Abstract
Aflatoxins are secondary toxic metabolites produced by some Aspergillus spp. particularly, Aspergillus flavus and A. parasiticus that contaminate food and feed. The objective of this study was to evaluate the contamination of feedstuffs with Aspergillus spp. and detect genes involved in the aflatoxin biosynthesis pathway of A. flavus and A. parasiticus isolates. A total of 110 cow feed samples (comprised of silage, concentrate, hay and total mixed ration) from 30 industrial and semi-industrial dairy farms of Khorasan Razavi province, northeastern Iran, were examined using cultural and PCR methods. 68 (61.82%) Aspergillus spp. were isolated from 110 samples of feedstuff. The predominant Aspergillus isolates were A. fumigates (21.81%), followed by A. flavus (17.27%), A. niger (10%), A. parasiticus (8.18%), and A. oryzae (4.54%). Fungal contamination levels of industrial and semi-industrial dairy farm samples were not significantly different (P>0.05). Using four sets of primers, a quadruplex PCR was developed to detect genes (nor1, ver1, omtA and aflR) at different loci coding enzymes in the aflatoxin biosynthesis pathway of A. flavus and A. parasiticus strains. Out of 28 strains of A. flavus and A. parasiticus, 10 isolates (35.71%) showed a quadruplet pattern indicating the important genes involved in the aflatoxin biosynthesis pathway, encoded for functional products. These isolates were confirmed to be aflatoxigenic by Thin Layer Chromatography. 18 isolates (64.29%) had three, two and single molecular patterns. The results obtained by this study show that rapid and specific detection of aflatoxigenic molds is important to ensure the microbiological safety of feedstuffs.
Key Words: Aflatoxin, Aspergillus spp., Feedstuff, Multiplex PCR
Introduction
Mold and mycotoxin contamination of animal feedstuffs is largely a feed management problem. Most species of Aspergillus are able to grow on wide ranges of feed. They are essentially saprophytic and particularly associated with stored moldy plant products (Shapira et al., 1996 ▶). Aspergillus genera is the most important aflatoxigenic fungi (Rodrigues et al., 2009 ▶). Aflatoxins, are highly toxic secondary metabolites produced predominately by A. flavus, A. parasiticus and A. nomius. Food and feed commodities are usually contaminated by a range of different fungi during growth, harvesting and storage. Local weather conditions as well as environmental conditions in storage facilities, especially temperature and relative humidity, contribute to the growth of A. flavus and A. parasiticus and are, therefore, potential risks for aflatoxin production (Michael et al., 1999; Rahimi et al., 2008 ▶). Aspergillus parasiticus are able to produce aflatoxins B1, B2, G1 and G2, whereas A. flavus is only able to produce aflatoxin B1 and B2 (Yu, 2012 ▶). Only about 40-50% of A. flavus strains are able to produce these mycotoxins. Aflatoxin B1 is widely regarded as the most potent liver carcinogen, affecting a large number of animal species and humans (Michael et al., 1999). Aflatoxin M1 and M2 are hydroxylate derivatives of AFB1 and AFB2, which are formed and excreted in the milk of lactating animals and humans that have consumed aflatoxin-contaminated foods (Michael et al., 1999). Aflatoxins are highly toxic and carcinogenic in animals and humans, leading to hepatotoxicity, immunotoxicity (Mehrzad et al., 2011 ▶), teratogenicity and even death (Erami et al., 2007 ▶; Ghiasian and Maghsood, 2011 ▶).
Traditional culture techniques for monitoring foods and animal feed for fungal contaminations are extremely labor-intensive and require several days to complete. During the past years, a number of molecular based detection methods have been developed to characterize aflatoxigenic and non-aflatoxigenic Aspergillus spp. isolates in human foods and animal feeds (van der Vossen, 1996 ▶; Konietzny and Greiner, 2003 ▶). Aflatoxin biosynthesis is a complex process involving many intermediates and enzymes. The regulation of aflatoxin gene expression occurs at multiple levels and by multiple regulatory components. Twenty seven enzymatic steps are estimated for an aflatoxin biosynthesis (Ehrlich and Yu, 2010 ▶), and as many as 30 genes are potentially involved in the process. The genes and corresponding enzymes have been extensively studied (Yu et al., 2004a ▶, b ▶). In A. flavus and A. parasiticus, aflatoxin pathway genes are clustered within a 75-kb region of the fungal genome on chromosome III, roughly 80 kb away from the telomere (Yu et al., 2004a ▶, b ▶; Chang et al., 2005 ▶). Nor-1, ver-1 and omt-A are three structural genes in the cluster genes of the biosynthesis aflatoxin pathway that are coded for key enzymes in the production of aflatoxin, thus they are essential for the production of aflatoxin (Erami et al., 2007 ▶). Norsolorinic acid (NOR) was confirmed to be the first stable aflatoxin precursor (Yu et al., 2004b ▶). The ver-1 gene was predicted to encode a ketoreductase, similar to nor-1 (Keller et al., 1994 ▶). Aflatoxin pathway genes were found to be clustered in the genome of A. flavus and A. parasiticus (Yu et al., 2004a ▶, b ▶). These genes are expressed concurrently except for the regulatory gene aflR. In this gene cluster, a positive-acting regulatory gene, aflR, is located in the middle of the gene cluster. The aflR gene, encoding a 47 kDa sequence-specific zinc-finger DNA-binding protein, is required for transcriptional activation of most, if not all, structural genes of the aflatoxin gene cluster (Ehrlich et al., 1998 ▶; Chang et al., 1999a ▶, b ▶). Several studies were carried out regarding the use of genes involved in aflatoxin biosynthesis (Shapira et al., 1996 ▶; Criseo et al., 2001 ▶) for the identification of aflatoxin-producing A. flavus and A. parasiticus.
The aims of this study were to determine Aspergillus spp. contamination of animal feed samples and to specifically detect genes involved in aflatoxin bio-synthesis in A. flavus and A. parasiticus isolated from feedstuffs of Khorasan dairy industries, Iran. To this end, a multiplex-PCR method was used to detect important genes involved in aflatoxin synthesis using four set of primers, namely aflR, omtA, ver1, nor1.
Materials and Methods
Feed samples
A total of 110 animal feed samples comprising silage (n=28), concentrate feed (n=30), total mixed ration (n=27) and hay (n=25) were randomly collected from 30 industrial and semi-industrial dairy farms of Khorasan Razavi province (Table 1). All samples were transported to the laboratory under cold (4°C) conditions. All samples were intended for animal consumption and none showed any visible sign of mold contamination.
Table 1.
The frequency of Aspergillus spp. isolated from feedstuffs of Khorasan dairy farms
| Feedstuff sample | No of samples |
Aspergillus spp. |
Total (%) | ||||
|---|---|---|---|---|---|---|---|
| A. flavus | A. parasiticus | A. fumigatus | A. niger | A. oryzae | |||
| Silage | 28 | 1 | 2 | 3 | 2 | 0 | 8 (28.57%) |
| Concentrate | 30 | 8 | 3 | 8 | 3 | 1 | 23 (76.66%) |
| TMR | 27 | 5 | 2 | 5 | 0 | 2 | 14 (51.85%) |
| Hay | 25 | 5 | 2 | 8 | 6 | 2 | 23 (92%) |
| Total (%) | 110 | 19 (17.27%) | 9 (8.18%) | 24 (21.81%) | 11 (10%) | 5 (4.54%) | 68 (61.82%) |
Identification of Asprgillus spp.
Samples were homogenized and stored at 4°C and protected against light until the day of analysis. A 10 g portion of each sample was homogenized in 90 ml of 0.1% peptone water solution for 30 min in an orbital shaker. Serial dilutions of up to 10-6 were made and 0.1 ml of each dilution was inoculated in duplicate onto potato dextrose agar (PDA, Merck). The plates were incubated at 25°C for 5-7 days. To identifyAspergillus species, the spores were transferred on the Czapek-Dox agar medium (CZA, Merck) after fungal colony formation and incubated for 5-7 days at 27°C. Wet mount smears and slide cultured colonies were stained with lactophenol cotton blue. Taxonomic fungi identification was made based on macroscopic and microscopic features according to appropriate keys proposed by Klich (2002) ▶.
Determination of aflatoxin production by chromatography
The isolates identified as A. flavus and A. parasiticus were tested for the production of aflatoxins based on the thin layer chromatography (TLC) method (Moubasher et al., 2013 ▶). The spore suspension (100 µL) of each strain containing 107 spores/ml prepared in 0.1% (vol/vol) Tween 20 was added to a 250 ml Erlenmeyer flask containing 100 ml yeast extract sucrose broth (YES, Merck), incubated for 7 days at 26°C and shook at 150 rpm. To extract aflatoxin from the mycelium, the cells were lysed by adding 100 ml chloroform to each flask and transferring the lower transparent phase to a new tube. Chloroform was evaporated at 100°C in a water bath and the remaining pellets were dissolved in 1 ml methanol. Silica gel TLC plates (Sigma) were used for the aflatoxin analysis. From each sample, 50 µL was spotted onto the TLC sheets, developed in a toluene-ethyl acetate-acetic acid (50:30:4) solvent system. Pure aflatoxins (Sigma) were used as standards. Aflatoxins were visualized under a UV lamp at 365 nm and their presence was chemically confirmed by spraying 50% H2SO4, and heating to charring. An aflatoxigenic strain was used as positive control.
DNA extraction
The isolation of fungal DNA was performed according to the method described by Yelton et al. (1984) ▶ with some modifications. The strains were grown for 72 h under continuous shaking conditions (150 rpm) in the PDA Broth. The mycelium was then harvested by filtration, transferred to a mortar, frozen in liquid nitrogen and ground to a powder which was resuspended in a lysis buffer (50 mmol/L EDTA, 0.2% SDS, pH =8.5) and heated immediately at 68°C for 15 min. After centrifugation for 15 min at 15000 × g, a 7-10 ml volume of the supernatant fluid was transferred to a new centrifuge tube and 1 ml 4 mol/L sodium acetate was added. This solution was placed on ice for 1 h and centrifuged for 15 min at 15000 × g. After centrifugation, the supernatant fluid was transferred to a fresh tube and extracted by AccuPrep® Genomic DNA extraction Kit (Bioneer, Korea).
Multiplex PCR reaction
In the present study, multiplex PCR was performed according to the method described by Criseo et al. (2001) ▶. All of the isolated A. flavus and A. parasiticus were examined for the presence of four important aflatoxin genes (aflR, omt-A, ver-1 and nor-1) enclosed in the aflatoxin biosynthesis pathway by multiplex PCR using four published primer sets (Table 2).
Table 2.
Primers used in this study, target gene, sequence and PCR product size
| Primer name | Target gene | Primer sequence (5´-3´) | PCR product size (bp) | Reference |
|---|---|---|---|---|
| NorF | nor-1 | ACCGCTACGCCGGCACTCTCGGCAC | 400 bp | Criseo et al., 2001 |
| NorR | GTTGGCCGCCAGCTTCGACACTCCG | |||
| VerF | ver-1 | GCCGCAGGCCGCGGAGAAAGTGGT | 537 bp | Criseo et al., 2001 |
| VerR | GGGGATATACTCCCGCGACACAGCC | |||
| OmtF | omt-A | GTGGACGGACCTAGTCCGACATCAC | 797 bp | Criseo et al., 2001 |
| OmtR | GTCGGCGCCACGCACTGGGTTGGGG | |||
| AflrF | aflR | TATCTCCCCCCGGGCATCTCCCGG | 1032 bp | Criseo et al., 2001 |
| AflrR | CCGTCAGACAGCCACTGGACACGG |
PCR reaction was performed in 25 µL containing 2.5 µL 1 X PCR buffer, 0.75 µL 50 mM Mgcl2, 0.5 µL 10 mM dNTPs, 2 µL of each primer, 0.2 µL Taq DNA polymerase (1 U/µL), 5 µL extracted DNA as template and 8.05 µL sterile distilled water. A total of 35 cycles was started with heating at 94°C for 5 min, and continued by denaturation for 30 s at 94°C, annealing for 30 s at 67°C, elongation for 30 s at 72°C and a final extension of 10 min at 72°C. Amplified products were visualized by UV illumination after electrophoresis on 1% agarose gel and ethidium bromdie staining.
Genomic DNA of the following organisms was used to test the sensitivity of primers listed in Table 2: A. parasiticus ATCC 15517, A. oryzae IMI 126842, Penicillium purpurogenome PTCC 5212, A. fumigates PTCC 5009, Fusarium oxysporum PTCC 5115, A. niger ATCC 9142, Alternaria alternata PTCC 5224.
Statistical analysis
Statistical analysis was performed using SPSS version 19. Chi-square and Fisher exact tests were used to assess the possible differences between fungi incidence in feeds and types of dairy farm. The significance level was set at P<0.05 for all tests.
Results
Among 110 feed samples, 68 (61.82%) were contaminated with Aspergillus species. The predominant isolate was A. fumigates (21.81%), followed by A. flavus (17.27%), A. niger (10%), A. parasiticus (8.18%) and A. oryzae (4.54%) (Table 1).
The most frequent isolated fungi were found in the hay samples (92%) and the lowest fungal contamination frequency was in the silage (28.57%). The incidence rates of fungal contamination in industrial and semi-industrial dairy farm samples were (62.5%) and (64.81%), respectively, with no significant differences between farm types (P>0.05).
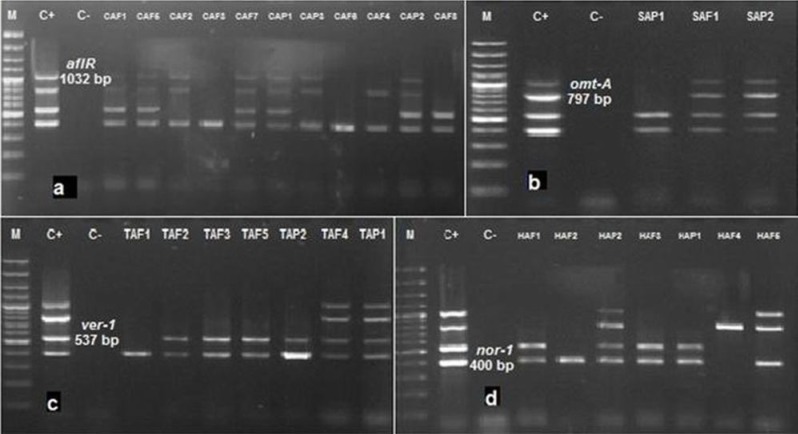
The molecular patterns obtained for 28 examined A. flavus and A. parasiticus isolates are shown in Fig. 1 and Table 3.
Fig. 1.
Agarose gel electrophoresis of multiplex PCR products amplified from A. flavus and A. parasiticus isolated from feedstuffs. M: 100 bp DNA molecular size marker, C+: Positive control (A. parasiticus ATCC 15517), and C-: Negative control (without genomic DNA). (a): DNA banding patterns of A. flavus and A. parasiticus isolates of concentrate samples, (b): DNA banding patterns of A. flavus and A. parasiticus isolates of silage samples, (c): DNA banding patterns of A. flavus and A. parasiticus isolates of TMR samples, and (d): DNA banding patterns of A. flavus and A. parasiticus isolates of hay samples
Table 3.
Results obtained by multiplex PCR and conventional methods (TLC and PCR)
| Isolate code | Aflatoxin biosynthesis gene |
Aflatoxin production by TLC method |
||||
|---|---|---|---|---|---|---|
| aflR | omt-A | ver-1 | nor-1 | Aflatoxin production | ||
| SAF1 | + | + | + | + | + | |
| SAP1 | - | - | + | + | - | |
| SAP2 | + | + | + | + | + | |
| CAF1 | + | + | + | + | + | |
| CAF2 | + | + | - | + | - | |
| CAF3 | - | - | - | + | - | |
| CAF4 | - | + | - | + | - | |
| CAF5 | + | + | + | + | + | |
| CAF6 | - | - | - | + | - | |
| CAF7 | + | + | + | + | + | |
| CAF8 | - | - | + | + | - | |
| CAP1 | + | + | + | + | + | |
| CAP2 | + | + | + | + | + | |
| CAP3 | + | + | - | + | - | |
| TAF1 | - | - | - | + | - | |
| TAF2 | - | - | + | + | - | |
| TAF3 | - | - | + | + | - | |
| TAF4 | + | + | + | + | + | |
| TAF5 | - | - | + | + | - | |
| TAP1 | + | + | + | + | + | |
| TAP2 | - | - | + | + | - | |
| HAF1 | - | - | + | + | - | |
| HAF2 | - | - | - | + | - | |
| HAF3 | - | - | + | + | - | |
| HAF4 | - | + | - | - | - | |
| HAF5 | + | + | - | + | - | |
| HAP1 | - | - | + | + | - | |
| HAP2 | + | + | + | + | + | |
| A. niger ATCC 9142 | - | - | - | - | - | |
| F. oxysporum PTCC 5115 | - | - | - | - | - | |
| A. parasiticus ATCC15517 | + | + | + | + | + | |
| P. purporogenome PTCC 5212 | - | - | - | - | - | |
S: Silage, C: Concentrate, T: TMR, H: Hay, AF: A. flavus, and AP: A. parasiticus
DNA fragments of aflR, omt-A, ver-1 and nor-1 genes were visualized at 1032, 797, 537 and 400 bp, respectively (Fig. 1). Five isolates (CAF1, CAF5, CAF7, CAP1 and CAP2) from the concentrate (Fig. 1a), 2 isolates (SAF1 and SAP2) from the silage (Fig. 1b), 2 isolates (TAF4 and TAP1) from the TMR (Fig. 1c) and 1 isolate (HAP2) from the hay (Fig. 1d) had a quadruplet pattern, indicating the presence of the four genes involved in the aflatoxin biosynthesis pathway. Other strains, however, showed different molecular patterns (Table 3). The results obtained by the TLC method indicated that all aflatoxigenic strains of A. flavus and A. parasiticus (10 strains) contained the four structural genes (Table 3).
No DNA amplification was observed with A. niger ATCC 9142, Penicillium purpurogenome PTCC 5212, Fusarium oxysporum PTCC 5115, A. oryzae IMI 126842, A. fumigates PTCC 5009 and Alternaria alternata PTCC 5224, even at the highest level, thus indicating the high specificity of the PCR. To determine the PCR’s sensitivity, lower concentrations of spores were tested. The DNA was only amplified in A. parasiticus ATCC 15517, even at the lowest spore level.
Discussion
In this study, A. fumigatus (21.81%) and A. flavus (17.27%) were the predominant Aspergillus spp. isolated from feedstuffs. These results differ from reports describing A. niger as the most predominant followed by A. flavus (Saleemi et al., 2010 ▶), and vice versa (Accensi et al., 2004 ▶; Somashekar et al., 2004 ▶). In a study conducted in Brazil on raw materials and finished cow feed samples by Rosa et al. (2008) ▶, eight fungal genera were isolated. The predominant Aspergillus isolated from finished cow feed samples were A. flavus (31.6%) followed by A. niger (22.4%). The predominance of A. flavus isolates showed that it can easily adapt itself to various geographical regions. Another important point to consider is that A. flavus can grow at low (11-14%) humidity levels (Pitt and Hocking, 1997 ▶; Macioro et al., 2007 ▶).
In the present study, similar to Khosravi et al. (2004) ▶, the most frequent isolated fungi were found in concentrate samples rather than other feedstuffs. In another study carried out in Iran by Ghiasian and Maghsood (2011) ▶, concentrate feed was reported to be the most contaminated, with mean colony counts of 7.25 × 102 and 7.50 × 102 CFU/g for A. flavus and A. parasiticus, respectively.
In our study, similar to Khosravi et al. (2004) ▶, the lowest fungal contamination frequency was found in silage. The incidence rate of fungal contamination in both dairy farm samples were (63.31%) and (67.18%), respectively, and no significant difference was detected between farm types (P>0.05). Similar results regarding Iranian farm types were reported by Ghiasian and Maghsood (2011) ▶.
PCR is a method of choice for the diagnosis of aflatoxigenic molds (Shapira et al., 1996 ▶; Erami et al., 2007 ▶). In the present work, using a set of four primers aflR, omtA, ver1 and nor1, a multiplex PCR procedure was used to detect genes involved in the aflatoxin biosynthesis pathway. Primers omt-1, nor-1, ver-1, are three structural genes used for the biosynthesis of aflatoxin. The aflR gene, that codes for a regulatory factor, seemed to be involved in the activation of the transcript of pathway genes (Woloshuk et al., 1994 ▶). It was also found to regulate aflatoxin biosynthesis. The omt gene was found to be involved in the conversion of sterigmatocystin to o-methylsterigmatocystin in the aflatoxin biosynthetic pathway. The results indicated that all aflatoxigenic strains of A. flavus (5 isolates) and A. parasiticus (5 isolates) contained the four tested: nor-1, ver-1, omtA, and aflR genes. In our study, 10 isolates (35.71%) of 28 strains of tested A. flavus and A. parasiticus showed a quadruplet pattern, indicating the presence of all genes which encode for functional products and their involvement in the aflatoxin biosynthetic pathway (Yu et al., 2004a ▶, b ▶). These strains had a quadruplet DNA banding pattern, indicating the presence of the four genes in the aflatoxin biosynthetic pathway. In contrast, non-aflatoxigenic strains showed different band patterns comprising of 1, 2 and 3 bands. These results complement those of the TLC aflatoxin detection method, and were in agreement with the findings of Rashid et al. (2008) ▶, who concluded that aflatoxigenic isolates of A. flavus and A. parasiticus had all four nor1, ver1, omtA, and aflR genes.
In our study, we found that conventional methods of using cultural media for aflatoxin production distinguished perfectly between aflatoxin-producing and non-producing strains. Unfortunately, these methods are time-consuming and labor-intensive, and can fail to detect some aflatoxin-producing strains due to the fact that aflatoxin production instability may occur in certain aflatoxigenic strains growing in culture media (Criseo et al., 2001 ▶). Using multiplex PCR utilizing primers targeting the aflR, nor-1, ver-1 and omt-A genes appears to offer some promise in detecting aflatoxigenic molds. This study confirms the importance of further surveillance of mycotoxigenic fungi and mycotoxin occurrence in feedstuffs in Iran.
Acknowledgements
The authors express their special thanks to Dr. J. Mehrzad for his kind comments on revising the paper and Mrs. S. Khajenasiri for technical help. Guidance and help on the aflatoxin measurement of the samples by TLC method and interpretation of the results in this study has been conducted by Dr. R. Rezaeian-Doloei. This research was supported by grant No. 16865 from the Research Council of the Ferdowsi University of Mashhad.
References
- Accensi, E, Abarca, ML, Cabanes, FJ. Occurrence of Aspergillus species in mixed feeds and component raw materials and their ability to produce ochratoxin A. Food Microb. 2004;21:623–627. [Google Scholar]
- Chang, PK, Horn, BW, Dorner, JW. Sequence breakpoints in the aflatoxin biosynthesis gene cluster and flanking regions in nonaflatoxigenic Aspergillusflavus isolates. Fungal Genet. Biol. 2005;42:914–923. doi: 10.1016/j.fgb.2005.07.004. [DOI] [PubMed] [Google Scholar]
- Chang, PK, Yu, J, Bhatnagar, D, Cleveland, TE. The carboxy-terminal portion of the aflatoxin pathway regulatory protein AFLR of Aspergillusparasiticus activates GAL1:lacZ gene expression in Saccharomycescerevisiae. Appl. Environ. Microbiol. 1999a;65:2508–2512. doi: 10.1128/aem.65.6.2508-2512.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, PK, Yu, J, Bhatnagar, D, Cleveland, TE. Repressor-AFLR interaction modulates aflatoxin biosynthesis in Aspergillusparasiticus. Mycopathologia. 1999b;147:105–112. doi: 10.1023/a:1007157309168. [DOI] [PubMed] [Google Scholar]
- Criseo, G, Bagnara, A, Bisignano, G. Differentiation of aflatoxin-producing and non-producing strain of Aspergillusflavus group. J. App. Microb. 2001;33:291–295. doi: 10.1046/j.1472-765x.2001.00998.x. [DOI] [PubMed] [Google Scholar]
- Ehrlich, KC, Montalbano, BG, Bhatnagar, D, Cleveland, TE. Alteration of different domains in AFLR affects aflatoxin pathway metabolism in Aspergillusparasiticus transformants. Fungal Genet. Biol. 1998;23:279–287. doi: 10.1006/fgbi.1998.1045. [DOI] [PubMed] [Google Scholar]
- Ehrlich, KC, Yu, J. Aflatoxin-like gene clusters and how they evolved. In: Rai, M, Varma, A, editors. Mycotoxins in food, feed, and bioweapons. New York: Springer; 2010. pp. 65–75. [Google Scholar]
- Erami, M, Hashemi, SJ, Pourbakhsh, SA, Shahsavandi, S, Mohammadi, S, Shooshtari, AH, Jahanshiri, Z. Application of PCR on detection of aflatoxinogenic fungi. Arch. Razi Inst. 2007;62:95–100. [Google Scholar]
- Ghiasian, SA, Maghsood, AH. Occurrence of aflatoxigenic fungi in cow feeds during the summer and winter season in Hamadan, Iran. Afr. J. Microbiol. Res. 2011;5:516–521. [Google Scholar]
- Keller, NP, Kantz, NJ, Adams, TH. Aspergillusnidulans verA is required for production of the mycotoxin sterigmatocystin. Appl. Environ. Microbiol. 1994;60:1444–1450. doi: 10.1128/aem.60.5.1444-1450.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosravi, AR, Shokri, H, Yahyaraeyat, R, Soltani, M. Isolation of toxigenic and nontoxigenic fungi from feedstuff referred to the center of mycology. J. Vet. Res. 2004;59:221–226. [Google Scholar]
- Klich, MA. Identification of common Aspergillus species. Utrecht, Netherlands: CBS; 2002. p. 116. [Google Scholar]
- Konietzny, U, Greiner, R. The application of PCR in the detection of mycotoxigenic fungi in foods. Braz. J. Microbiol. 2003;34:283–300. [Google Scholar]
- Macioro, KG, Herrera, P, Jones, FT, Pillai, SD, Ricke, SC. Effect on poultry and livestock of feed contamination with bacteria and fungi. Anim. Feed Sci. Tech. 2007;133:109–136. [Google Scholar]
- Mehrzad, J, Klein, G, Kamphues, J, Wolf, P, Grabowski, N, Schuberth, HJ. In vitro effects of very low levels of aflatoxin B1 on free radicals production and bactericidal activity of bovine blood neutrophils. Vet. Immunol. Immunopatol. 2011;141:16–25. doi: 10.1016/j.vetimm.2011.01.010. [DOI] [PubMed] [Google Scholar]
- Moubasher, H, Abutaleb, A, Senousy, HH. Molecular differentiation between aflatoxinogenic and non-aflatoxinogenic strains of Aspergillusflavus and Aspergillusparasiticus. Microbiology. 2013;82:642–646. [Google Scholar]
- Pitt, JI, Hocking, AD. 3rd Edn. New York: Springer-Verlag; 2009. Fungi and food spoilage; pp. 3–9. [Google Scholar]
- Rahimi, P, Sharifnabi, B, Bahar, M. Detection of aflatoxin in Aspergillus species isolated from pistachio in Iran. Phytopathology. 2008;156:15–20. [Google Scholar]
- Rashid, M, Khalil, S, Ayub, N, Ahmed, W, Ghaffarkhan, A. Categorization of Aspergillus flavus and Aspergillus parasiticus isolates of stored wheat grains in to aflatoxinogenics and non-aflatoxinogenics. Pak. J. Bot. 2008;40:2177–2192. [Google Scholar]
- Rodrigues, P, Armando, V, Kozakiewicz, Z, Lima, N. A polyphasic approach to the identification of aflatoxigenic and non-aflatoxigenic strain of Aspergillus section flavi isolation from Portuguesa almond. Food Microbiol. 2009;129:187–193. doi: 10.1016/j.ijfoodmicro.2008.11.023. [DOI] [PubMed] [Google Scholar]
- Rosa, CAR, Cavaglieri, LR, Ribeiro, JMM, Keller, KM, Alonso, VA, Chiacchiera, SM, Dalcero, AM. Mycobiota and naturally-occurring ochratoxin A in dairy cattle feed from Rio de Janeiro state, Brazil. World Mycotoxin J. 2008;1:195–201. [Google Scholar]
- Saleemi, MK, Khan, MZ, Ahrar, K, Javed, I. Mycoflora of poultry feeds and mycotoxins producing potential of Aspergillus species. Pak. J. Bot. 2010;42:427–434. [Google Scholar]
- Shapira, R, Paster, N, Eyal, O, Menashero, M, Mett, A, Salomon, R. Detection of aflatoxigenic mold in grain by PCR. App. Environ. Microbiol. 1996;62:3270–3273. doi: 10.1128/aem.62.9.3270-3273.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somashekar, D, Rati, ER, Anad, S, Chandrashekar, A. Isolation, enumeration and PCR characterization of aflatoxigenic fungi from food and feed samples in India. Food Microb. 2004;21:809–813. [Google Scholar]
- van der Vossen, JM, Hofstra, H. DNA based typing, identification and detection systems for food spoilage microorganisms: development and implementa-tion. Int. J. Food Microbiol. 1996;33:35–49. doi: 10.1016/0168-1605(96)01136-1. [DOI] [PubMed] [Google Scholar]
- Woloshuk, CP, Foutz, KR, Brewer, JF, Bhatnagar, D, Cleveland, TE, Payne, GA. Molecular characterization of aflR, a regulatory locus for aflatoxin biosynthesis. App. Environ. Microbiol. 1994;60:2408–2414. doi: 10.1128/aem.60.7.2408-2414.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelton, MM, Hamer, JE, Timberlake, WE. Transformation of Aspergillus nidulans by using a trpC plasmid. National Academy of Sciences of the USA; 1984. pp. 1470–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J. Current understanding on aflatoxin biosynthesis and future perspective in reducing aflatoxin contamination. Toxins. 2012;4:1024–1057. doi: 10.3390/toxins4111024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J, Bhatnagar, D, Cleveland, TE. Completed sequence of aflatoxin pathway gene cluster in Aspergillus parasiticus. FEBS Lett. 2004a;564:126–130. doi: 10.1016/S0014-5793(04)00327-8. [DOI] [PubMed] [Google Scholar]
- Yu, J, Chang, PK, Ehrlich, KC, Cary, JW, Bhatnagar, D, Cleveland, TE, Payne, GA, Linz, JE, Woloshuk, CP, Bennett, JW. Clustered pathway genes in aflatoxin biosynthesis. Appl. Environ. Microbiol. 2004b;70:1253–1262. doi: 10.1128/AEM.70.3.1253-1262.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]