Abstract
Dietary proteins influence colorectal cancer (CRC) risk, depending on their quantity and quality. Here, using pyrosequencing, we compared the fecal microbiota composition in Balb/c mice fed either a normal protein/carbohydrate diet (ND, 20% casein and 68% carbohydrate) or a high-protein/low-carbohydrate diet (HPLCD, 30% casein and 57% carbohydrate). The results showed that HPLCD feeding for 2 weeks reduced the diversity and altered the composition of the microbiota compared with the ND mice, which included a decrease in the proportion of the family Lachnospiraceae and Ruminococcaceae and increases in the proportions of the genus Bacteroides and Parabacteroides, especially the species EF09600_s and EF604598_s. Similar changes were reported in patients with inflammatory bowel disease, and in mouse models of CRC and colitis, respectively. This suggests that HPLCD may lead to a deleterious luminal environment and may have adverse effects on the intestinal health of individuals consuming such a diet.
Keywords: gut microbiota, high protein diet, low carbohydrate diet, pyrosequencing, mouse
INTRODUCTION
The gastrointestinal microbiota has become a major focus of interest to investigate the etiology of many diseases including inflammatory bowel disease (IBD), obesity, and colorectal cancer (CRC). Accumulating evidence shows that the gut microbiota is involved in gastrointestinal immune stimulation, essential nutrient production, control of pathogenic microorganism, modulation of gastrointestinal epithelial cell proliferation and differentiation, and management of bioactive foods and chemical components (1). Diets, especially of the 3 major macronutrients (carbohydrates, proteins, and fats), can significantly affect the composition and metabolic activities of the microbiota, which adapts according to the new colonic luminal environment and substrate availability. In contrast to carbohydrate fermentation, microbial metabolism of dietary proteins can result in the formation of deleterious metabolites such as phenols, indoles, amines, sulfides, ammonia, and monocarboxylic acids, which are cytotoxic and mutagenic to colonic epithelial cells (2). Whereas the influence of carbohydrates, fibers, and fat-rich diets on the repartition of species and functional groups of the microbiota have been well documented (3–5), few studies have addressed the effect of a high-protein/low-carbohydrate diet (HPLCD) on the composition and activity of the gut microbiota (6). Recently, Liu et al. (7) reported changes of the colonic microbiota and luminal environment by a HPLCD in the rat model. In their study, however, they used a HPLCD composed of 53% protein, which far exceeds the normal amount of daily protein intake. The recommended dietary allowance for protein is 7~20% and 10~35% of the total food energy in Korea and in the United States, respectively. According to the National Health and Nutrition Examination Survey, the average protein intake of Koreans is 14.7% of the total energy (Korea Health Statistics 2013). Considering that our diet style is more westernized, it is therefore needed to investigate the influence of a 30% protein diet on the mouse gut microbiota composition by pyrosequencing.
MATERIALS AND METHODS
Animals and diets
Six week old female Balb/c mice were obtained from KOATECH Bio Inc. (Busan, Korea). Animals were acclimated for 1 week and separated randomly into two groups: normal diet group (ND, 20% protein/68% carbohydrate, n=2), and high-protein/low-carbohydrate diet group (HPLCD, 30% protein/57% carbohydrate, n=2). The experimental diet composition based on AIN-76A is shown in Table 1. The diet was fed to the animals ad libitum for 2 weeks. Animals were kept in controlled conditions of temperature (23±2°C), humidity (50±10%), and light (12-h light/dark cycle). All animal experimentation was approved by the Animal Care and Use Committee at the Catholic University of Daegu (IACUC-2014-042).
Table 1.
Composition of experimental diets
| Ingredients | ND | HPLCD |
|---|---|---|
| Corn starch | 150 | 150 |
| Casein | 200 | 300 |
| D,L-methionine | 3 | 3 |
| Sucrose | 500 | 400 |
| Cellulose powder | 50 | 50 |
| Corn oil | 50 | 50 |
| Mineral mixture | 35 | 35 |
| Vitamin mixture | 10 | 10 |
| Choline bitartrate | 2 | 2 |
| t-BHQ | 0.01 | 0.01 |
| Total (g) | 1,000 | 1,000 |
| Protein [kcal (% energy)] | 800 (20.8%) | 1,200 (31.2%) |
| Carbohydrate [kcal (% energy)] | 2,600 (67.5%) | 2,200 (57.1%) |
| Fat [kcal (% energy)] | 450 (11.7%) | 450 (11.7%) |
| Total energy (kcal/kg) | 3,850 | 3,850 |
ND, normal diet; HPLCD, high-protein/low-carbohydrate diet.
DNA isolation from fecal samples of mice
For the pyrosequencing analysis, feces were collected for 3 consecutive days before the animals were euthanized with CO2. The fecal samples were stored at −70°C until analysis. Genomic DNA was extracted from pooled fecal samples using the FastDNA™ SPIN kit for Feces (MP Biomedical, Aurora, OH, USA) according to the manufacture’s protocol, except that a Mini-BeadBeater-8™ Cell Disrupter (Biospec Products, Bartlesville, OK, USA) was used for the cell disruption.
Pyrosequencing and data analysis
For pyrosequencing, amplification of genomic DNA was performed using barcoded primers that targeted the V1 to V3 region of the bacterial 16S rRNA gene. The amplification, sequencing, and basic analysis were performed according to the methods described by Chun et al. (8) and were completed by Chunlab Inc. (Seoul, Korea), using a 454 GS FLX Titanium Sequencing Systems (Roche, Branford, CT, USA). Sequence reads were identified using the EzTaxon-e database (http://eztaxon-e.ezbiocloud.net) (9) on the basis of 16S rRNA sequence data. We analyzed the number of sequences, observed the diversity richness [operational taxonomic units (OTUs)], and estimated OTU richness (abundance-based coverage estimator and Chao1 indices). Bacterial community abundance and composition were generated using the CLcommunity software (Chunlab Inc.).
Statistical analysis
The data are presented as means±standard error (SE). All statistical analyses were performed by the SPSS Program ver. 19.0 (IBM Co., Armonk, NY, USA). The data were analyzed by Student’s t-test and differences between experimental groups were evaluated at the P< 0.05.
RESULTS AND DISCUSSION
Effect of HPLCD on the diversity of mouse gut microbiota
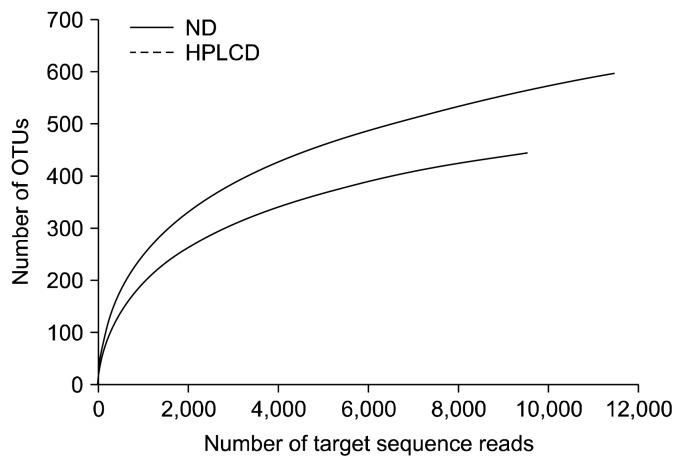
As demonstrated by the rarefaction curves shown in Fig. 1, the gut microbiotas of ND mice were more diverse than those of HPLCD mice. In a rat model, HPLCD consumption was found to decrease the microbiome diversity in the colon (7). Decreased diversity was also seen in mice bearing tumors developed from dextran sodium sulfate administration (10), in infants with atopic eczema (11), in patients with Crohn’s disease (12), and in colorectal cancerous tissue samples from diagnosed colorectal carcinoma (CRC) patients (13). This suggests that a low diversity of the gut microbiota is associated with some health deterioration and would be an indicator for unhealthy conditions. Collectively, a HPLCD may also have adverse effects on intestinal health by reducing the diversity of the gut microbiota.
Fig. 1.
CD-HIT rarefaction curves. Rarefaction analysis of V1–V3 region pyrosequencing tags of the 16S rRNA gene in fecal microbiota from mice. ND, normal diet; HPLCD, high protein low carbohydrate diet.
Changes of gut microbiota composition by HPLCD
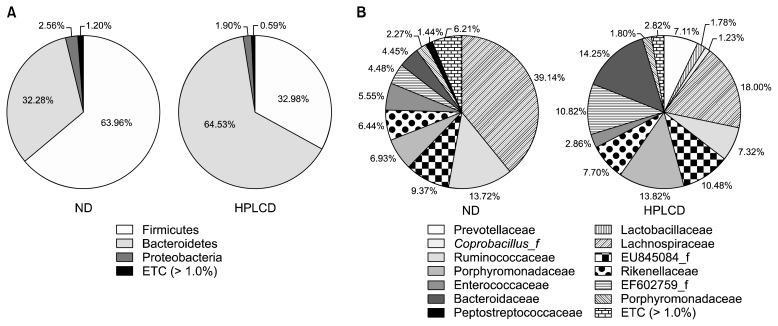
Taxonomy-based analysis showed that Firmicutes and Bacteroidetes were the dominant taxa at the phylum level (ND, 96.24%; HPLCD, 97.51%), but the composition of these two phyla was dramatically different between the 2 mouse groups (Fig. 2A). The ND group showed a high Firmicutes level (63.96%) and a relatively low Bacteroidetes level (32.28%), whereas the HPLCD group had a low Firmicutes level (32.98%) and a high Bacteroidetes level (64.53%). Ley et al. (14) have demonstrated low proportions of Bacterioidetes/Firmicutes in ob/ob mice and in obese individuals (15). This suggests that a HPLCD may lead to weight loss, caused by the high proportions of Bacterioidetes/Firmicutes. In fact, HPLCDs are widely used for weight loss (16). In this study, we did not observe weight differences between the ND and HPLCD groups (Table 2). Daily food intake was also similar between the 2 groups. Although we cannot exclude the possibility that long term HPLCD feeding may lead to weight loss, 2 weeks of such a diet may be too short to cause body weight changes.
Fig. 2.
The composition of gut microbiota in phylum and family levels. Phylum (A) and family (B) levels. Genomic DNA was extracted from the fecal samples taken from mice maintained for 2 weeks on diets described in Table 1. Samples were analyzed for the bacterial composition by pyrosequecing of the bacterial 16S rRNA V1–V3 region. ND, normal diet; HPLCD, high-protein/low-carbohydrate diet.
Table 2.
Daily food intake and body weight
| Daily food intake (g/d) | Final body weight (g) | |
|---|---|---|
| ND | 3.3±0.14NS | 18.4±0.3NS |
| HPLCD | 3.2±0.12 | 18.1±0.2 |
Food intake (measured daily) and body weight (measured once a week) of mice in each group [ND (n=2) and HPLCD (n=2)]. The value is expressed as mean±SE.
ND, normal diet; HPLCD, high-protein/low-carbohydrate diet.
NS: not significant.
With regard to the microbiota composition at the family level, Lachnospiraceae (phylum Firmicutes, class Clostridia) was the most abundant taxon, but the HPLCD group had less than half of that of the ND group (ND, 39.14%; HPLCD, 18.04%) (Fig. 2B). The members of Lachnospiraceae family are associated with obesity and protection from CRC in humans, and these roles are linked to butyrate production (17). This family also has been found to be of significantly low proportion in the colon of human IBD patients (18). HPLCD mice also showed lower levels of Ruminococcaceae proportion compared with ND mice (ND, 13.72%; HPLCD, 7.32%). Prevotellaceae, Lactobacillaceae, and Coprobacillus were detected in the feces of HPLCD mice (7.11%, 1.78%, and 1.23%, respectively) but not in the feces of ND mice. The proportions of Bacteroidaceae and Porphyromonadaceae were higher in the HPLCD mouse fecal samples. CRC animal models and cancerous tissues have shown higher levels of the families Bacteroidaceae, Streptococcaceae, Fusobacteriaceae, Peptostreptococcaceae, Veillonellaceae, and Pateurellaceae and significantly lower levels of Lanchnospriaecae, Ruminococcaceae, and Lactobacillaceae family Prevotellaceae compared with healthy animals and the intestinal lumen (10,19–21). It is hard to conclude that the shift of microbiota caused by HPLCD would be harmful, but some taxonomic changes such as decreasing Lachnospriaecae and Ruminococcaceae, and increasing Bacteroidaceae indicate the possibility of aggravating the gut environment.
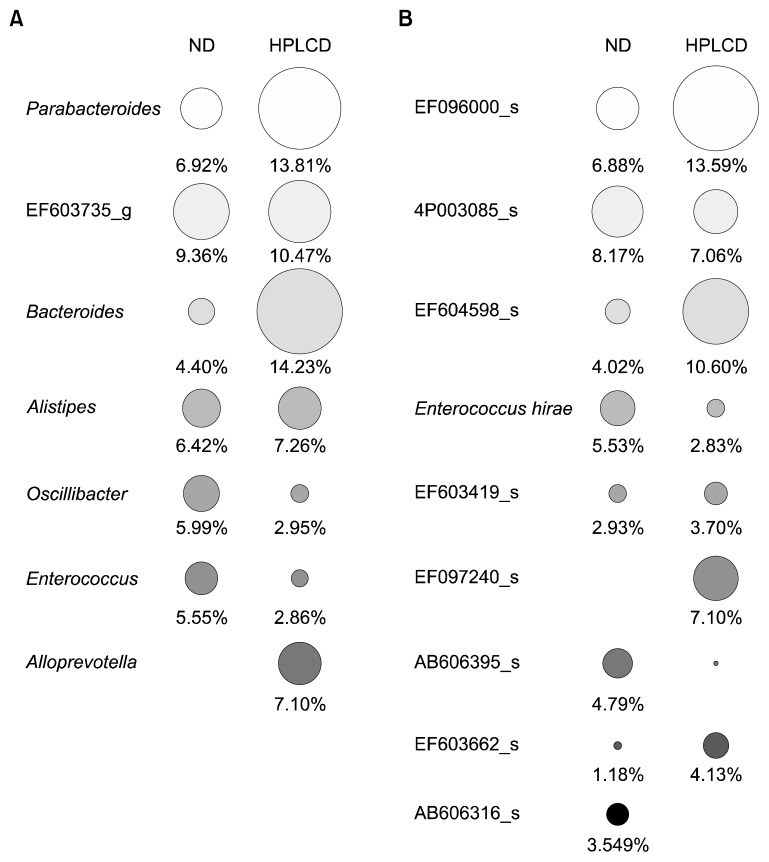
At the genus level, the proportions of Bacteroides and Parabacteroides were significantly higher in the feces of HPLCD mice (14.23% and 13.81%, respectively) than in the samples from ND mice (4.40% and 6.92%), whereas Oscillibacter and Enterococcus levels were higher in HPLCD mice (2.95% and 2.86%, respectively) than in ND mice (5.99% and 5.55%) (Fig. 3A). Alloprevotella comprised a 7.10% portion of the fecal microbiome in HPLCD mice, but it was not detected in the feces of ND mice. Zackular et al. (10) showed that manipulation of the microbiome using antibiotics could dramatically reduce tumor formation in a colitis-associated CRC animal model. This clearly suggests the importance of bacterium-driven factors in tumorigenesis. In this study, mice showed a significant decrease in microbial diversity in the gut microbiome during tumor development, and Bacteroides and Parabacteroides in particular were associated with increased tumorigenesis. In the CRC rat model, Bacteroides and Parabacteroides also showed higher levels compared with healthy rats (19). This shows the possibility of harmful changes in the gut environment of HPLCD mice.
Fig. 3.
Circle type heat map of relative percent abundance of major taxa in gut microbiota of mice. Genus (A) and species (B) levels. Circle size indicates relative composition of each taxon [full size of circles of (A) and (B) indicates 30.27% and 28.39%, respectively]. Minor taxa that individual composition was less than 5% and 3% were excluded in genus and species level, respectively. ND, normal diet; HPLCD, high-protein/low-carbohydrate diet.
At the species level, EF09600_s (Pararbacteroides genus), EF604598_s (Bacteroides genus), and EF097240_s (Alloprevotella genus) were increased in HPLCD mice to 13.59%, 10.60%, and 7.10%, respectively (Fig. 3B). EF 09600_s and EF604598_s were detected to be in high proportions in the stools of 2,3,4-trinitrobenzene sulfonic acid (TNBS)-treated colitis mice, whereas they were not detected in normal mice and doenjang-treated mice administered TNBS (22). This suggests that these 2 bacteria are associated with colon colitis, and a HPLCD might be associated with this condition.
In conclusion, the gut microbiomes were significantly changed in HPLCD-fed mice in this study. In particular, a decrease in the microbial diversity, a decrease in the proportion of the family Lachnospiraceae and Ruminoococcaceae, the increased level of the family Bacteroidaceae, the genus Bacteroides, and parabacteroides, and the increased levels of the species EF09600_s and EF604598 _s were similar to changes found in IBD patients, a CRC mouse model, and a colitis mouse model, respectively. Even though our observations have some limitations including the short-term diet period (2 weeks) and a low number of samples (n=2/group), our research showed that a HPLCD could decrease the diversity of the mouse gut microbiota and alter its composition. Further studies are needed to examine the effects of a HPLCD during a long-term diet, and the effects of a HPLCD in CRC animal models.
ACKNOWLEDGEMENTS
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIP) (No. NRF-2010-0012980 and 2015 R1C1A2A01054514).
Footnotes
AUTHOR DISCLOSURE STATEMENT
The authors declare no conflict of interest.
REFERENCES
- 1.Akin H, Tözün N. Diet, microbiota, and colorectal cancer. J Clin Gastroenterol. 2014;48:S67–S69. doi: 10.1097/MCG.0000000000000252. [DOI] [PubMed] [Google Scholar]
- 2.Kim E, Coelho D, Blachier F. Review of the association between meat consumption and risk of colorectal cancer. Nutr Res. 2013;33:983–994. doi: 10.1016/j.nutres.2013.07.018. [DOI] [PubMed] [Google Scholar]
- 3.Hooda S, Boler Vester BM, Serao Rossoni MC, Brulc JM, Staeger MA, Boileau TW, Dowd SE, Fahey GC, Jr, Swanson KS. 454 pyrosequencing reveals a shift in fecal microbiota of healthy adult men consuming polydextrose or soluble corn fiber. J Nutr. 2012;142:1259–1265. doi: 10.3945/jn.112.158766. [DOI] [PubMed] [Google Scholar]
- 4.Martinez-Medina M, Denizot J, Dreux N, Robin F, Billard E, Bonnet R, Darfeuille-Michaud A, Barnich N. Western diet induces dysbiosis with increased E. coli in CEABAC10 mice, alters host barrier function favouring AIEC colonisation. Gut. 2014;63:116–124. doi: 10.1136/gutjnl-2012-304119. [DOI] [PubMed] [Google Scholar]
- 5.Walker AW, Ince J, Duncan SH, Webster LM, Holtrop G, Ze X, Brown D, Stares MD, Scott P, Bergerat A, Louis P, McIntosh F, Johnstone AM, Lobley GE, Parkhill J, Flint HJ. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J. 2011;5:220–230. doi: 10.1038/ismej.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blachier F, Mariotti F, Huneau JF, Tomé D. Effects of amino acid-derived luminal metabolites on the colonic epithelium and physiopathological consequences. Amino Acids. 2007;33:547–562. doi: 10.1007/s00726-006-0477-9. [DOI] [PubMed] [Google Scholar]
- 7.Liu X, Blouin JM, Santacruz A, Lan A, Andriamihaja M, Wilkanowicz S, Benetti PH, Tomé D, Sanz Y, Blachier F, Davila AM. High-protein diet modifies colonic microbiota and luminal environment but not colonocyte metabolism in the rat model: the increased luminal bulk connection. Am J Physiol Gastrointest Liver Physiol. 2014;307:G459–G470. doi: 10.1152/ajpgi.00400.2013. [DOI] [PubMed] [Google Scholar]
- 8.Chun J, Kim KY, Lee JH, Choi Y. The analysis of oral microbial communities of wild-type and toll-like receptor 2-deficient mice using a 454 GS FLX Titanium pyrosequencer. BMC Microbiol. 2010;10:101. doi: 10.1186/1471-2180-10-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J. Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol. 2012;62:716–721. doi: 10.1099/ijs.0.038075-0. [DOI] [PubMed] [Google Scholar]
- 10.Zackular JP, Bzxter NT, Iverson KD, Sadler WD, Petrosino JF, Chen GY, Schloss PD. The gut microbiome modulates colon tumorigenesis. mBio. 2013;4:e00692–13. doi: 10.1128/mBio.00692-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abrahamsson TR, Jakobsson HE, Andersson AF, Björkstén B, Engstrand L, Jenmalm MC. Low diversity of the gut microbiota in infants with atopic eczema. J Allergy Clin Immunol. 2012;129:434–440. doi: 10.1016/j.jaci.2011.10.025. [DOI] [PubMed] [Google Scholar]
- 12.Dicksved J, Halfvarson J, Rosenquist M, Järnerot G, Tysk C, Apajalahti J, Engstrand L, Jansson JK. Molecular analysis of the gut microbiota of identical twins with Crohn’s disease. ISME J. 2008;2:716–727. doi: 10.1038/ismej.2008.37. [DOI] [PubMed] [Google Scholar]
- 13.Gao Z, Guo B, Gao R, Zhu Q, Qin H. Microbiota disbiosis is associated with colorectal cancer. Front Microbiol. 2015;6:20. doi: 10.3389/fmicb.2015.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 16.Johnstone AM, Horgan GW, Murison SD, Bremner DM, Lobley GE. Effects of a high-protein ketogenic diet on hunger, appetite, and weight loss in obese men feeding ad libitum. Am J Clin Nutr. 2008;87:44–55. doi: 10.1093/ajcn/87.1.44. [DOI] [PubMed] [Google Scholar]
- 17.Meehan CJ, Beiko RG. A phylogenomic view of ecological specialization in the Lachnospiraceae, a family of digestive tract-associated bacteria. Genome Biol Evol. 2014;6:703–713. doi: 10.1093/gbe/evu050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frank DN, St. Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu Q, Jin Z, Wu W, Gao R, Guo B, Gao Z, Yang Y, Qin H. Analysis of the intestinal lumen microbiota in an animal model of colorectal cancer. PLoS One. 2014;9:e90849. doi: 10.1371/journal.pone.0090849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen W, Liu F, Ling Z, Tong X, Xiang C. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS One. 2012;7:e39743. doi: 10.1371/journal.pone.0039743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu Q, Gao R, Wu W, Qin H. The role of gut microbiota in the pathogenesis of colorectal cancer. Tumour Biol. 2013;34:1285–1300. doi: 10.1007/s13277-013-0684-4. [DOI] [PubMed] [Google Scholar]
- 22.Kim KA, Jang SE, Jeong JJ, Yu DH, Han MJ, Kim DH. Doenjang, a Korean soybean paste, ameliorates TNBS-induced colitis in mice by suppressing gut microbial lipopolysaccharide production and NF-κB activation. J Funct Foods. 2014;11:417–427. doi: 10.1016/j.jff.2014.09.021. [DOI] [Google Scholar]