication fork as opposed to the longer homology tracts that are used in BIR92 (FIG. 3b). MMBIR has been proposed as the major formative mechanism that underlies nonrecurrent structural variants in genomic disorders92 on the basis that three main observations could not be readily explained by NAHR, NHEJ or BIR alone: first, the presence of microhomology in most of the breakpoint junctions, which may reflect priming DNA replication; second, template-driven juxtaposition of DNA sequences separated by large genomic distances, that is, long-range template switching; and third, iterative template switches that generate breakpoint complexity and CGRs77,92.
RBMs are error prone
Microhomology mediates long-range template switching
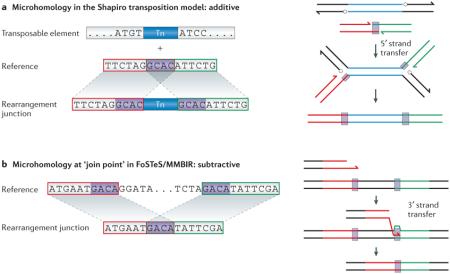
Template switching refers to a change of the single-stranded DNA template during replication within the same replication fork (short-distance template switch) or between distinct replication forks (long-distance template switch). Long-range template switching that produces tandem amplification was observed in stress-induced Escherichia coli between segments sharing microhomology81. The presence of microhomology indicates a recombination process that does not require homology for strand invasion and DNA synthesis81. In humans, structural variants that are causative of PMD have revealed evidence for long-distance template switching that is mediated by microhomology, which led to the proposal of the FoSTeS mechanistic model in genomic disorders77. A key observation during the formation of the structural variants at the PMD locus was the occurrence of multiple iterative template switches (FoSTeSX2, FoSTeSX3, and so on) mediated by microhomology that generated CGR, such as interspersed duplications and triplications77. This study also showed that the use of microhomology to assist repair can contribute to the high error rate that is associated with mechanisms that rely on a short stretch of homology to achieve strand transfer76 (BOX 1).
Box 1. Microhomology can be added or removed during structural variant formation.
A replicative mechanism was proposed by James Shapiro in 1979 to explain the semi-conservative transposition of the bacteriophage Mu149, in which short nucleotide sequences are duplicated flanking the target site of transposition as a result of the replication step that follows the 5′ strand transfer (microhomology additive mode; see the figure, part a). By contrast, the fork stalling and template switching (FoSTeS)/microhomology-mediated break-induced replication (MMBIR) model proposes that single-ended, double-stranded DNA (seDNA) is processed by a nuclease. A single strand end of the 3′ tail is then generated upon resection of the 5′ end anneals by virtue of Watson–Crick base pairing to a single-strand DNA sharing microhomology (that is, template switching) and primes DNA replication. This process has the opposite effect to the Shapiro model and results in the deletion of a short nucleotide stretch that is in common or shared between the segments that are involved in generating the rearrangement junction, essentially reducing it from two copies to one (microhomology subtractive mode; see the figure, part b).
Template switches can generate complex structural variants
Replicative mechanisms can undergo multiple rounds of strand invasion; microhomology can be used to resume DNA synthesis. If strand dissociation occurs and is followed by template switches it can lead to the generation of complex structural variants81,92–96. Intrachromosomal and interchromosomal template switches have the potential to generate different types of CGRs, such as the insertion of short genomic segments at the repair site, large-scale copy number alterations (for example, duplications, triplications and higher-order amplifications) and inversions (FIG. 4). There seems to be a bias towards intrachromosomal template switching in human rearrangements, although interchromosomal rearrangements do also occur7,10,97. Importantly, if nonallelic, homologous segments are used as templates and replication extends for several kilobases78,94, template switches will not lead to a change in copy number but the resulting genomic segment can show loss of heterozygosity, or observed AOH94.
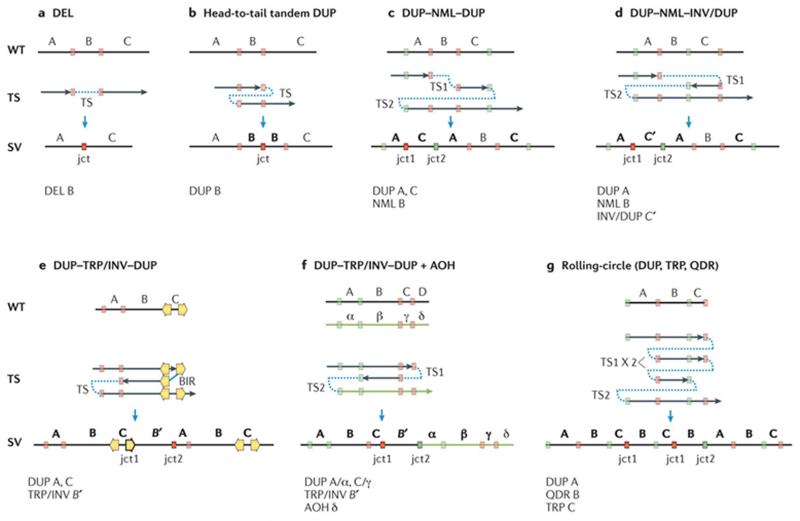
Figure 4. Template switches can generate distinct patterns of ‘simple’ or ‘complex’ structural variants.
All displayed structural patterns have been identified in one or more genomic disorders (TABLE 1). Top panel: black vertical line indicates a wild-type (WT) genomic segment. Middle panel: black arrows represent segments of DNA generated upon template switches (TSs). Red and green colour-matched boxes represent microhomology regions in WT genomic segment further involved in annealing and resumption of replication during TS. Orange arrows represent inverted low-copy repeats (LCRs) or regions of extended homology that can also be used during TS. Note that formation of any homology/microhomology breakpoint junctions (represented by outlined colour-matched boxes) is accompanied by a relative net reduction of the copy number of those homology/microhomology regions compared to WT regardless of whether the rearrangement results in gain or loss of the flanking genomic material (see microhomology subtractive mode (BOX 1)). Bottom panel: resulting structural variant (SV). Letters (A, B, C, D) represent chromosome alleles. Alleles subject to copy number gains in the resulting SVs are marked as bold letters. Primed letters represent an inverted-oriented genomic segment that originates upon a TS between complementary strands. α, β, γ, δ, represent corresponding homologue alleles. A single TS can generate deletion (DEL) (part a) or a head to tail tandem duplication (DUP) with one junction (jct) (part b). Two TSs can generate distinct complex end products presenting with two breakpoint junctions (jct1 and jct2). DUP–NML–DUP: copy number neutral segment (normal; NML) interspersed between duplications (DUP) (part c). DUP–NML–INV/DUP: inverted interspersed duplications (part d). DUP–TRP/INV–DUP: inverted triplication interspersed with duplications (part e). DUP–TRP/INV–DUP can be generated by break-induced replication (BIR), if inverted repeats are used as substrates for TS, or microhomology-mediated BIR if microhomology is used for TS. DUP–TRP/INV–DUP (part f) can be associated with extended regions of absence of heterozygosity (AOH) if TS occurs between homologous chromosomes (interchromosomal TS). Rolling-circle amplification (part g) can be generated by re-replication (for instance, the generation of two copies of jct1) culminating in the formation of multiple copies of a given segment.
The molecular characterization of structural variants in genomic disorders has revealed that apparently ‘simple’ nonrecurrent rearrangements may actually consist of complex breakpoint junctions. Insertions of short templated segments (<100 nucleotides) can be found in up to 35% of simple CNV junctions10,98–103. Most short templated insertions originate from nearby segments (within 300 bp), probably reflecting short-distance template switches that are due to misalignment or to replication slippage within the same replication fork10,13,93. Long-distance template switches introducing short template segments at the junctions occurred within <27 kb (REF. 10). By contrast, long-range template switches that produced large-scale CGRs, such as CNVs that are visible by high-resolution microarrays, vary in breakpoint junction distance from a few kilobases to megabases apart39,77,99,101–105. These observations may also be relevant for human genomic variability, as 27% of copy number losses analysed in samples from the 1,000 Genomes Project show insertions varying from 1 to 96 nucleotides at their breakpoint junctions; approximately 50% could be characterized as a templated insertion106.
Complex rearrangements arise due to reduced processivity of DNA polymerases
The presence of de novo indels, as well as short- and long-distance trans and cis template switches that occur concomitantly, strongly support a faulty DNA replication process during repair by RBMs6,10,13,39,77,82,104,105. In fact, the properties observed from studies of breakpoint junction formation suggest that the polymerase used for RBM has reduced processivity and fidelity relative to intergenerational DNA polymerases107. Distinct polymerases may contribute to template switching via different mechanisms. For example, in mammalian cell culture, Polθ introduces short templated insertions that are synthesized from mismatched primer termini during DSB repair by microhomology-mediated end joining (MMEJ)108. Long-range template switching, particularly involving the replication of long DNA segments, may involve replicative polymerases; Polδ is a prominent candidate due to its lower processivity compared with Polε. Supporting this argument, BIR microhomology-mediated events depend on the Polδ subunit Pol32 (in yeast) or POLD3 (in humans) to initiate the repair of broken forks; in humans this repair can produce duplications as large as 200 kb (REFS 5,89). The participation of translesion synthesis (TLS) polymerases such as Polζ in short-range template switching is supported by evidence that fork stalling uses Polζ to overcome the impairment in progression109,110. Importantly, recent studies in yeast have revealed that the TLS polymerases Polζ and Rev1 mediate template switches in BIR-defective cells using microhomology to prime replication111. This finding suggests that MMBIR can actually result from an interrupted BIR111. In aggregate, these data support the idea that template switching events that generate CGR may result from the disruption of replication, dissociation of replicative polymerases and switching to lower processivity polymerases that are error prone.
RBMs show a high mutational rate
In addition to having a high risk of undergoing template switching, DNA synthesis that is associated with repair can also lead to an increased local mutational rate10,107. The involvement of DNA synthesis during repair in humans is accompanied by increased de novo SNV and indel mutation rates at or near the breakpoint junction of duplicated segments (~2.1 × 10−4 mutations per base pair and ~1.7 × 10−3 events per base pair, respectively)10. Furthermore, kataegis, which is frequently associated with genomic rearrangements in some types of cancer such as breast cancer112,113, is hypothesized to result from susceptible BIR-generated single-stranded DNA (ssDNA)4.
Consistent with human data, BIR in yeast is accompanied by a 1,000-fold increase in the SNV mutation rate compared with S phase replication107. A 5′ extensive resection generates a 3′ ssDNA end that will precede the strand invasion mediated by Rad51 (REF. 114). In BIR, the combination of an extensive ssDNA in addition to leading and lagging strands that are synthesized in an asynchronous way115–117 results in the accumulation of ssDNA that is highly susceptible to unrepaired DNA lesions, the formation of secondary structures and cleavage by endonucleases. The conservative mode of DNA synthesis of newly synthesized strands in BIR via a migrating DNA bubble (D-loop) may contribute to the propagation of these orientation-dependent acquired mutations116,117 (FIG. 3a). Importantly, the extent of replication inaccuracy resulting from BIR seems to be limited by the distance of the break-induced event to an efficient convergent fork as well as to the activity of the endonuclease Mus81. Mus81 cleaves the D-loop and converts conservative DNA synthesis into a canonical replication fork and W-C semi-conservative synthesis118. Therefore, Mus81 limits the high mutational rate of RBMs, including template switches, to regions that are near the breakpoint junctions of the structural variants formed by this mechanism118. Switching to a semi-conservative synthesis mode is likely to have a selective advantage by limiting the burst of mutations in the long genomic regions that would otherwise follow.
The role of genomic architecture
Formation of nonrecurrent structural variants is non-random
Breakpoint junction mapping of nonrecurrent rearrangements in genomic disorders indicates that the occurrence of structural variants in certain regions of the genome is unlikely to be random. Particular genomic architectures, such as repeated sequences and repetitive elements, with the potential to form non-B DNA structures (for example, A-T rich palindromes, G-quadruplexes, short inverted repeats and retrotransposable elements) are frequently observed in association with the location of a template switch or structural variant breakpoint45,119–121. How such elements contribute to structural variant formation is currently being debated: one possibility is that they render certain regions susceptible to the transient formation of secondary DNA structures that can lead to fork collapse and, in some cases, to the formation of DSBs119,120. DSBs can contribute to the primary cause of instability in a particular genomic region; however, DSBs may not even be required to trigger genomic stability. For example, common fragile sites (CFSs) that harbour [A]n and [TA]n repeats and quasi-palindrome sequences such as short inverted repeats (4–6 nt) have been shown to perturb the progression of S phase Polδ replication upon which the polymerase may pause and dissociate from the replication machinery122. This event potentially causes an error-prone polymerase to take over to more efficiently replicate such complex sites, thus leading to genomic instability that is conducive to nonrecurrent genomic rearrangements122,123.
Nonrandom formation of nonrecurrent rearrangements is supported by analyses of polymorphic structural variants in human populations15,106 in which distinct signatures of the underlying structural variant mechanism can be associated with the local genomic features where they occurred106. For example, DNA replication timing differences across the genome are associated with distinct types of CNVs. Recurrent CNVs are more frequently observed in early replicating regions, whereas nonrecurrent CNVs are more frequently observed in late replicating regions106,124. Further analysis has indicated that regions of reduced rates of replication, which are consistent with polymerase pausing, seem to associate with breakpoint junctions of short homology-mediated CNVs125, supporting the contention that genomic regions that are prone to polymerase pausing may be more susceptible to genomic instability. Nearby repeated sequences may themselves facilitate replicative repair that can lead to nonrecurrent structural variants.
The role of repeat sequences in the formation of non-recurrent structural variants
Genomic architecture can stimulate the occurrence of template switches near a DNA break. In yeast, the presence of repeats close to an induced break increases the frequency of the formation of CGRs, whereas diverged human-derived Alu elements inserted near seDSBs lead to increased template switching118. Interestingly, a recent study in budding yeast showed that the position of the strand invasion during repair by RBMs is influenced by ‘microhomology islands’ that flank the junctions of resulting rearrangements126. It remains unclear how frequently microhomology islands are found in nonrecurrent human rearrangements but it is tempting to speculate that repetitive elements such as Alus may provide such microhomology islands that could assist strand transfer or stimulate template switching during repair by RBM. Moreover, a strong association between the locations of the breakpoint junctions of nonrecurrent rearrangements within LCRs has been documented in locus-specific studies (TABLE 1). This association has also been observed genome-wide in polymorphic structural variants127; in fact, Kidd et al.128 indicate that ~20% of the breakpoints of certain types of structural variants map within 5 kb of LCRs.
Mechanistically, how LCRs contribute to the formation of nonrecurrent rearrangements is not entirely clear and probably differs in nature from the homologous recombination role that LCRs have in mediating recurrent rearrangements. This is evident from a genome-wide study of polymorphic structural variants that showed that the location of nonrecurrent rearrangements hotspots differs from those of recurrent rearrangements129. However, contrary to the challenge of predicting structural variant breakpoints using sequence motifs, LCRs do allow some degree of prediction of the occurrence of nonrecurrent rearrangements and support the hypothesis that the genomic architecture exerts multiple but distinct influences on structural variant formation genome-wide. This is exemplified by specific CGR structures such as DUP–TRP/INV–DUP that can be formed if highly identical inverted LCRs are used as substrates for BIR7,33,34,130. The presence of inverted repeats is hypothesized to provide a high degree of nucleotide sequence identity, which renders the region susceptible to undergoing ectopic template switching. Such ectopic template switching during a replicative repair process may result in an inverted segment being formed concomitantly with a copy number gain (FIG. 4e). DUP–TRP/INV–DUP is frequently observed in patients with PMD due to PLP1 duplication33 and is also present in ~20% of individuals with MECP2 duplication syndrome7,39. Importantly, DUP–TRP/INV–DUP can cause a much more severe clinical phenotype if the dosage-sensitive gene, that is, PLP1 or MECP2, maps within the triplicated segment7,33,131,132. Therefore, the presence of inverted repeats near dosage-sensitive genes can indicate regions in the human genome that are susceptible to DUP–TRP/INV–DUP formation53, which has been proved to be of clinical relevance at different genomic loci34,35,130 (TABLE 1).
RBMs underlie some human translocations
Extensive homology present at the telomere and subtelomere of most chromosomes29 can trigger breakage–fusion–bridge (BFB) cycles133,134 by forming dicentric chromosomes, intrachromosomal or interchromosomal ectopic recombination135 and secondary structures that can render these regions prone to breaks136. NHEJ is likely to be a predominant mechanism of repair137, although recurrent NAHR between interchromosomal LCRs also occurs51,135,138. Recent data have implicated RBMs in the repair of broken telomeres on which homology, homeology or microhomology is used during repair37,139–141, including telomeric short inverted repeats142. Therefore, LCRs and other types of repeats probably contribute to the rearrangement events involving telomeres, supporting the conclusion that genomic architecture can have multiple roles in the formation of structural variants genome-wide and that RBMs might contribute to increasing an individual’s genome complexity.
Conclusions
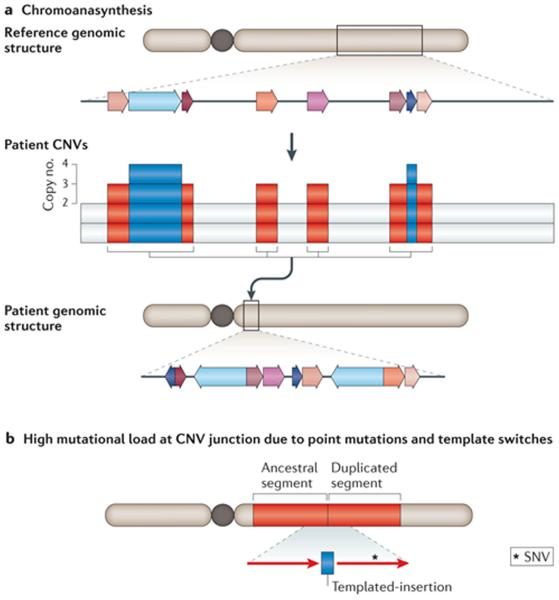
Studies of structural variation in genomic disorders have resulted in mechanistic findings that apply to model organisms and have wide implications for fields as diverse as human biology, genomics, molecular diagnostics, disease understanding and therapeutic interventions based on gene dosage correction. The emerging picture is a perplexing fractal: large genomic-scale structural variants, that is, alterations involving megabases of DNA, can be associated with additional complexities, such as other structural variants that are generated in a one-off event (chromoanasynthesis and chromothripsis-like events (FIG. 5a))8,9 or a CGR accompanied by AOH11,12. Further alterations might be revealed when these structural variants are scrutinized down to the base-pair sequence level owing to the presence of small-scale changes that can act as mutational signatures, for example, insertions or deletions of short DNA segments and/or de novo point mutations that are present adjacent to the junction of a structural variant10,106,128,143 (FIG. 5b). This picture may further complicate the interpretation of potential functional consequences of a mutagenic event for an a priori apparently simple structural variant.
Figure 5. Replication-based mechanisms are error prone.
a | Schematic example of copy number variants (CNVs) in multiple, interspersed segments within the same chromosome arm. Chromoanasynthesis in constitutional genomic disorders resembles the phenomenon of chromosome catastrophes (that is, chromothripsis) initially described based on the analyses of hundreds of cancer genome sequences and in different types of cancers83. The chromosomal region involved in chromoanasynthesis is denoted by a grey box; local genomic segments involved are represented by coloured arrows. Array comparative genomic hybridization (aCGH) reveals genomic segments with copy number variation. The rearranged chromosome is shown on which the extra copy number segments (represented by colour-matched arrows) are inserted together in a new chromosomal position with respect to the reference genome. This clustering of CNVs in a new position is a hallmark of chromoanasynthesis. b | Increased mutational load can be observed at the breakpoint junctions of CNVs generated by replication-based mechanisms (RBMs). SNVs include point mutations and short insertions/short deletions (indels). Red rectangles represent the duplicated segment and red arrows represent the orientation of the duplicated segments.
The genomic architecture at a particular locus contributes to an increase in structural variant complexity, with a prominent role for replicative mechanisms. LCRs can mediate recurrent structural variants by providing substrate sequences for NAHR, and also stimulate non-recurrent structural variants by RBMs, where ectopic repeats can assist DNA repair by providing homology, homeology or microhomology sequences. RBMs underlying the formation of a ‘simple’ or a ‘complex’ structural variant have important implications for human disease and transmission genetics. First, the occurrence of triplications can lead to more severe clinical phenotypes; chromoanasynthesis events can cause multiple malformations and congenital diseases, whereas segmental AOH may enable the expression of recessive traits and regional uniparental disomy. Second, point mutations and indels that are derived from error-prone repair that is likely to arise concomitant with a disease-associated structural variant may affect gene expression and contribute to the variability of the associated genomic disorder. Third, RBM-generated nonrecurrent rearrangements are hypothesized to be formed during mitosis. Therefore, these mutations may contribute to diseases that are caused by somatic mutations, including cancer, developmental and neurological disorders144 and to an increased probability of parental low-level mosaicism6,145. Germline mosaicism can alter the recurrence risk for future pregnancies19.
Importantly, how genomic architecture contributes to altered gene expression must be broadly investigated. The impact of a particular structural variant on the expression of overlapping genes seems to go beyond simply the direct effect of the altered copy number. For example, lymphoblastoid cells from patients with deletions of the Williams–Beuren region (7q11.23) show dysregulation of flanking genes, reflecting long-range cis-regulatory elements within the structural variant146 that can also potentially change the timing of gene expression147. Recently, chromosome conformation capture (4C-seq) and ChIP-seq revealed that modifications of local chromatin were strongly associated with the altered expression of particular genes in 7q11.23 (REF. 16). The contribution of structural variants to the regulation of gene expression in cis or in trans needs to be further explored as it may help to explain phenotypic variability in patients who carry overlapping nonrecurrent rearrangements148. For example, new discoveries of mechanisms for structural variant formation in humans are elucidating how such events can re-structure a specific region of the genome to change the expression of transcripts locally or genome-wide16,17, including through altering topologically associating domains (TADs), with pathological consequences for carriers18.
In summary, the contribution of genomic architecture to structural variant formation goes beyond the definition and expansion of a group of human diseases designated as genomic disorders. It implicates particular mechanisms for rearrangement formation and helps to guide studies about the potential causes of human disease, including germline and somatic mutagenesis events.
Acknowledgements
The authors thank C. R. Beck, S. Gu, P. Stankiewicz and P. J. Hastings for thoughtful comments and helpful discussions. The authors apologize to colleagues and the authors of relevant papers who could not be cited owing to space limitations. The research conducted by the authors was supported in part by the US National Institutes of Neurologic Disorders and Stroke (RO1NS058529), National Human Genome Research Institute/National Heart Blood Lung Institute jointly funded Ba ylor Hopkins Center f or Mendelian Gen omics (U54HG006542), National Institute of General Medical Sciences (RO1 GM106373), the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) (476217/2013-0) and the Young Investigator fellowship (Science without Borders Program) grant 402520/2012-2 to C.M.B.C. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NINDS, NHGRI/NHBLI, NIGMS or NIH.
Glossary
- Genomic disorders
Conditions that result from rearrangements of the genome rather than base pair changes of DNA, and in which genomic instability results from the endogenous genome architecture.
- Structural variants
Variants that include copy number variants and copy number neutral inversions, insertions and translocations in a personal genome compared with a reference genome.
- Copy number variants (CNVs)
Alteration in copy number (gain or loss) of a locus resulting in deviation from the normal diploid state.
- Single nucleotide variants (SNVs)
A single site in a DNA sequence that differs among individuals.
- Chromothripsis “Shattering of chromosomes”
A single catastrophic event affecting one chromosome and leading to complex rearrangements in cancer.
- Absence of heterozygosity (AOH)
Refers to copy number neutral genomic segments that lack heterozygosity for assayed polymorphic markers.
- Template switching
Refers to a transient dissociation of the primer and template followed by a re-association to a distinct template during DNA replication. It can occur within the same replication fork (short-distance template switch) or between distinct replication forks (long-distance template switch).
- Array comparative genomic hybridization (aCGH)
Microarray-based technique that measures the relative copy number of DNA segments.
- Replication-based mechanisms (RBMs)
Replicative non-homologous DNA repair mechanism of single-ended, double-stranded DNA (seDNA).
- Rearrangement susceptibility
Regions of the genome prone to structural variation formation.
- Mobile elements
A segment of DNA capable of moving into a new genomic position.
- Paralogous sequences
Homologous sequences that arose by duplication.
- Nonallelic homologous recombination
Nonallelic pairing of paralogous sequences and crossover leading to deletions, duplications and inversions.
- Ectopic synapsis
Chromosomal homologue synapses at a nonallelic position.
- Fosmid paired-end sequencing
A clone-based method to sequence the ends of fragments with a known size range.
- Homeologous sequences
Imperfectly matched paralogous genomic segments.
- Microhomologies
Short stretches of shared nucleotide identity present at the junctions of rearranged genomic segments.
- Non-homologous end joining (NHEJ)
Double-stranded break (DSB) mechanism of repair that processes the broken DNA ends and joins non-homologous sequences. It repairs the programmed DSBs created in the immune system.
- Break-induced replication (BIR)
Homologous recombination pathway that repairs single-ended double-stranded breaks (seDSBs) through the establishment of a unidirectional replication fork.
- Microhomology-mediated break-induced replication (MMBIR)
RAD51-independent break-induced replication that relies on microhomology to resume replication.
- Serial replication slippage (SRS)
Multiple rounds of slipped strand mispairing at the replication fork.
- Fork stalling and template switching (FoSTeS)
Mechanism of template switching between different replication forks.
- Chromoanasynthesis
Chromosome reconstitution or chromosome re-assortment. Constitutive complex rearrangements resulting from multiple template switches.
- Microhomology-mediated end joining (MMEJ)
An alternative non-homologous end joining mechanism that repairs broken double-stranded breaks using sequence microhomology to join and stabilize DNA end intermediates.
- Kataegis
Somatic single-nucleotide mutation clusters or mutation showers in cis.
- Breakage–fusion–bridge (BFB) cycles
Processes by which sister chromatids that lack a telomere (breakage) can retrieve them by fusion and the creation of an unstable dicentric chromosome that will be pulled apart during anaphase (bridge). Eventually, the bridge breaks and the cycle starts again until the chromosome is stabilized.
Footnotes
Competing interests statement
The authors declare competing interests: see Web version for details.
References
- 1.Lupski JR. Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 1998;14:417–422. doi: 10.1016/s0168-9525(98)01555-8. [DOI] [PubMed] [Google Scholar]
- 2.Zhang F, Gu W, Hurles ME, Lupski JR. Copy number variation in human health, disease, and evolution. Annu. Rev. Genom. Hum. Genet. 2009;10:451–481. doi: 10.1146/annurev.genom.9.081307.164217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mills RE, et al. An initial map of insertion and deletion (INDEL) variation in the human genome. Genome Res. 2006;16:1182–1190. doi: 10.1101/gr.4565806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sakofsky CJ, et al. Break-induced replication is a source of mutation clusters underlying kataegis. Cell Rep. 2014;7:1640–1648. doi: 10.1016/j.celrep.2014.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Costantino L, et al. Break-induced replication repair of damaged forks induces genomic duplications in human cells. Science. 2014;343:88–91. doi: 10.1126/science.1243211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang F, et al. The DNA replication FoSTeS/MMBIR mechanism can generate genomic, genic and exonic complex rearrangements in humans. Nat. Genet. 2009;41:849–853. doi: 10.1038/ng.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carvalho CM, et al. Inverted genomic segments and complex triplication rearrangements are mediated by inverted repeats in the human genome. Nat. Genet. 2011;43:1074–1081. doi: 10.1038/ng.944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu P, et al. Chromosome catastrophes involve replication mechanisms generating complex genomic rearrangements. Cell. 2011;146:889–903. doi: 10.1016/j.cell.2011.07.042. Very complex rearrangements with multiple template switches can be formed constitutively in a one-off event by RBM that is reminiscent of the chromothripsis events that were first described in cancer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kloosterman WP, et al. Constitutional chromothripsis rearrangements involve clustered double-stranded DNA breaks and nonhomologous repair mechanisms. Cell Rep. 2012;1:648–655. doi: 10.1016/j.celrep.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 10.Carvalho CM, et al. Replicative mechanisms for CNV formation are error prone. Nat. Genet. 2013;45:1319–1326. doi: 10.1038/ng.2768. This work revealed how apparently simple structural variants can actually be highly complex and the complexity revealed by applying multiple experimental techniques to deduce structure and understand the resultant end product of mutation. An unexpectedly high mutational spectrum represented by both SNVs and template switches can be detected in up to 52% of the CNVs at the locus studied. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carvalho CM, et al. Absence of heterozygosity due to template switching during replicative rearrangements. Am. J. Hum. Genet. 2015;96:555–564. doi: 10.1016/j.ajhg.2015.01.021. CNVs generated post-zygotically by replication-based mechanisms can produce triplications that are associated with inversion and long regions of AOH. The importance of this observation relies on the potential implication for human diseases that may include not only dosage-sensitive genes but also unmasking of recessive traits due to the extensive AOH, distorting transmission genetics leading to disease in a family with only a single carrier parent, as well as imprinting disease due to the presence of uniparental disomy (UPD) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sahoo T, et al. Concurrent triplication and uniparental isodisomy: evidence for microhomology-mediated break-induced replication model for genomic rearrangements. Eur. J. Hum. Genet. 2015;23:61–66. doi: 10.1038/ejhg.2014.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, et al. Characterization of 26 deletion CNVs reveals the frequent occurrence of micro-mutations within the breakpoint-flanking regions and frequent repair of double-strand breaks by templated insertions derived from remote genomic regions. Hum. Genet. 2015;134:589–603. doi: 10.1007/s00439-015-1539-4. [DOI] [PubMed] [Google Scholar]
- 14.Coe BP, et al. Refining analyses of copy number variation identifies specific genes associated with developmental delay. Nat. Genet. 2014;46:1063–1071. doi: 10.1038/ng.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sudmant PH, et al. An integrated map of structural variation in 2,504 human genomes. Nature. 2015;526:75–81. doi: 10.1038/nature15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gheldof N, et al. Structural variation-associated expression changes are paralleled by chromatin architecture modifications. PLoS ONE. 2013;8:e79973. doi: 10.1371/journal.pone.0079973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ricard G, et al. Phenotypic consequences of copy number variation: insights from Smith-Magenis and Potocki-Lupski syndrome mouse models. PLoS Biol. 2010;8:e1000543. doi: 10.1371/journal.pbio.1000543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lupianez DG, et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 2015;161:1012–1025. doi: 10.1016/j.cell.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Campbell IM, Shaw CA, Stankiewicz P, Lupski JR. Somatic mosaicism: implications for disease and transmission genetics. Trends Genet. 2015;31:382–392. doi: 10.1016/j.tig.2015.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weischenfeldt J, Symmons O, Spitz F, Korbel JO. Phenotypic impact of genomic structural variation: insights from and for human disease. Nat. Rev. Genet. 2013;14:125–138. doi: 10.1038/nrg3373. [DOI] [PubMed] [Google Scholar]
- 21.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 22.Bailey JA, et al. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- 23.Sebat J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 24.Sharp AJ, et al. Segmental duplications and copy-number variation in the human genome. Am. J. Hum. Genet. 2005;77:78–88. doi: 10.1086/431652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kurotaki N, Stankiewicz P, Wakui K, Niikawa N, Lupski JR. Sotos syndrome common deletion is mediated by directly oriented subunits within inverted Sos-REP low-copy repeats. Hum. Mol. Genet. 2005;14:535–542. doi: 10.1093/hmg/ddi050. [DOI] [PubMed] [Google Scholar]
- 26.Park SS, et al. Structure and evolution of the Smith-Magenis syndrome repeat gene clusters, SMS-REPs. Genome Res. 2002;12:729–738. doi: 10.1101/gr.82802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang Z, et al. Ancestral reconstruction of segmental duplications reveals punctuated cores of human genome evolution. Nat. Genet. 2007;39:1361–1368. doi: 10.1038/ng.2007.9. [DOI] [PubMed] [Google Scholar]
- 28.Dittwald P, et al. NAHR-mediated copy-number variants in a clinical population: mechanistic insights into both genomic disorders and Mendelizing traits. Genome Res. 2013;23:1395–1409. doi: 10.1101/gr.152454.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Linardopoulou EV, et al. Human subtelomeres are hot spots of interchromosomal recombination and segmental duplication. Nature. 2005;437:94–100. doi: 10.1038/nature04029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stankiewicz P, Lupski JR. Genome architecture, rearrangements and genomic disorders. Trends Genet. 2002;18:74–82. doi: 10.1016/s0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- 31.Sharp AJ, et al. Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat. Genet. 2006;38:1038–1042. doi: 10.1038/ng1862. The authors apply the conceptual mechanistic understanding of NAHR to predict genomic instability regions and define five novel genomic disorders. This article provides evidence that comprehending the rules underlying structural variation formation in the human genome is important and provides insights enabling predictions of rearrangement-prone genomic regions. [DOI] [PubMed] [Google Scholar]
- 32.Nathans J, Piantanida TP, Eddy RL, Shows TB, Hogness DS. Molecular genetics of inherited variation in human color vision. Science. 1986;232:203–210. doi: 10.1126/science.3485310. [DOI] [PubMed] [Google Scholar]
- 33.Beck CR, et al. Complex genomic rearrangements at the PLP1 locus include triplication and quadruplication. PLoS Genet. 2015;11:e1005050. doi: 10.1371/journal.pgen.1005050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beri S, Bonaglia MC, Giorda R. Low-copy repeats at the human VIPR2 gene predispose to recurrent and nonrecurrent rearrangements. Eur. J. Hum. Genet. 2013;21:757–761. doi: 10.1038/ejhg.2012.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ishmukhametova A, et al. Dissecting the structure and mechanism of a complex duplication-triplication rearrangement in the DMD gene. Hum. Mutat. 2013;34:1080–1084. doi: 10.1002/humu.22353. [DOI] [PubMed] [Google Scholar]
- 36.Soler-Alfonso C, et al. CHRNA7 triplication associated with cognitive impairment and neuropsychiatric phenotypes in a three-generation pedigree. Eur. J. Hum. Genet. 2014;22:1071–1076. doi: 10.1038/ejhg.2013.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gu S, et al. Alu-mediated diverse and complex pathogenic copy-number variants within human chromosome 17 at p13.3. Hum. Mol. Genet. 2015;24:4061–4077. doi: 10.1093/hmg/ddv146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bauters M, et al. Nonrecurrent MECP2 duplications mediated by genomic architecture-driven DNA breaks and break-induced replication repair. Genome Res. 2008;18:847–858. doi: 10.1101/gr.075903.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carvalho CM, et al. Complex rearrangements in patients with duplications of MECP2 can occur by fork stalling and template switching. Hum. Mol. Genet. 2009;18:2188–2203. doi: 10.1093/hmg/ddp151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Inoue K, et al. Genomic rearrangements resulting in PLP1 deletion occur by nonhomologous end joining and cause different dysmyelinating phenotypes in males and females. Am. J. Hum. Genet. 2002;71:838–853. doi: 10.1086/342728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Small K, Warren ST. Emerin deletions occurring on both Xq28 inversion backgrounds. Hum. Mol. Genet. 1998;7:135–139. doi: 10.1093/hmg/7.1.135. [DOI] [PubMed] [Google Scholar]
- 42.Woodward KJ, et al. Heterogeneous duplications in patients with Pelizaeus-Merzbacher disease suggest a mechanism of coupled homologous and nonhomologous recombination. Am. J. Hum. Genet. 2005;77:966–987. doi: 10.1086/498048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boone PM, et al. The Alu-rich genomic architecture of SPAST predisposes to diverse and functionally distinct disease-associated CNV alleles. Am. J. Hum. Genet. 2014;95:143–161. doi: 10.1016/j.ajhg.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stankiewicz P, et al. Genomic and genic deletions of the FOX gene cluster on 16q24.1 and inactivating mutations of FOXF1 cause alveolar capillary dysplasia and other malformations. Am. J. Hum. Genet. 2009;84:780–791. doi: 10.1016/j.ajhg.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vissers LE, et al. Rare pathogenic microdeletions and tandem duplications are microhomology-mediated and stimulated by local genomic architecture. Hum. Mol. Genet. 2009;18:3579–3593. doi: 10.1093/hmg/ddp306. [DOI] [PubMed] [Google Scholar]
- 46.Liu P, et al. Frequency of nonallelic homologous recombination is correlated with length of homology: evidence that ectopic synapsis precedes ectopic crossing-over. Am. J. Hum. Genet. 2011;89:580–588. doi: 10.1016/j.ajhg.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lichten M, Borts RH, Haber JE. Meiotic gene conversion and crossing over between dispersed homologous sequences occurs frequently in Saccharomyces cerevisiae. Genetics. 1987;115:233–246. doi: 10.1093/genetics/115.2.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McKusick VA. Human genetics. Annu. Rev. Genet. 1970;4:1–46. doi: 10.1146/annurev.ge.04.120170.000245. [DOI] [PubMed] [Google Scholar]
- 49.Vissers LE, Stankiewicz P. Microdeletion and microduplication syndromes. Methods Mol. Biol. 2012;838:29–75. doi: 10.1007/978-1-61779-507-7_2. [DOI] [PubMed] [Google Scholar]
- 50.Liu P, et al. Mechanism, prevalence, and more severe neuropathy phenotype of the Charcot-Marie-Tooth type 1A triplication. Am. J. Hum. Genet. 2014;94:462–469. doi: 10.1016/j.ajhg.2014.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ou Z, et al. Observation and prediction of recurrent human translocations mediated by NAHR between nonhomologous chromosomes. Genome Res. 2011;21:33–46. doi: 10.1101/gr.111609.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robberecht C, Voet T, Zamani Esteki M, Nowakowska BA, Vermeesch JR. Nonallelic homologous recombination between retrotransposable elements is a driver of de novo unbalanced translocations. Genome Res. 2013;23:411–418. doi: 10.1101/gr.145631.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dittwald P, et al. Inverted low-copy repeats and genome instability—a genome-wide analysis. Hum. Mutat. 2013;34:210–220. doi: 10.1002/humu.22217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Golzio C, Katsanis N. Genetic architecture of reciprocal CNVs. Curr. Opin. Genet. Dev. 2013;23:240–248. doi: 10.1016/j.gde.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Turner DJ, et al. Germline rates of de novo meiotic deletions and duplications causing several genomic disorders. Nat. Genet. 2008;40:90–95. doi: 10.1038/ng.2007.40. In this paper the authors calculate the locus-specific ratio of deletions and duplications by NAHR in male meiosis. The observed higher ratio of deletions versus duplications, 2/1 for autosomes, correlates well with theoretical predictions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Myers S, Freeman C, Auton A, Donnelly P, McVean G. A common sequence motif associated with recombination hot spots and genome instability in humans. Nat. Genet. 2008;40:1124–1129. doi: 10.1038/ng.213. [DOI] [PubMed] [Google Scholar]
- 57.Berg IL, et al. PRDM9 variation strongly influences recombination hot-spot activity and meiotic instability in humans. Nat. Genet. 2010;42:859–863. doi: 10.1038/ng.658. Variation within the genomic structure of PRDM9 was reported to correlate with the frequency of meiotic recombination in individuals, providing direct evidence for PRDM9 involvement in HR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang F, et al. Identification of uncommon recurrent Potocki-Lupski syndrome-associated duplications and the distribution of rearrangement types and mechanisms in PTLS. Am. J. Hum. Genet. 2010;86:462–470. doi: 10.1016/j.ajhg.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cooper GM, et al. A copy number variation morbidity map of developmental delay. Nat. Genet. 2011;43:838–846. doi: 10.1038/ng.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lam KW, Jeffreys AJ. Processes of de novo duplication of human alpha-globin genes. Proc. Natl Acad. Sci. USA. 2007;104:10950–10955. doi: 10.1073/pnas.0703856104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.MacArthur JA, et al. The rate of nonallelic homologous recombination in males is highly variable, correlated between monozygotic twins and independent of age. PLoS Genet. 2014;10:e1004195. doi: 10.1371/journal.pgen.1004195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Flores M, et al. Recurrent DNA inversion rearrangements in the human genome. Proc. Natl Acad. Sci. USA. 2007;104:6099–6106. doi: 10.1073/pnas.0701631104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kidd JM, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Waldman AS, Liskay RM. Dependence of intrachromosomal recombination in mammalian cells on uninterrupted homology. Mol. Cell. Biol. 1988;8:5350–5357. doi: 10.1128/mcb.8.12.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sun C, et al. Deletion of azoospermia factor a (AZFa) region of human Y chromosome caused by recombination between HERV15 proviruses. Hum. Mol. Genet. 2000;9:2291–2296. doi: 10.1093/oxfordjournals.hmg.a018920. [DOI] [PubMed] [Google Scholar]
- 66.Shuvarikov A, et al. Recurrent HERV-H-mediated 3q13.2-q13.31 deletions cause a syndrome of hypotonia and motor, language, and cognitive delays. Hum. Mutat. 2013;34:1415–1423. doi: 10.1002/humu.22384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Campbell IM, et al. Human endogenous retroviral elements promote genome instability via non-allelic homologous recombination. BMC Biol. 2014;12:74. doi: 10.1186/s12915-014-0074-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Steinmann K, et al. Type 2 NF1 deletions are highly unusual by virtue of the absence of nonallelic homologous recombination hotspots and an apparent preference for female mitotic recombination. Am. J. Hum. Genet. 2007;81:1201–1220. doi: 10.1086/522089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lam KW, Jeffreys AJ. Processes of copy-number change in human DNA: the dynamics of α-globin gene deletion. Proc. Natl Acad. Sci. USA. 2006;103:8921–8927. doi: 10.1073/pnas.0602690103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mezard C, Pompon D, Nicolas A. Recombination between similar but not identical DNA sequences during yeast transformation occurs within short stretches of identity. Cell. 1992;70:659–670. doi: 10.1016/0092-8674(92)90434-e. [DOI] [PubMed] [Google Scholar]
- 71.Callinan PA, Batzer MA. Retrotransposable elements and human disease. Genome Dyn. 2006;1:104–115. doi: 10.1159/000092503. [DOI] [PubMed] [Google Scholar]
- 72.Boone PM, et al. Alu-specific microhomology-mediated deletion of the final exon of SPAST in three unrelated subjects with hereditary spastic paraplegia. Genet. Med. 2011;13:582–592. doi: 10.1097/GIM.0b013e3182106775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hsiao MC, et al. Decoding NF1 intragenic copy-number variations. Am. J. Hum. Genet. 2015;97:238–249. doi: 10.1016/j.ajhg.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Deininger PL, Batzer MA. Alu repeats and human disease. Mol. Genet. Metab. 1999;67:183–193. doi: 10.1006/mgme.1999.2864. [DOI] [PubMed] [Google Scholar]
- 75.Stankiewicz P, Pursley AN, Cheung SW. Challenges in clinical interpretation of microduplications detected by array CGH analysis. Am. J. Med. Genet. A. 2010;152A:1089–1100. doi: 10.1002/ajmg.a.33216. [DOI] [PubMed] [Google Scholar]
- 76.Ottaviani D, LeCain M, Sheer D. The role of microhomology in genomic structural variation. Trends Genet. 2014;30:85–94. doi: 10.1016/j.tig.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 77.Lee JA, Carvalho CM, Lupski JR. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131:1235–1247. doi: 10.1016/j.cell.2007.11.037. Long-range template switching leading to complex genomic rearrangements and causing disease was reported. Fork stalling and template switching mechanisms were proposed for the first time to explain the observed unusual breakpoint complexity. [DOI] [PubMed] [Google Scholar]
- 78.Hastings PJ, Lupski JR, Rosenberg SM, Ira G. Mechanisms of change in gene copy number. Nat. Rev. Genet. 2009;10:551–564. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pannunzio NR, Li S, Watanabe G, Lieber MR. Non-homologous end joining often uses microhomology: implications for alternative end joining. DNA Repair (Amst.) 2014;17:74–80. doi: 10.1016/j.dnarep.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu P, Carvalho CM, Hastings PJ, Lupski JR. Mechanisms for recurrent and complex human genomic rearrangements. Curr. Opin. Genet. Dev. 2012;22:211–220. doi: 10.1016/j.gde.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Slack A, Thornton PC, Magner DB, Rosenberg SM, Hastings PJ. On the mechanism of gene amplification induced under stress in Escherichia coli. PLoS Genet. 2006;2:e48. doi: 10.1371/journal.pgen.0020048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen JM, Chuzhanova N, Stenson PD, Ferec C, Cooper DN. Complex gene rearrangements caused by serial replication slippage. Hum. Mutat. 2005;26:125–134. doi: 10.1002/humu.20202. [DOI] [PubMed] [Google Scholar]
- 83.Stephens PJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. Chromothripsis was defined and recognized as a one-off event in cancer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Malkova A, Ivanov EL, Haber JE. Double-strand break repair in the absence of RAD51 in yeast: a possible role for break-induced DNA replication. Proc. Natl Acad. Sci. USA. 1996;93:7131–7136. doi: 10.1073/pnas.93.14.7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Morrow DM, Connelly C, Hieter P. “Break copy” duplication: a model for chromosome fragment formation in Saccharomyces cerevisiae. Genetics. 1997;147:371–382. doi: 10.1093/genetics/147.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Malkova A, Ira G. Break-induced replication: functions and molecular mechanism. Curr. Opin. Genet. Dev. 2013;23:271–279. doi: 10.1016/j.gde.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lydeard JR, Jain S, Yamaguchi M, Haber JE. Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature. 2007;448:820–823. doi: 10.1038/nature06047. [DOI] [PubMed] [Google Scholar]
- 88.Lydeard JR, et al. Break-induced replication requires all essential DNA replication factors except those specific for pre-RC assembly. Genes Dev. 2010;24:1133–1144. doi: 10.1101/gad.1922610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Payen C, Koszul R, Dujon B, Fischer G. Segmental duplications arise from Pol32-dependent repair of broken forks through two alternative replication-based mechanisms. PLoS Genet. 2008;4:e1000175. doi: 10.1371/journal.pgen.1000175. This paper shows that segmental duplications are formed due to DNA synthesis rather than to ectopic homologous recombination and that the underlying mechanism is dependent on the nonessential Polδ subunit, Pol32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Arlt MF, Rajendran S, Birkeland SR, Wilson TE, Glover TW. De novo CNV formation in mouse embryonic stem cells occurs in the absence of Xrcc4-dependent nonhomologous end joining. PLoS Genet. 2012;8:e1002981. doi: 10.1371/journal.pgen.1002981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ira G, Haber JE. Characterization of RAD51-independent break-induced replication that acts preferentially with short homologous sequences. Mol. Cell. Biol. 2002;22:6384–6392. doi: 10.1128/MCB.22.18.6384-6392.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hastings PJ, Ira G, Lupski JR. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 2009;5:e1000327. doi: 10.1371/journal.pgen.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chen JM, Chuzhanova N, Stenson PD, Ferec C, Cooper DN. Meta-analysis of gross insertions causing human genetic disease: novel mutational mechanisms and the role of replication slippage. Hum. Mutat. 2005;25:207–221. doi: 10.1002/humu.20133. [DOI] [PubMed] [Google Scholar]
- 94.Smith CE, Llorente B, Symington LS. Template switching during break-induced replication. Nature. 2007;447:102–105. doi: 10.1038/nature05723. Another important aspect of the mutagenic nature of BIR was revealed in this work. BIR can undergo multiple rounds of template switching during DSB repair; dispersed repetitive sequences were observed to lead to non-reciprocal translocations. [DOI] [PubMed] [Google Scholar]
- 95.Tsaponina O, Haber JE. Frequent interchromosomal template switches during gene conversion in S. cerevisiae. Mol. Cell. 2014;55:615–625. doi: 10.1016/j.molcel.2014.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Iraqui I, et al. Recovery of arrested replication forks by homologous recombination is error-prone. PLoS Genet. 2012;8:e1002976. doi: 10.1371/journal.pgen.1002976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sun Z, et al. Replicative mechanisms of CNV formation preferentially occur as intrachromosomal events: evidence from Potocki-Lupski duplication syndrome. Hum. Mol. Genet. 2013;22:749–756. doi: 10.1093/hmg/dds482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang F, Carvalho CM, Lupski JR. Complex human chromosomal and genomic rearrangements. Trends Genet. 2009;25:298–307. doi: 10.1016/j.tig.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chanda B, et al. A novel mechanistic spectrum underlies glaucoma-associated chromosome 6p25 copy number variation. Hum. Mol. Genet. 2008;17:3446–3458. doi: 10.1093/hmg/ddn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chauvin A, et al. Elucidation of the complex structure and origin of the human trypsinogen locus triplication. Hum. Mol. Genet. 2009;18:3605–3614. doi: 10.1093/hmg/ddp308. [DOI] [PubMed] [Google Scholar]
- 101.Coccia M, et al. X-linked cataract and Nance-Horan syndrome are allelic disorders. Hum. Mol. Genet. 2009;18:2643–2655. doi: 10.1093/hmg/ddp206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Giorgio E, et al. Analysis of LMNB1 duplications in autosomal dominant leukodystrophy provides insights into duplication mechanisms and allele-specific expression. Hum. Mutat. 2013;34:1160–1171. doi: 10.1002/humu.22348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rugless MJ, et al. A large deletion in the human α-globin cluster caused by a replication error is associated with an unexpectedly mild phenotype. Hum. Mol. Genet. 2008;17:3084–3093. doi: 10.1093/hmg/ddn205. [DOI] [PubMed] [Google Scholar]
- 104.Bi W, et al. Increased LIS1 expression affects human and mouse brain development. Nat. Genet. 2009;41:168–177. doi: 10.1038/ng.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Liu P, et al. Copy number gain at Xp22.31 includes complex duplication rearrangements and recurrent triplications. Hum. Mol. Genet. 2011;20:1975–1988. doi: 10.1093/hmg/ddr078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Abyzov A, et al. Analysis of deletion breakpoints from 1,092 humans reveals details of mutation mechanisms. Nat. Commun. 2015;6:7256. doi: 10.1038/ncomms8256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Deem A, et al. Break-induced replication is highly inaccurate. PLoS Biol. 2011;9:e1000594. doi: 10.1371/journal.pbio.1000594. The mutagenic nature of BIR was shown in this yeast system; Polδ was shown to contribute in part to these errors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yousefzadeh MJ, et al. Mechanism of suppression of chromosomal instability by DNA polymerase POLQ. PLoS Genet. 2014;10:e1004654. doi: 10.1371/journal.pgen.1004654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Northam MR, Garg P, Baitin DM, Burgers PM, Shcherbakova PV. A novel function of DNA polymerase ζ regulated by PCNA. EMBO J. 2006;25:4316–4325. doi: 10.1038/sj.emboj.7601320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cherng N, et al. Expansions, contractions, and fragility of the spinocerebellar ataxia type 10 pentanucleotide repeat in yeast. Proc. Natl Acad. Sci. USA. 2011;108:2843–2848. doi: 10.1073/pnas.1009409108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sakofsky CJ, et al. Translesion polymerases drive microhomology-mediated break induced replication leading to complex chromosomal rearrangements. Mol. Cell. 2015;60:860–872. doi: 10.1016/j.molcel.2015.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nik-Zainal S, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. Kataegis, the phenomenon of localized hypermutational segments in cis, was defined in cancer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chung WH, Zhu Z, Papusha A, Malkova A, Ira G. Defective resection at DNA double-strand breaks leads to de novo telomere formation and enhances gene targeting. PLoS Genet. 2010;6:e1000948. doi: 10.1371/journal.pgen.1000948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wilson MA, et al. Pif1 helicase and Polδ promote recombination-coupled DNA synthesis via bubble migration. Nature. 2013;502:393–396. doi: 10.1038/nature12585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Saini N, et al. Migrating bubble during break-induced replication drives conservative DNA synthesis. Nature. 2013;502:389–392. doi: 10.1038/nature12584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Donnianni RA, Symington LS. Break-induced replication occurs by conservative DNA synthesis. Proc. Natl Acad. Sci. USA. 2013;110:13475–13480. doi: 10.1073/pnas.1309800110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mayle R, et al. Mus81 and converging forks limit the mutagenicity of replication fork breakage. Science. 2015;349:742–747. doi: 10.1126/science.aaa8391. Endonuclease Mus81 was shown to limit the mutagenic synthesis associated with BIR during DNA repair. It also inhibits template switches between interspersed homeologous repeats, including human Alu. This work importantly adds to our understanding of how genomic architecture contributes to increased instability and which factors have evolved to avoid this. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bacolla A, et al. Breakpoints of gross deletions coincide with non-B DNA conformations. Proc. Natl Acad. Sci. USA. 2004;101:14162–14167. doi: 10.1073/pnas.0405974101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chen JM, Chuzhanova N, Stenson PD, Ferec C, Cooper DN. Intrachromosomal serial replication slippage in trans gives rise to diverse genomic rearrangements involving inversions. Hum. Mutat. 2005;26:362–373. doi: 10.1002/humu.20230. [DOI] [PubMed] [Google Scholar]
- 121.Bose P, Hermetz KE, Conneely KN, Rudd MK. Tandem repeats and G-rich sequences are enriched at human CNV breakpoints. PLoS ONE. 2014;9:e101607. doi: 10.1371/journal.pone.0101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Walsh E, Wang X, Lee MY, Eckert KA. Mechanism of replicative DNA polymerase delta pausing and a potential role for DNA polymerase kappa in common fragile site replication. J. Mol. Biol. 2013;425:232–243. doi: 10.1016/j.jmb.2012.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Northam MR, et al. DNA polymerases ζ and Rev1 mediate error-prone bypass of non-B DNA structures. Nucleic Acids Res. 2014;42:290–306. doi: 10.1093/nar/gkt830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Koren A, et al. Differential relationship of DNA replication timing to different forms of human mutation and variation. Am. J. Hum. Genet. 2012;91:1033–1040. doi: 10.1016/j.ajhg.2012.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Chen L, et al. CNV instability associated with DNA replication dynamics: evidence for replicative mechanisms in CNV mutagenesis. Hum. Mol. Genet. 2015;24:1574–1583. doi: 10.1093/hmg/ddu572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Anand RP, et al. Chromosome rearrangements via template switching between diverged repeated sequences. Genes Dev. 2014;28:2394–2406. doi: 10.1101/gad.250258.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Korbel JO, et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kidd JM, et al. A human genome structural variation sequencing resource reveals insights into mutational mechanisms. Cell. 2010;143:837–847. doi: 10.1016/j.cell.2010.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lam HY, et al. Nucleotide-resolution analysis of structural variants using BreakSeq and a breakpoint library. Nat. Biotechnol. 2010;28:47–55. doi: 10.1038/nbt.1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Shimojima K, et al. Pelizaeus-Merzbacher disease caused by a duplication-inverted triplication-duplication in chromosomal segments including the PLP1 region. Eur. J. Med. Genet. 2012;55:400–403. doi: 10.1016/j.ejmg.2012.02.013. [DOI] [PubMed] [Google Scholar]
- 131.del Gaudio D, et al. Increased MECP2 gene copy number as the result of genomic duplication in neurodevelopmentally delayed males. Genet. Med. 2006;8:784–792. doi: 10.1097/01.gim.0000250502.28516.3c. [DOI] [PubMed] [Google Scholar]
- 132.Wolf NI, et al. Three or more copies of the proteolipid protein gene PLP1 cause severe elizaeus–Merzbacher disease. Brain. 2005;128:743–751. doi: 10.1093/brain/awh409. [DOI] [PubMed] [Google Scholar]
- 133.McClintock B. The behavior in successive nuclear divisions of a chromosome broken at meiosis. Proc. Natl Acad. Sci. USA. 1939;25:405–416. doi: 10.1073/pnas.25.8.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.McClintock B. The stability of broken ends of chromosomes in Zea Mays. Genetics. 1941;26:234–282. doi: 10.1093/genetics/26.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zuffardi O, Bonaglia M, Ciccone R, Giorda R. Inverted duplications deletions: underdiagnosed rearrangements?? Clin. Genet. 2009;75:505–513. doi: 10.1111/j.1399-0004.2009.01187.x. [DOI] [PubMed] [Google Scholar]
- 136.Hannes F, et al. Telomere healing following DNA polymerase arrest-induced breakages is likely the main mechanism generating chromosome 4p terminal deletions. Hum. Mutat. 2010;31:1343–1351. doi: 10.1002/humu.21368. [DOI] [PubMed] [Google Scholar]
- 137.Ghezraoui H, et al. Chromosomal translocations in human cells are generated by canonical nonhomologous end-joining. Mol. Cell. 2014;55:829–842. doi: 10.1016/j.molcel.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ballif BC, Yu W, Shaw CA, Kashork CD, Shaffer LG. Monosomy 1p36 breakpoint junctions suggest pre-meiotic breakage-fusion-bridge cycles are involved in generating terminal deletions. Hum. Mol. Genet. 2003;12:2153–2165. doi: 10.1093/hmg/ddg231. [DOI] [PubMed] [Google Scholar]
- 139.Luo Y, et al. Diverse mutational mechanisms cause pathogenic subtelomeric rearrangements. Hum. Mol. Genet. 2011;20:3769–3778. doi: 10.1093/hmg/ddr293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yatsenko SA, et al. Human subtelomeric copy number gains suggest a DNA replication mechanism for formation: beyond breakage-fusion-bridge for telomere stabilization. Hum. Genet. 2012;131:1895–1910. doi: 10.1007/s00439-012-1216-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lowden MR, Flibotte S, Moerman DG, Ahmed S. DNA synthesis generates terminal duplications that seal end-to-end chromosome fusions. Science. 2011;332:468–471. doi: 10.1126/science.1199022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hermetz KE, et al. Large inverted duplications in the human genome form via a fold-back mechanism. PLoS Genet. 2014;10:e1004139. doi: 10.1371/journal.pgen.1004139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Conrad DF, et al. Mutation spectrum revealed by breakpoint sequencing of human germline CNVs. Nat. Genet. 2010;42:385–391. doi: 10.1038/ng.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.King DA, et al. Mosaic structural variation in children with developmental disorders. Hum. Mol. Genet. 2015;24:2733–2745. doi: 10.1093/hmg/ddv033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Poduri A, Evrony GD, Cai X, Walsh CA. Somatic mutation, genomic variation, and neurological disease. Science. 2013;341:1237758. doi: 10.1126/science.1237758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Merla G, et al. Submicroscopic deletion in patients with Williams-Beuren syndrome influences expression levels of the nonhemizygous flanking genes. Am. J. Hum. Genet. 2006;79:332–341. doi: 10.1086/506371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Chaignat E, et al. Copy number variation modifies expression time courses. Genome Res. 2011;21:106–113. doi: 10.1101/gr.112748.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Stranger BE, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Shapiro JA. Molecular model for the transposition and replication of bacteriophage Mu and other transposable elements. Proc. Natl Acad. Sci. USA. 1979;76:1933–1937. doi: 10.1073/pnas.76.4.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Koolen DA, et al. A new chromosome 17q21.31 microdeletion syndrome associated with a common inversion polymorphism. Nat. Genet. 2006;38:999–1001. doi: 10.1038/ng1853. [DOI] [PubMed] [Google Scholar]
- 151.Ballif BC, et al. Expanding the clinical phenotype of the 3q29 microdeletion syndrome and characterization of the reciprocal microduplication. Mol. Cytogenet. 2008;1:8. doi: 10.1186/1755-8166-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Mochizuki J, et al. Alu-related 5q35 microdeletions in Sotos syndrome. Clin. Genet. 2008;74:384–391. doi: 10.1111/j.1399-0004.2008.01032.x. [DOI] [PubMed] [Google Scholar]
- 153.Rosenfeld JA, et al. Further evidence of contrasting phenotypes caused by reciprocal deletions and duplications: duplication of NSD1 causes growth retardation and microcephaly. Mol. Syndromol. 2013;3:247–254. doi: 10.1159/000345578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Antonell A, et al. Partial 7q11.23 deletions further implicate GTF2I and GTF2IRD1 as the main genes responsible for the Williams–Beuren syndrome neurocognitive profile. J. Med. Genet. 2010;47:312–320. doi: 10.1136/jmg.2009.071712. [DOI] [PubMed] [Google Scholar]
- 155.Berg JS, et al. Speech delay and autism spectrum behaviors are frequently associated with duplication of the 7q11.23 Williams-Beuren syndrome region. Genet. Med. 2007;9:427–441. doi: 10.1097/gim.0b013e3180986192. [DOI] [PubMed] [Google Scholar]
- 156.Beunders G, et al. A triplication of the Williams–Beuren syndrome region in a patient with mental retardation, a severe expressive language delay, behavioural problems and dysmorphisms. J. Med. Genet. 2010;47:271–275. doi: 10.1136/jmg.2009.070490. [DOI] [PubMed] [Google Scholar]
- 157.Cheroki C, et al. Genomic imbalances associated with mullerian aplasia. J. Med. Genet. 2008;45:228–232. doi: 10.1136/jmg.2007.051839. [DOI] [PubMed] [Google Scholar]
- 158.Nogueira SI, et al. Atypical 22q11.2 deletion in a patient with DGS/VCFS spectrum. Eur. J. Med. Genet. 2008;51:226–230. doi: 10.1016/j.ejmg.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Yamagishi H, Garg V, Matsuoka R, Thomas T, Srivastava D. A molecular pathway revealing a genetic basis for human cardiac and craniofacial defects. Science. 1999;283:1158–1161. doi: 10.1126/science.283.5405.1158. [DOI] [PubMed] [Google Scholar]
- 160.Potocki L, et al. Characterization of Potocki-Lupski syndrome (dup(17)(p11.2p11.2)) and delineation of a dosage-sensitive critical interval that can convey an autism phenotype. Am. J. Hum. Genet. 2007;80:633–649. doi: 10.1086/512864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zhang F, et al. Mechanisms for nonrecurrent genomic rearrangements associated with CMT1A or HNPP: rare CNVs as a cause for missing heritability. Am. J. Hum. Genet. 2010;86:892–903. doi: 10.1016/j.ajhg.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Venturin M, et al. Evidence for non-homologous end joining and non-allelic homologous recombination in atypical NF1 microdeletions. Hum. Genet. 2004;115:69–80. doi: 10.1007/s00439-004-1101-2. [DOI] [PubMed] [Google Scholar]
- 163.Small K, Iber J, Warren ST. Emerin deletion reveals a common X-chromosome inversion mediated by inverted repeats. Nat. Genet. 1997;16:96–99. doi: 10.1038/ng0597-96. [DOI] [PubMed] [Google Scholar]
- 164.Fusco F, et al. Genomic architecture at the Incontinentia Pigmenti locus favours de novo pathological alleles through different mechanisms. Hum. Mol. Genet. 2012;21:1260–1271. doi: 10.1093/hmg/ddr556. [DOI] [PubMed] [Google Scholar]