Abstract
Canine distemper virus (CDV) is the cause of canine distemper (CD) which is a severe and highly contagious disease in dogs. In the present study, a duplex reverse transcription polymerase chain reaction (RT-PCR) method was developed for the detection and differentiation of wild-type and vaccine strains of CDV. Four primers were designed to detect and discriminate the two viruses by generating 638- and 781-bp cDNA products, respectively. Furthermore, the duplex RT-PCR method was used to detect 67 field samples suspected of CD from Guangdong province in China. Results showed that, 33 samples were to be wild-type-like. The duplex RT-PCR method exhibited high specificity and sensitivity which could be used to effectively detect and differentiate wild-type and vaccine CDV, indicating its use for clinical detection and epidemiological surveillance.
Key Words: Canine distemper virus, Differentiation, Duplex RT-PCR, Sensitivity, Specificity
Introduction
Canine distemper (CD), a highly contagious and fatal disease of dogs caused by the canine distemper virus (CDV), is a single-stranded negative RNA virus belonging to the Morbillivirus genus within the Paramyxoviridae family (Wiezorek and Harder, 1998 ▶). Canine distemper virus has a very broad host-range, including dogs, ferrets, foxes, jackals, coyotes, hyenas, tigers, lions, leopards, cheetahs, seals, sea lions and dolphins (Appel et al., 1994 ▶; van de Bildt et al., 2002 ▶). The virus infects many cell types such as hematopoietic, epithelial, mesenchymal and neuroendocrine cells from various organs and tissues. Main clinical manifestations include respiratory and gastrointestinal signs, immuno-suppression, and demyelinating leukoencephalitis (Carvalho et al., 2012 ▶).
Canine distemper virus is a highly effective prophylactic disease and leads a large number of dogs to death every year, causing significant economic losses in China. Therefore, accurate vaccination, good sur-veillance programs and most importantly, reliable detection methods to differentiate CDV wild-type and vaccine strains are needed to control this disease. In the recent years, with the use of methods based on molecular biological techniques such as nucleic acid hybridization and PCR, accurate, specific and sensitive diagnoses of CDV infection have been made (Gaedke et al., 1997 ▶; Yoshida et al., 1998 ▶; Hoyland et al., 2003 ▶). Real-time RT-PCR targeting the hypervariable C-terminal domain of the established nucleocapsid (N) gene, was shown to be more sensitive and effective (Grant et al., 2009 ▶). In addition, RT-PCR-RFLP could be used to detect the hemagglutinin (H) gene of CDV (Calderon et al., 2007 ▶). Di Francesco et al., (2012) ▶ also applied a new highly sensitive and specific hemi-nested RT-PCR-RFLP assay to detect the nucleoprotein (NP) gene of CDV.
Harder et al. (1996) ▶ reported marked differences between wild-type and vaccine strains of CDV, thus raising the question of whether CDV vaccine strains are able to provide protection from the current strains of CDV or not. Furthermore, since the attenuated CDV vaccine is used widely in China, it is both difficult and necessary to discriminate wild-type from vaccine strains. Thus, a method of specifically detecting wild-type CDV strains is necessary. A previous study found that a multiplex reverse transcription-nested polymerase chain reaction (RT-nPCR) method could be used to effectively detect and differentiate wild-type CDV-infected dogs from those vaccinated with CDV vaccine (Si et al., 2010 ▶). An RT-PCR-RFLP method was established based on the CDV N gene, which effectively differentiated the vaccine and field strains of CDV (Wang et al., 2011 ▶; Fischer et al., 2013 ▶). A hemi-nested RT-PCR-RFLP assay was applied to differentiate among vaccine and wild-type CDV strains and to characterize the field viral strains (Di Francesco et al., 2012 ▶). In this study, a duplex RT-PCR method was established to effectively differentiate the vaccine and wild-type strains of CDV. Being sensitive and specific, this method not only meets the need for veterinary technology, but also sets a theoretical foundation for the early differential diagnosis of clinical samples.
Materials and Methods
Viruses and cells
Canine distemper virus vaccine and wild-type strains, canine parvovirus (CPV), rabies virus (RV), Escherichia coli and Pasteurella multocida were preserved in our laboratory (Laboratory of Microbiology and Immuno-logy, College of Veterinary Medicine, South China Agricultural University). Canine parainfluenza virus (CPIV) and canine influenza virus (CIV) were provided by the Laboratory of Clinical Surgery in Veterinary Medicine (College of Veterinary Medicine, South China Agricultural University). Vero E6 cells preserved in our laboratory were used in this study.
Optimization of duplex RT-PCR
To optimize reaction conditions, preliminary assays were performed to test different concentrations of each primer set, TaqDNA polymerase, dNTPs and Mg2+ to establish the optimal reaction protocol for the duplex RT-PCR assay. Furthermore, the appropriate annealing temperature was selected to yield the best results for the target CDV.
Primer design
With the help of DNAStar 5.07, DNAMAN 6.0.3.99 and premier 5.0 softwares, four primers were designed based on the genomic sequences of CDV strains published in GenBank. Primers P1 (5´-AGG AGC AAT AAG AGG GAT AAA GC-3´) and P2 (5´-CCC GAG AGC CGG ATA CAT AGT-3´) were used as common primers, designed based on N gene conserved fragments in virulent and attenuated viruses. Primers P3 (5´-GGG CAA CAC CTA TGG ATC GAG-3´) and P4 (5´-ATA AAC AAT TGC ATG ATC GCC C-3´) were used as the primers specific for wild-type strains.
Duplex RT-PCR
Viral RNA samples of CDV were isolated from all specimens using a commercial kit (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s instructions. Using a ReverTra Ace kit (TOYOBO Bio-Technology Co., Ltd., Shanghai), cDNA synthesis was conducted. Briefly, a 20 μL reaction system containing 4.0 µL 5 × Ace Buffer, 2.0 µL dNTPs (10 mM), 0.5 µL RNase inhibitor, 1.0 µL Ace, 2.0 μL random primers and 10.5 µL RNA samples was bathed in water at 42°C for 1 h and stored at -20°C for further study. The PCR amplification cycle was optimized as follows: 94°C 45 s, 55°C 45 s and 72°C 45 s for 30 cycles with a final extension step at 72°C for 10 min. A total of 5 µL of the amplified products was visualized using gel electro-phoresis in a 2.0% agarose gel, stained with ethidium bromide. Bands were visualized by ultraviolet light transillumination and compared with a 100 bp Ladder (TakaRa, Osaka, Japan).
Specificity, sensitivity, and repeatability tests
Duplex RT-PCR was used to detect Vero E6 cells infected with CDV wild-type strain, CDV vaccine strain, mixed CDV wild-type and vaccine strains, CPV, CPIV, CIV, RV, E. coli, P. multocida and uninfected cells in order to test its specificity. Extracted RNA was serially diluted (101, 100, 10-1, 10-2, 10-3, 10-4, and 10-5) and assayed by duplex RT-PCR to determine its sensitivity. To validate the repeatability of the test, duplex RT-PCR was also performed three times to identify cells infected with CDV wild-type strain, CDV vaccine strain, mixed CDV wild-type and vaccine strains, and uninfected cells.
Application of duplex RT-PCR
A total of 67 samples were taken from dogs clinically suspected of having CDV infection. All samples including 5 serum samples, 7 tissue samples and 55 swab samples were obtained from dogs of the Guangdong province in China and assayed by duplex RT-PCR. All positive field samples of the wild-type strain were confirmed by this method.
Results
Determination of the application conditions of the duplex RT-PCR
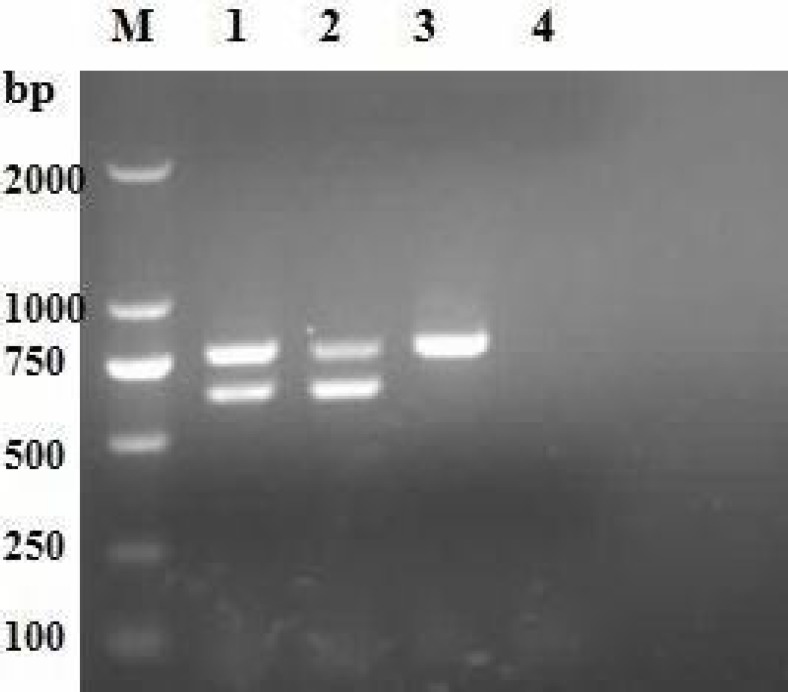
Designed primers (P1 and P2) specific to CDV corresponding to the highly conserved region of the CDV genome were used as a common primer pair and primers (P3 and P4) specific to the CDV wild-type strain were used in PCR. According to the results, when the anneal temperature was 55°C, and the concentrations of 5 pmol primers, 1.5 U TaqDNA polymerase, 0.20 mM dNTPs and 3.0 mM Mg2+ were set, the most distinct band appeared. These results were thus chosen for the duplex RT-PCR. Using the best selected incubation profile conditions for amplification tests, the results in Fig. 1, show that a fragment of 638 bp, together with a fragment of 781 bp, were amplified from the wild-type strain genomic RNA, a fragment of 781 bp from the vaccine strain genomic RNA in the RT-PCR, and two fragments of 781 bp and 638 bp from the wild-type strain or mixed samples of vaccine and wild-type strains.
Fig. 1.
Amplification of genomes of canine viruses by duplex reverse transcription PCR. Lane M: DL2000 DNA marker. Lane 1: Wild-type and vaccine canine distemper viruses, Lane 2: Wild-type canine distemper virus, Lane 3: Vaccine canine distemper virus, and Lane 4: Negative control
Sensitivity of the duplex RT-PCR
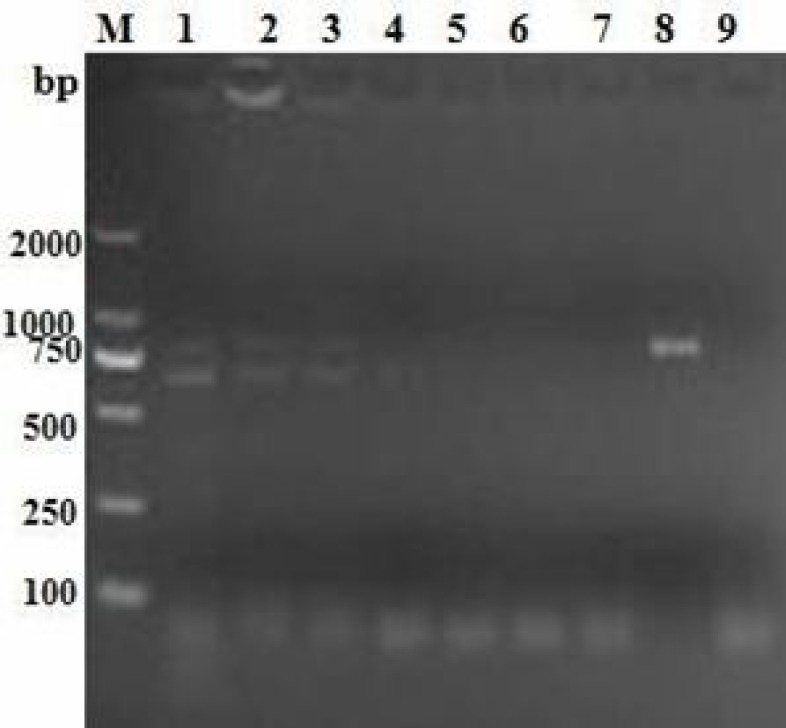
To evaluate the sensitivity of this method, serial 10-fold dilutions of the CDV wild-type strain were subjected to amplification by three separated duplex RT-PCRs. As Fig. 2 shows, the lowest limit of detection with this method was shown to be 8 pg/ml.
Fig. 2.
Sensitivity of duplex reverse transcription PCR. Lane M: DL2000 DNA marker. Lane 1: 80 ng, Lane 2: 8 ng, Lane 3: 0.8 ng, Lane 4: 8 × 10-2 ng, Lane 5: 8 × 10-3 ng, Lane 6: 8 × 10-4 ng, Lane 7: 8 × 10-5 ng, Lane 8: Positive control, and Lane 9: Negative control
Specificity of the multiplex RT-PCR
To test whether the amplified PCR fragment corresponded to the expected virus, the PCR product was run on a gel and the band was visualized. The data confirmed that a fragment of 781 bp was amplified from the CDV vaccine strain, and two bands of 638 and 781 bp were detected simultaneously from the CDV wild-type strain. No amplification was achieved for uninfected cells, or cells infected with CPV, RV, CAV, CPIV and CIV, E. coli and P. multocida by duplex RT-PCR. This indicates that the assay was completely specific for CDV (Fig. 3).
Fig. 3.
Specificity of duplex reverse transcription PCR. Lane M: DL2000 DNA marker. Lane 1: Wild-type canine distemper viruses, Lane 2: Vaccine canine distemper virus, Lane 3: Canine adenovirus (CAV), Lane 4: Canine parvovirus (CPV), Lane 5: Canine parainfluenza virus (CPIV), Lane 6: Canine influenza virus (CIV), Lane 7: Rabies virus (RV), Lane 8: Escherichia coli, Lane 9: Pasteurella multocida, and Lane 10: Negative control
Repeatability and applicability of the duplex RT-PCR
Three inter- and intra-assay replicates of the duplex RT-PCR in eight independent experiments gave con-sistent results, indicating the repeatability of the assay. The applicability of the duplex RT-PCR was also investigated. From the 67 field samples, 33 were tested positive for CDV, all showing the presence of wild-type strains.
Discussion
Canine distemper, caused by CDV, is a disease of dogs that should be included in the list of differentials for clinical diagnosis of any multi-systemic condition characterized by fever, respiratory, enteric, and neurological signs (Martella et al., 2008 ▶). Laboratory tests are essential to confirm CDV infection, and molecular assays are useful for monitoring and differentiating CDV wild-type from vaccine strains. Molecular detection by RT-PCR is an ideal detection method of the viruses in hosts, since it is rapid, specific, sensitive, repeatable, and amenable to automation. RT-PCR has been used for the efficient detection of CDV (Shin et al., 1995 ▶; Elia et al., 2006 ▶; Calderon et al., 2007 ▶; Yi et al., 2012a ▶). In the present study, we reported on the development of a duplex RT-PCR suitable for the rapid and sensitive discrimination of wild-type from vaccine strains of CDV.
With the RT-PCR, optimal primers are critically important to obtain distinct and specific amplifications of virus genome parts. The selection of suboptimal primers often causes undesirable results such as hairpin formation, primer dimer formation, false-negative or false-positive results and the generation of spurious products (Yi et al., 2012b ▶). In this study, primer sequences with similar lengths, hybridization kinetics, and G-C contents were selected. Additionally, some important parameters were optimized. For example, 5 pmol was chosen as the best primer concentration to obtain uniform amplification signals of the fragments, and 55°C was selected for an optimal annealing temperature to obtain a desired specific product. Furthermore, other factors such as 1.5 U TaqDNA polymerase, 0.20 mM dNTPs and 3.0 mM Mg2+ were set to avoid affecting the quality of the duplex RT-PCR assays. A distinct band appeared using these optical conditions for amplification tests.
Sensitivity of the duplex RT-PCR assay was suitable, allowing for the CDV detection at 8 pg/ml. Specificity confirmed that the assay was completely specific for CDV but not for CPV, RV, CAV, CPIV and CIV, E. coli and P. multocida. Repeatability and applicability of the assay was also investigated and it was found that this method was repeatable and offered the potential for very rapid detection with a single clinical sample against a large number of potential pathogens.
In conclusion, the duplex RT-PCR developed in this study is a highly specific and sensitive assay for the rapid detection and differentiation of wild-type and vaccine strains of CDV. Therefore, this assay has the potential to be used in clinical diagnoses and epidemiological surveillance.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (No. 31072137 and 31172321), the Key Project of Natural Science Foundation of Guangdong province, China (No. S2011020001037), the Special Project for Scientific and Technological Innovation in Higher Education of Guangdong, China (No. 2012CXZD0013), the Special Fund for Agro-Scientific Research in the Public Interest (No. 201203056) and the Research Fund for the Doctoral Program of Higher Education of China (No. 20114404110015).
Conflict of interest
The authors declare that they have no competing interests.
References
- Appel, MJ, Yates, RA, Foley, GL, Bernstein, JJ, Santinelli, S, Spelman, LH, Miller, LD, Arp, LH, Anderson, M, Barr, M, Et, A. Canine distemper epizootic in lions, tigers, and leopards in North America. J. Vet. Diagn. Invest. 1994;6:277–288. doi: 10.1177/104063879400600301. [DOI] [PubMed] [Google Scholar]
- Calderon, MG, Remorini, P, Periolo, O, Iglesias, M, Mattion, N, La Torre, J. Detection by RT-PCR and genetic characterization of canine distemper virus from vaccinated and non-vaccinated dogs in Argentina. Vet. Microbiol. 2007;125:341–349. doi: 10.1016/j.vetmic.2007.05.020. [DOI] [PubMed] [Google Scholar]
- Carvalho, OV, Botelho, CV, Ferreira, CG, Scherer, PO, Soares-Martins, JA, Almeida, MR, Silva, JA. Immunopathogenic and neurological mechanisms of canine distemper virus. Adv. Virol. 2012;2012 doi: 10.1155/2012/163860. Article ID 163860, 10 pages. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Francesco, CE, Di Francesco, D, Di Martino, B, Speranza, R, Santori, D, Boari, A, Marsilio, F. Detection by hemi-nested reverse transcription polymerase chain reaction and genetic characterization of wild type strains of canine distemper virus in suspected infected dogs. J. Vet. Diagn. Invest. 2012;24:107–115. doi: 10.1177/1040638711425700. [DOI] [PubMed] [Google Scholar]
- Elia, G, Decaro, N, Martella, V, Cirone, F, Lucente, MS, Lorusso, E, Di Trani, L, Buonavoglia, C. Detection of canine distemper virus in dogs by real-time RT-PCR. J. Virol. Methods. 2006;136:171–176. doi: 10.1016/j.jviromet.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Fischer, CD, Ikuta, N, Canal, CW, Makiejczuk, A, Allgayer, MD, Cardoso, CH, Lehmann, FK, Fonseca, AS, Lunge, VR. Detection and differentiation of field and vaccine strains of canine distemper virus using reverse transcription followed by nested real time PCR (RT-nqPCR) and RFLP analysis. J. Virol. Methods. 2013;194:39–45. doi: 10.1016/j.jviromet.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaedke, K, Zurbriggen, A, Baumgartner, W. In vivo and in vitro detection of canine distemper virus nucleoprotein gene with digoxigenin-labelled RNA, double-stranded DNA probes and oligonucleotides by in situ hybridization. Zentralbl. Veterinarmed. B. 1997;44:329–340. doi: 10.1111/j.1439-0450.1997.tb00983.x. [DOI] [PubMed] [Google Scholar]
- Grant, RJ, Banyard, AC, Barrett, T, Saliki, JT, Romero, CH. Real-time RT-PCR assays for the rapid and differential detection of dolphin and porpoise morbilliviruses. J. Virol. Methods. 2009;156:117–123. doi: 10.1016/j.jviromet.2008.11.008. [DOI] [PubMed] [Google Scholar]
- Harder, TC, Kenter, M, Vos, H, Siebelink, K, Huisman, W, van Amerongen, G, Orvell, C, Barrett, T, Appel, MJ, Osterhaus, AD. Canine distemper virus from diseased large felids: biological properties and phylogenetic relationships. J. Gen. Virol. 1996;77:397–405. doi: 10.1099/0022-1317-77-3-397. [DOI] [PubMed] [Google Scholar]
- Hoyland, JA, Dixon, JA, Berry, JL, Davies, M, Selby, PL, Mee, AP. A comparison of in situ hybridisation, reverse transcriptase-polymerase chain reaction (RT-PCR) and in situ-RT-PCR for the detection of canine distemper virus RNA in Paget’s disease. J. Virol. Methods. 2003;109:253–259. doi: 10.1016/s0166-0934(03)00079-x. [DOI] [PubMed] [Google Scholar]
- Martella, V, Elia, G, Buonavoglia, C. Canine distemper virus. Vet. Clin. N. Am. Small Anim. Pract. 2008;38:787–797. doi: 10.1016/j.cvsm.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Shin, Y, Mori, T, Okita, M, Gemma, T, Kai, C, Mikami, T. Detection of canine distemper virus nucleocapsid protein gene in canine peripheral blood mononuclear cells by RT-PCR. J. Vet. Med. Sci. 1995;57:439–445. doi: 10.1292/jvms.57.439. [DOI] [PubMed] [Google Scholar]
- Si, W, Zhou, S, Wang, Z, Cui, SJ. A multiplex reverse transcription-nested polymerase chain reaction for detection and differentiation of wild-type and vaccine strains of canine distemper virus. Virol. J. 2010;7 doi: 10.1186/1743-422X-7-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Bildt, MW, Kuiken, T, Visee, AM, Lema, S, Fitzjohn, TR, Osterhaus, AD. Distemper outbreak and its effect on African wild dog conservation. Emerg. Infect. Dis. 2002;8:211–213. doi: 10.3201/eid0802.010314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, F, Yan, X, Chai, X, Zhang, H, Zhao, J, Wen, Y, Wu, W. Differentiation of canine distemper virus isolates in fur animals from various vaccine strains by reverse transcription-polymerase chain reaction-restriction fragment length polymorphism according to phylogenetic relations in china. Virol. J. 2011;8:85. doi: 10.1186/1743-422X-8-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiezorek, T, Harder, D. Dosimetric aspects of physical and dynamic wedges of the Clinac 2100C Linear Accelerator”. Comment on the publication of Avadhani, JS et al. in Strahlenther. Onkol. (1997), 173: 524-528. Strahlenther. Onkol. 1998;174:105. doi: 10.1007/BF03038483. [DOI] [PubMed] [Google Scholar]
- Yi, L, Cheng, S, Xu, H, Wang, J, Cheng, Y, Yang, S, Luo, B. Development of a combined canine distemper virus specific RT-PCR protocol for the differentiation of infected and vaccinated animals (DIVA) and genetic characterization of the hemagglutinin gene of seven Chinese strains demonstrated in dogs. J. Virol. Methods. 2012;179:281–287. doi: 10.1016/j.jviromet.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, E, Iwatsuki, K, Miyashita, N, Gemma, T, Kai, Cand Mikami, T. Molecular analysis of the nucleocapsid protein of recent isolates of canine distemper virus in Japan. Vet. Microbiol. 1998;59:237–244. doi: 10.1016/s0378-1135(97)00194-6. [DOI] [PubMed] [Google Scholar]