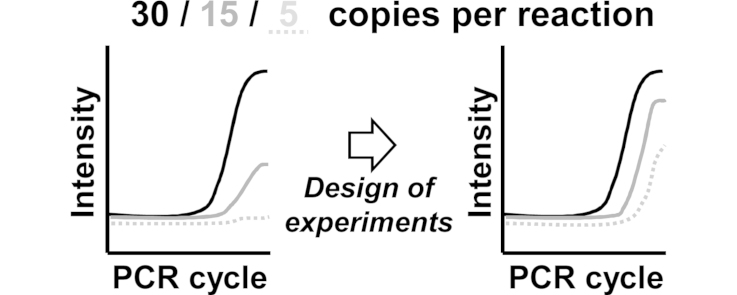
Graphical abstract
Keywords: Design of experiments, Mediator probe PCR, Universal reporter, Real-time PCR, PCR optimization
Highlights
-
•
Design of experiments (DOE) is an appropriate approach for probe sequence optimization in real-time PCR.
-
•
Nine MP designs led to maximum information for the 3 input factors.
-
•
Using DOE the maximum number of individual reactions performed was 180.
-
•
A one-factor-at-a-time approach in contrast would require 320 individual reactions.
Abstract
Primer and probe sequence designs are among the most critical input factors in real-time polymerase chain reaction (PCR) assay optimization. In this study, we present the use of statistical design of experiments (DOE) approach as a general guideline for probe optimization and more specifically focus on design optimization of label-free hydrolysis probes that are designated as mediator probes (MPs), which are used in reverse transcription MP PCR (RT-MP PCR). The effect of three input factors on assay performance was investigated: distance between primer and mediator probe cleavage site; dimer stability of MP and target sequence (influenza B virus); and dimer stability of the mediator and universal reporter (UR). The results indicated that the latter dimer stability had the greatest influence on assay performance, with RT-MP PCR efficiency increased by up to 10% with changes to this input factor. With an optimal design configuration, a detection limit of 3–14 target copies/10 μl reaction could be achieved. This improved detection limit was confirmed for another UR design and for a second target sequence, human metapneumovirus, with 7–11 copies/10 μl reaction detected in an optimum case. The DOE approach for improving oligonucleotide designs for real-time PCR not only produces excellent results but may also reduce the number of experiments that need to be performed, thus reducing costs and experimental times.
These definitions of the following terms have been taken from the specified references
Analytical performance characteristics (of real-time PCR): express the potential power of the analytical method (the real-time PCR assay) [1], [2]. Here we employ:
-
○
Selectivity (also expressed as specificity): the ability of a measurement procedure to solely assess the measurand [3].
-
○
Accuracy: closeness of agreement between a test result and the accepted reference value [4].
-
○
Precision: degree of agreement between independent test results obtained under stipulated conditions [4].
-
○
Limit of detection: lowest amount of analyte in a sample that can be detected but not quantified as an exact value [5].
-
○
Linear dynamic range: quantity of the measurand to be determined by a linear calibration curve [6].
-
○
Real-time PCR efficiency: measure of the power of product formation in one particular PCR cycle [6].
Statistical design of experiments (DOE): aims to optimize a method with minimized costs and time by lowering the extent of the experiments to a statistically relevant level [7]. The following are important terms from the DOE approach used within this article:
-
○
Target value: single or a combination of values, such as different performance characteristics, that can be measured and that are representative of the performance of the method to be optimized [7].
-
○
Input factor: parameter that can systematically vary in magnitude (=input factor level) during a DOE study to determine its effect on one or more target values [7].
-
○
Screening: experimental design that enables the evaluation of whether one input factor, a set of input factors, or a certain input factor combination effectively alters one or several target values [7].
-
○
Full factorial design: a design of experiment in which for each input factor and input factor combination, all input factor levels are present [7].
Gibbs free energy (ΔG): expresses the spontaneity of a product formation out of reactants at a given temperature and reactant concentrations [8]. Here negative values indicate spontaneous formation of a dimer of two different oligonucleotides with a partial reverse complementary. The lower the ΔG the higher the stability of the dimer.
1. Introduction
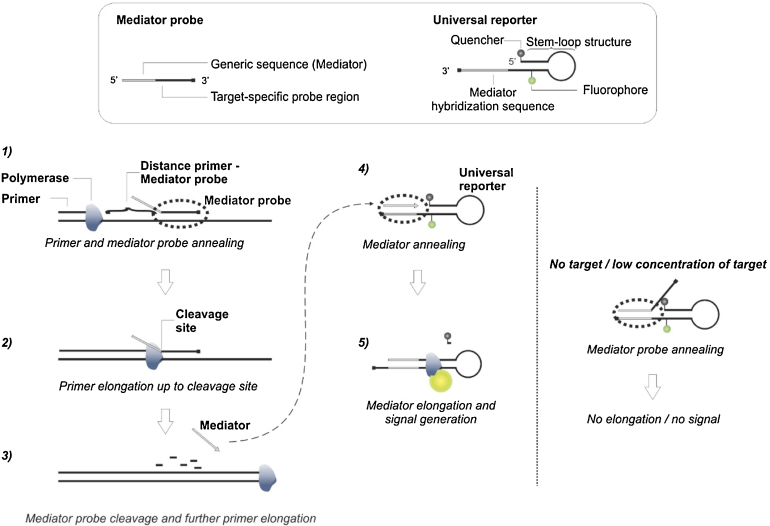
Real-time polymerase chain reaction (PCR) has become the gold standard for detecting nucleic acids because of its robustness, low analytical detection limits, equipment access, and ease of performance due to the numerous publications describing experimental setup guidelines [9], [10], [11], [12]. The most commonly used probe-based real-time PCR methods are hydrolysis probes, molecular beacons, and scorpion primers [13], all of which are labeled with both a fluorescence reporter dye and fluorescence quencher. In contrast, the more novel detection method of mediator probe PCR (MP PCR, Fig. 2) [14], [15], [16] incorporates the use of unlabeled, sequence-specific MPs in conjunction with a fluorogenic universal reporter (UR) oligonucleotide. Signal generation is initiated by releasing the mediator during the amplification of the target sequence and its hybridization to a dual-labeled UR. The same UR can be used to detect many different target sequences. This flexibility facilitates cost reductions, improved homogeneity in fluorescence signals, and easier optimization of fluorescence signal generation, which is primarily determined by UR quenching and fluorescence signal emission efficiencies [17].
Fig. 2.
Mediator probe PCR cycle with mediator probe sequence interactions (dotted ovals). The mediator probe (MP) and universal reporter (UR) with corresponding sequence modifications are presented in the box. The different steps of MP PCR are given below (1–5). In the presence of its complementary target DNA sequence, MP anneals to the sequence (1) and is subsequently cleaved during primer extension (2 + 3). The mediator is set free to hybridize to the UR mediator hybridization site (4). Through the subsequent extension of the mediator at UR, the fluorophore and quencher are separated and a fluorescence signal is generated (5). If no target DNA or an abundant amount (e.g., initial PCR cycles) of target DNA is present, the uncleaved mediator probe hybridizes to UR but is not extended. (Modified from [14]).
Independent of the probing method used to detect nucleic acid target sequences, a suitable probe design is crucial to achieve good analytical performance. The analytical performance characteristics of a real-time PCR assay are referred to as “target values” throughout this article. These target values mainly comprise selectivity, precision, accuracy, detection limits, linear dynamic range, and efficiency of the real-time PCR assay [2], [6], [18], [19], [20], [21]. In an experiment designed for probe optimization, not all of these target values can be determined because of the cost involved and time restrictions. However, at a minimum, the most important target values with respect to practical application should be identified.
Previous studies reported that the statistical design of experiments (DOE) approach as described by G. Taguchi [18], [22], [23], [24], [25], [26], which originated from optimization studies for engineering processes, demonstrates distinct advantages over approaches that optimize one factor at a time in improving PCR-based assay performance. In general, the DOE approach guides the investigator through the experimental setup and data analysis, thus simplifying the analysis of the complex interactions between the input factors. Furthermore, the number of experiments required for optimization can be reduced because of the maximization of information on the effects of certain input factors on the basis of the tests performed [7], [27]. In addition, the DOE approach can improve input factor differentiation between those that influence local target value optimum and those that lead to a more general target value optimization.
Taguchi-based optimization has been previously used to improve PCR target values by modifying the input factors of the primer, probe, and magnesium chloride concentrations or changing the temperature profile [28], [29], [30]. To the best of our knowledge, there are no reports regarding DOE application to real-time PCR probe sequence optimization, despite this input factor exerting a substantial influence on real-time PCR performance [8], [31], [32].
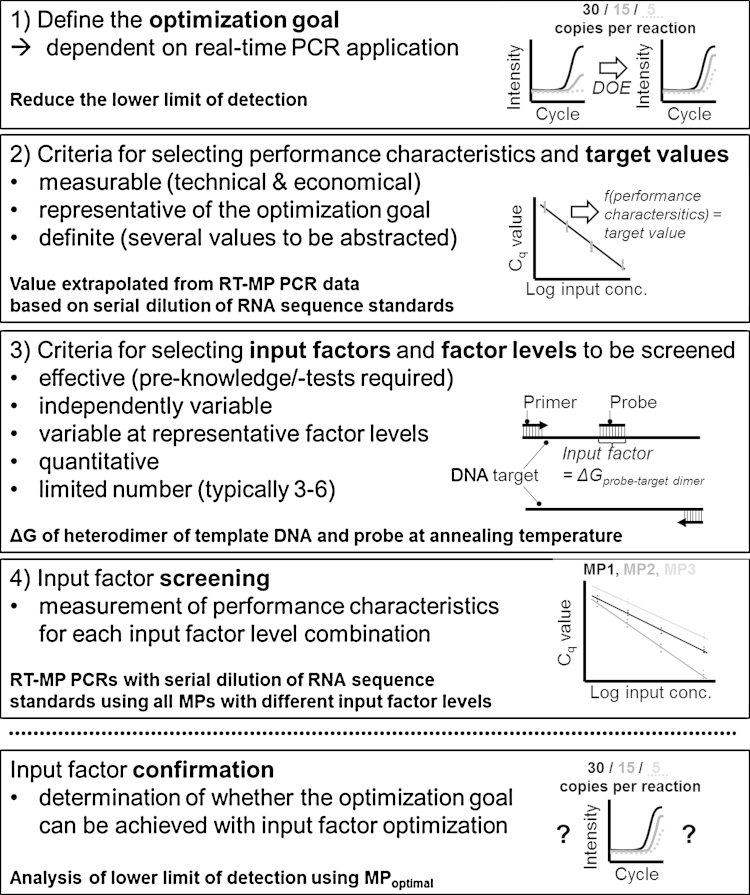
With MP PCR, the target value depends on MP sequence interaction with both the target sequence itself and UR. Other relevant input factors include the polymerase’s MP cleavage efficiency, or the UR’s signal generation efficiency. The latter depends on the polymerase-mediated extension of the released mediator and the ratio between the initial quenching efficiency and fluorescence signal emission following the introduction of spatial separation between the fluorophore and quencher [17]. In this study, we first used DOE to determine the dominant input factors that had the greatest effect on MP-PCR target values. Second, we optimized RT-MP PCR performance and particularly the analytical detection limit by optimizing the level of each determined input factor. Using RT-MP PCR, the detection limit must be at a minimum, i.e., as low as with hydrolysis probes, which are the gold standard references for real-time PCR. For this DOE-based optimization, we selected two clinically relevant targets that cause acute respiratory tract infections, namely influenza B virus (InfB) and human metapneumovirus (hMPV) [17]. In this manner, a guideline for mediator probe sequence optimization has been established that can be extended to other probe formats. This DOE approach is outlined in Fig. 1 and described in detail below.
Fig. 1.
DOE-based process optimization. The examples (bold text) were developed on the basis of the presented optimization of RT-MP PCR.
2. Materials and methods
This DOE-based input factor screening study for RT-MP PCR optimization followed the four steps described in Fig. 1. The following section describes the principle approach and actual experiments performed.
2.1. Definition of optimization goal
Selected RT-MP PCRs were used to detect InfB and hMPV viral RNA. In clinical studies the viral load of these targets ranged between 103and108 RNA copies per ml sample [33]. Therefore, the optimization goal was defined as follows: The assay must enable the detection of 10–100 RNA copies per μl RNA input volume per reaction.
2.2. Selection of performance characteristics and target value
To determine the lower limit of detection, many replicates need to be analyzed per concentration (nine in this study). Comparing all possible MP designs at this level of replication would not be economically feasible. Other performance characteristics that at least influence the limit of detection were, thus, selected and measured using fewer replicates as previously proposed [34]. A target value was calculated (Eq. (1)) from data using an RNA dilution series (InfB) to determine as many performance characteristics as possible: PCR efficiency (calculated from the standard curve), quantitative correlation between RNA input and output concentrations (R2 from the standard curve), mean signal increase over the fluorescence background signal as a measure for detecting sensitivity, and Cq value of reactions with 104 target RNA copies/reaction, which is a measure for the reaction velocity. The abstracted target value was, thus, calculated as follows:
| (1) |
Coefficients a–d were used to balance the performance characteristic value influence on the target value. Coefficient values were determined according to the mean values of the four different performance characteristics for the nine different MPs (refer to Section 4).
2.3. Selection of input factors and factor levels for screening
It is necessary to select an appropriate number of input factors to evaluate their real effectiveness on the target value. All possible input factors were collected on the basis of the MP interactions present during an MP PCR cycle (Fig. 2).
These input factors were collectively assessed (Table 1) according to the interactions illustrated in Fig. 2 using important DOE criteria [7]. Most input factors [(D)–(G)] could be excluded from the screening study because of poor variability, i.e., no sufficiently different MP designs could be created on the basis of these input factors. The qualitative assumption of input factor effectiveness was based on different previous (RT-)MP PCR assays. In these previous experiments, the effects of input factors were not systematically investigated, as was the case in this study. However, input factors (A)–(D) reproducibly demonstrated a strong effect in different assays and different experimental conditions, e.g., a strong effect on PCR efficiency.
Table 1.
Assessment of input factors. Qualitative assessment ranged from +++ (very good) to + (poor). The input factors (A)–(C) were ultimately selected (bold text).
| Input factor | Assumed effectiveness | Independent variability | Variability at representative factor levels | Quantitative variability |
|---|---|---|---|---|
| (A) ΔG of the MP-target heterodimer | +++ | + | +++ | +++ |
| (B) ΔG of the mediator-UR heterodimer | +++ | + | +++ | +++ |
| (C) Distance between primer and MP cleavage site | +++ | + | +++ | +++ |
| (D) Mediator length | +++ | + | ++ | +++ |
| (E) 3′-terminal base at mediator | + | + | + | + |
| (F) Mediator binding position at UR | ++ | + | + | +++ |
| (G) Sequence around MP cleavage site | ++ | + | + | + |
Three input factors were finally selected: ΔG of the MP-target heterodimer, ΔG of the mediator-UR heterodimer, and distance of the sense primer 3′-terminal to the MP-target heterodimer cleavage site. Next, two levels were established for each factor that spanned an appropriate range according to the pre-test results. This resulted in 23 = 8 MP designs. In addition to these eight designs, a ninth design was selected that incorporated all input factors at a central level. The latter design enabled screening for non-linear relationships between factor levels and the target value. Moreover, this MP design was used for data calibration between several RT-PCR runs conducted during the screening experiment.
3. Materials
Concentration standards of RNA target sequences for use with input factor screening and confirmation studies comprised in vitro transcripts from synthetic DNA sequences encoding InfB (hemagglutinin (HA) gene; transcript length: 170 nt) and hMPV (fusion protein gene; transcript length: 70 nt). RNA concentration standards were provided by the University Medical Center Göttingen Virology Department and were prepared as previously described [35]. The transcript sequences are listed as supplementary information. Both InfB and hMPV RNA concentration standards were diluted in 100 μg/ml baker’s yeast tRNA, which served as a blocking agent (Roche, Germany, cat. no. 10109495001).
All primers, MP and UR sequence designs, and reference hydrolysis probe (HP) sequence designs are listed in Table 2. All sequences were designed using VisualOMP™ (DNA Software®, USA, Version 7.8.42.0) [8], [11], [36].
Table 2.
Oligonucleotide sequences (supplier: biomers.net).
| Oligonucleotide or target name | Description | Sequence (5′–3′) | Probe input factor and value |
||
|---|---|---|---|---|---|
| Factor A/kcal × mol−1 | Factor B/kcal × mol−1 | Factor C/bp | |||
| Universal reporter A | UR Aa | Q2-CACGCG°A°A°GATGAGATCGCG(dT-Cy5)GTGTTGGTCGTAGAGCCCAGAACGAT-C3 | |||
| Universal reporter B | UR Ba | Q2-CACCGG °C°T°AAGACGCGCCGG(dT-Cy5)GTGTTGCACCTGGGACATCGACTAT-C3 | |||
| Influenza B virus (InfB) HA gene (amplicon length 160 bp) | Forward primer (Fw InfB) | GGATTAAATAAAAGCAAGCCTTAC | |||
| Reverse primer (Rv InfB) | CAGCAATAGCTCCGAAGAAAC | ||||
| Hydrolysis probe Aa (HP InfB A)c | 6F-CACCCATATTGGGCAATTTCCTATGGC-Q1 | −18.0 | |||
| Hydrolysis probe Ba (HP InfB B) | 6F-CCCATATTGGGCAATTTCCTATGGC-Q1 | −15.5 | |||
| Mediator probeb A1 (MP InfB A1) | TCGTTCTGGGCTCTACGACC|AGGAGGCCTATATTTGGTTCCATTGG-PH | −16.9 | −13.9 | 21 | |
| Mediator probeb A2 (MP InfB A2) | TCGTTCTGGGCTCTACGACC|ACCCATATTGGGCAATTTCCTATGGC-PH | −17.5 | −14.0 | 67 | |
| Mediator probeb A3 (MP InfB A3) | TCGTTCTGGGCTCTACGACC|AGGAGGCCTATATTTGGTTCCA-PH | −13.9 | −13.9 | 21 | |
| Mediator probeb A4 (MP InfB A4) | TCGTTCTGGGCTCTACGACC|ACCCATATTGGGCAATTTCCTAT-PH | −13.8 | −14.0 | 67 | |
| Mediator probeb A5 (MP InfB A5) | ATCGTTCTGGGCTCTACGAC|ATTGGCAAGCTTCAAAGGTGTTT-PH | −15.2 | −13.1 | 37 | |
| Mediator probeb A6 (MP InfB A6) | TTCTGGGCTCTACGACC|AGGAGGCCTATATTTGGTTCCATTGG-PH | −16.9 | −11.3 | 21 | |
| Mediator probeb A7 (MP InfB A7) | TTCTGGGCTCTACGACC|ACCCATATTGGGCAATTTCCTATGGC-PH | −17.5 | −11.3 | 67 | |
| Mediator probeb A8 (MP InfB A8) | TTCTGGGCTCTACGACC|AGGAGGCCTATATTTGGTTCCA-PH | −13.9 | −11.3 | 21 | |
| Mediator probeb A9 (MP InfB A9) | TTCTGGGCTCTACGACC|ACCCATATTGGGCAATTTCCTAT-PH | −13.8 | −11.3 | 67 | |
| Mediator probeb B1 (MP InfB B1) | TCGATGTCCCAGGTGC|CCATATTGGGCAATTTCCTATGGC-PH | −15.9 | −11.0 | 70 | |
| Mediator probeb B2 (MP InfB B2) | ATAGTCGATGTCCCAGGTGC|CCATATTGGGCAATTTCCTATGGC-PH | −15.9 | −13.1 | 70 | |
| Forward primer (Fw hMPV) | GCTTCAGTCAATTCAACAGAAG | ||||
| Reverse primer (Rv hMPV) | TGGTGTTATYCCRGCATTGTCTG | ||||
| Hydrolysis probea (HP hMPV) | 6F-CTAAATGTTGTGCGGCARTTTTCAG-Q1 | −16.5 | |||
| Human Metapneumovirus F gene (amplicon length 70 bp) | Mediator probeb A1 (MP hMPV A1) | TTCTGGGCTCTACGACC|TAAATGTTGTGCGGCARTTTTCAG-PH | −15.0 | −11.9 | 2 |
| Mediator probeb A2 (MP hMPV A2) | TCGTTCTGGGCTCTACGACC|TAAATGTTGTGCGGCARTTTTCAG-PH | −15.9 | −13.2 | 2 | |
| Mediator probeb B1 (MP hMPV A1) | TCGATGTCCCAGGTGC|TAAATGTTGTGCGGCARTTTTCAG-PH | −16.1 | −11.0 | 2 | |
| Mediator probeb B2 (MP hMPV A2) | ATAGTCGATGTCCCAGGTGC|TAAATGTTGTGCGGCARTTTTCAG-PH | −16.1 | −13.1 | 2 | |
“°” = phosphorothioate, underlining indicates stem, text in italics and bold indicates a mediator hybridization site; 6F = 6-Carboxyfluoresceine; Cy5 = Cyanine 5; Q1 = BHQ-1; Q2 = BHQ-2; C3 = C3-Spacer; R = A or G; Y = C or T.
Text in italics and bold indicates a mediator region, underlining indicates a target-specific probe region; PH = phosphate moiety, a dashed vertical line indicates an MP cleavage site.
The Qiagen QuantiTect Multiplex RT-PCR NR Kit (Qiagen, Germany, cat. no. 204843) was used for one-step reverse transcription–amplification reactions. Each 10 μl reaction was performed with final concentrations of primers at 300 nM, MPs or HPs at 150 nM, and UR at 100 nM.
3.1. Setup and analysis of input factor screening
All nine InfB MP designs (Table 2 A1–A9) were tested in the amplification reactions with different InfB target RNA concentrations spanning 105–102 InfB copies/10 μl reaction. All reactions were performed in quadruplicate. For each RT-MP PCR run, four 10 μl reactions for amplification and detection of InfB target RNA (104 copies/10 μl reaction) using MP InfB A5 were used as an inter-run calibrator and were co-analyzed with all other InfB amplifications. The RT-real-time MP PCR were performed in a Rotor-Gene Q (Qiagen, Hilden, Germany) using the following reaction protocol: 20 min at 50 °C (reverse transcription), 15 min at 95 °C (hot start), 50 × (10 s at 94 °C and 45 s at 60 °C). For fluorescence signal generation, UR A was used in all reactions. Fluorescence signals were acquired at the end of each 60 °C-cycling step with signal acquisition using the Cy5-readout gain for UR and the FAM-readout gain for InfB HP and hMPV HP (refer to input factor confirmation study).
For data analysis, the maximum fluorescence signal achieved during data acquisition was divided by the mean of the fluorescence background signal during cycles 3–10 for each test at 102 copies/reaction. For establishing the other target values, the Cq values for all setups were determined using the Rotor-Gene Q analysis software Q 2.1.0.9 with slope correction for normalized fluorescence data and Cq value determination using a fluorescence threshold set to 0.02, which was in the exponential phase of the sigmoidal fluorescence curves for all setups. The Cq values for the separated experimental runs (Cqi) were normalized according to the mean of the Cq values of the inter-run calibrator (four technical replicates) during the specific run [Cq(calibrator run)] and in all runs [Cq(calibrator all runs)] using the equation:
| (2) |
Standard curves were generated using normalized Cq values as an input into the Origin 9.0 software (OriginLab Coorp., USA). Using the standard curve, the efficiency of each RT-real-time MP PCR and the target copy number of each test were calculated. The back-calculated copy numbers were plotted against the theoretical copy number input and the R2 value for this correlation was determined. Mean values for the four technical replicates were determined for all four values to establish one discrete target value per MP design. JMP® 8.0.1 software (SAS Institute Inc., USA) was used for the experimental setup design and statistical analysis. A full factorial screening design was selected to cover all combinations of input factor levels. This experimental setup permits the analysis of significant effects for single input factors and programmed factor combinations on the target value. In addition, one center point was added to the experimental design to verify whether these input factors affect the target value in a linear or non-linear manner. Next, the statistical significance of these effects was analyzed using the JMP screening function. This function refers to Lenth’s method for statistical analysis, where a significance level of 95% is applied. This means that the effect of an input factor or input factor combination on the target value was significant if the analysis produced a p value of <0.05.
3.2. Setup and analysis of input factor confirmation
This test was performed to assess the appropriateness of the DOE-based approach for MP design optimization. The input factor with the largest effect on the target value was selected according to the results of the screening study (refer to input factor with highest value in Fig. 4). For this screening, two factor levels, one resulting in relatively low target value magnitudes (MP InfB A2, Table 2) and one resulting in higher magnitudes (MP InfB A7, Table 2) were used for MP design. The influence of this input factor on the lower limit of detection (optimization goal) was tested for both InfB and hMPV detections. Two different UR designs (A and B, Table 2) were used for detecting both the targets. InfB was diluted to the final concentrations of 150, 50, 17, 6, 2, and 0.6 copies/10 μl reaction, while hMPV was diluted to 150, 75, 38, 19, 9, and 5 copies/10 μl reaction. All setups were performed in nine technical replicates. In addition, nine NTCs were analyzed for each setup. RT-real-time MP PCR was performed as previously described. Sample amplification was considered positive if the following conditions were met: 1) a sigmoidal curve shape, 2) the normalized fluorescence signal in the corresponding sample (cycles, 40–50) was of >10-fold greater than the standard deviation of the fluorescence signal from nucleic acid negative samples (cycles, 5–15) [9]. The 95% confidence intervals around the detection limit were determined using the SPSS software version 19 Probit analysis (IBM, USA) [37].
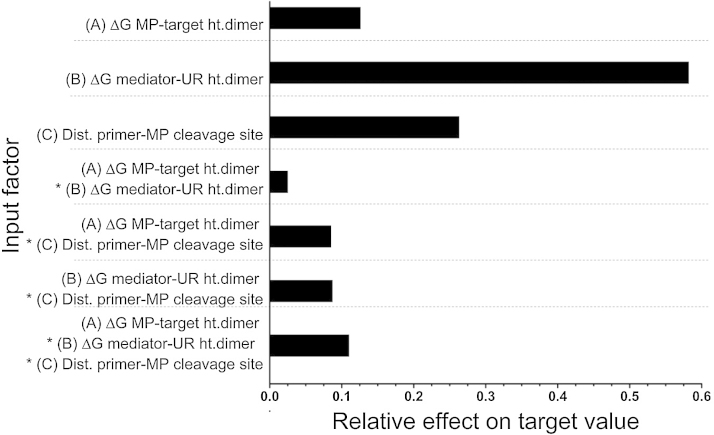
Fig. 4.
The relative effect of all input factors and input factor level combinations on target values. All input factors and input factor level combinations (depicted by multiplication sign “*”) had a significant effect on their target values (significance level, 95%) as shown using the JMP software analysis. ΔG mediator-UR ht. dimer was found as dominant factor. Dist. = distance.
4. Results and discussion
4.1. Input factor screening
Using a DOE-based approach, the effects of different MP design input factors on a target value describing RT-MP PCR performance were evaluated. Different combinations of three MP design input factors at two factor levels each were screened: (A) ΔG of the MP-target heterodimer; (B) ΔG of the mediator-UR heterodimer; and (C) distance of the sense primer 3′-terminal to the MP-target heterodimer cleavage site. For calculating the target value, the coefficients included in Equation 1 were determined. The target value was based on four single performance characteristics. To analyze these performance characteristics, all MPs were evaluated for detectable amplification with the serial dilution of InfB input RNA (102–105 copies per reaction). The single performance characteristics demonstrated the following ranges of values: R2, 0.997–0.996; PCR efficiency, 0.89–0.99 (i.e., 89%–99%); signal increase, 2.4–16.3; and Cq value, 19–23. Different coefficients were used for calculating the target value so that all single performance characteristics were balanced according to the mean value for all nine individual MP designs.
| (1.2) |
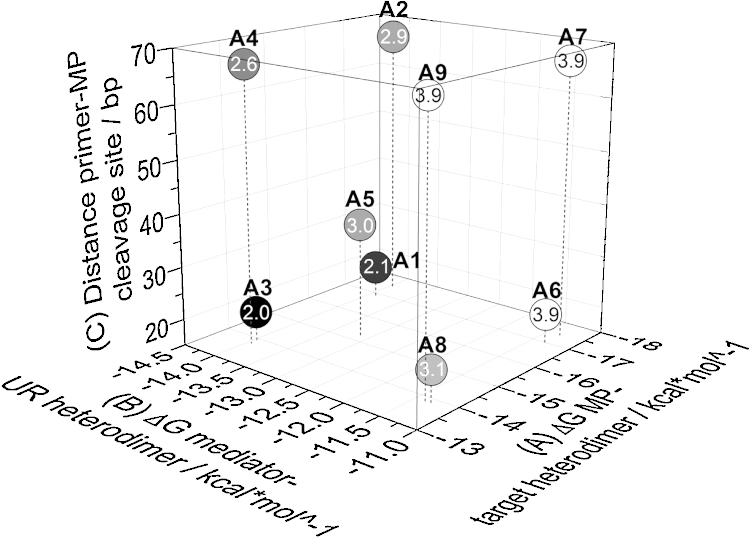
Fig. 3 illustrates the different input factor level combinations together with the recorded target value from Eq. (1.2.)
Fig. 3.
Screening results. Orthogonal view of MP input factors and target values. Three MP input factors with two factor levels each were screened for potential influence on the target values. Low numbers and dark shading indicate a low magnitude target value, while large numbers and bright shading indicate a high magnitude target value. Furthermore, a center point (A5) was added to check for non-linear interactions.
The results clearly revealed that MP designs with higher levels of input factor (B) ΔG of the mediator-UR heterodimer consistently produced higher target values (brighter points) compared with those with lower values for the same input factor. Thus, the lower the mediator-UR heterodimer stability at a given annealing temperature, the higher the RT-real-time MP PCR performance. Because an increase in the (B) value was accomplished by reducing the mediator sequence length at its 5′-terminal, the following disturbance variable (input variables that can cause the controlled variables to deviate from their respective set points) had to be taken into consideration. All MPs that achieved higher target values [MPs with higher levels of input factor (B)] had a mediator sequence length of only 16–18 nt compared with the 19–21 nt length for the mediator sequences from the designs that achieved lower target values [MPs with lower levels of input factor (B)].
Changing either (A) or (C), the two other input factors did not significantly change the target value at least if factor (B) was already at the optimal level. When factor (C) was reduced, all target values also declined. However, this effect was only significant in cases that included non-optimal levels of input factor (B). Factor (A) had the smallest impact on the target value. In contrast to (B) or (C), changing (A) in one direction did not impact the target value. Similar to (C), a strong effect was only found for (A) in the range of non-optimal values for (B). Misinterpreting these locally optimal target values was prevented by the design of the DOE-based experimental setup: i.e., input factors (A) and (C) could be identified as input factors that do not have the largest effect on the target value.
From the single performance characteristics that produced the target value (supplementary information), it is apparent that MP with the highest target values (MP InfB A6, A7, and A9) demonstrated better performance in a minimum of two out of the four single performance characteristics measured as compared with the designs with the lowest target values (MP InfB A1-A4).
As already derived from the data space presented in Fig. 3 the results of the significance analysis demonstrated that all input factors and input factor combinations did not have a significant effect on the target value (Fig. 4). However, input factor (B) demonstrated the greatest effect on target value and was, therefore, regarded as the dominant input factor. The relatively large effect of the primer-MP distance [input factor (C)] can be explained by the large drop in the target value using MP design A1 and A3. In all other setups, the dominance of the effect of factor (C) is clear for both factors (B) and (A).
RT-real-time MP PCR efficiency was evaluated as one of the single performance values leading to the target values (Fig. 3). In summary, ΔG of the mediator-UR heterodimer was less than −11.29 kcal/mol (disturbance variable mediator length: 19–21 nt); thus, the minimum PCR efficiency achieved (MP design A3) was 89% ± 2%. In comparison, MP with ΔG of the mediator-UR heterodimer was more than −11.29 kcal/mol (disturbance variable mediator length: 15–17 nt), and the minimum efficiency was 95% ± 1% (MP designs A8 & A7).
Two effects were regarded as being responsible for the superior effects of input factor (B) on the target value: the efficiency of MP cleavage increases with decreasing mediator length [38], and the equilibrium between MP-UR heterodimers and MP-target sequence heterodimers shifts as well.
Although it was not evaluated using sequence lengths of <20 nt, the cleavage efficiency of Taq polymerases toward fork-like structures, as was present in the case of the MP-target heterodimer, was shown as being dependent on the length of the 5′-unhibridized flap (in MP PCR, i.e., the length of the mediator sequence) [38]. It is assumed that this is because of the mechanism of flap processing by the polymerase nuclease domain as previously described for the flap endonuclease 1, which is structurally and mechanistically related to the nuclease domain in Taq polymerases [39], [40]: Prior to cleavage, the flap is threaded into the enzyme's active site. Thus, shorter flaps might be threaded and cleaved with a higher efficiency.
Concerning the observed influence of the equilibrium between MP-UR heterodimers and MP-target sequence heterodimers, Holland et al. first investigated the 5′-3′ nuclease activity of Taq polymerases when different probes were used during primer extension [41]. The experiments proved that the probe binding must be favored toward primer binding to enable probe binding before the primer extension reaches the probe binding site.
Target values achieved using the homologous probes MP InfB A2 (ΔG mediator UR heterodimer = −13.95 kcal mol−1; mediator length, 19 nt) and A7 (ΔG mediator UR heterodimer = −11.33 kcal mol−1; mediator length, 16 nt) were compared with the corresponding target value using dual-labeled hydrolysis probes (HP InfB A) as the gold standard of the probe design in real-time PCR. Therefore, the same target-specific probe sequence was used; however, the mediator was replaced by a fluorophore and the 3′-terminal phosphate by a quencher. The following mean target values were achieved using four replicates of each experiment at each RNA dilution step: MP InfB A2: 2.93 ± 0.15; MP InfB A7: 3.85 ± 0.11; and HP InfB A: 2.22 ± 0.02. Differences in the target value are because of the different values for the single performance characteristics of Cq value for reactions with an RNA-input concentration of 104 copies/μl and because of the signal increase measured for the reactions with an RNA-input concentration of 102 copies/μl. For both characteristics, MP InfB A7 significantly performed better than both others, with MP InfB A2 containing a longer mediator (higher ΔG mediator UR heterodimer) along with the hydrolysis probe HP InfB A. This observation becomes obvious on observing the RT-real-time PCR fluorescence curves of the three different setups (Supplementary information).
Since the variability of the efficiency of the reverse transcription step was not evaluated, results are prone to an uncertainty [42], [43]. However, to account for the statistical variability of the RT efficiency we performed technical replicates (4 replicates per RNA input concentration). As can be derived from the R2 values of the standard curve (Supplementary information), the Cq-variation between these replicates and thus the variability of the reverse transcription efficiency is considered to be not significant in the case of the presented study.
4.2. Input factor confirmation
The actual performance characteristic to be optimized was the assay detection limit. This important assay performance characteristic was selected to test the relevance of results gathered in the input factor screening. The InfB assay detection limit using a non-optimal MP design (MP InfB A2) was compared with the one that used an optimal MP design (MP InfB A7) and compared with a corresponding hydrolysis probe. Furthermore, the InfB assay detection was also analyzed using an alternative UR. The same analyses were performed for the hMPV detection assay.
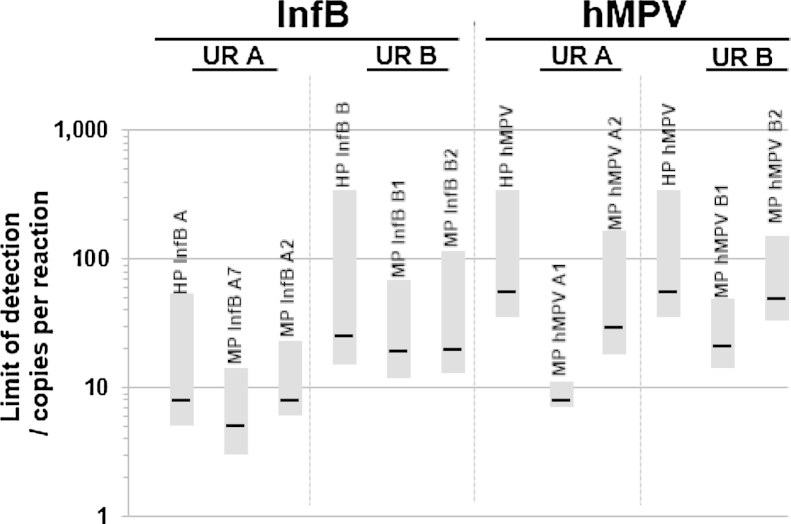
As shown in Fig. 5, for two different InfB and hMPV targets using two different URs (different mediator hybridization sequences), the optimal MP design according to the target value magnitude demonstrated reduced detection limits compared with the corresponding HP RT-PCR. As expected from the input factor screening, MPs with higher ΔG mediator-UR heterodimer levels (MP InfB A7:−11.33 kcal mol−1; MP InfB B1: −11.0 kcal mol−1; MP hMPV A1: −11.9 kcal mol−1; and MP hMPV B1: −11.0 kcal mol−1) demonstrated lower limits of detection (95% probability of detection) with the corresponding 95% confidence interval than MPs with lower levels of this input factor (B) (MP InfB A2: −13.95 kcal mol−1; MP InfB B2: −13.1 kcal mol−1; MP hMPV A2: − 13.2 kcal mol−1; and MP hMPV B2: −13.1 kcal mol−1). Compared with corresponding HP PCRs, detection limits achieved with MP PCR are significantly reduced. This is because of the higher signal-to-noise ratio of the universal reporter with a low fluorophore-quencher distance achieved by the hairpin structure compared with the dual-labeled HPs with larger fluorophore-quencher distances, depending on the actual probe sequence length. The analytical detection limits of <50 copies/reaction of the improved RT-MP PCR assays demonstrated a good agreement with commercially available assays for InfB and hMPV detection [33], [44].
Fig. 5.
Detection limits of InfB and hMPV obtained by HP and RT-MP PCRs. For RT-MP PCRs, the universal reporters A and B with different mediator hybridization sequences oligonucleotides were used. The gray areas span the lower and upper bounds of the detection limit estimate within a 95% confidence interval.
For the reverse transcription efficiency variability the same considerations as demonstrated before apply for the input factor confirmation.
5. Conclusion
In a DOE-based MP design screening, three different design input factors were selected from a group of candidates. These factors were tested and had a significant effect on RT-MP PCR performance in pre-tests. We determined that the input factor ΔG of the mediator-UR heterodimer has the highest effect on RT-MP PCR performance. Easy adaption of MPs leading to improved RT-MP PCR results could thus be realized just by changing the mediator sequence length. At the same time, these mediator probes also demonstrated superior performances in RT real-time PCR compared with homologous hydrolysis probes. Thus, the DOE method appears to be an appropriate approach for MP sequence optimization for use with real-time PCR. Only nine MP designs led to maximum information for the three input factors and their combinatorial effects. The nine designs were screened across four technical replicates at each of the five different RNA input concentrations, resulting in 180 individual performances. In contrast, a one-factor-at-a-time approach would require 320 individual reactions [eight designs (no center point), 16 technical replicates, five different RNA concentrations] to receive information for the effect of an input factor on the target value at the same accuracy. Second, there is the risk associated with a local minimum in data collection and assay optimization [24]. On the basis of these results, we can, thus, recommend DOE as a guideline for MP sequence optimization. However, the DOE approach may also be applicable to other detection formats for real-time PCR (e.g., hydrolysis probes or molecular beacons) or to PCR primer sequence design. This approach may be particularly relevant with diagnostic applications in which the best performing primers and probes must be defined for the detection of pathogenic nucleic acid sequences.
Acknowledgments
This study was funded by the German research society as part of the project Mediatorsonden Technologie (FKZ STE 1937/1-1). We gratefully acknowledge Prof. Dr. Manfred Weidmann and Prof. Dr. Frank Hufert for providing the RNA target material and corresponding primer- and hydrolysis probe-binding regions. Moreover, we thank Christian Köhler for assisting with the analysis software and data interpretation and Dr. Günter Roth for helpful advice during input factor collection.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.bdq.2015.12.002.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- 1.Danzer K. Springer; Berlin, Heidelberg: 2007. Analytical Chemistry: Theoretical and Metrological Fundamentals; with 31 Tables. [Google Scholar]
- 2.Wilson J.A. Wayne; PA: 2008. Verification and Validation of Multiplex Nucleic Acid Assays: Approved Guideline, Clinical and Laboratory Standards Institute. [Google Scholar]
- 3.International Organization for Standardization, In vitro diagnostic medical devices – Measurement of quantities in biological samples – Metrological traceability of values assigned to calibrators and control materials, http://www.iso.org/iso/iso_catalogue/catalogue_tc/catalogue_detail.htm?csnumber=30716 (last accessed: 03.2015).
- 4.International Organization for Standardization, Statistics – Vocabulary and symbols – Part 1: General statistical terms and terms used in probability, http://www.iso.org/iso/catalogue_detail.htm?csnumber=40145 (last accessed: 03.2015).
- 5.World Health Organization . World Health Organization; Geneva: 1998. WHO Expert Committee on Biological Standardization. Fortx-Sixth Report. [PubMed] [Google Scholar]
- 6.Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L., Vandesompele J., Wittwer C.T. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 7.W. Kleppmann, Taschenbuch Versuchsplanung: Produkte und Prozesse optimieren, 7., aktualisierte und erw Aufl Hanser München (2011).
- 8.SantaLucia J., Jr. Methods Mol. Biol. (Clifton, NJ) 2007;402:3–34. doi: 10.1007/978-1-59745-528-2_1. [DOI] [PubMed] [Google Scholar]
- 9.Heid C.A., Stevens J., Livak K.J., Williams P.M. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 10.Mackay I.M. Clin. Microbiol. Infect. 2004;10:190–212. doi: 10.1111/j.1198-743x.2004.00722.x. [DOI] [PubMed] [Google Scholar]
- 11.Mackay I.M., Arden K.E., Nitsche A. Nucleic Acids Res. 2002;30:1292–1305. doi: 10.1093/nar/30.6.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thornton B., Basu C. Biochem. Mol. Biol. Educ. 2011;39:145–154. doi: 10.1002/bmb.20461. [DOI] [PubMed] [Google Scholar]
- 13.Juskowiak B. Anal. Bioanal. Chem. 2011;399:3157–3176. doi: 10.1007/s00216-010-4304-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faltin B., Wadle S., Roth G., Zengerle R., von Stetten F. Clin. Chem. 2012;58:1546–1556. doi: 10.1373/clinchem.2012.186734. [DOI] [PubMed] [Google Scholar]
- 15.Faltin B., Zengerle R., von Stetten F. Clin. Chem. 2013;59:1567–1582. doi: 10.1373/clinchem.2013.205211. [DOI] [PubMed] [Google Scholar]
- 16.S. Wadle, S. Dr. Rubenwolf, M. Dr. Lehnert, B. Dr. Faltin, M. Dr. Weidmann, F. Dr. Hufert, R. Prof. Dr. Zengerle, F. Dr. habil. von Stetten, Quantitative Real-Time PCR, pp. 55-73
- 17.Marras S.A.E., Kramer F.R., Tyagi S. Nucleic Acids Res. 2002;30:122e–122. doi: 10.1093/nar/gnf121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.S. Stanzel, Optimale statistische Versuchsplanung dreifaktorieller Zwei-Farben-cDNA-Microarray-Experimente, Dortmund Dissertation (2007).
- 19.Kitchen R.R., Kubista M., Tichopad A. Methods(San Diego, Calif.) 2010;50:231–236. doi: 10.1016/j.ymeth.2010.01.025. [DOI] [PubMed] [Google Scholar]
- 20.U.S. Department of Health and Human Services/Food and Drug Administration, Bioanalytical Method Validation (2001) http://www.fda.gov/downloads/Drugs/Guidances/ucm070107.pdf. (last accessed: 03.2015).
- 21.European Medicines Agancy, Quality guidelines, http://www.ema.europa.eu/ema/index.jsp?curl=pages/regulation/general/general_content_000081.jsp&mid=WC0b01ac0580027546. (last accessed: 12.2015).
- 22.Wrobel G., Schlingemann J., Hummerich L., Kramer H., Lichter P., Hahn M. Nucleic Acids Res. 2003;31:67e–67. doi: 10.1093/nar/gng067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wildsmith S.E., Archer G.E., Winkley A.J., Lane P.W., Bugelski P.J. Biotechniques. 2001;30:202–206. doi: 10.2144/01301dd04. 208. [DOI] [PubMed] [Google Scholar]
- 24.Luo W., Pla-Roca M., Juncker D. Anal. Chem. 2011;83:5767–5774. doi: 10.1021/ac103239f. [DOI] [PubMed] [Google Scholar]
- 25.Rao R.S., Kumar C.G., Prakasham R.S., Hobbs P.J. Biotechnol. J. 2008;3:510–523. doi: 10.1002/biot.200700201. [DOI] [PubMed] [Google Scholar]
- 26.Heyden Y.V. Lc Gc Eur. 2015;19:469–475. [Google Scholar]
- 27.Derringer G.C., Suich R. J. Qual. Technol. 1980;12:214–219. [Google Scholar]
- 28.Altekar M., Homon C.A., Kashem M.A., Mason S.W., Nelson R.M., Patnaude L.A., Yingling J., Taylor P.B. Clin. Lab. Med. 2007;27:139–154. doi: 10.1016/j.cll.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 29.de Souza Celani, Jose Helder, Moyses C.B., Pontes F.J., Duarte R.N., da Silva Sanches, Eduardo C., Alberto F.L., Ferreira U.R., Silva M.B. Mol. Cell. Probes. 2011;25:231–237. doi: 10.1016/j.mcp.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 30.Ballantyne K.N., van Oorschot R.A.H., Mitchell R.J. Genomics. 2008;91:301–305. doi: 10.1016/j.ygeno.2007.10.016. [DOI] [PubMed] [Google Scholar]
- 31.Lunge V., Miller B., Livak K., Batt C. J. Microbiol. Methods. 2002;51:361–368. doi: 10.1016/s0167-7012(02)00113-6. [DOI] [PubMed] [Google Scholar]
- 32.Livak K.J. Genet. Anal. Biomol. Eng. 1999;14:143–149. doi: 10.1016/s1050-3862(98)00019-9. [DOI] [PubMed] [Google Scholar]
- 33.van Wesenbeeck L., Meeuws H., van Immerseel A., Ispas G., Schmidt K., Houspie L., van Ranst M., Stuyver L. J. Clin. Microbiol. 2013;51:2977–2985. doi: 10.1128/JCM.00911-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Köppel R., Zimmerli F. Eur. Food Res. Technol. 2010;231:727–732. [Google Scholar]
- 35.Weidmann M., Mühlberger E., Hufert F.T. J. Clin. Virol. 2004;30:94–99. doi: 10.1016/j.jcv.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 36.Visual OMP DNA Software, http://www.dnasoftware.com/our-products/visual-omp/ (last accessed: 12.2015).
- 37.Finney D.J. 3rd edition. Cambridge University Press; New York: 1971. Probit Analysis. [Google Scholar]
- 38.Lyamichev V., Brow M.A., Dahlberg J.E. Science (New York, NY) 1993;260:778–783. doi: 10.1126/science.7683443. [DOI] [PubMed] [Google Scholar]
- 39.Allawi H.T., Kaiser M.W., Onufriev A.V., Ma W.-P., Brogaard A.E., Case D.A., Neri B.P., Lyamichev V.I. J. Mol. Biol. 2003;328:537–554. doi: 10.1016/s0022-2836(03)00351-6. [DOI] [PubMed] [Google Scholar]
- 40.Liu Y., Kao H.-I., Bambara R.A. Ann. Rev. Biochem. 2004;73:589–615. doi: 10.1146/annurev.biochem.73.012803.092453. [DOI] [PubMed] [Google Scholar]
- 41.Holland P.M., Abramson R.D., Watson R., Gelfand D.H. Proc. Natl. Acad. Sci. U. S. A. 1991;88:7276–7280. doi: 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bustin S., Dhillon H.S., Kirvell S., Greenwood C., Parker M., Shipley G.L., Nolan T. Clin. Chem. 2015;61:202–212. doi: 10.1373/clinchem.2014.230615. [DOI] [PubMed] [Google Scholar]
- 43.Bustin S.A., Nolan T. J. Biomol. Tech. 2004;15:155–166. [PMC free article] [PubMed] [Google Scholar]
- 44.Altona Diagnostics, RealStar® hMPV RT-PCR Kit, http://www.altona-diagnostics.com/realstar-mpv-rt-pcr-kit.html (last accessed: 03.2015).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.